Abstract
We have identified a distal point mutation in streptavidin that causes a 1000-fold reduction in biotin binding affinity without disrupting the equilibrium complex structure. The F130L mutation creates a small cavity occupied by a water molecule, but all neighboring side chain positions are preserved and protein-biotin hydrogen bonds are unperturbed. Molecular dynamics simulations reveal reduced mobility of biotin binding residues but no observable destabilization of protein-ligand interactions. Our combined structural and computational studies suggest that the additional water molecule may affect binding affinity through an electronic polarization effect that impacts the highly cooperative hydrogen-bonding network in the biotin binding pocket.
High resolution crystal structure information is crucial for the prediction and design of protein-protein and protein-ligand interactions, and has been particularly useful in explaining how point mutations or ligand modifications impact ligand recognition and binding affinity. However, there are a growing number of examples of point mutations, often quite far from the ligand binding site, that affect ligand binding affinities even when no structural changes are observed.
Protein residues distant from recognition sites can have a dramatic impact on protein activity through long-range effects on the structure or dynamics of the active site. Long-range effects on active site structure through propagation of conformational changes are well documented in allosteric proteins (1, 2), and similar “dynamically-driven allostery” by propagation of fluctuations has also been observed (3). Long-range effects of distal residues on the electronic properties of active sites are less well characterized, though long-range effects such as electrostatic steering and electrostatic effects on protein-ligand association rates are well known (4, 5).
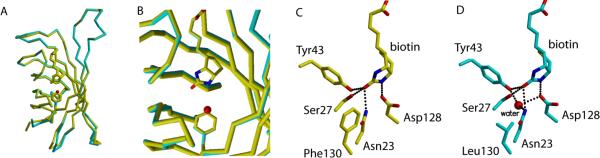
One source of binding free energy in the extremely high affinity (Ka = 1013–1014 M−1) streptavidin-biotin interaction is a highly cooperative hydrogen bond network that polarizes the biotin ureido group and extends into the second contact shell of streptavidin, i.e., the residues next to the first shell of residues in contact with biotin (6–8). Of the five hydrogen bonds to the biotin ureido group, the D128-ureido nitrogen interaction makes one of the largest contributions to binding energy (9) and is the most critical to the cooperative effect (10). Here we describe a mutation in the second contact shell of streptavidin that introduces additional hydrogen bonds to D128 and other biotin-contacting residues and diminishes binding affinity 1000-fold through a large increase in dissociation rate. This mutation, F130L, causes no discernable change to the bound equilibrium structure of the active site (Figure 1A shows the superposition with the WT1-biotin complex, Figure 1B shows details of the binding site, and Figure S1 in the Supporting Information shows a superposition of WT and F130L binding sites), and no destabilizing effect in terms of increased fluctuations of streptavidin-biotin bonds in molecular dynamics simulations.
FIGURE 1.
Bound WT (yellow) and F130L (blue) streptavidin structures. (A) Superposition of the overall structures. (B) Close-up of the superimposed binding pocket and mutation site. The additional water molecule in F130L is shown as a red sphere. (C) Details of the WT streptavidin binding pocket. (D) Details of the F130L binding pocket described in the text.
The crystal structure of the F130L mutant with bound biotin (1.3 Å resolution, Figure 1) reveals that a water molecule occupies the pocket adjacent to L130 which is formed when the larger phenylalanine side chain is removed. However, there are no observable changes in side chain positions or hydrogen bonds in the biotin binding pocket that would explain the large effect on affinity. Moreover, molecular dynamics simulations exhibit reduced mobility of side chains in the binding pocket which appear to increase rather than decrease the structural stability of hydrogen bonds formed with biotin when compared to reference simulations for the WT complex. Our simulations indicate that the additional water molecule forms hydrogen bonds with several key binding pocket residues, including N23, Y43 and D128. While the water molecule does not cause any observable structural perturbations in the streptavidin-biotin hydrogen bonding network, it does reduce fluctuations of the N23 and D128 side chains, apparently stabilizing the hydrogen bonding interactions these residues make with biotin, as compared to simulations results for the WT complex (11).
The overall structure of biotin-bound F130L is very similar to that of biotin-bound WT streptavidin (Figure 1A; a stereoview version of this figure and crystallographic data are included in the Supporting Information). Superimposing the A subunits of the two structures using 98 Cα atoms of the subunit core gives an RMSD value of 0.377 Å; similar values were obtained for other subunit superpositions (Supporting Information). Figure 1B–D depicts an enlarged view of the region of the mutation and binding site. The mutation has no effect on main-chain atom positions for residue 130 (Figure 1B), and the leucine side chain partially fills the space occupied by the aromatic side chain in WT; a water molecule fills the remaining cavity. This water is hydrogen-bonded to ND2 of Asn23, OH of Tyr43, and OD2 of Asp128 (Figure 1D), but these changes do not affect surrounding side chain positions in the binding pocket or hydrogen bonding distances to biotin.
We performed a 500 nanosecond (ns) molecular dynamics simulation to probe the impact of the F130L point mutation on biotin binding site structure and dynamics. Our simulation yields an average structure that compares favorably with the crystal structure. The backbone RMSD fluctuation for all core residues (all residues except those in the three large surface loops) relative to the crystal structure over the final 400 ns of simulation is ~ 0.8 Å, and all core residue side chain positions are maintained relative to the crystal structure. All protein-biotin hydrogen bonds are also well maintained with the exception of the D128-biotin interaction. This hydrogen bond fluctuates more dramatically in WT-biotin simulations (11), but due to the interaction of D128 with the additional water molecule in the F130L mutant, D128 fluctuations are diminished, somewhat increasing the stability of the D128-biotin hydrogen bond.
The additional water molecule in F130L also forms stable hydrogen bonds with biotin-binding residue Y43, and more sporadically with residue N23, during our simulation, and these hydrogen bond contacts dramatically reduce side chain fluctuations for N23 in the mutant relative to WT-biotin simulations. We calculate a residency value of 0.998 for the water molecule, averaged over all four subunits during the trajectory, in good agreement with the experimental value of 1, and an average residence time on the order of tens of nanoseconds. The water molecule occasionally exchanges with bulk solvent via interaction with water molecules that enter the biotin binding site through the streptavidin subunit interface water channel (12). The water molecule exhibits considerable rotational mobility within the small cavity, leading to fluctuations in hydrogen bonding contacts with residues N23, Y43 and D128. The most consistent hydrogen bond is with Y43, based on measurements of hydrogen bond distance and angle as a function of time. Though the reduction of side chain fluctuations relative to WT is largest in N23, this reduced mobility has no statistically measurable impact on N23-biotin interactions. The additional water molecule appears to exert the most dramatic effect on the D128-biotin interaction.
We have performed preliminary quantum mechanical geometry optimization calculations for a simplified binding site model (biotin, the water molecule, and residues N23, S27, Y43, S45, and D128, with amino acid Cα carbons restrained at their relative crystallographic positions to maintain general binding site geometry). Like the MD simulations, these calculations suggest that the water molecule forms favorable hydrogen bonds with the binding site residues, and causes no structural disruption of side chain-biotin interactions. These calculations also suggest that the water molecule interacts most prominently with D128.
The F130L mutation causes a loss of binding affinity of 975 ± 79 relative to WT streptavidin at 37 °C, corresponding to a decrease in the free energy of binding of 4.2 ± 0.1 kcal/mol (Table 1; Figure S2 in the Supporting Information contains competitive binding, calorimetric, and kinetic data). This ΔΔG° is enthalpically driven, with a large 5.5 kcal/mol loss of binding enthalpy, as measured using isothermal titration calorimetry. The unfavorable change in the enthalpy of binding is partially compensated for by a more favorable entropy of binding than WT (TΔΔS°) of 1.3 kcal/mol (calculated from ΔΔG° = ΔΔH° − TΔΔS°). Biotin dissociation for F130L was measured at 0, 3.3, 8 and 12 °C; dissociation was too fast to measure above 12 °C. Activation thermodynamic parameters were calculated by fitting all kinetic data to an Eyring model and using ΔH≠ and ΔS≠ as the only adjustable parameters. An increase in dissociation rate (Δkoff) of 7600 for F130L relative to WT was observed at 12 °C, and a Δkoff of 2700 at 37 °C is predicted based on the fit values of ΔH≠ and ΔS≠, 19.8 kcal/mol and 4.1 cal/mol·K, respectively. All Δ(activation parameter) values are based on previously published values for WT streptavidin (13).
Table 1.
Thermodynamic parameters for F130L versus WT at 37 °C
| Parameter | Value |
|---|---|
| Δ K d | 975 ± 79 |
| ΔΔG° (kcal/mol) | 4.2 ± 0.1 |
| ΔΔH° (kcal/mol) | 5.5 ± 0.2 |
| TΔΔS° (kcal/mol) | 1.3 ± 0.2 |
| Δ k off | 2700 ± 500 |
| ΔΔG≠ (kcal/mol) | 4.9 ± 0.3 |
| ΔΔH≠ (kcal/mol) | 10.6 ± 0.8 |
| TΔΔS≠ (kcal/mol) | 5.7 ± 0.2 |
These results demonstrate a large thermodynamic consequence for a mutation outside the streptavidin-biotin binding pocket with no observable structural or destabilizing dynamical changes in the binding site. In the F130L mutant crystal structure, a water molecule occupies a small pocket that is created when the native phenylalanine residue is replaced with the smaller leucine side chain. While several specific nonbonded contacts are lost around residue 130 when phenylalanine is replaced, several new hydrogen bonds are formed, and there are no other structural changes that would easily explain this large enthalpy loss. The additional water molecule appears to effectively fill the void generated by the phenylalanine to leucine substitution. Our molecular dynamics simulations do suggest that there are local changes in binding pocket side chain fluctuations as a consequence of the point mutation, but the calculations indicate that the side chain fluctuations are reduced relative to simulation results for the WT complex (11). This reduced motion is due to specific hydrogen bonds the additional water molecule makes with binding site residues, especially N23, Y43 and D128. The reduced motion actually stabilizes the hydrogen bonds these residues make with biotin in the simulations, and thus cannot be used to easily rationalize the diminished binding affinity observed experimentally.
The current study was inspired by ongoing crystallographic and calorimetric experiments which demonstrate that some streptavidin point mutations can cause large changes in binding energetics with minimal changes in structure, suggesting that other mechanisms must be responsible. Dynamical regulation of ligand binding has often been invoked in the case of allosterically regulated proteins. However, neither structural changes nor changes in structural fluctuations alone appear to provide a satisfactory explanation for the considerable reduction in biotin binding affinity we measure for the F130L mutant. Instead, our results suggest that the additional water molecule influences the behavior of the cooperative hydrogen bonding network in the biotin binding pocket, likely via a polarization mechanism, and that the impact on residue D128 is particularly pronounced. This hydrogen bonding network makes a major contribution to biotin binding affinity in WT streptavidin (6, 7). We have reported previously that D128 has a significant impact on biotin binding thermodynamics (9) and hydrogen bond cooperativity (10), and DeChancie and Houk have also proposed that this residue is a crucial component of the cooperative hydrogen bonding network and in polarizing the biotin ureido group (6). The importance of polarization-induced stabilization of hydrogen bonds in avidin-biotin binding was also recently described by Tong et al. (14). It is intriguing to note that the decreased binding free energy we measure for the F130L mutant (4.2 kcal/mol at 37 °C) is comparable to the result we obtained previously for the D128A mutation (4.3 kcal/mol at 37 °C) (9). We are currently performing coupled QM/MM calculations and additional experimental studies to characterize the impact of this crucial structural water molecule in F130L in more detail, including comparison with unliganded F130L. Characterization of D128N, a structurally conservative mutation replacing the side chain carboxyl with a neutral amide, will also help illuminate the role of D128 polarization on biotin binding.
The results reported here have important implications for applications of high resolution structural information such as structure-based drug design. We know of no scoring functions typically used in molecular docking studies or empirical free energy binding calculations that could correctly predict the dramatically reduced biotin binding affinity we measure for the F130L streptavidin mutant. Indeed, standard pair-additive molecular mechanics potential functions typically used for protein molecular dynamics simulations cannot provide much insight in this case. Our current results illustrate that localized water molecules may exert a dramatic impact in ligand binding thermodynamics, even when these water molecules are outside the binding pocket and produce no structural perturbations in the active site.
Supplementary Material
ACKNOWLEDGMENT
We thank Richard To for help with mutagenesis.
This work was supported by National Institutes of Health Grant GM080214 (T.P.L.). Portions of this work were performed at the Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland.
Footnotes
SUPPORTING INFORMATION AVAILABLE Detailed experimental procedures, crystallographic data and refinement statistics, stereoview of superimposed WT and F130L streptavidin structures, biophysical data including competitive binding, calorimetry and off-rate data. This material is available free of charge via the Internet at http://pubs.acs.org.
Abbreviations: WT, wild type; RMSD, root mean square deviation
REFERENCES
- 1.Monod J, Wyman J, Changeux J-P. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 2.Perutz M. Cooperativity and allosteric regulation in proteins. Cambridge University Press; Cambridge, UK: 1990. [Google Scholar]
- 3.Popovych N, Sun S, Ebright R, Kalodimos C. Nat. Struct. Mol. Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meltzer R, Thompson E, Soman K, Song X, Ebalunode J, Wensel T, Briggs J, Pedersen S. Biophys. J. 2006;91:1302–1314. doi: 10.1529/biophysj.106.081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myles T, Le Bonniec B, Betz A, Stone S. Biochemistry. 2001;40:4972–4979. doi: 10.1021/bi0023549. [DOI] [PubMed] [Google Scholar]
- 6.DeChancie J, Houk K. J. Am. Chem. Soc. 2007;129:5419–5429. doi: 10.1021/ja066950n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klumb L, Chu V, Stayton PS. Biochemistry. 1998;37:7657–7663. doi: 10.1021/bi9803123. [DOI] [PubMed] [Google Scholar]
- 8.Le Trong I, Freitag S, Klumb L, Chu V, Stayton PS, Stenkamp RE. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1567–1573. doi: 10.1107/s0907444903014562. [DOI] [PubMed] [Google Scholar]
- 9.Freitag S, Chu V, Penzotti J, Klumb L, To R, Hyre D, Le Trong I, Lybrand TP, Stenkamp RE, Stayton PS. Proc. Natl. Acad. Sci. U S A. 1999;96:8384–8389. doi: 10.1073/pnas.96.15.8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyre D, Le Trong I, Merritt E, Eccleston J, Green N, Stenkamp RE, Stayton PS. Protein Sci. 2006;15:459–467. doi: 10.1110/ps.051970306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerutti DS, Le Trong I, Stenkamp RE, Lybrand TP. J. Phys. Chem. B. 2009;113:6971–6985. doi: 10.1021/jp9010372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyre D, Amon L, Penzotti J, Le Trong I, Stenkamp RE, Lybrand TP, Stayton PS. Nat. Struct. Biol. 2002;9:582–585. doi: 10.1038/nsb825. [DOI] [PubMed] [Google Scholar]
- 13.Hyre D, Le Trong I, Freitag S, Stenkamp RE, Stayton PS. Protein Sci. 2000;9:878–885. doi: 10.1110/ps.9.5.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong Y, Mei Y, Li Y, Ji C, Zhang J. J. Am. Chem. Soc. 2010;132:5137–5142. doi: 10.1021/ja909575j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.