Abstract
Recent studies have identified several amino acid sequences that interact with the ribosomal interior components and arrest their own elongation. Whereas stalling of the inducible class depends on specific low-molecular weight compounds, that of the intrinsic class is released when the nascent chain is transported across or inserted into the membrane. The stalled ribosome alters messenger RNA secondary structure and thereby contributes to regulation of the cis-located target gene expression at different levels. The stalling sequences are divergent but likely to utilize non-uniform nature of the peptide bond formation reactions and are recruited relatively recently to different biological systems, possibly including those to be identified in forthcoming studies.
Keywords: Ribosome stalling, Translation regulation, Elongation arrest, Nascent polypeptide, Protein localization, Peptidyl transferase center, Exit tunnel
The ribosomal peptidyl transferase center (PTC) catalyzes peptide bond formation by allowing nascent peptidyl-tRNA in the P-site to receive a nucleophilic attack by the A-site aminoacyl-tRNA [1]. Translation is concluded at a termination codon by the PTC- and a release factor-mediated hydrolysis of the P-site peptidyl-tRNA [2]. The growing nascent polypeptide chain moves through the exit tunnel, an interior conduit that bridges PTC and the cytosol, which is about 100-Å long and ~15-Å wide and capable of accommodating 30–40 amino acid residues of an extended to α-helical polypeptide [3,4]. The tunnel is largely composed of 23S rRNA, having a constriction at about 1/3 away from PTC, where tips of rod-shaped parts of r-proteins L22 and L4 are located [4]. The tunnel wall is charged negatively [5], and originally thought not to interact with the polypeptide product [4]. However recent studies have identified a number of amino acid sequences that interact with the exit tunnel and arrest their own translation. This mini-review focuses on bacterial regulatory systems that use ribosome-stalling sequences.
Non-uniformity in translation
Although in vitro examinations suggest that translation proceeds uniformly [6], in vivo rates of polypeptide elongation show some heterogeneity [7,8]. In particular, prolyl-tRNA at the P-site only inefficiently participates in termination [9,10] or transfer to puromycin [11,12]. Also, proline at the A-site is a poor attacker against the P-site peptidyl-tRNA [13]. More generally, peptidyl transfer reactions are affected by nature of the side-chains of participating amino acids [14]. An array of positively charged amino acids lead to retarded elongation due presumably to their electrostatic interaction with the tunnel wall [15,16]. Thus, elongation of nascent chains is not absolutely uniform but could be a target of biological regulation.
Regulation by stalling sequences
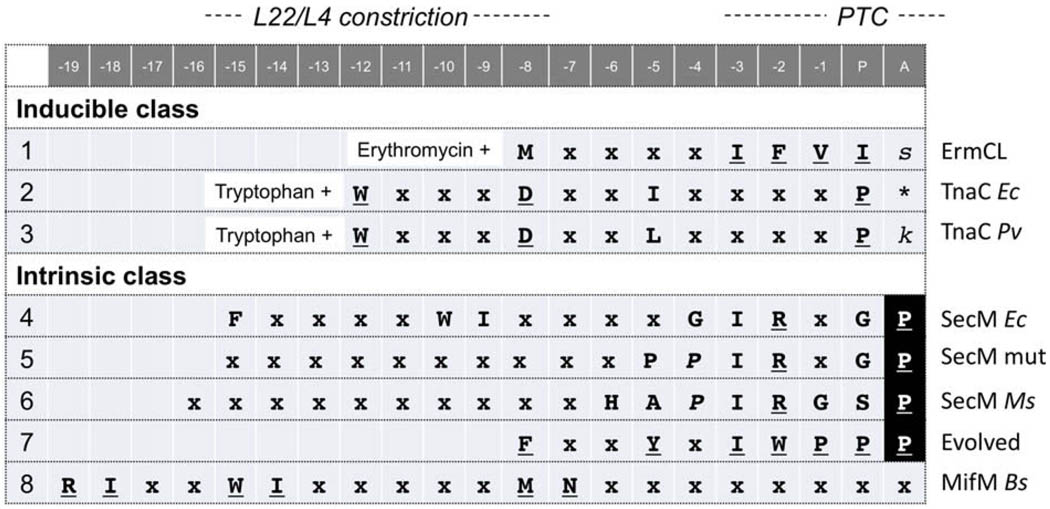
A class of amino acid sequences, some 9–21 residues long, interact with ribosomal interior components and lead to PTC dysfunction. Fig. 1 aligns some well-characterized stalling sequences with their likely occupancy within the ribosome (numbered downward from the P-site residue). They span segments from the PTC-proximal arrest point to the region proximal to the tunnel constriction (Fig. 1), although the exact positioning should differ between extended [17] and compacted [18] nascent chains. The stalling sequences act as a conditional brake upon translation. The inducible class stalls only in the presence of a specific effector, such as an antibiotic or a metabolite. The intrinsic class stalls without any inducing molecule until the arrest is released as the nascent chain interacts with specific cellular machineries. Therefore, the latter class of stalling sequences are preceded by N-terminal regions long enough to reach other cellular components (Fig. 2).
Fig. 1.
Stalling sequences that have been subjected to comprehensive mutational analysis. Sequences that arrest translation elongation are aligned based on their likely positions in the stalled ribosome, with numbering starting inversely from −1 for the position immediately preceding the P-site amino acid. Approximate locations of amino acids in the ribosome are indicated at the top, on the basis of the structure of the extended TnaC peptide–ribosome complex [17]. Note that in the cases of SecM and others, the intraribosomal peptide may be more compacted. Translation ends with the P-site amino acid as the last amino acid of the nascent peptidyl-tRNA (note, however, that ribosomal occupancy has not been determined for MifM). Residues essential for the elongation arrest are underlined. Residues denoted×are less important as they can be changed to one or more different amino acid(s) without affecting the arrest. In all cases that have been examined, the arrest-essential amino acids need to be separated with the exact spacings shown. The A-site amino acids shown in lower case italics are not required for the arrest. The A-site prolines shown in reverse upper case are essential for the arrest. In the case of E. coli TnaC, the A-site codon is a UGA stop (shown by asterisk). 1, Leader peptide of the erythromycin resistance gene, ermC [21]; 2, and 3, arrest sequence of the tryptophanase operon of E. coli [26] and Proteus vulgaris [30], respectively; 4, arrest sequence of SecM from E. coli [38]; 5, a mutant form of the SecM arrest sequence having proline at −4 and −5 positions, of which the one at −4 (italicized) alleviates the specificity of the constriction-proximal residues [39]; 6, arrest sequence of SecM from Mannheimia succiniciproducens [39]; 7, an experimentally evolved arrest sequence obtained by genetic screening [47]; 8, arrest sequence of MifM from B. subtilis [40].
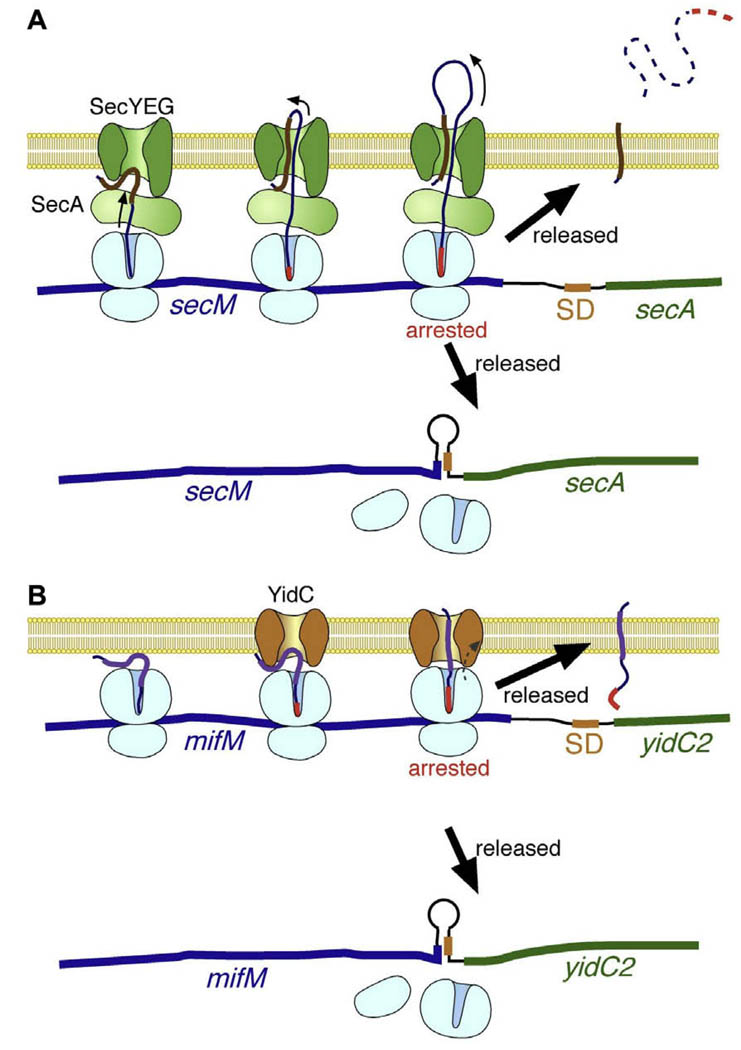
Fig. 2.
Arrest regulation. (A) SecM. As translation proceeds, the N-terminal region of the SecM nascent peptide, including the signal sequence (shown by purple thick line), engages in SecA-SecYEG-dependent translocation across the cytoplasmic membrane. When the ribosome attempts to translate the arrest sequence (shown in red thick line), it stalls on the messenger RNA. The stalled ribosome disrupts the secondary structure formed at the secM-secA intergenic region (shown in thin black part in the bottom line, representing the messenger RNA) and consequently exposes the SD sequence (shown in orange) required for translation of secA. Although the elongation arrest is transient in wild-type cells as it is released by active translocation reaction, it is prolonged when the Sec translocation activity is compromised by mutation or at low temperature, exposing the SD sequence for prolonged lengths of time and allowing for enhanced levels of secA translation. It should be noted that the released, completed product of SecM is rapidly proteolyzed in the periplasmic space [57] and that it is not well understood how SecA is able to participate in translocation of ribosome-tethered nascent polypeptide, which might be expected to spatially occlude SecA binding. (B) MifM. Although the situation is similar to the case of SecM shown in (A), there are important differences. MifM contains a predicted Nout–Cin transmembrane sequence (shown by purple thick line), which inserts into the membrane with the aid of the YidC membrane protein integration/folding factor (known as SpoIIIJ in B. subtilis). This mode of MifM membrane insertion is different from the conventional protein export, in which the C-terminus crosses the membrane. The ultimate fate of the released, completed MifM product is unknown.
The translating ribosome prevents secondary structure formation in the ribosome-covered and some 3′ regions of the messenger RNA [19,20]. Therefore, a typical consequence of ribosomal stalling is to uncover the ribosome-binding SD sequence of the downstream target gene by directly or indirectly disrupting the SD-sequestering secondary structure, facilitating the entry of active ribosomes that up-regulate translation. Stalling can also regulate transcription, as exemplified by the TnaC system, in which altered RNA secondary structure affects Rho-dependent transcription termination in front of the target gene. Still another level of regulation is provided by stalling sequences that prolong co-translational interactions of the nascent regulatory peptide (see “cis-chaperone” function of SecM discussed below).
Antibiotic-sensing leader peptides
Genes conferring resistance to antibiotics are often preceded by a short ORF. ErmCL (19 amino acids) [21] and CrbCmlA (9 amino acids) [22] regulate erythromycin resistance and chloramphenicol resistance, respectively. They contain a sequence that arrests translation in the presence of the respective antibiotic and consequently up-regulates translation of the resistance gene. In the ErmCL-stalled ribosome, Ile9 is located at the P-site without further elongation. This and the three preceding amino acids comprise the essential arrest element, IFVI, pointing to the importance of the PTC-proximal side-chains. More N-terminal 5 residues are substitutable but cannot be deleted, suggesting that some promiscuous molecular interaction involving the tunnel constriction contributes to the elongation arrest. In support of this notion, alterations in L22 and an rRNA residue A2062 alleviate the elongation arrest [21]. These polypeptides show an arrest induced by antibiotics that bind specifically to each nascent chain, thereby acting on the ribosome in manners different from their general translation inhibitory mechanism [23].
Tryptophan-sensing TnaC
TnaC ORF (24 amino acids) precedes the tryptophanase gene in Escherichia coli [24]. Its stalling sequence spans PTC-proximal Pro24 (position P) to constriction-proximal Trp12 (position −12) (Fig. 1, line 2). Pro24 is followed by a UAG stop codon, suggesting that the stalling utilizes the peculiarity of proline in translation termination. In the presence of tryptophan, TnaC-Pro24-tRNA is refractory to the PTC-RF2-catalyzed hydrolysis [25]. In addition, stalling requires Asp16 (position −8) and Trp12 (−12) with the exact spacing [26]. Position −12 and its neighbors interact with the constriction components [27]. Free tryptophan was proposed to bind to a site near the A-site of the TnaC-bearing ribosome, to induce structural changes of 23S rRNA that interfere with the PTC-RF2 actions [28,29]. Consistent with the PTC dysfunction, stalling occurs also when the following codon is not a stop but an Ile codon in an E. coli mutant [28] or a Lys codon in Proteus vulgaris (Fig. 1, line 3) [30]. It was proposed that a PTC residue 2585 is involved in the tryptophan to PTC transmission of a stalling signal [31].
Secretion-monitoring SecM
SecM ORF (170 amino acids) precedes secA for the secretion-driving ATPase. Having a signal sequence, it is designed to be exported to the periplasmic space (Fig. 2A). While its stalling-sequence (17 amino acids; Fig. 1, line 4) halts translation, its N-terminal region engages in SecA-SecYEG-mediated translocation (Fig. 2A). Cellular attempts to export SecM as a nascent polypeptide leads to release of the elongation arrest. Thus, the duration of ribosome stalling is inversely correlated with cellular protein (more precisely, SecM) export activity. While the transient arrest in normal cells is essential for the basal-level secA expression, prolonged arrest in export-compromised cells leads to the secretion defect-responding up-regulation of SecA biosynthesis. This regulation is particularly important at low temperature, a secretion-retarding physiological condition in E. coli [32,33]. SecM may have an another level of role, facilitating folding of newly synthesized SecA [34]. In this “cis-chaperone” function, translocon-targeting of nascent SecM brings the secM-secA messenger RNA to the membrane, where localized biosynthesis of SecA, a multi-conformation protein [35], enhances its folding into the working structure.
Translation of SecM stops at Gly165, producing peptidyl-glycyl-tRNA bound to the P-site. The next codon Pro166 is essential for the arrest but not incorporated into the peptide [36,37]. This prolyl-tRNA seems to act as an A-site effector of arrest, like free tryptophan in the TnaC system. However, whereas the intracellular concentration of tryptophan varies according to nutritional conditions, prolyl-tRNA is genetically encoded, constitutive and integrated into the intrinsic arrest function of SecM; the regulatory cue here is the translocation status of the nascent SecM polypeptide. Pro166 and Arg163 (at −2 position) are absolutely required while Gly165 (P), Ile156 (−9) and Trp155 (−10) contribute substantially to the arrest. The importance of the constricted region was inferred from arrest-alleviating mutations affecting it [38]. However, introduction of proline at −4 makes the constriction-proximal SecM residues less important (Fig. 1, line 5) [39]. Interestingly, SecM from Mannheimia succiniciproducens and some other bacteria naturally contains proline at −4 [39]. The constriction-proximal parts of this version of SecM may still participates in the arrest, as its −24 and 21 residues are crosslinkable with L22 and a chemical modification of −25 impairs the arrest [39]. Taken together, the PTC-adjacent arginine–proline combination has a primary role in the arrest of SecM, while molecular interactions at the tunnel constriction make secondary contributions.
Membrane integration-monitoring MifM
MifM (95 amino acids) is encoded upstream of the secondary YidC paralog in Bacillus subtilis (yidC2) and has an N-terminal transmembrane segment and a stalling sequence of at least ~21 amino acids near the C-terminus [40] (Fig. 2B). Presumably, MifM is integrated into the membrane with amino-terminus-out orientation by SpoIIIJ (YidC1), the primary YidC in B. subtilis [41,42]. Thus, MifM up-regulates translation of YidC2 by stalling the ribosome for extended time when MifM, as a monitor, is not efficiently integrated into the membrane [40]. In contrast to the other stalling sequences, arrest-important amino acids of MifM reside at position −7 and further upstream of the arrest site (Fig. 1, line 6), suggesting that interaction with the constriction region provides the main mechanism of stall. Consistent with this notion, L22 alterations strongly alleviate the arrest [40]. Note, however, that the arrest point has not precisely been determined for MifM and that its PTC proximal residues, mostly acidic, could still have a role in the elongation arrest.
Nascent chain-induced ribosomal stall is not interfered with trans-translation
The SecM-translating, stalled ribosome is refractory to the action of the SsrA trans-translation/tagging system [37], which likely acts on the A-site-vacant ribosome after cleavage of the messenger RNA [43,44]. The inability of the tmRNA system to rescue the stalled ribosome should ensure stalling to continue when needed and, hence, stalling-based regulations to be operational in vivo. SsrA-tagging of SecM was nevertheless observed upon its overproduction [45,46], because nascent chain–ribosome complexes having vacant A-site are generated by SecM-overproduction-provoked depletion of the tRNAPro, which is indeed antagonized by additional overproduction of this tRNA [37,47].
Experimental evolution of stalling sequences
Overproduction of a SecM-type stalling peptide, having genetically encoded A-site effector, will lead to its SsrA-tagging. Thus, Tanner et al. utilized an engineered tmRNA-coding sequence to select new stalling sequences. In addition to the known tagging-enhancers (Pro-stop and its derivatives), a new elongation-arresting sequence FxxYxIWPPP (Fig. 1, line 7) has been identified. As expected from the selection design, the final proline acts as a non-polypeptide effector at the A-site. Interestingly, Asp-tRNA and Trp-tRNA can also work at this position as an arrest effector for this sequence [47], whereas Pro166 of SecM cannot be replaced by these amino acids [38]. The other two prolines may contribute to the stalling through the peculiarity of this imino acid.
Divergence of stalling sequences
Stalling sequences are quite divergent [23,38–40,47] (Fig. 1). They interact with the ribosome in distinct ways, as shown by the distinct spectrum of effects they receive from mutations of ribosomal components [21,22,27,29,31,38–40,47]. Homologs of SecM and MifM are found only in limited subclasses of bacterial species [40,48], suggesting that these regulatory peptides were recruited relatively recently in evolution for the purpose of fine tuning of the already established translation and protein delivery systems. Notably, these regulatory systems nevertheless work under strikingly similar workflows.
Structures and mechanisms of stall
FRET measurements suggest that the stalled SecM peptide assumes some compacted structure, possibly an α-helix, which is required but not sufficient for the arrest [18]. The compacted structure of SecM contrasts with the extended TnaC peptide [17], but agrees with both the extended distribution of arrest-important SecM residues (Fig. 1) and crosslinking results [27,39]. This suggests that that Pro166 and the ribosomal PTC/tunnel components may induce secondary structure formation in SecM-Gly165-tRNA. It is possible that the MifM nascent chain is similarly compact, since it has a similarly broad distribution of arrest-important residues. Reconstructed cryo-electron microscopy images of the SecM-stalled ribosome revealed extensive structural rearrangements as compared to structures of the pretranslocational state of the normal ribosome [49]. It was proposed that SecM arrest produced an elongation-incompatible form of the ribosome by a cascade of internal signal transduction events.
More recently, Seidelt et al. visualized the TnaC nascent chain in a ~5.8 Å resolution image of the ribosome–TnaC complex [17]. The overall structure of the TnaC-bearing 70S ribosome was similar to that of the empty 70S ribosome. However, a PTC residue A2585, which assumes variable configurations in other nascent chain complexes, shows a robust contact with Pro24. Another PTC residue, A2602, which is otherwise flexible, is in a distinct and rigid structure in the TnaC complex. Such “frozen” structure of PTC appears to preclude functional accommodation of RF2. Puzzlingly, A-site bound tryptophan was not detected by this analysis. The TnaC peptide was largely extended but assumed a fixed configuration by contacting the tunnel wall at multiple (~10) sites, confirming previous biochemical and genetic inferences. Signal transduction from the tunnel to the PTC does not appear to involve large-scale conformational changes in the ribosome. Instead, the TnaC nascent chain itself and/or subtle conformational changes of the tunnel components might generate a stalling signal. Given this result, it is important to establish whether SecM [49] and any other nascent chain sequences induce more global conformational changes of the ribosome.
Taken together with a structure of another nascent polypeptide [50], Seidelt et al. argue that nascent peptides generally assume distinct configurations in the exit tunnel. A nascent chain might experience a series of programmed changes in its spatial configurations as it passes through the exit tunnel. The countless and changing molecular interactions between the ribosome and nascent chains may have provided the translation system with ample sampling opportunities to evolve divergent stalling sequences.
Arrest release by nascent chain dynamics
Two general possibilities, which are not mutually exclusive, can be considered for the mechanism by which ribosomal stalling is relieved by nascent chain engagement in protein translocation or membrane integration [51]. First, the dynamism of the nascent chain outside the ribosome may generate a physical pulling-force that disrupts the peptide–tunnel molecular interactions, leading to the resumption of elongation (Fig. 2A). Secondly, interaction of the nascent chain with the machinery for membrane translocation or integration might induce a ribosomal conformational change and elicit a signal to circumvent the PTC inhibition. The pulling model was suggested for SecM, as a placement of a stop-transfer hydrophobic sequence after its signal sequence antagonized the arrest release [52]. The predicted disposition of the transmembrane region of MifM is similar to that of a stop-transfer sequence, in that both of them assume an Nout–Cin configuration that prevents continued export of the C-terminus (Fig. 2B), raising the question about the relevance of the pulling model for the regulation of MifM [40]. It is conceivable that even the membrane integration process could generate a subtle physical force, which leads to cooperative configuration changes of the nascent chain, canceling the peptide–tunnel molecular interactions. As a signal transduction model is equally possible, the actual mechanisms of arrest release are important issues to be investigated further by various approaches, including theoretical calculations [53–55].
The knowledge we have gained so far raises a tempting possibility that there may be many more sequences that interact with the exit tunnel with a range of affinities and thereby contribute to fine tuning of translational elongation speed. Temporary pauses of elongation could not only contribute to translation/transcription of cis-located genes, as in the systems described thus far, but also to co-translational events of the protein itself, such as subcellular targeting, folding and assembly [56]. Such systems are inherently feedback regulated, allowing judicious use of cellular translational capacity. Following the process of nascent chain completion in relation to its folding or targeting with sufficient time resolution will be a challenging subject left for us.
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voss NR, Gerstein M, Steitz TA, Moore PB. The geometry of the ribosomal polypeptide exit tunnel. J. Mol. Biol. 2006;360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 4.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Kobertz WR, Deutsch C. Mapping the electrostatic potential within the ribosomal exit tunnel. J. Mol. Biol. 2007;371:1378–1391. doi: 10.1016/j.jmb.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 6.Ledoux S, Uhlenbeck OC. Different aa-tRNAs are selected uniformly on the ribosome. Mol. Cell. 2008;31:114–123. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorensen MA, Kurland CG, Pedersen S. Codon usage determines translation rate in Escherichia coli. J. Mol. Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 8.Zamora-Romo E, Cruz-Vera LR, Vivanco-Dominguez S, Magos-Castro MA, Guarneros G. Efficient expression of gene variants that harbour AGA codons next to the initiation codon. Nucleic Acids Res. 2007;35:5966–5974. doi: 10.1093/nar/gkm643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J. Biol. Chem. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- 10.Sunohara T, Abo T, Inada T, Aiba H. The C-terminal amino acid sequence of nascent peptide is a major determinant of SsrA tagging at all three stop codons. RNA. 2002;8:1416–1427. doi: 10.1017/s1355838202020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muto H, Ito K. Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem. Biophys. Res. Commun. 2008;366:1043–1047. doi: 10.1016/j.bbrc.2007.12.072. [DOI] [PubMed] [Google Scholar]
- 12.Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J. Biol. Chem. 2008;283:32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- 13.Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. USA. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rychlik I, Cerna J, Chladek S, Pulkrabek P, Zemlicka J. Substrate specificity of ribosomal peptidyl transferase. The effect of the nature of the amino acid side chain on the acceptor activity of 2′(3′)-O-aminoacyladenosines. Eur. J. Biochem. 1970;16:136–142. doi: 10.1111/j.1432-1033.1970.tb01064.x. [DOI] [PubMed] [Google Scholar]
- 15.Lu J, Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache J-P, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woolhead CA, Johnson AE, Bernstein HD. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol. Cell. 2006;22:587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 20.Takyar S, Hickerson RP, Noller HF. RNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 21.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence MG, Lindahl L, Zengel JM. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J. Bacteriol. 2008;190:5862–5869. doi: 10.1128/JB.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol. Microbiol. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 24.Gong F, Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 25.Gong F, Ito K, Nakamura Y, Yanofsky C. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro) Proc. Natl. Acad. Sci. USA. 2001;98:8997–9001. doi: 10.1073/pnas.171299298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruz-Vera LR, Yanofsky C. Conserved residues Asp16 and Pro24 of TnaCtRNAPro participate in tryptophan induction of Tna operon expression. J. Bacteriol. 2008;190:4791–4797. doi: 10.1128/JB.00290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome–peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell. 2005;19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Cruz-Vera LR, Gong M, Yanofsky C. Changes produced by bound tryptophan in the ribosome-peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. USA. 2006;103:3598–3603. doi: 10.1073/pnas.0600082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruz-Vera LR, New A, Squires C, Yanofsky C. Ribosomal features essential for tna operon induction: tryptophan binding at the peptidyl transferase center. J. Bacteriol. 2007;189:3140–3146. doi: 10.1128/JB.01869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruz-Vera LR, Yang R, Yanofsky C. Tryptophan inhibits Proteus vulgaris TnaC leader peptide elongation, activating tna operon expression. J. Bacteriol. 2009;191:7001–7006. doi: 10.1128/JB.01002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang R, Cruz-Vera LR, Yanofsky C. 23S rRNA nucleotides in the peptidyl transferase center are essential for tryptophanase operon induction. J. Bacteriol. 2009;191:3445–3450. doi: 10.1128/JB.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami A, Nakatogawa H, Ito K. Translation arrest of SecM is essential for the basal and regulated expression of SecA. Proc. Natl. Acad. Sci. USA. 2004;101:12330–12335. doi: 10.1073/pnas.0404907101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pogliano KJ, Beckwith J. The Cs sec mutants of Escherichia coli reflect the cold sensitivity of protein export itself. Genetics. 1993;133:763–773. doi: 10.1093/genetics/133.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakatogawa H, Murakami A, Mori H, Ito K. SecM facilitates translocase function of SecA by localizing its biosynthesis. Genes Dev. 2005;19:436–444. doi: 10.1101/gad.1259505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol. Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 37.Garza-Sanchez F, Janssen BD, Hayes CS. Prolyl-tRNA(Pro) in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J. Biol. Chem. 2006;281:34258–34268. doi: 10.1074/jbc.M608052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 39.Yap MN, Bernstein HD. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol. Cell. 2009;34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiba S, Lamsa A, Pogliano K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 2009;28:3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saller MJ, Fusetti F, Driessen AJ. Bacillus subtilis SpoIIIJ and YqjG function in membrane protein biogenesis. J. Bacteriol. 2009;191:6749–6757. doi: 10.1128/JB.00853-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie K, Dalbey RE. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat. Rev. Microbiol. 2008;6:234–244. doi: 10.1038/nrmicro3595. [DOI] [PubMed] [Google Scholar]
- 43.Janssen BD, Hayes CS. Kinetics of paused ribosome recycling in Escherichia coli. J. Mol. Biol. 2009;394:251–267. doi: 10.1016/j.jmb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Yagi M, Morita T, Aiba H. Cleavage of mRNAs and role of tmRNA system underaminoacidstarvationin Escherichia coli. Mol.Microbiol. 2008;68:462–473. doi: 10.1111/j.1365-2958.2008.06167.x. [DOI] [PubMed] [Google Scholar]
- 45.Collier J, Bohn C, Bouloc P. SsrA tagging of Escherichia coli SecM at its translation arrest sequence. J. Biol. Chem. 2004;279:54193–54201. doi: 10.1074/jbc.M314012200. [DOI] [PubMed] [Google Scholar]
- 46.Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J. Biol. Chem. 2004;279:15368–15375. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- 47.Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR. Genetic identification of nascent peptides that induce ribosome stalling. J. Biol. Chem. 2009;284:34809–34818. doi: 10.1074/jbc.M109.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Sluis EO, Driessen AJ. Stepwise evolution of the Sec machinery in Proteobacteria. Trends Microbiol. 2006;14:105–108. doi: 10.1016/j.tim.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 49.Mitra K, Schaffitzel C, Fabiola F, Chapman MS, Ban N, Frank J. Elongation arrest by SecM via a cascade of ribosomal RNA rearrangements. Mol. Cell. 2006;22:533–543. doi: 10.1016/j.molcel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Becker T, Bhushan S, Jarasch A, Armache J-P, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakatogawa H, Murakami A, Ito K. Control of SecA and SecM translation by protein secretion. Curr. Opin. Microbiol. 2004;7:145–150. doi: 10.1016/j.mib.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Butkus ME, Prundeanu LB, Oliver DB. Translocon “pulling” of nascent SecM controls the duration of its translational pause and secretion-responsive secA regulation. J. Bacteriol. 2003;185:6719–6722. doi: 10.1128/JB.185.22.6719-6722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fulle S, Gohlke H. Statics of the ribosomal exit tunnel: implications for cotranslational peptide folding, elongation regulation, and antibiotics binding. J. Mol. Biol. 2009;387:502–517. doi: 10.1016/j.jmb.2009.01.037. [DOI] [PubMed] [Google Scholar]
- 54.Voelz VA, Petrone P, Pande VS. A multiscale approach to sampling nascent peptide chains in the ribosomal exit tunnel. Pac. Symp. Biocomput. 2009:340–352. doi: 10.1142/9789812836939_0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrone PM, Snow CD, Lucent D, Pande VS. Side-chain recognition and gating in the ribosome exit tunnel. Proc. Natl. Acad. Sci. USA. 2008;105:16549–16554. doi: 10.1073/pnas.0801795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 57.Nakatogawa H, Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol. Cell. 2001;7:185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]