Summary
During Bacillus subtilis sporulation, the engulfment checkpoint is thought to directly regulate late fore-spore transcription but to indirectly regulate late mother cell transcription, via the σG-produced pro-tease SpoIVB. We here demonstrate that SpoIIQ is subject to σG-independent, but engulfment-dependent, proteolysis that depends on SpoIVB. Thus, SpoIVB produced before engulfment supports some SpoIVB-dependent events, suggesting that its activity or access to substrates must be regulated by engulfment. Furthermore, a mutation (bofA) that allows σK to be active without σG does not allow σK activity in engulfment mutants, although the pro-σK processing enzyme (SpoIVFB) is localized to the septum in engulfment mutants, suggesting that engulfment comprises a second checkpoint for σK Finally, we find that SpoIIQ and another protein required for σG activity (SpoIIIAH), which directly interact and assemble helical structures around the forespore, recruit the σK-processing enzyme SpoIVFB to the forespore and these structures. We suggest that these foci serve a synapse-like role, allowing engulfment to simultaneously control both σG and σK, and integrating multiple checkpoints and signalling pathways.
Introduction
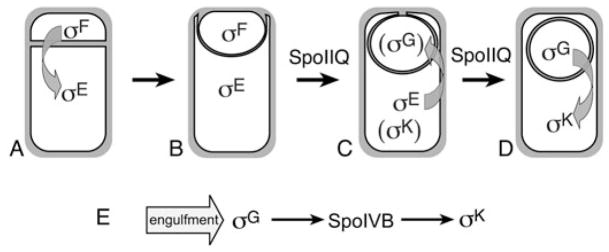
Transcription in the two cells required to make a Bacillus subtilis spore is governed both by intracellular signal transduction cascades, which ensure the coordinated development of the two cells, and by morphological checkpoints, which ensure that landmark morphological events are complete before the onset of transcription that could interfere with these events (reviewed by Rudner and Losick, 2001; Errington, 2003). The first morphological checkpoint of sporulation is polar septation (Fig. 1A), which generates the smaller forespore (the future spore), and the larger mother cell, which lyses to release the mature spore. The second is engulfment, a phagocytosis-like process during which the mother cell membrane migrates around the smaller forespore (Fig. 1B), until the engulfing membrane meets (Fig. 1C) and fuses to release the forespore into the mother cell cytoplasm (Fig. 1D). Engulfment is essential for spore assembly, which occurs within the mother cell cytoplasm. Uncoupling transcription from either checkpoint reduces the number of viable spores produced, and can directly block completion of these key morphological events (Cutting et al., 1990; Coppolecchia et al., 1991; Eichenberger et al., 2001; Fujita and Losick, 2002). For example, immediately after polar septation, a second polar division event commences in the mother cell that if unchecked will give rise to a defective sporangium with two forespores and an anucleate mother cell (Setlow et al., 1991; Lewis et al., 1994; Piggot et al., 1994). The early mother cell transcription factor σE therefore directs the synthesis of proteins that inhibit this division event (Pogliano et al., 1999; Eichenberger et al., 2001); premature synthesis of these proteins inhibits polar septation (Eichenberger et al., 2001). Thus, coupling σE activity to the morphological checkpoint of septation is likely essential for asymmetric division and the generation of daughter cells with distinct developmental cell fates.
Fig. 1.
Engulfment pathway of B. subtilis and cell-specific gene expression.
A. Asymmetric division allows activation of σF in the smaller forespore, followed by activation of sE in the larger mother cell.
B. During engulfment, the mother cell membrane migrates around the forespore, meets (C) and fuses (D) to release the forespore into the mother cell cytoplasm. The forespore protein SpoIIQ is required to synthesize the second forespore transcription factor (σG) and for σG activation, which also requires the completion of engulfment and the mother cell proteins encoded by the spoIIIA operon (arrow in C). The σG factor then produces the secreted signal transduction protein SpoIVB (arrow in D), allowing σK activation.
E. Summary of checkpoints controlling late gene expression: engulfment directly controls σG activity, which produces SpoIVB, allowing σK activity.
The engulfment-regulated activation of the mother cell transcription factor σK occurs by regulated intramembrane proteolysis (RIP), resulting in the removal of a membrane associated N-terminal leader sequence from pro-σK (Zhou and Kroos, 2004). The pro-σK protease, SpoIVFB is a zinc metallopeptidase related to bacterial and eukaryotic proteases involved in RIP (Rudner et al., 1999; Yu and Kroos, 2000), including those involved in bacterial mating (Antiporta and Dunny, 2002), extracellular stress (Ades, 2004; Alba and Gross, 2004), eukaryotic development (Ebinu and Yankner, 2002), sterol biogenesis (Brown and Goldstein, 1997) and the unfolded protein response (Liu and Kaufman, 2003). In these pathways, signal transduction culminates in the cleavage of a protein within a membrane bilayer, often with an initiating cleavage event outside the membrane. These pathways can release signalling peptides or proteins that govern transcription, and in bacteria, the substrate is often an anti-sigma factor whose elimination allows transcription (Brown et al., 2000; Gottesman, 2003). For example, the extracytoplasmic stress response of Escherichia coli responds to elevated levels of unfolded outer membrane proteins, which bind to the PDZ domain of a secreted serine protease (Walsh et al., 2003). This protease then cleaves the extracellular domain of an anti-sigma factor (Walsh et al., 2003), allowing an intramembrane protease to inactivate the anti-sigma factor and release the active sigma factor (Ades et al., 1999). In the case of B. subtilis σK, the hydrophobic leader sequence of pro-σK acts as a covalently attached anti-sigma factor (Zhang et al., 1998), and there is no evidence for an initiating extracellular cleavage event.
One mechanism by which the activation of pro-σK is coupled to engulfment is by its dependence on the prior activation of the late forespore transcription factor σG, whose activity depends on engulfment (Cutting et al., 1990). The σG factor produces a secreted serine protease, SpoIVB, which activates the pro-σK processing machinery (Cutting et al., 1991a), comprised of three mother cell membrane proteins SpoIVFA, SpoIVFB and BofA (Resnekov et al., 1996; Rudner and Losick, 2002). SpoIVFA mediates the septal localization of SpoIVFB and BofA (Rudner and Losick, 2002) while SpoIVFB processes pro-σK (Rudner et al., 1999; Yu and Kroos, 2000; Zhou and Kroos, 2004). BofA is an inhibitor of SpoIVFB protease activity (Resnekov and Losick, 1998; Zhou and Kroos, 2004), whose elimination uncouples σK activity from forespore transcription and from SpoIVB (Cutting et al., 1990; Ricca et al., 1992). One model for the onset of pro-σK processing is that engulfment allows activation of σG, and the consequent production of the SpoIVB protease, which cleaves SpoIVFA in vitro, dissociating BofA from SpoIVFB and allowing processing (Dong and Cutting, 2003). However, SpoIVB is also produced before engulfment, because its expression is also mediated by the early transcription factor σF (Gomez and Cutting, 1996), suggesting that its activity or that of the SpoIVFB intramembrane protease might be regulated by engulfment. Indeed, given the detrimental effect of premature σK activation on sporulation (Cutting et al., 1990), one might expect its activity to be directly coupled both to the completion of engulfment and to σG activity.
A promising candidate for a protein that senses the completion of engulfment is the forespore-expressed membrane protein SpoIIQ, which is conserved in all endospore-forming bacteria (Stragier, 2002) and required for σG synthesis and activation (Londono-Vallejo et al., 1997; Sun et al., 2000). SpoIIQ assembles helical arcs surrounding the forespore, is degraded after the membrane fusion event that is the final step of engulfment (Rubio and Pogliano, 2004), and directly interacts with the mother cell protein SpoIIIAH, which is also involved in σG activation (Blaylock et al., 2004; Doan et al., 2005). We here demonstrate that SpoIIQ is subject to an engulfment-dependent but σG-independent proteolytic event involving at least two cleavage sites, one on each side of its transmembrane domain. Extracellular proteolysis requires the SpoIVB serine protease, suggesting that the activity of this protease or its access to substrates is regulated by engulfment. We also provide preliminary evidence that σK activation is separately regulated by both the completion of engulfment and by σG activity. Finally, we find that SpoIIQ and its mother cell ligand SpoIIIAH direct the localization of the pro-σK processing enzyme, SpoIVFB, to the forespore and mediate its assembly into helical arcs and foci. This finding is in keeping with those of Doan et al. (2005), who noted that localization of the SpoIVFB interacting protein SpoIVFA depended on SpoIIQ and SpoIIIAH. We propose that the coassembly of proteins required for activation of post-engulfment transcription factors in both the forespore and the mother cell allows engulfment to simultaneously govern transcription in both cells.
Results
SpoIIQ is degraded after engulfment
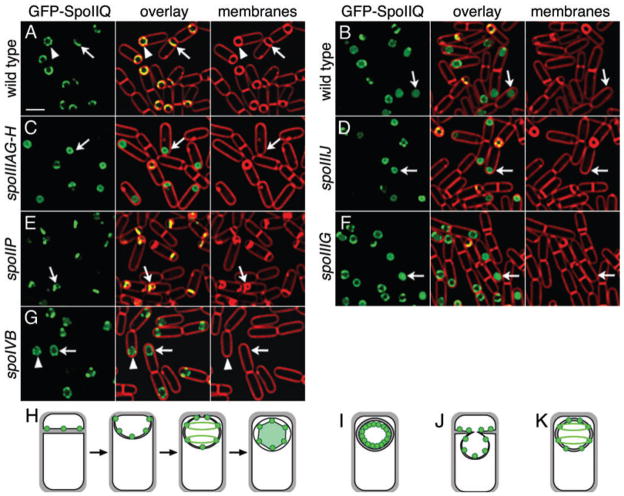
SpoIIQ is a forespore-expressed protein with a single membrane spanning segment and a C-terminal extracellular domain (Londono-Vallejo et al., 1997). We previously noted that GFP-SpoIIQ assembles a complex structure of helical arcs and foci surrounding the forespore (Fig. 2A) and is degraded after engulfment to release soluble GFP, resulting in cytoplasmic fluorescence (Figs 2B and 3B; Rubio and Pogliano, 2004). Immunoblot analysis with antibodies to the C-terminus of SpoIIQ showed that native SpoIIQ was also degraded to release a product corresponding in size to the extracellular C-terminal domain (Fig. 3A). Fractionation analysis demonstrated that while full-length GFP-SpoIIQ and SpoIIQ-His6 was in the insoluble membrane fraction, both the N-terminal (GFP-containing) and C-terminal (His-tagged) products released from the proteins were in the soluble fractions (Fig. 3C). The size and solubility of the degradation products suggest that GFP-SpoIIQ is subject to two proteolytic events, one on each side of the transmembrane domain (Fig. 3D), releasing the extracellular domain from native SpoIIQ and cytoplasmic GFP from the N-terminus of GFP-SpoIIQ.
Fig. 2.
Localization and proteolysis of GFP-SpoIIQ. Live cells expressing either GFP-SpoIIQ (A–G) or MalF-GFP (H) (green), with FM 4–64 stained membranes (red).
A. Wild type (KP845) at t2. Arrow shows GFP-SpoIIQ early in engulfment. Arrowhead, the punctate structure assembled prior to membrane fusion. Bar = 2 μm.
B. GFP-SpoIIQ (KP845) at t3. After membrane fusion (arrow), FM 4–64 is excluded from the forespore and soluble GFP is observed (C) spoIIIAG-H (KP872) at t3. GFP-SpoIIQ remains membrane associated after fusion (arrow).
D. spoIIIJ (KP873)at t3. GFP-SpoIIQ remains membrane associated after fusion (arrow).
E. In spoIIP::tet (KP848) at t3, engulfment is blocked, and the forespore bulges into the mother cell, forming bulges to which GFP-SpoIIQ localizes (arrow).
F. A mutant lacking σG (KP850; spoIIIG::neo) at t3, showing cytoplasmic GFP after fusion (arrow).
G. spoIVB::Tn917Ωmls (KP949) at t3. No cytoplasmic GFP is observed (arrow, arrowhead).
H. SpoIIQ (green circles) localization and degradation (green shading).
I. GFP-SpoIIQ in spoIIIA and spoIIIJ mutants.
J. Cartoon of SpoIIQ in a spoIIP mutant.
K. GFP-SpoIIQ remains punctate and membrane associated after fusion in the spoIVB mutant.
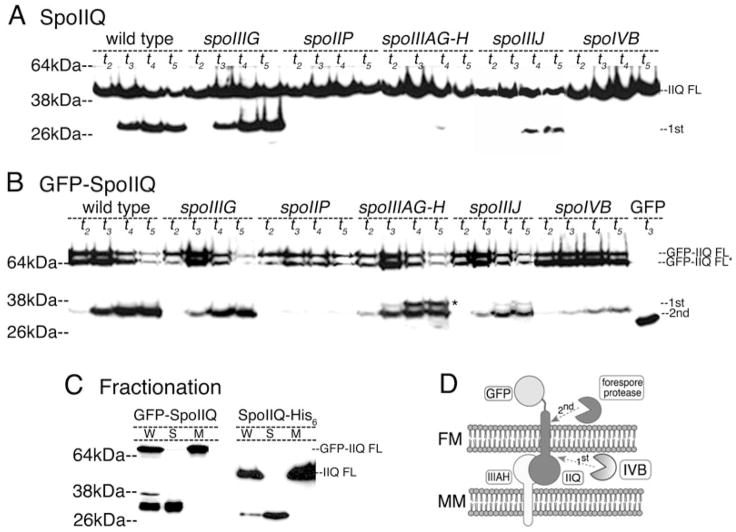
Fig. 3.
Regulated proteolysis of SpoIIQ.
A. Immunoblot and fractionation analysis of GFP-SpoIIQ and native SpoIIQ.
A. Native SpoIIQ in wild type (PY79), spoIIIG (KP85), spoIIP (KP719), spoIIIAG-H (KP896), spoIIIJ (KP901) and spoIVB (KP468). The ~30 kDa product does not accumulate in spoIIP or spoIVB and is reduced in spoIIIAG-H and spoIIIJ mutants.
B. GFP-SpoIIQ in wild type (KP845), spoIIIG (KP850), spoIIP (KP848), spoIIIAG-AH (KP872), spoIIIJ (KP873) and KP949 (spoIVB) and a strain expressing soluble GFP from the spoIIQ promoter (KP835; labelled GFP). Full length GFP-SpoIIQ often migrates as a doublet (~68 kDa, GFP-IIQ FL and GFP-IIQ FL*), although the use of additional protease inhibitors eliminates the smaller product, suggesting proteolysis during sample preparation. By t3, a ~32 kDa N-terminal fragment of GFP-SpoIIQ is produced. spoIIIAG-H and spoIIIJ mutant strains produce the ~32 kDa product and a 38 kDa product (*).
C. Fractionation analysis of GFP-SpoIIQ at t4 (KP845), and SpoIIQ-His6 at t2.5 (KP980). W = whole cells, S = soluble, M = insoluble membrane fraction. Full length GFP-SpoIIQ and SpoIIQ-His6 are in the membrane fraction, while the N-terminal, GFP-containing product of GFP-SpoIIQ and the C-terminal, His6-containing product of SpoIIQ-His6 are soluble.
D. Model for SpoIIQ proteolysis. GFP-SpoIIQ in the forespore membrane (FM) interacts with SpoIIIAH in the mother cell membrane (MM) (Blaylock et al., 2004). Extracellular proteolysis depends on the secreted serine protease SpoIVB (light pac-man), allowing a second cleavage within the cytoplasm by an unknown protease (dark pac-man).
SpoIIQ degradation commenced after the final step of engulfment, membrane fusion (Rubio and Pogliano, 2004), suggesting it was either directly regulated by engulfment or that it required the post-engulfment transcription factors σG or σK We therefore compared the localization and degradation of GFP-SpoIIQ in engulfment mutants and in a strain with a mutation in the gene encoding σG, spoIIIG, which also abolishes activity of the downstream transcription factor σK. GFP-SpoIIQ localized normally in the spoIIIG mutant, with cytoplasmic GFP fluorescence observed after membrane fusion (Fig. 2F, arrow), suggesting that neither σG nor σK directed gene expression is required for SpoIIQ proteolysis. Immunoblot analysis confirmed that in the spoIIIG mutant, both native SpoIIQ and GFP-SpoIIQ were degraded as in wild type (Fig. 3A and B). In the spoIIP mutant, engulfment is blocked at the first step, septal thinning, and the growing forespore pushes into the mother cell, forming a bulge (Frandsen and Stragier, 1995; Abanes-De Mello et al., 2002). In this strain, GFP-SpoIIQ stayed at the septum, sometimes localizing to the bulge, with no obvious release of soluble GFP (Fig. 2E, arrow; J). Immunoblot analysis showed that the spoIIP mutation abolished degradation of both SpoIIQ (Fig. 3A) and GFP-SpoIIQ (Fig. 3B). Thus, engulfment is a morphological checkpoint for SpoIIQ proteolysis.
Altered SpoIIQ proteolysis in spoIIIA and spoIIIJ mutants
A key event that also depends on engulfment is activation of σG, which requires SpoIIQ (Sun et al., 2000), SpoIIIJ (an Oxa1p/YidC homologue) (Errington et al., 1992; Yen et al., 2001; Serrano et al., 2003) and the eight mother cell proteins encoded by the spoIIIA operon (Kellner et al., 1996), one of which (SpoIIIAH) directly interacts with SpoIIQ (Blaylock et al., 2004). We were interested in determining if these proteins were involved in SpoIIQ proteoysis and therefore assessed GFP-SpoIIQ localization and degradation in a spoIIIJ mutant and a mutant in which the last two genes in the spoIIIA operon were deleted, spoIIIAG and spoIIIAH (hereafter designated spoIIIAG-H). In both cases GFP-SpoIIQ localized to the septum, tracked the engulfing membrane and assembled foci early in sporulation, but remained membrane-bound even late in sporulation, with no GFP fluorescence in the forespore cytoplasm (Fig. 2C and D, arrows; I). Immunoblot analysis demonstrated an altered GFP-SpoIIQ degradation pattern in the spoIIIA and spoIIIJ mutants (Fig. 3B), with the accumulation of a product (Fig. 3B, asterisk) less abundant in wild type. Surprisingly, although fluorescence microscopy showed little cytoplasmic GFP fluorescence in spoIIIA and spoIIIJ mutants (Fig. 2C and D, arrows), a significant amount of soluble GFP-SpoIIQ degradation product was detected (Fig. 3B). This discrepancy might be due to cleavage of the insoluble product during sample preparation.
The absence of SpoIIIJ or SpoIIIAGH caused a more notable difference in the degradation pattern of native SpoIIQ. In the spoIIIAG-H mutant, the soluble extracellular domain was almost undetectable, although the level of full-length SpoIIQ decreased during sporulation (Fig. 3A). A somewhat weaker, but similar effect was seen in the spoIIIJ null mutant, where a reduced amount of the soluble extracellular domain was visualized (Fig. 3A). It is therefore possible that SpoIIIJ and SpoIIIAGH are required for both proteolysis events, and that the GFP-SpoIIQ fusion protein alters the proteolytic pathway (although providing a useful tag for cytoplasmic proteolysis). Alternatively, it is possible that the interaction between SpoIIQ and SpoIIIAH (Blaylock et al., 2004) stabilizes the SpoIIQ extracellular domain, so that in the absence of SpoIIIAH, this domain is subject to further degradation and undetectable.
In summary, after engulfment SpoIIQ is subject to at least two proteolysis events, one outside the forespore membrane that releases the extracellular domain (extracellular proteolysis) and a second inside the forespore membrane that releases GFP into the cytoplasm (intracellular proteolysis). Two proteins required for σG activation are required for either proteolysis or for the accumulation of proteolytic products, SpoIIIJ and SpoIIIAGH, suggesting that SpoIIQ proteolysis might play a role in activating σG after engulfment.
SpoIVB serine protease is required for extracellular proteolysis of SpoIIQ
While examining the lab collection of spoIIIA and spoIIIJ mutant strains, we found that one ‘spoIIIA’ Tn917 insertion mutation completely inhibited GFP-SpoIIQ degradation in immunoblots, unlike the other spoIIIA mutations. The insertion site was identified using random primed polymerase chain reaction (PCR) and DNA sequencing, and we found that the transposon was actually within the spoIVB coding region. Indeed, a spoIVB deletion mutation had a phenotype identical to that of the spoIVB::Tn917 mutation, strongly inhibiting extracellular proteolysis of native SpoIIQ (Fig. 3A) and intracellular proteolysis of GFP-SpoIIQ (Fig. 3B), with no decrease in the amount of full length protein. Fluorescence microscopy demonstrated that GFP-SpoIIQ localized normally in the spoIVB mutant, but no cytoplasmic GFP fluorescence was observed after membrane fusion (Fig. 2G, arrow, K). Thus, SpoIIQ degradation depends, directly or indirectly, on the forespore protein SpoIVB, a serine protease required for σK activation in the mother cell. The extracellular location of SpoIVB suggests that it is required for the extracellular proteolysis of SpoIIQ; the striking effect that the spoIVB null mutation also has on the intracellular proteolysis event could indicate that either SpoIVB is involved in both proteolysis events, or that intracellular proteolysis normally occurs only after extracellular proteolysis.
SpoIVB is a secreted signalling molecule that triggers σK activation by interacting with the σK processing complex in the mother cell membrane (Cutting et al., 1991a; Wakeley et al., 2000). SpoIVB cleaves SpoIVFA (Dong and Cutting, 2003), which has an extracellular domain with sequence similarity to that of SpoIIQ; it is possible that cleavage of SpoIIQ represents the second essential function of SpoIVB identified by genetic studies (Oke et al., 1997). SpoIVFA is a component of the σK processing machinery and is required to localize the pro-σK protease SpoIVFB to the septum (Rudner and Losick, 2002). It is generally reported that σK activation is indirectly coupled to engulfment by its dependence on the production of SpoIVB by σG, whose transcriptional activity is regulated by engulfment by an unknown mechanism (Fig. 1E). However, SpoIVB is initially expressed by the early forespore transcription factor σF, although it is produced in higher levels after the engulfment-dependent activation of σG (Gomez and Cutting, 1996). Our results demonstrate that the initial, σF-dependent production of SpoIVB in the spoIIIG mutant is sufficient to mediate degradation of SpoIIQ at rates identical to wild type, which together with our observation that engulfment defective mutants block SpoIIQ degradation, suggests that SpoIVB activity might be regulated by engulfment.
A σG -independent role for SpoIIQ, SpoIIIA and engulfment proteins in σK activation
SpoIVB-dependent proteolysis of SpoIIQ occurred in the absence of σG activity, so we were interested in testing the possibility that engulfment and σG activity comprised two independent checkpoints for σK activation, and that SpoIIQ might be required both for σG and σK activity. To assess the possibility that SpoIIQ has a separate role in σK activation, we used a bofA mutant, in which σK is activated in the absence of σG activity and SpoIVB-mediated signalling (Cutting et al., 1990, 1991b; Ricca et al., 1992). If activating σG is the only role that SpoIIQ plays in σK activation, then the bofA mutation should completely rescue σK activity in the spoIIQ mutant. Surprisingly, in the spoIIQ bofA double mutant, no σK activity was detected (Fig. 4B, filled circle), suggesting that SpoIIQ has independent roles in the activation of σG and σK.
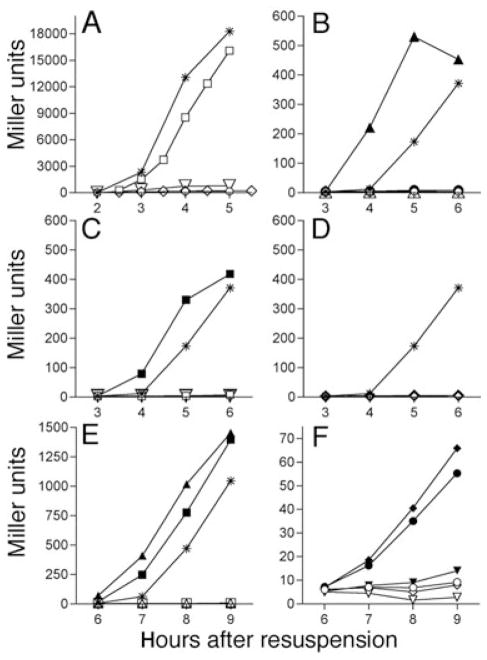
Fig. 4.
Effect of various spo mutations on σG and σK activity. Strains were induced to sporulate via resuspension with samples removed and β-galactosidase activity measured at hourly intervals.
A. Activity of σG-dependent sspB-lacZ at 37°C. Wild type (*; KP6); spoIIQ (○ KP950); spoIVB (□; KP957); spoIIIAG-H (◇; KP952); spoIIP (▽; KP951).
B–D. Activity of the σK-dependent cotD–lacZ fusion at 37°C.
B. Wild type (*; KP946), spoIIIG (△; KP947), spoIIIG bofA (▲; KP948), spoIIQ (○; KP953), spoIIQ bofA (●; KP955).
C. Wild type (*; KP946), spoIIP (▽; KP958), spoIIP bofA (▼; KP959), spoIVB (□; KP954); spoIVB bofA (■; KP956).
D. Wild type (*; KP946), spoIIIAG-H (◇; KP962); spoIIIAG-H bofA (◆; KP963), spoIVB (□; KP954); spoIVB bofA (■; KP956).
E–F. Activity of the σK-dependent cotD–lacZ fusion at 30°C.
E. Wild type (*; KP946), spoIIIG (△; KP947), spoIIIG bofA (▲; KP948), spoIVB (□; KP954); spoIVB bofA (■; KP956).
F. spoIIQ (○; KP953), spoIIQ bofA (●; KP955), spoIIP (▽; KP958), spoIIP bofA (▼; KP959), spoIIIAA-H (◇; KP960); spoIIIAA-H bofA (◆; KP961). Identical results were obtained with other engulfment mutants (spoIID or spoIIM).
We next assayed σG and σK activity in engulfment mutants (spoIID, spoIIM and spoIIP) and in spoIIIA mutants. Similar to the spoIIQ mutant, no σG activity (Fig. 4A, inverted triangles and diamonds) consequently, no σK activity was detected in these mutants (Fig. 4C and D, inverted triangles and diamonds) and the bofA mutation failed to rescue σK activity (Fig. 4C and D, filled inverted triangles, diamonds). Thus, at 37°C, the bofA mutation fails to rescue engulfment or spoIIQ or spoIIIA mutants, although the engulfment and spoIIIA mutants both produce pro-σK (Lu et al., 1990), and spoIIQ mutants have normal σE-directed gene expression (Sun et al., 2000). We next collected samples from cultures at 30°C, a more permissive temperature for many mutants, and performed β-galactosidase assays. Consistent with the results from 37°C experiments, spoIIIG and spoIVB mutants were bypassed by the bofA mutant, producing high levels of σK activity, while the engulfment mutants were not bypassed (Fig. 4E, filled triangle and square; Fig. 4F, filled inverted triangle). However, spoIIQ bofA and spoIIIA bofA double mutants showed small but reproducible increases in σK activity to ~5% of wild type levels (Fig. 4F, filled circle and filled diamond), perhaps explaining why previous publications observed that bofA could suppress the requirement for spoIIIA (Cutting et al., 1990). Because the bofA mutation completely bypasses the σG checkpoint for σK activation at both 37°C and 30°C, we conclude that SpoIIQ and SpoIIIA are required for both σG and σK activation and we speculate that engulfment might serve as a separate checkpoint for σK activation.
SpoIVFB localization requires SpoIIQ, SpoIIIA and engulfment
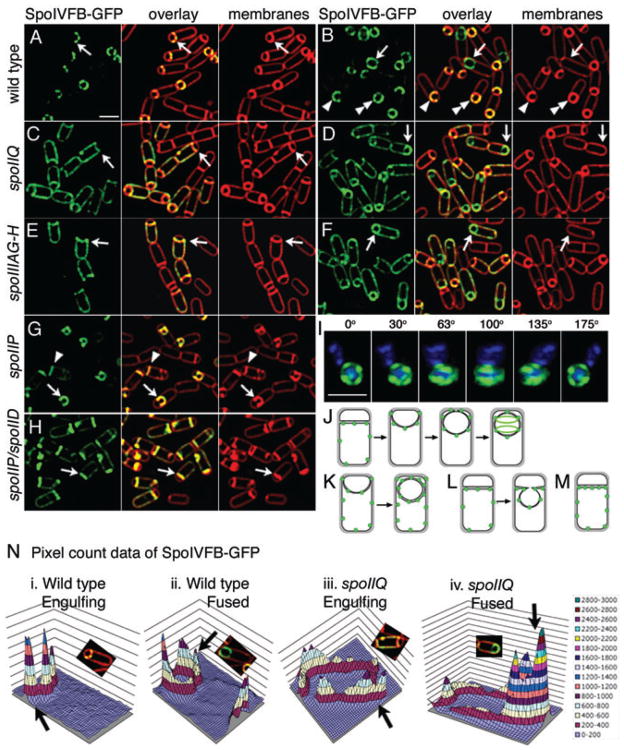
A simple mechanism by which both SpoIIQ and SpoIIIA could be involved in σK activation is that the SpoIIQ-SpoIIIAH zipper (Blaylock et al., 2004) might recruit the σK processing complex (SpoIVFA, SpoIVFB and BofA) to the sporulation septum. We tested this hypothesis by investigating the subcellular distribution of SpoIVFB-GFP in wild type and in spoIIQ and spoIIIA mutants. Early in engulfment, SpoIVFB-GFP localized to the septum, with discrete foci at the septum and very faint fluorescence in the mother cell cytoplasmic membrane (Fig. 5A, arrow), tracking the engulfing membrane (Fig. 5B, arrowhead) and assembling foci around the forespore before and after fusion (Fig. 5B, double arrowhead and arrow respectively; Fig. 5J). After three-dimensional reconstuction, SpoIVFB-GFP appeared as helical arcs surrounding the forespore (Fig. 5I). The localization and structure formed by SpoIVFB-GFP is very similar to that of GFP-SpoIIQ and SpoIIIAH-flag, except GFP-SpoIIQ is degraded while the others persist as foci around the forespore (Blaylock et al., 2004; Rubio and Pogliano, 2004).
Fig. 5.
Localization of mother cell-expressed SpoIVFB-GFP (green). Live cells expressing SpoIVFB-GFP (green) at t2 (A, C, E) or t3 (B, D, F, G, H). FM 4–64 stained membranes are red.
A. Wild type (KP969) at t2. Arrow shows punctate localization early in engulfment. Bar = 2 μm.
B. Wild type (KP969) at t3. Sporangia before (arrowhead, double arrowhead) and after (arrow) membrane fusion.
C. spoIIQ (KP970) at t2. SpoIVFB-GFP is randomly distributed in mother cell membrane (arrow).
D. spoIIQ (KP970) at t3. SpoIVFB-GFP enriched at the forespore after fusion (arrow).
E. spoIIIAG-H (KP971) at t2. SpoIVFB-GFP randomly distributed (arrow).
F. spoIIIAG-H (KP971) at t3. SpoIVFB-GFP enriched at the forespore after fusion (arrow).
G. spoIIP (KP972) is engulfment defective. SpoIVFB localizes to flat septa (arrowhead) and bulges (arrow).
H. spoIIP spoIID double mutant (KP973).
I. Three dimensional reconstruction of SpoIVFB-GFP from 12 focal planes (z = 0.15 μm); rotated around the Y-axis. DAPI-stained DNA (blue). Bar = 2 μm.
J. SpoIVFB-GFP (green) localization during engulfment.
K. SpoIVFB-GFP (green) in the absence of either SpoIIQ or SpoIIIA.
L. SpoIVFB-GFP (green) in the absence of either SpoIIP, M or D or (M) both SpoIIP and SpoIID.
N. Quantification of SpoIVFB-GFP fluorescence intensity in wild type or spoIIQ. Deconvolved pixel count data from cells indicated by arrows in panels A (i), B (ii), C (iii) and D (iv) were imported into Microsoft Excel and three-dimensional graphs created. Scale on right.
Similar effects were noted in both the spoIIQ and spoIIIAG-H mutants. In spoIIQ mutant sporangia early in engulfment, SpoIVFB-GFP showed a somewhat patchy distribution throughout the mother cell membrane (Fig. 5C, arrow) with a twofold enrichment at the leading edge of the engulfing membrane, which contains two layers of the mother cell membrane (Fig. 1). However, after membrane fusion, SpoIVFB-GFP fluorescence was more intense in the outer forespore membrane than in the cytoplasmic membrane (Fig. 5D, arrow). A similar result was obtained in the spoIIIAG-H mutant, with little septal localization of SpoIVFB-GFP during engulfment (Fig. 5E, arrow), but significant localization to the outer forespore membrane after membrane fusion (Fig. 5F, arrow). To quantitatively view the cytological data, we exported the deconvolved but unadjusted pixel data (photons/pixel) of select cells from SoftWoRx to Excel and visualized the pixel intensities as three-dimensional graphs (Fig. 5N). In wild type before and after fusion, little SpoIVFB-GFP fluorescence was visible in the mother cell cytoplasmic membrane, and peaks corresponding to foci were seen at the forespore. However, in spoIIQ before membrane fusion, SpoIVFB-GFP was uniformly distributed in the cytoplasmic and septal membrane domains. After fusion, the GFP intensity in the mother cell cytoplasmic membrane remained the same, but that in the outer forespore membrane increased about sevenfold (Fig. 5N), suggesting a continued accumulation of SpoIVFB-GFP in the outer forespore membrane after fusion. The requirement for SpoIIQ and SpoIIIAGH for localization of the pro-σK processing enzyme SpoIVFB to the forespore during engulfment could explain why they are required for σK activation.
Engulfment mutants showed only a slight increase in non-localized SpoIVFB in sporangia with flat septa (Fig. 5G, arrowhead), and in sporangia with a bulge, SpoIVFB clearly localized to the bulge, which is peptidoglycan-free (Fig. 5G, arrow). The spoIID spoIIP double mutant blocked bulge formation (Pogliano et al., 1999) and had a somewhat more severe defect in SpoIVFB-GFP localization (Fig. 5H, arrow). Thus, SpoIVFB appears to localize to the septum of engulfment mutants better than does SpoIIIAH, whose localization is strongly inhibited by the absence of septal thinning (Blaylock et al., 2004). This suggests the existence of an alternate mechanism for SpoIVFB localization, as is also supported by the SpoIIQ and SpoIIIAG-H independent recruitment of SpoIVFB to the outer forespore membrane after membrane fusion.
While this manuscript was in preparation, Doan et al. (2005) made similar observations regarding the dependence of the SpoIVFB interacting protein SpoIVFA to the septum, which they found to be dependent on SpoIIQ, SpoIIIAH and engulfment proteins. However, while we found that SpoIVFB (Fig. 5C) and SpoIIIAH (Blaylock et al., 2004) are completely dependent on SpoIIQ for septal localization during engulfment (Fig. 5N, iii), Doan et al. (2005) found substantial localization of SpoIVFA in the absence of either SpoIIQ or SpoIIIAH. These differences might be due to either the localization of different proteins (SpoIVFA vs. SpoIVFB) or to subtle differences in the ways the various fusion proteins were constructed.
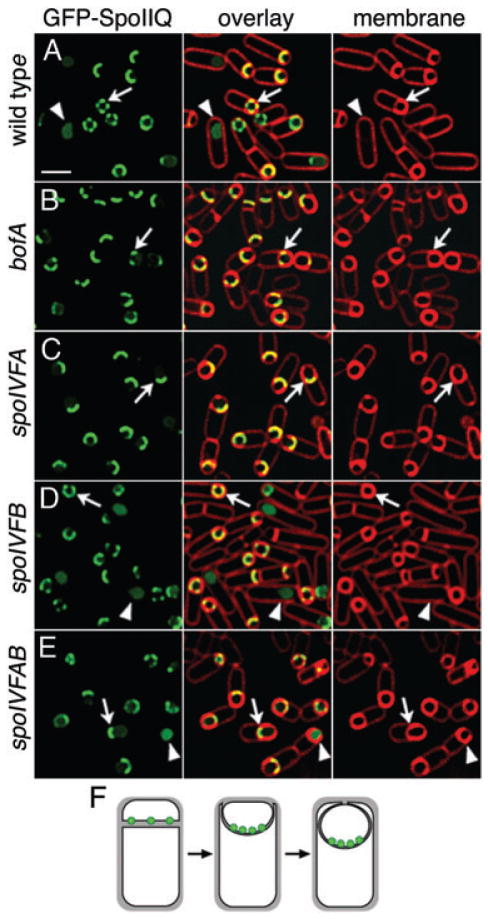
GFP-SpoIIQ has a tracking defect in bofA and spoIVFA mutants
GFP-SpoIIQ tracks the engulfing mother cell membrane during engulfment, moving around the forespore together with the engulfing mother cell membrane. In the absence of mother cell gene expression, GFP-SpoIIQ moves around the forespore ahead of the engulfing membrane (Rubio and Pogliano, 2004), suggesting that an interaction between SpoIIQ and a mother cell membrane protein tethers SpoIIQ to the septum. We were interested in determining if the σK processing machinery (SpoIVFA, SpoIVFB or BofA) could be the SpoIIQ tether or involved in SpoIIQ degradation. We therefore investigated the localization of GFP-SpoIIQ in bofA, spoIVFA, spoIVFAB and spoIVFB mutants. In each of these mutants, GFP-SpoIIQ initially localized to the septum, with discrete foci similar to wild type, so neither BofA, SpoIVFA nor SpoIVFB are required for SpoIIQ tethering. However, in bofA, spoIVFA and spoIVFAB mutants, GFP-SpoIIQ remained at the septum, failing to track the engulfing mother cell membrane around the forespore (Fig. 6B, C and E, arrows; Fig. 6F). Surprisingly, the bofA, spoIVFA and spoIVFAB sporangia failed to complete engulfment, remaining unfused (Fig. 6B, C and E, arrows). This unanticipated engulfment defect was observed in bofA, spoIVFA and spoIVFAB mutant strains expressing GFP-SpoIIQ (data not shown), but not in strains lacking GFP-SpoIIQ, suggesting a synergistic effect between GFP-SpoIIQ and the absence of either SpoIVFA or BofA. In these mutants, soluble GFP was observed even in sporangia that had not completed engulfment (Fig. 6E, arrowhead), demonstrating that the absence of these proteins partially uncouples SpoIIQ proteolysis from engulfment. In the mutant missing only spoIVFB, GFP-SpoIIQ localized normally and was degraded to release soluble GFP after engulfment (Fig. 6D, arrow and arrowhead). In summary, BofA and SpoIVFA but not SpoIVFB are required to allow GFP-SpoIIQ to track the engulfing mother cell membrane, but they are not required for SpoIIQ tethering or proteolysis.
Fig. 6.
GFP-SpoIIQ tracking defect in spoIVF and bofA mutants. GFP-SpoIIQ (green) at t3 with FM 4–64 stained membranes (red).
A. Wild type (KP845). Cells before fusion show punctate localization (arrow), while after fusion, soluble GFP (arrowhead).
B. bofA (KP964).
C. spoIVFA (KP966).
D. spoIVFB (KP976).
E. spoIVFAB (KP965). In the absence of BofA or SpoIVFA, GFP-SpoIIQ fails to track the engulfing mother cell membrane around the forespore, remaining at the septum (arrows in B, C, E or F). In the absence of only SpoIVFB, GFP-SpoIIQ localizes normally (arrow and arrowhead in D). Scale, 2 μm.
F. Localization of GFP-SpoIIQ in the absence of BofA or SpoIVFA.
Discussion
We here demonstrate that the forespore membrane protein SpoIIQ is subject to a proteolysis event regulated by the phagocytosis-like process of engulfment. Complete SpoIIQ proteolysis requires SpoIVB, a protease secreted from the forespore, SpoIIIAH, a mother cell membrane protein with which SpoIIQ directly interacts (Blaylock et al., 2004), and SpoIIIJ, a homologue of the YidC translocase subunit. SpoIIQ proteolysis involves at least two proteolysis events, one on each side its transmembrane domain. Our results suggest that extracellular proteolysis requires the SpoIVB serine protease, which is also essential for the engulfment-dependent activation of the mother cell transcription factor σK Although it remains possible that SpoIVB activates a second extracellular protease that cleaves SpoIIQ, we favour the hypothesis that it directly cleaves SpoIIQ, which is in the same protein family as the only other SpoIVB substrate described to date, SpoIVFA (Dong and Cutting, 2003).
While the role of SpoIIQ proteolysis remains unclear, our studies impact our understanding of the regulated RIP event that activates the mother cell transcription factor σK by removing the inhibitory leader sequence from pro-σK (Rudner et al., 1999; Yu and Kroos, 2000). Proteolytic activation of pro-σK by the SpoIVFB protease depends on SpoIVB, which is produced in high levels after engulfment by the late forespore transcription factor σG(Cutting et al., 1991a). Thus, σK activity is considered to be indirectly coupled to engulfment by its dependence on σG for production of the SpoIVB signal transduction protein (Fig. 1E). However, SpoIVB is also produced before engulfment by σF (Gomez and Cutting, 1996) so the simple view that activation of σG produces, for the first time, this key signal transduction protein, is unlikely to be correct. Indeed two of our results lead us to speculate that σK activation might be directly regulated by engulfment. First, SpoIVB-dependent proteolysis of SpoIIQ does not depend on σG, demonstrating that the SpoIVB protein produced by σF before engulfment is sufficient to support certain SpoIVB-dependent events (Fig. 7A). Further, SpoIIQ degradation commences after engulfment and is blocked by engulfment defective mutants, suggesting that SpoIVB activity or access to its substrates might be regulated by engulfment. Second, the bofA mutation, which completely bypasses the requirement for σG activity and for SpoIVB to activate σK, does not bypass the requirement for engulfment to activate σK Thus, while engulfment-defective mutants (spoIID, spoIIM and spoIIP) lack σG activity, this is unlikely to be the only reason they lack σK activity. We therefore speculate that the ability of the SpoIVFB intramembrane protease to activate σK is separately coupled to both the completion of engulfment and σG activity (Fig. 7C). The efficiency of SpoIVFB-mediated pro-σK processing in E. coli cells expressing SpoIVFB and pro-σK (Zhou and Kroos, 2004) suggests that the engulfment checkpoint might be governed by an inhibitor of σK processing, rather than an activator.
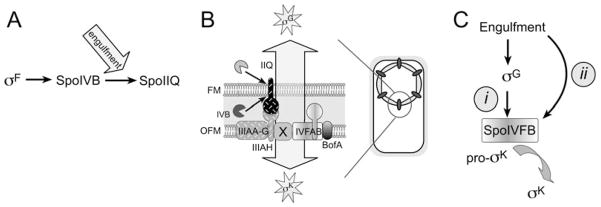
Fig. 7.
Summary and model of engulfment regulated events.
A. SpoIIQ degradation depends on the SpoIVB serine protease produced before engulfment by σF, but commences after engulfment, which must regulate either protease activity or access to its substrate.
B. The forespore protein SpoIIQ recruits the mother cell proteins SpoIIIAH and SpoIVFB to the outer forespore membrane (OFM) thereby assembling a protein complex containing proteins required for the engulfment-dependent activation of transcription in both cells (arrow). We propose that this synapse-like complex serves as a dedicated location for cell-cell communication.
C. We speculate that σK activity is governed by two checkpoints, the previously described σG checkpoint (i), in which the production of high levels of SpoIVB by σG relieves BofA-mediated inhibition of SpoIVFB and indirectly couples σK to engulfment, and a separate checkpoint (ii) that directly couples σK to engulfment, which could be comprised of a second inhibitory pathway or an activator.
Our results further emphasize the importance that the SpoIIQ membrane protein plays directing mother cell membrane proteins to the sporulation septum. We previously demonstrated that the extracellular domain of SpoIIQ directly interacts with that of the mother cell protein SpoIIIAH within the septal space, thereby tethering SpoIIIAH to the sporulation septum (Blaylock et al., 2004). Doan et al. (2005) also made similar findings regarding the interaction of SpoIIQ with SpoIIIAH and observed that the absence of these proteins reduced, but did not abolish, septal localization of SpoIVFA (which forms a complex with SpoIVFB). We here demonstrate that SpoIIQ/SpoIIIAH are essential for the localization of the pro-σK protease, SpoIVFB to the sporulation septum during engulfment, as in the absence of this complex SpoIVFB-GFP is randomly distributed within the mother cell membrane. However, after membrane fusion, SpoIVFB-GFP shows improved localization in the absence of SpoIIIAH or SpoIIQ, suggesting that after fusion, a SpoIIQ and SpoIIIAH independent mechanism recruits SpoIVFB to the outer forespore membrane. This proposal would require SpoIVFB to be directly inserted into this location, as membrane fusion generates a mother cell with two separate membrane systems, the mother cell cytoplasmic membrane and the outer forespore membrane. Thus, SpoIIQ recruits the SpoIIIAH membrane protein to the sporulation septum by virtue of a direct interaction between their extracellular domains, and SpoIIIAH in turn recruits SpoIVFA and SpoIVFB, perhaps via another protein since thus far, no interaction has been demonstrated between SpoIIIAH and either SpoIVFB (our data not shown) or SpoIVFA (Doan et al., 2005).
The SpoIIQ-SpoIIIAH-SpoIVFB localization pathway involves key proteins required for activation of engulfment-dependent transcription factors in both the forespore and the mother cell (Fig. 7B). We propose that the coassembly of these proteins into a single protein complex allows the coordinated regulation of both σG and σK activity by engulfment. Remarkably, each of these signal transduction proteins assemble foci in what appears to be a roughly icosahedral arrangement around the forespore (Blaylock et al., 2004; Rubio and Pogliano, 2004; Fig. 2A, arrowhead, Fig. 5B, double arrowhead), suggesting that these forespore and mother cell membrane proteins interact only at discrete contact sites (Fig. 7B). The restriction of membrane-localized signal transduction proteins to discrete points of contact between two cells also occurs during the assembly of neurological and immunological synapses, which are thought to allow prolonged signalling between the cells and the integration of multiple signalling pathways and checkpoints (Davis and Dustin, 2004; Jacobelli et al., 2004). It is tempting to speculate that the SpoIIQ/SpoIIIAH/SpoIVFB complex plays a similar role during B. subtilis development.
Experimental procedures
Bacterial strains
Bacillus subtilis strains (Table 1) are PY79 derivatives (Youngman et al., 1984), constructed by transformation (Dubnau and Davidoff-Abelson, 1971). EZ::TN(kan) insertions were isolated as described (Blaylock et al., 2004). Sporulation was induced by resuspension at 30°C or 37°C with tn representing the hours after the onset of sporulation (Sterlini and Mandelstam, 1969). Plasmids were constructed by PCR (using primers listed below) in E. coli DH5α selecting ampicillin resistance (100 μg ml−1) and sequenced by the Shared Source UCSD Cancer Center (funded in part by NCI Cancer Center Support Grant #2 P30 CA23100-18). Sporulation efficiency was determined as described (Rubio and Pogliano, 2004).
Table 1.
Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| PY79 | Wild type | Youngman et al. (1984) |
| KP6 | sspB-lacZΩcat | Sun et al. (1991a) |
| KP85 | spoIIIGΔ1 | Sun et al. (1991b) |
| KP265 | spoIVFB152, Tn917ΩHU144 | Cutting et al. (1991b) |
| KP270 | spoIVFAΔ91 | Cutting et al. (1991b) |
| KP468 | ΔspoIVB::spc | Oke et al. (1997) |
| KP575 | ΔspoIIQ::spc | Sun et al. (2000) |
| KP719 | ΔspoIIP::tet | Frandsen and Stragier (1995) |
| KP835 | amyE::PspoIIQ-gfpΩcat | Rubio and Pogliano (2004) |
| KP845 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat | Rubio and Pogliano (2004) |
| KP848 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat ΔspoIIP::tet | Rubio and Pogliano (2004) |
| KP850 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat ΔspoIIIG::neo | Rubio and Pogliano (2004) |
| KP872 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat ΔspoIIIAG-H::kan | Blaylock et al. (2004) |
| KP873 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat ΔspoIIIJ-jag::cat::tet | Blaylock et al. (2004) |
| KP896 | ΔspoIIIAG-H::kan | Blaylock et al. (2004) |
| KP901 | ΔspoIIIJ-jag::cat::tet | Blaylock et al. (2004) |
| KP946 | thrC::cotD-lacZΩmls | Sharp and Pogliano (1999) |
| KP947 | thrC::cotD-lacZΩmls spoIIIGΔ1 | This study |
| KP948 | thrC::cotD-lacZΩmls spoIIIGΔ1 ΔbofA::cat | This study |
| KP949 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat spoIVB::Tn917Ωmls | This study |
| KP950 | sspB-lacZΩcat ΔspoIIQ::spc | This study |
| KP951 | sspB-lacZΩcat ΔspoIIP::tet | This study |
| KP952 | sspB-lacZΩcat ΔspoIIIAG-H::kan | This study |
| KP953 | thrC::cotD-lacZΩmls ΔspoIIQ::spc | This study |
| KP954 | thrC::cotD-lacZΩmls ΔspoIVB::spc | This study |
| KP955 | thrC::cotD-lacZΩmls ΔspoIIQ::spc ΔbofA::cat | This study |
| KP956 | thrC::cotD-lacZΩmls ΔspoIVB::spc ΔbofA::cat | This study |
| KP957 | sspB-lacZΩcat spoIVB::Tn917 | This study |
| KP958 | thrC::cotD-lacZΩmls ΔspoIIP::tet | This study |
| KP959 | thrC::cotD-lacZΩmls ΔspoIIP::tet ΔbofA::cat | This study |
| KP960 | thrC::cotD-lacZΩmls ΔspoIIIAA-H::kan | This study |
| KP961 | thrC::cotD-lacZΩmls ΔspoIIIAA-H::kan ΔbofA::cat | This study |
| KP962 | thrC::cotD-lacZΩmls ΔspoIIIAG-H::kan | This study |
| KP963 | thrC::cotD-lacZΩmls ΔspoIIIAG-H::kan ΔbofA::cat | This study |
| KP964 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat bofA::erm | This study |
| KP965 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat ΔspoIVFAB::cat::tet | This study |
| KP966 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat spoIVFAΔ91 | This study |
| KP967 | ΔbofA::cat | Ricca et al. (1992) |
| KP969 | spoIVFB-gfpΩcat | This study |
| KP970 | spoIVFB-gfpΩcat ΔspoIIQ::spc | This study |
| KP971 | spoIVFB-gfpΩcat ΔspoIIIAA-H::kan | This study |
| KP972 | spoIVFB-gfpΩcat ΔspoIIP::tet | This study |
| KP973 | spoIVFB-gfpΩcat ΔspoIIP::tet spoIID::Tn917ΩHU298 | This study |
| KP975 | spoIVFB::pAR98(kan) | This study |
| KP976 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat spoIVFB::pAR98(kan) | This study |
| KP978 | ΔspoIIQ::spc amyE::PspoIIQgfp-spoIIQΩcat spoIVFB::pAR98(kan) ΔbofA::cat::tet | This study |
| KP979 | ΔspoIIP::tet amyE::PspoIIDgfp-spoIIPΩcat ΔspoIIQ::spc ΔbofA::erm | This study |
| KP980 | ΔspoIIQ::spc, amyE::PspoIIQhis6-spoIIQΩcat | This study |
| AR391 | BL21 (λDE3)/pAR92 | This study |
Microscopy and image analysis
Live cells were stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (0.2 μg ml−1; Molecular Probes) and Mitotracker Red (0.1 μg ml−1; Molecular Probes) or FM 4–64 (5 μg ml−1; Molecular Probes) (Pogliano et al., 1999; Sharp and Pogliano, 1999). Images were collected with a Spectris optical sectioning microscope with 15 iterations of the Delta Vision constrained-iterative deconvolution program (Applied Precision, Issaquah, WA). The three-dimensional model of SpoIVFB was constructed using 12 sections (z = 0.15 μm) and the volume builder function of SoftWoRx (v. 3.3).
SpoIIQ antibody production, membrane fractionation and Western blot analysis
Polyclonal rabbit anti-SpoIIQ antibodies were raised against purified C-terminal SpoIIQ (amino acids 63–283), expressed from a clone constructed by PCR using primers 5′-GGAAACTCCGCATATGGACAACAAC and 5′-TTTTCTCTCGAGAGACTGTTCAGT (NdeI and XhoI in bold) cloned into pET28b (Novagen; Madison, WI). This plasmid (pAR92) was electroporated into BL21 (λDE3) by selecting on kanamycin (50 μg ml−1). A growing culture was induced with 1 mM IPTG for 24 h at 25°C. Subsequent purification of His6-SpoIIQ63–283-His6 was performed by nickel affinity chromatography (HIS-Select Nickel Affinity Gel, Sigma; St. Louis, MO) yielding ~2.7 mg ml−1 purified protein which was sent to Antibodies (Davis, CA).
Cultures of KP845 and KP980 were induced to sporulate by resuspension, 4 ml harvested at t2.5 (KP980) and 25 ml at t4 (KP845) and fractionated (Blaylock et al., 2004). Samples were prepared (Pogliano et al., 1997), heated for 10 min at 50°C, loaded on a 12.5% SDS-polyacrylamide gel, transferred to PVDF (Perez et al., 2000) and probed with 0.4 μg ml−1 mouse monoclonal anti-GFP antibodies (Roche) or 1:10 000 dilution of rabbit polyclonal anti-SpoIIQ antibodies, followed by 1:1500 HRP-labelled anti-mouse or anti-rabbit antibodies and visualized with enhanced chemiluminescence (Amersham).
β-Galactosidase assay
Cultures were sporulated at 37°C or 30°C (with the 30°C cultures approximately 2 h slower), and β-galactosidase assays performed (Miller, 1972; Pogliano et al., 1997).
Plasmid construction
The spoIIQ-his6 gene was constructed by cloning a EcoRI-HindIII-digested PCR product amplified from PY79 chromosome using primers 5′-ATGTCATGAATTCACGTTTTTGGCACTCCTCTC and 5′-CGTACGTAAGCTTATGTAACTACATAGACGGTA (bold, EcoRI and HindIII sites, respectively) into pDG1662 (Guerout-Fleury et al., 1996) to yield pCH505 (amyE::PspoIIQspoIIQΩcat). The hexa-histidine tag was introduced at the spoIIQ termination codon by site-directed mutagenesis (Sawano and Miyawaki, 2000) using the primer (5′-CAGAAGACACTGAACAGTCTCATCACCATCACCATCACTAATGAAGAAAACGTCTATC-3′; bold, hexa-histidine sequence).
SpoIVFB-GFP was constructed using pPL51 (Levin et al., 1999), containing gfp downstream and in frame with EcoRI and XhoI restriction sites with a chloramphenicol resistance gene for selection in B. subtilis. The 3′ ~500 bp of spoIVFB was amplified using 5′-CCCAAGCTTCTGCCGATCTGGCCGCTG and 5′-CCGCTCGAGGTAGGGCAGAAGCAGTTC (HindIII and XhoI sites in bold), digested with HindIII and filled in using T4 polymerase. pPL51 was digested with EcoRI and filled in with T4 polymerase, and the PCR product and plasmid digested with XhoI and ligated to yield pXJ58, which was integrated at spoIVFB, selecting CmR.
The spoIVFB::pAR98(kan) insertion mutant was constructed using a derivative of pMOD2 (Epicenter; Madison, WI), pXJ52, containing a kanamycin resistance gene (aphIII) amplified from pUK19 and cloned into the BamHI and XbaI restriction sites of pMOD2 (Trieu-Cout and Courvalin, 1983). The 5′ end of spoIVFB was amplified using 5′-ACCCTGCAGTCAGATTAAACCCGCCGT and 5′-AAGGTCGACCCATTTATTCAAATGAAA, digested with PstI and SalI (bold) and ligated to pXJ52 to yield pAR98. The plasmid contained the 3′ end of spoIVFA and the first four codons of spoIVFB and was integrated at spoIVFA selecting KanR, producing a phenotype identical to that of spoIVFB152.
Acknowledgments
We thank Ruanbao Zhou and Lee Kroos for their helpful comments. This research was supported by the National Science Foundation (NSF 0135955 to K.P. and DBI 0109229 to A.R.) and the National Institute of Health (GM57045). S.C. was supported by a Japan Society for the Promotion of Science Research Fellowship for Young Scientists.
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ades SE. Control of the alternative sigma factor sigmaE in Escherichia coli. Curr Opin Microbiol. 2004;7:157–162. doi: 10.1016/j.mib.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- Antiporta MH, Dunny GM. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J Bacteriol. 2002;184:1155–1162. doi: 10.1128/jb.184.4.1155-1162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP, Jr, Pogliano K. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Coppolecchia R, DeGrazia H, Moran J. Deletion of spoIIAB blocks endospore formation in Bacillus subtilis at an early stage. J Bacteriol. 1991;173:6678–6685. doi: 10.1128/jb.173.21.6678-6685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs Pro-sigma K processing in Bacillus subtilis. Genes Dev. 1991a;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J Mol Biol. 1991b;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- Davis DM, Dustin ML. What is the importance of the immunological synapse? Trends Immunol. 2004;25:323–327. doi: 10.1016/j.it.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Dong TC, Cutting SM. SpoIVB-mediated cleavage of SpoIVFA could provide the intercellular signal to activate processing of Pro-sigmaK in Bacillus subtilis. Mol Microbiol. 2003;49:1425–1434. doi: 10.1046/j.1365-2958.2003.03651.x. [DOI] [PubMed] [Google Scholar]
- Dubnau D, Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971;56:209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Ebinu JO, Yankner BA. A RIP tide in neuronal signal transduction. Neuron. 2002;34:499–502. doi: 10.1016/s0896-6273(02)00704-3. [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Fawcett P, Losick R. A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol Microbiol. 2001;42:1147–1162. doi: 10.1046/j.1365-2958.2001.02660.x. [DOI] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Errington J, Appleby L, Daniel RA, Goodfellow H, Partridge SR, Yudkin MD. Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for sigma G activity at an intermediate stage of sporulation. J Gen Microbiol. 1992;138:2609–2618. doi: 10.1099/00221287-138-12-2609. [DOI] [PubMed] [Google Scholar]
- Frandsen N, Stragier P. Identification and characterization of the Bacillus subtilis spoIIP locus. J Bacteriol. 1995;177:716–722. doi: 10.1128/jb.177.3.716-722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis. Mol Microbiol. 2002;43:27–38. doi: 10.1046/j.1365-2958.2002.02732.x. [DOI] [PubMed] [Google Scholar]
- Gomez M, Cutting SM. Expression of the Bacillus subtilis spoIVB gene is under dual sigma F/sigma G control. Microbiology. 1996;142:3453–3457. doi: 10.1099/13500872-142-12-3453. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Jacobelli J, Andres PG, Boisvert J, Krummel MF. New views of the immunological synapse: variations in assembly and function. Curr Opin Immunol. 2004;16:345–352. doi: 10.1016/j.coi.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Kellner EM, Decatur A, Moran CP., Jr Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol Microbiol. 1996;21:913–924. doi: 10.1046/j.1365-2958.1996.461408.x. [DOI] [PubMed] [Google Scholar]
- Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci USA. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PJ, Partridge SR, Errington J. Sigma factors, asymmetry, and the determination of cell fate in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91:3849–3853. doi: 10.1073/pnas.91.9.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Kaufman RJ. The unfolded protein response. J Cell Sci. 2003;116:1861–1862. doi: 10.1242/jcs.00408. [DOI] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Frehel C, Stragier P. SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- Lu S, Halberg R, Kroos L. Processing of the mother-cell sigma factor, sigma K, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci USA. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Oke V, Shchepetov M, Cutting S. SpoIVB has two distinct functions during spore formation in Bacillus subtilis. Mol Microbiol. 1997;23:223–230. doi: 10.1046/j.1365-2958.1997.2091573.x. [DOI] [PubMed] [Google Scholar]
- Perez AR, Abanes-De Mello A, Pogliano K. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ, Bylund JE, Higgins ML. Morphogenesis and gene expression during sporulation. In: Piggot PJ, Moran CP, Youngman P, editors. Regulation of Bacterial Differentiation. Washington, DC: American Society for Microbiology; 1994. pp. 113–137. [Google Scholar]
- Pogliano K, Hofmeister AE, Losick R. Disappearance of the sigma E transcription factor from the fore-spore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnekov O, Losick R. Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Proc Natl Acad Sci USA. 1998;95:3162–3167. doi: 10.1073/pnas.95.6.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnekov O, Alper S, Losick R. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- Ricca E, Cutting S, Losick R. Characterization of bofA, a gene involved in intercompartmental regulation of pro-sigma K processing during sporulation in Bacillus subtilis. J Bacteriol. 1992;174:3177–3184. doi: 10.1128/jb.174.10.3177-3184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A, Pogliano K. Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis. EMBO J. 2004;23:1636–1646. doi: 10.1038/sj.emboj.7600171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. Morphological coupling in development: lessons from prokaryotes. Dev Cell. 2001;1:733–742. doi: 10.1016/s1534-5807(01)00094-6. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. A sporulation membrane protein tethers the pro-sigmaK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 2002;16:1007–1018. doi: 10.1101/gad.977702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Corte L, Opdyke J, Moran CP, Jr, Henriques AO. Expression of spoIIIJ in the pre-spore is sufficient for activation of sigma G and for sporulation in Bacillus subtilis. J Bacteriol. 2003;185:3905–3917. doi: 10.1128/JB.185.13.3905-3917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow B, Magill N, Febbroriello P, Nakhimousky L, Koppel DE, Setlow P. Condensation of the forespore nucleoid early in sporulation of Bacillus species. J Bacteriol. 1991;173:6270–6278. doi: 10.1128/jb.173.19.6270-6278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P. A gene odyssey: exploring the genomes of endospore-forming bacteria. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus Subtilis and its Relatives: From Genes to Cells. Washington, DC: American Society for Microbiology; 2002. pp. 519–526. [Google Scholar]
- Sun D, Fajardo-Cavazos P, Sussman MD, Tovar-Roja F, Cabrera-Martinez RM, Setlow P. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by EσF: identification of features of good EσF-dependent promoters. J Bacteriol. 1991a;173:7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Cabrera-Martinez RM, Setlow P. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore specific transcription factor σG. J Bacteriol. 1991b;173:2977–2984. doi: 10.1128/jb.173.9.2977-2984.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YL, Sharp MD, Pogliano K. A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J Bacteriol. 2000;182:2919–2927. doi: 10.1128/jb.182.10.2919-2927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cout P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5′-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- Wakeley PR, Dorazi R, Hoa NT, Bowyer JR, Cutting SM. Proteolysis of SpolVB is a critical determinant in signalling of Pro-sigmaK processing in Bacillus subtilis. Mol Microbiol. 2000;36:1336–1348. doi: 10.1046/j.1365-2958.2000.01946.x. [DOI] [PubMed] [Google Scholar]
- Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- Yen MR, Harley KT, Tseng YH, Saier MH., Jr Phylogenetic and structural analyses of the oxa1 family of protein translocases. FEMS Microbiol Lett. 2001;204:223–231. doi: 10.1111/j.1574-6968.2001.tb10889.x. [DOI] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, Losick R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet. 1984;195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
- Yu YT, Kroos L. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing inter-compartmental communication during Bacillus subtilis sporulation. J Bacteriol. 2000;182:3305–3309. doi: 10.1128/jb.182.11.3305-3309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Hofmeister A, Kroos L. The prosequence of pro–sigmaK promotes membrane association and inhibits RNA polymerase core binding. J Bacteriol. 1998;180:92434–92441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Kroos L. BofA protein inhibits intramembrane proteolysis of pro-sigmaK in an intercompartmental signaling pathway during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 2004;101:6385–6390. doi: 10.1073/pnas.0307709101. [DOI] [PMC free article] [PubMed] [Google Scholar]