Abstract
The onset of engulfment-dependent gene expression during Bacillus subtilis sporulation requires the forespore membrane protein SpoIIQ, which recruits mother cell proteins involved in late gene expression to the outer forespore membrane. Engulfment activates the late forespore transcription factor σG, which produces high levels of the secreted SpoIVB protease that is required for activation of the late mother cell transcription factor σK. Engulfment also triggers the proteolytic cleavage of SpoIIQ, an event that depends on the SpoIVB protease but not on σG activity. To determine if SpoIVB directly cleaves SpoIIQ and to determine if this event participates in the onset of late gene expression, we purified SpoIVB, SpoIIQ, and SpoIVFA (another SpoIVB substrate). SpoIVB directly cleaved SpoIIQ at the same site in vitro and in vivo and cleaved SpoIVFA in at least three different locations. SpoIIQ cleavage depends on membrane fusion, but not on σG activity, suggesting that the ability of SpoIVB to cleave substrates is regulated by membrane fusion. We isolated SpoIVB-resistant SpoIIQ proteins by random mutagenesis of codons at the cleavage site and demonstrated that SpoIIQ processing is dispensable for spore formation and for activation of late forespore and mother cell gene expression. Fluorescence recovery after photobleaching analysis demonstrated that membrane fusion releases SpoIIQ from an immobile complex, an event that could allow SpoIVB to cleave SpoIIQ. We propose that this membrane fusion-dependent reorganization in the complex, rather than SpoIIQ proteolysis itself, is necessary for the onset of late transcription.
Endospore formation in Bacillus subtilis and its relatives depends on engulfment, a phagocytosis-like process that mediates a dramatic rearrangement of the sporangium from two adjacent daughter cells, to an endospore in which the forespore lies within the cytoplasm of the larger mother cell (see Fig. 1; reviewed by Refs. 1 and 2). Engulfment is critical for sporulation, because it allows spore assembly to occur in a protected environment. It also serves as a morphological checkpoint for activation of the late forespore and mother cell transcription factors σG and σK, respectively (reviewed by Refs. 1–4). The endospore-forming bacteria must therefore have some mechanism to sense the completion of engulfment and to couple this morphological event to the onset of late gene expression.
FIGURE 1. B. subtilis engulfment, SpoIIQ proteolysis, and σG and σK activation.
A, after septation, the larger mother cell engulfs the smaller forespore, which is ultimately completely enclosed in the mother cell cytoplasm. GFP-SpoIIQ (dark gray) assembles foci at the septal midpoint and migrates around the forespore with the engulfing membrane, assembling a helical or hexagonal structure in the forespore (19). After engulfment, SpoIIQ is cleaved to release the N-terminal GFP fragment into the forespore cytoplasm (17, 19). The forespore transcription factor σG and the mother cell transcription factor σK are synthesized during engulfment and activated after engulfment (reviewed by Ref. 1). B, the interacting proteins SpoIIQ (Q) and SpoIIIAH (AH) are required for activation of σG and recruit the BofA-SpoIVFA-SpoIVFB complex necessary for activation of σK to the outer forespore membrane (OFM) (17, 24). Activation of σK is governed by two checkpoints: the forespore checkpoint (i) in which σG activation allows the production of high levels of SpoIVB (11), which cleaves SpoIVFA to relieve SpoIVFB from inhibition by BofA, and the engulfment checkpoint (ii) in which σK activity is coupled to engulfment by a BofA-independent mechanism (17). SpoIVB is also required to process SpoIIQ (17). C, SpoIIQ traverses the forespore membrane (FM), with a cytoplasmic N terminus that is the site of the GFP fusion. The extracellular C-terminal domain was used as the antigen for polyclonal antibodies (anti-IIQ) (17) while His6 and FLAG tags were fused to the extreme C terminus. Three proteolysis events are indicated by roman numerals (I–III). The first initiating proteolysis event (I) requires SpoIVB serine protease (lightning bolt), and allows a subsequent cleavage within the membrane or in the cytoplasm (II). There might also be a third cleavage event (III?) near the C terminus of the protein that releases the His6 and FLAG tags. D and E, models for the role of SpoIIQ in engulfment-dependent gene expression. D, the “pre-proteolytic activation model,” in which full-length SpoIIQ is required for σG and σK activity; E, “post-proteolytic activation model,” in which SpoIIQ proteolysis mediates intracellular signal transduction.
The forespore transcription factor σG is the first to become active after engulfment, but it remains unclear how σG is held inactive during engulfment or activated after engulfment. More is known about the regulation of the second late transcription factor, σK, which becomes active in the mother cell (summarized in Fig. 1B and reviewed by Refs. 1–4). The σK factor is initially synthesized as an inactive pro-protein containing a hydrophobic leader sequence, which functions as a covalently attached anti-sigma factor (5). This leader sequence is removed by the intramembrane protease SpoIVFB, which cleaves pro-σK within the membrane to release active σK (6–8). This processing event shares many characteristics with Regulated Intramembrane Proteolysis (RIP),3 a widespread signal transduction mechanism in which extracellular signals are transduced to a protease that cleaves its substrate within the plane of the membrane to release an active transcription factor (reviewed by Refs. 9 and 10). As is typical of RIP systems, the intramembrane protease SpoIVFB is inactive until it receives an extracellular signal. In the case of SpoIVFB, the signal is the prior activation of the late forespore-specific transcription factor σG (11), which produces higher levels of the SpoIVB protease (12). This protease disrupts an inhibitory complex between the SpoIVFB intramembrane protease and two other proteins, SpoIVFA, which is necessary for complex assembly and localization (13), and BofA, which inhibits SpoIVFB activity (Fig. 1B) (7, 11, 13). BofA is likely the main inhibitor of the SpoIVFB intramembrane protease, because it can inhibit SpoIVFB activity in an ectopic expression system (7), and because its genetic inactivation allows σK to become active in the absence of σG activity and in the absence of the SpoIVB signal transduction protease (11). The SpoIVB protease cleaves SpoIVFA and relieves BofA-mediated inhibition of SpoIVFB (12, 14 –16). This pathway results in the indirect coupling of σK activity to engulfment, by virtue of its dependence on the prior activation of σG.
Recent evidence suggests that activation of σK is also directly coupled to the completion of engulfment in two distinct manners (17). First, although the genetic elimination of BofA allows σK to become active in the absence of σG and SpoIVB, it does not allow σK activity in engulfment defective mutants (11, 17). Thus, the failure of these mutants to activate σG is not the only reason for the failure to activate σK. This suggests that σK activation is directly governed by engulfment via a pathway that does not depend on BofA, the known inhibitor of SpoIVFB intramembrane protease activity. We therefore previously proposed the existence of two distinct checkpoints for σK activity (17): the previously described forespore checkpoint that couples σK activity to σG activity via the BofA inhibitor (11) and the engulfment checkpoint that couples σK activity to the completion of engulfment via a BofA-independent mechanism (Fig. 1B) (17). Second, engulfment also appears likely to govern the activity of the SpoIVB protease that participates in the forespore checkpoint. This was first suggested by the observation that SpoIVB is synthesized both before engulfment by the early forespore transcription factor σF and after engulfment by the late forespore transcription factor σG (18). Additional support for the regulation of SpoIVB protease activity came from observations that the forespore membrane protein SpoIIQ is subject to a proteolytic processing event that depends on both engulfment and on the SpoIVB protease (17, 19). However, SpoIIQ processing occurs with apparently identical kinetics in wild-type strain or in a strain with a mutation in the gene encoding σG (spoIIIG). Thus, if SpoIVB directly cleaves SpoIIQ then its activity or access to substrates must be regulated by engulfment.
SpoIIQ proteolysis could be involved in engulfment-dependent gene expression, because this forespore membrane protein plays several key roles in this stage of sporulation. First, SpoIIQ is essential for the engulfment-dependent activation of σG (20, 21), although its precise role in this process remains unclear. Second, SpoIIQ participates in engulfment, providing a secondary mechanism that is necessary for membrane migration when the activity of the primary engulfment machinery is compromised (22). This secondary engulfment mechanism requires the interaction between the extracellular domains of SpoIIQ and that of the mother cell membrane protein SpoIIIAH (Fig. 1B) (22, 23). This interaction can readily be detected by several biochemical methods such as co-immunoprecipitation (23), affinity chromatography (23, 24), and sucrose density gradient analysis (Fig. 5). Third, SpoIIQ is required for the localization of mother cell membrane proteins needed for both σG and σK activation to the outer forespore membrane that is the site of intracellular signal transduction (17, 23, 24). Specifically, the interaction between SpoIIQ and SpoIIIAH prevents SpoIIIAH from diffusing away from the outer forespore membrane (23, 24), where it is required for σG activity. SpoIIIAH and SpoIIQ together are needed to localize the σK-processing machinery, SpoIVFA and SpoIVFB, to the outer forespore membrane, although it is unclear if SpoIIQ or SpoIIIAH directly interact with SpoIVFA or SpoIVFB (17, 24). Interestingly, SpoIIQ, SpoIIIAH, and SpoIVFB all localize to foci surrounding the forespore (Fig. 4B) (17, 19, 23). These foci might represent synapse-like sites for intracellular signal transduction, perhaps allowing transcription in both cells to be coordinately regulated by engulfment. It remains unclear how the completion of engulfment is sensed, and if SpoIIQ serves only as a scaffold to localize these signal transduction proteins or if it also participates in signal transduction.
FIGURE 5. The C-terminal fragment of SpoIIQ shows reduced interaction with SpoIIIAH.

A and B, sucrose density gradient analysis to assess the apparent molecular masses of proteins in whole cell lysates from t2 (A) and t3.5 (B). Fractions were collected from the bottom (lane 1) to the top (lane 17) of the gradient. SpoIIIAH-FLAG (AH-flag) and full-length SpoIIQ (IIQ-FL) and the C-terminal cleavage product (C-term) were visualized by Western blot with anti-FLAG or anti-SpoIIQ antibodies, respectively. Arrowheads indicate positions of proteins used as size standards. A, at early times of sporulation (t2), full-length SpoIIQ and SpoIIIAH-FLAG are present in the same fractions (~100 kDa). In the absence of one, the other migrates at a lower apparent molecular mass (~40 kDa). B, at later times (t3.5), full-length SpoIIQ migrates at ~100 kDa, whereas the C-terminal proteolytic product migrates at a lower apparent molecular mass. C, co-immunoprecipitation of SpoIIQ with SpoIIIAH-FLAG. Whole cell lysates from strains PY79 (spoIIIAH; lanes 1–3) and KP856 (spoIIIAH-flag; lanes 4 – 6) were immunoprecipitated with anti-FLAG M2 antibody. W, B, and U indicate whole cell lysate, bound, and unbound protein fractions, respectively.
FIGURE 4. Altered proteolysis, localization, and dynamics of cleavage-defective GFP-SpoIIQ.
A, proteolysis of GFP-SpoIIQ (SCB6) V72Y (SCB138) and V72E (SCB139), analyzed by Western blots with anti-GFP. “GFP-Q” and “GFP” indicate full-length and N-terminal GFP degradation product of GFP-SpoIIQ. B, localization of GFP-SpoIIQ (wt) and GFP-SpoIIQ (V72Y) 4 h after the initiation of sporulation (t4). Membranes were stained with FM4-64 (red). Arrowheads indicate septal localization of GFP-SpoIIQ (green); double arrows, GFP-SpoIIQ migrating with mother cell membrane; double arrowheads, helical structure. Membrane fusion (indicated by exclusion of FM4-64 from the forespore membranes, arrow and double arrowhead) occurs before proteolysis, which releases GFP into the cytoplasm (arrow). After fusion, V72Y remains membrane-bound but localizes smoothly around the forespore (arrow). C, FRAP analysis of GFP-SpoIIQV72Y at t3 performed and quantified as described under “Experimental Procedures.” Images of GFP (green) and FM4-64 (red)-stained membranes of cells before bleaching are to the right of each graph. Images below each plot show the GFP fluorescence during the experiment; the second panel shows the cell just after bleaching. Recovery kinetics were quantified and plotted to show the mean pixel intensity of the bleached (filled square) and unbleached (empty circle) regions and the theoretical pixel intensity value following equilibration between these regions (dashed line). Wild-type GFP-SpoIIQ shows very little recovery (see supplemental Fig. S2 and Ref. 22).
SpoIIQ proteolysis could affect its interaction with proteins involved in σG and σK activation, and therefore provides a potential mechanism by which late transcription might be coupled to engulfment. To determine if this is the case, and to determine if SpoIVB directly cleaves SpoIIQ, we further characterized SpoIIQ proteolysis. Specifically, we demonstrated that purified SpoIVB directly cleaves SpoIIQ, mapped the sites at which SpoIIQ is cleaved in vivo and in vitro, and isolated protease-resistant mutants. These mutants had no effect on σG or σK activity or spore formation, demonstrating that SpoIIQ processing is dispensable for sporulation. SpoIIQ proteolysis requires the membrane fusion event that is the final step of engulfment. Finally, fluorescence recovery after photobleaching (FRAP) demonstrates that the mobility of SpoIIQ dramatically increases after membrane fusion (in the absence of proteolysis), indicating that SpoIIQ is released from an essentially immobile complex after fusion. This reorganization in the SpoIIQ complex might control both proteolysis and signal transduction.
EXPERIMENTAL PROCEDURES
Strain and Plasmid Construction
B. subtilis strains (Table 1) were constructed by transformation (25). Plasmids (supplemental Table S1) were constructed by standard cloning methods and site-directed mutagenesis (26). Following introduction into the B. subtilis chromosome, recombinants were checked for their antibiotic resistance, the inactivation of amyE, and the loss of any additional drug resistance markers on the plasmid backbone. Primers used for plasmid construction are in supplemental Table S2. Plasmid pCH507 (amyE::PspoIIQgfp(Δ2–6)-spoIIQΩcat) was constructed by deleting the five codons that are derived from the spoIIQ gene and redundantly exist at the 5′ region of gfp on pAR47 (amyE::PspoIIQgfp-spoIIQΩcat (19)) by site-directed mutagenesis (26) using a primer, SP1. Plasmids encoding either spoIIQ, gfp-spoIIQ, or gst-spoIIQ derivatives were constructed by site-directed mutagenesis using either pCH505 (amyE::PspoIIQspoIIQ), pCH507 (amyE::PspoIIQgfp(Δ2–6)-spoIIQ), or pGEX-SpoIIQ (gst-spoIIQ43–283 (23)) as the template. Primer SP2 was used for pCH510 (amyE::PspoIIQspoIIQ-his6Ωcat). The plasmids encoding spoIIQ cleavage site mutants (V72, G73, or K74) were constructed by using either SP3, SP4, or SP5 (respectively), which had a random mixture of bases in the target codon. pCH550 (spoIIQ1–72) was also isolated by this random mutagenesis. The DNA sequence of the resulting products was confirmed prior to transformation into B. subtilis. Plasmids encoding mono-Cys mutants of spoIIQ were constructed by site-directed mutagenesis using the template plasmid pCH505 (spoIIQ) and primers either SP10 (pCH528; A62C), SP11 (pCH530; D64C), SP12 (pCH531; D68C), SP13 (pCH532; V70C), SP14 (pCH533; E71C), SP15 (pCH534; V72C), SP16 (pCH535; G73C), or SP17 (pCH536; K74C), respectively.
TABLE 1.
B. subtilis strains and plasmids used in this study
| Strain | Genotype | Source |
|---|---|---|
| PY79 | Wild type | (39) |
| KP575 | ΔspoIIQ::spc | (21) |
| KP701 | sspB-lacZΩcat::tet | This study |
| KP856 | spoIIIAH-flagΩcat | (23) |
| KP857 | spoIIIAH-flagΩcat, ΔspoIIQ::spc | (23) |
| KP953 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls | (17) |
| KP6012 | amyE::spoIIIE-gfpΩcat, ΔspoIIIE::spc | (33) |
| KP6111 | amyE::spoIIIE124::i31-gfpΩcat, ΔspoIIIE::spc | (33) |
| AR232 | ΔspoIIQ::spc, sspB-lacZΩcat::tet | This study |
| XJ220 | thrC::cotD-lacZΩmls | This study |
| XJ401 | ΔspoIIIAH, trpC2 | This study |
| XJ412 | spoIIQ-flagΩcat | This study |
| SCB3 | ΔspoIIQ::spc, amyEΩcat | This study |
| SCB4 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQΩcat | This study |
| SCB6 | ΔspoIIQ::spc, amyE::PspoIIQgfp(Δ2–6)-spoIIQΩcat | This study |
| SCB9 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQ-his6Ωcat | (17) |
| SCB15 | ΔspoIIQ::spc, spoIIIGΔ1, spoIIIG-lacZΩcat::erm, amyE::PspoIIQspoIIQΩcat | This study |
| SCB18 | ΔspoIIQ::spc, sspB-lacZΩcat::tet, amyE::PspoIIQspoIIQΩcat | This study |
| SCB21 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE::PspoIIQspoIIQΩcat | This study |
| SCB43 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQA62CΩcat | This study |
| SCB45 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQD64CΩcat | This study |
| SCB46 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQD68CΩcat | This study |
| SCB47 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV70CΩcat | This study |
| SCB48 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQE71CΩcat | This study |
| SCB49 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72CΩcat | This study |
| SCB50 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQG73CΩcat | This study |
| SCB51 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQK74CΩcat | This study |
| SCB62 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72RΩcat | This study |
| SCB63 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72YΩcat | This study |
| SCB64 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72NΩcat | This study |
| SCB65 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72SΩcat | This study |
| SCB66 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72AΩcat | This study |
| SCB67 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72IΩcat | This study |
| SCB68 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72HΩcat | This study |
| SCB69 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72KΩcat | This study |
| SCB70 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72EΩcat | This study |
| SCB71 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQV72MΩcat | This study |
| SCB72 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQG73VΩcat | This study |
| SCB73 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQG73EΩcat | This study |
| SCB74 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQ1–72Ωcat | This study |
| SCB75 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQG73LΩcat | This study |
| SCB76 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQG73PΩcat | This study |
| SCB77 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQG73TΩcat | This study |
| SCB78 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQG73IΩcat | This study |
| SCB79 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQG73SΩcat | This study |
| SCB80 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQK74EΩcat | This study |
| SCB81 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQK74IΩcat | This study |
| SCB83 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQK74RΩcat | This study |
| SCB84 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQK74AΩcat | This study |
| SCB85 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQK74NΩcat | This study |
| SCB86 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQK74SΩcat | This study |
| SCB87 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQK74PΩcat | This study |
| SCB88 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQK74DΩcat | This study |
| SCB89 | ΔspoIIQ::spc, amyE::PspoIIQspoIIQK74GΩcat | This study |
| SCB108 | ΔspoIIQ::spc, sspB-lacZΩcat::tet, amyE::PspoIIQspoIIQV72CΩcat | This study |
| SCB109 | ΔspoIIQ::spc, sspB-lacZΩcat::tet, amyE::PspoIIQspoIIQV72MΩcat | This study |
| SCB110 | ΔspoIIQ::spc, sspB-lacZΩcat::tet, amyE::PspoIIQspoIIQG73VΩcat | This study |
| SCB111 | ΔspoIIQ::spc, sspB-lacZΩcat::tet, amyE::PspoIIQspoIIQG73EΩcat | This study |
| SCB114 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE::PspoIIQspoIIQV72CΩcat | This study |
| SCB115 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE::PspoIIQspoIIQV72MΩcat | This study |
| SCB116 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE::PspoIIQspoIIQG73VΩcat | This study |
| SCB117 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE::PspoIIQspoIIQV73EΩcat | This study |
| SCB138 | ΔspoIIQ::spc, amyE::PspoIIQgfp(Δ2–6)-spoIIQV72YΩcat | This study |
| SCB139 | ΔspoIIQ::spc, amyE::PspoIIQgfp(Δ2–6)-spoIIQV72EΩcat | This study |
| SCB223 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE:: Ωcat, bofA::cat::tet, spoIIIG::neo | This study |
| SCB224 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE::PspoIIQspoIIQ, bofA::cat::tet, spoIIIG::neo | This study |
| SCB225 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE::PspoIIQspoIIQV72CΩcat, bofA::cat::tet, spoIIIG::neo | This study |
| SCB227 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE::PspoIIQspoIIQV72MΩcat, bofA::cat::tet, spoIIIG::neo | This study |
| SCB228 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE::PspoIIQspoIIQG73VΩcat, bofA::cat::tet, spoIIIG::neo | This study |
| SCB229 | ΔspoIIQ::spc, thrC::cotD-lacZΩmls, amyE::PspoIIQspoIIQV73EΩcat, bofA::cat::tet, spoIIIG::neo | This study |
pCH687 (gst-spoIVFA101–264-FLAG) was constructed as follows: spoIVFA was amplified from PY79 chromosomal DNA using primers SP6 and SP7, digested by BamHI, and then cloned into pDG1662. A FLAG tag was introduced at the C-terminal end of spoIVFA by site-directed mutagenesis using SP8. A BamHI site was further introduced between the 100th and 101st codons by site-directed mutagenesis using primer SP9. A BamHI fragment encoding the C-terminal extracytoplasmic region was cloned into the same site of pGEX4T-3 (Amersham Biosciences).
Sporulation Conditions
Sporulation was induced by resuspension at 37 °C (27), with tn being the hours after the onset of sporulation. For small scale sporulations, the cells were resuspended in 2 ml of the sporulation media and cultured with rotating in test tubes (small scale culture for Fig. 3A). For larger scale sporulations, the cells were resuspended in 15 ml (for other Western blotting, β-galactosidase activity assay, and microscopy) or 200 ml (for immunopurification of SpoIIQ-FLAG derivatives in Fig. 2B) and shaken in flasks. Sporulation efficiency was determined as described (28). β-Galactosidase assays were performed as described (29, 30).
FIGURE 3. Protease sensitivity and σG and σK activity of SpoIIQ cleavage site-mutants.
A, proteolysis in small-scale cultures. Sporulation was induced by resuspension in a 2-ml culture in test tubes, and samples were prepared after 5 h at 37 °C for Western blot analysis with anti-SpoIIQ. Amino acids introduced at Val-72 (lanes 2–11), Gly-73 (lanes 12–18), and Lys-74 (lanes 19 –27) are indicated. B, time course of proteolysis in large scale cultures. IIQ-FL and C-term indicate full-length SpoIIQ and C-terminal cleavage products. C, alignment of predicted cleavage sites of SpoIIQ from various Bacillus sp. The arrowhead indicates cleavage site of B. subtilis SpoIIQ. D–F, affect of various spoIIQ mutations on σG and σK activity. All strains contained the lacZ fusion indicated in each panel, and the spoIIQ mutation indicated by the following symbols (circles, spoIIQ+; squares, ΔspoIIQ; triangles, wt; inverted triangles, V72C; solid diamonds, V72M; open diamonds, G73V; solid circles, G73E). In addition, all strains with a spoIIQ derivative at amyE also contained a spoIIQ null mutation. D, to assay σG activity, strains KP701, AR232, SCB18, SCB108, SCB109, SCB110, and SCB111 were used. E, for σK activity, XJ220, KP953, SCB21, SCB114, SCB115, SCB116, and SCB117 were used. F, the cleavage site mutants also supported σK activity in strains lacking spoIIIG and bofA. Strains contained the indicated spoIIQ mutations plus bofA and spoIIIG (SCB223, SCB224, SCB225, SCB227, SCB228, and SCB229).
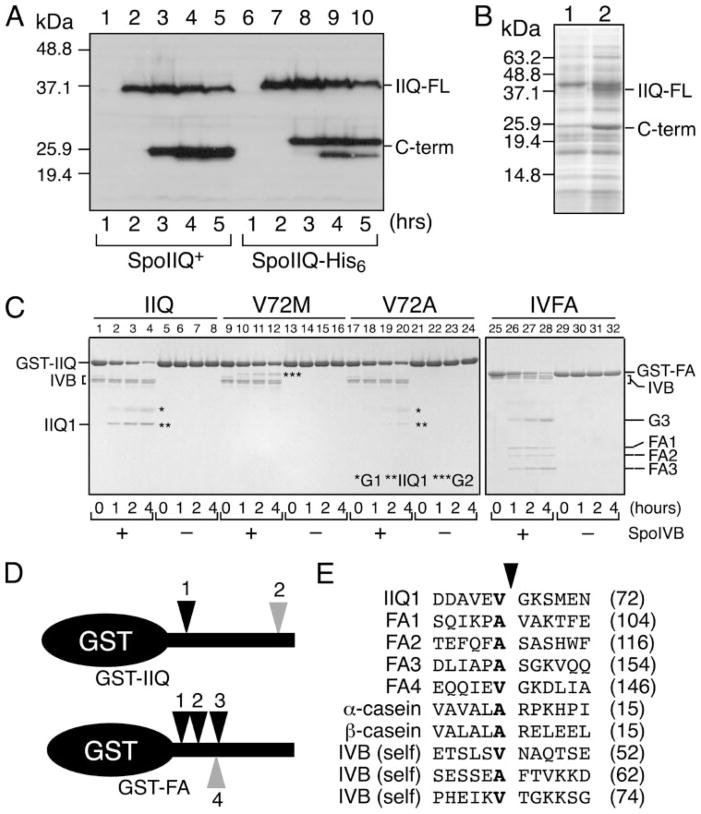
FIGURE 2. Identification of SpoIVB cleavage sites in SpoIIQ and SpoIVFA.
A, in vivo proteolysis of SpoIIQ (PY79) and SpoIIQ-His6 (SCB9) analyzed by Western blot using anti-SpoIIQ polyclonal antiserum at various times of sporulation. Full-length SpoIIQ (IIQ-FL) migrates at ~37 kDa, the C-terminal product (C-term) at ~26 kDa. B, SpoIIQ-FLAG (lane 2, XJ412) and the negative control strain PY79 (lane 1) were immunoprecipitated with anti-FLAG antibodies, and visualized by Coomassie staining. The bands indicated by C-term in lane 2 and lane 1 were extracted and subject to N-terminal amino acid sequencing. C, in vitro proteolysis of GST-SpoIIQ, various mutant derivatives, and GST-SpoIVFA101–264-FLAG by purified SpoIVB-His6. Purified GST-SpoIIQ43–283 and GST-SpoIVFA were incubated at 37 °C for 4 h in the presence or absence of purified SpoIVB-His6 and visualized by Coomassie staining. IIQ1 and FA1–3 are C-terminal cleavage products of SpoIIQ and SpoIVFA, respectively. G1–3 are fragments containing GST released from the N terminus. D, cleavage sites are indicated by arrowheads. Gray arrowhead 2 indicates the estimated position of the second C-terminal cleavage in SpoIIQ; gray arrowhead 4 indicates cleavage at FA4 (15). E, amino acid alignment of SpoIVB-dependent cleavage sites in SpoIIQ, SpoIVFA, SpoIVB (22), and α- and β-casein. Arrowhead indicates position of cleavage. The numbers of amino acids before the cleavage site are in parentheses.
Co-immunoprecipitation and Western Blotting
Sporulating cells were harvested and washed with SMM buffer (0.5 M sucrose, 20 mM MgCl2, and 20 mM maleic acid, pH 6.5), resuspended in the same buffer and treated with 1 mg/ml lysozyme at 37 °C for 10 min. The spheroplasts were harvested by centrifugation and resuspended in ice-cold buffer A (20 mM HEPES-NaOH, 150 mM NaCl, and 1 mM EDTA, pH 7.6) with 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml pepstatin and then treated with 0.5% n-dodecyl β-D-maltoside (Sigma) on ice for 30 min. The insoluble fraction was removed by ultracentrifugation with a Beckman TLA120-2d rotor (40,000 rpm, 30 min), and the supernatant was incubated with anti-FLAG M2 affinity gel (Sigma) overnight at 4 °C with gentle rolling. The affinity gel was washed twice with buffer A containing 0.5% n-dodecyl β-D-maltoside, and the bound proteins were eluted by SDS-loading buffer (without reducing agent) at 42 °C. Proteins in the cell lysate and in the flow-through containing unbound protein were precipitated by 5% trichloroacetic acid for 20 min on ice, washed by acetone, and then solubilized in SDS loading buffer. Proteins were analyzed by SDS-PAGE or Western blotting as described previously (17, 23). 1:5,000 dilutions of anti-SpoIIQ (17), anti-SpoIIIAH (see below), anti-GFP (Roche Applied Science), and anti-FLAG M2 antibodies (Sigma) were used to probe derivatives of SpoIIQ, GFP-SpoIIQ, and SpoIIIAH-FLAG, respectively.
Sucrose Density Gradient Ultracentrifugation
Sucrose density gradient ultracentrifugation was performed essentially as described (31), with the following modifications. Sporulating cell lysates were prepared by the same procedure as described above for co-immunoprecipitation, except that buffer B (20 mM HEPES-NaOH, 300 mM KCl, and 1 mM EDTA, pH 7.6) was used instead of buffer A. The lysates were loaded on top of 5-ml sucrose gradient beds (5–20% sucrose, 20 mM HEPES-NaOH, 300 mM KCl, and 1 mM EDTA, pH7.6). Proteins were separated by ultracentrifugation in a Beckman MLS50 rotor (4 °C, 43,000 rpm, 16 h) and collected into 17–18 fractions. Purified proteins catalase (226 kDa), adolase (146 kDa, Amersham Biosciences), bovine serum albumin (68 kDa), and lysozyme (14.3 kDa, Sigma) were used as the protein standards.
AMS Modification of Monocysteine Derivatives of SpoIIQ
For AMS treatment, whole cell proteins were solubilized in SDS-loading buffer, containing 1 mM Tris(2-carboxyethyl) phosphine instead of 2 mM dithiothreitol. The pH of the SDS-loading buffer changed by addition of Tris(2-carboxyethyl) phosphine was adjusted to pH 6.8 by adding the proper volume of 0.5 M Tris-HCl (pH 6.8). Samples were treated with 2 mM AMS (4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid) at 37 °C for 1 h and then analyzed by Western blotting.
In Vivo Proteolysis Assay of SpoIIQ Derivatives
A whole cell trichloroacetic acid precipitation was prepared from 1 ml of culture to which trichloroacetic acid was added (to 5% final concentration). The cells were collected by centrifugation, and the pellet was washed with 0.75 ml of 1 M Tris-HCl (pH 8) and treated with 1 mg/ml lysozyme in 60 μl of buffer C (33 mM Tris-HCl, 40% sucrose, 1 mM EDTA, pH 8). Proteins were then solubilized by SDS-loading buffer with 2 mM dithiothreitol and analyzed by Western blotting.
Purification of SpoIVB, SpoIIQ, and SpoIVFA
BL21(DE3)/pZR53 (SpoIVB-His6) was grown in LB ampicillin (100 μg/ml) media, and expression of SpoIVB-His6 was induced by 1 mM isopropyl 1-thio-β-D-galactopyranoside for 1 h at 30 °C. Cells were washed by buffer D (20 mM Tris-HCl (pH 8.0), 150 mM NaCl) and disrupted by sonication. After removing debris by centrifugation, cell lysate was subjected to nickel affinity column (Sigma) equilibrated in buffer D. The column was washed with buffer D plus 20 mM imidazole and eluted with 300 mM imidazole in the same buffer. Eluted protein was then dialyzed in buffer D, loaded on Hi-Trap Q (Amersham Biosciences) anion-exchange chromatography column previously equilibrated in buffer D. Flow-through fractions that include SpoIVB-His6 were collected and then loaded onto a Hi-Trap SP (Amersham Biosciences) cation-exchange chromatography column previously equilibrated in buffer D. SpoIVB-His6 was eluted by a sodium chloride gradient (0–1 M) in buffer D.
GST-SpoIVFA and GST-SpoIIQ were expressed in BL21 (DE3). Expression was induced by the addition of 1 mM isopropyl 1-thio-β-D-galactopyranoside for 2–3 h at 37 °C. Cells were washed with phosphate-buffered saline (PBS), suspended in PBS/1 mM dithiothreitol/1 mM EDTA, and disrupted by sonication. Cell lysate was loaded on a glutathione-Sepharose column (Amersham Biosciences) previously equilibrated in PBS, washed with PBS, and eluted with 50 mM Tris-HCl (pH 8.0) containing 10 mM glutathione. GST-SpoIVFA101–264-FLAG was dialyzed by buffer D and further purified by using a Sephacryl S-200 column (Amersham Biosciences). All proteins were dialyzed in buffer D, and concentration was determined by a Bradford protein assay (Sigma).
In Vitro Protease Assay and Amino Acid Sequence Analysis
Purified substrates (240 ng/μl) were incubated in buffer D at 37 °C in 0–4 h in the presence or absence of 30 ng/μl purified SpoIVB-His6. The reaction was stopped with SDS-loading buffer, and the products were analyzed by SDS-PAGE and Coomassie staining. Bands were excised from polyvinylidene difluoride membrane and subjected to N-terminal sequencing.
Microscopy and Image Analysis
For GFP visualization, live cells were stained with 4′,6′-diamidino-2-phenylindole (0.2 μg/ml, Molecular Probes) and FM4-64 (5 μg/ml, Molecular Probes) as described previously (32). Images were collected with an Applied Precision Spectris microscope with a QLM laser module (described in Ref. 33). Photobleaching was performed and quantified as described (22), using a 0.02- to 0.05-s pulse of a 488 nm argon laser at 50% power. Subsequent GFP images were collected at 30-s intervals for 5 min for GFP-SpoIIQ and V72Y or as quickly as possible for smoothly localized GFP-SpoIIQV72Y. Exposure times were limited to 1.5–2.5 s. These experiments were quantified as previously described (22). Briefly, the images were corrected for photobleaching during image collection, and the fluorescence intensity of the bleached and unbleached regions were quantified throughout the experiment, and corrected for bleaching during image acquisition. The small size of the forespore results in a small and variably sized pool of unbleached GFP-SpoIIQ from which recovery can occur. We therefore calculated the theoretical equilibration point between the bleached and unbleached regions, which is represented as a dashed line.
Immunofluorescence and Preparation of Anti-SpoIIIAH Antibody
Anti-SpoIIIAH polyclonal antiserum was made by injection of the KLH-conjugated chemically synthesized antigen peptide, CDLFTTYRLDLEDARSKEREE, into rabbits (subcontracted to Sigma Genosys). This peptide was chosen for its hydrophilicity and corresponds to amino acids 104–123 of SpoIIIAH plus an N-terminal Cys for conjugation to KLH. Immunofluorescence microscopy was performed as described (34).
RESULTS
Identification of in Vivo Cleavage Site of SpoIIQ
Previous results demonstrated that both N- and C-terminal proteolytic products of SpoIIQ, GFP-SpoIIQ, and SpoIIQ-His6 were soluble, suggesting that the protein was cleaved on both sides of the membrane (Fig. 1C) (17). The ability to completely block SpoIIQ proteolysis with spoIVB mutations suggested that SpoIIQ proteolysis is initiated by the secreted SpoIVB protease, which releases the extracellular domain from the membrane, and allows a subsequent cytoplasmic (or perhaps intramembrane) proteolysis that releases the N-terminal GFP tag into the cytoplasm (Fig. 1C) (17). There might also be a third proteolysis event near the C terminus of SpoIIQ (Fig. 1C), based on our analysis of SpoIIQ-His6. This protein shows two proteolytic products (Fig. 2A) in immunoblot experiments with anti-SpoIIQ antibodies, one slightly larger than that of native SpoIIQ (which reacts with His6-specific antibodies) and one identical in size to that of native SpoIIQ (and that does not react with His6-specific antibodies).
To further characterize the SpoIVB-dependent proteolysis event that appears to initiate SpoIIQ degradation we used two independent methods to identify the site at which SpoIIQ was cleaved in intact cells. First, we immunoprecipitated SpoIIQ-FLAG from whole cell lysates and extracted a band of ~25 kDa that appeared only when SpoIIQ-FLAG was expressed (Fig. 2B, lane 2). The major signal from the amino acid sequencing analysis was GKSMEN (corresponding to amino acids 73–78 of SpoIIQ), suggesting that extracellular cleavage occurs between Val-72 and Gly-73. Second, we constructed a series of SpoIIQ derivatives with individual cysteines introduced between codons 62 and 74 and used AMS modification (35) to determine if the cysteine residue was present in the C-terminal degradation product, which confirmed that in intact cells proteolysis occurred between Val-72 and Gly-73 (supplemental Fig. S1).
Purified SpoIVB Cleaves SpoIIQ
To determine if SpoIVB (a serine protease) directly cleaves SpoIIQ, we tested if purified SpoIVB cleaved purified GST-SpoIIQ in vitro, using GST-SpoIVFA (a known SpoIVB substrate), GST and α- and β-casein as control proteins. SpoIVB was able to cleave GST-SpoIIQ and GST-SpoIVFA with similar kinetics (Fig. 2C), whereas GST was stable during the incubation (data not shown). The degradation products of GST-SpoIIQ and GST-SpoIVFA were excised and subjected to N-terminal amino acid sequencing. GST-SpoIIQ was cleaved once to yield a GST-containing product (G1) and a product with an N-terminal sequence starting at Gly-73 of SpoIIQ (IIQ1). Thus, purified SpoIVB cleaves SpoIIQ between Val-72 and Gly-73, the same position at which SpoIIQ is cut in intact cells. SpoIVB cleaved GST-SpoIVFA and itself in three locations (Fig. 2E) (36). A comparison of the SpoIVB cleavage sites of these three proteins and α- and β-casein demonstrated that in each case SpoIVB cut after either alanine (in the case of SpoIVFA, caseins, and one site in SpoIVB) or valine (in the case of SpoIIQ and two sites in SpoIVB), with little other primary sequence similarity in the surrounding region. A similar analysis by another group also identified these three sites in SpoIVFA (15). Using mass spectrometry analysis, they also observed cleavage at an EVGK motif within SpoIVFA that is identical to the site of proteolysis in SpoIIQ (15). Cleavage at this site depended on cleavage at the most C-terminal cleavage site. This explains why our N-terminal sequencing analysis failed to detect cleavage at EVGK, because we sequenced only the larger C-terminal products, which will have the same N terminus with or without cleavage at EVGK. The 8-amino acid internal product of cleavage at these sites is too small to be detected by SDS-PAGE.
Isolation of Cleavage-defective SpoIIQ Proteins
The above results demonstrate that SpoIIQ is directly cleaved by the SpoIVB serine protease that is essential for σK activation. To determine if SpoIIQ proteolysis was essential for sporulation, we used site-directed mutagenesis to randomize codons 72, 73, and 74 of wild-type spoIIQ (“Experimental Procedures”), isolating 26 different amino acid substitutions at these sites. The mutations were introduced into B. subtilis and screened for defects in SpoIIQ proteolysis using small scale sporulations (in test tubes) and anti-SpoIIQ Western blots from extracts prepared 5 h after the initiation of sporulation (t5, Fig. 3A). Under these conditions, sporulation is slowed, likely due to decreased aeration, but wild-type SpoIIQ was cleaved by t5 (Fig. 3A, lanes 1 and 28). Of the 29 mutants tested (including three Cys substitutions, supplemental Fig. S1), 14 were cleavage-defective, and 15 were permissive. At amino acid 72 only V72A and V72C allowed proteolysis (Fig. 3A, lanes 2–11, supplemental Fig. S1), demonstrating that Val-72 plays a crucial role in substrate recognition. In contrast, many substitutions at amino acids 73 and 74 allowed proteolysis (Fig. 3A, lanes 12–27), although introduction of negatively charged amino acids (Glu or Asp) or proline inhibited proteolysis. Alignment of the N-terminal regions of SpoIIQ from various Bacillus species (Fig. 3C) demonstrated that each had valine at the predicted SpoIVB cleavage site and permissive substitutions at positions corresponding to Gly-73 and Lys-74 of B. subtilis SpoIIQ (aside from Geobacter and Oceanobacillus iheyensis, which had a non-permissive substitution at position 74). Thus, valine appears to be conserved at the cleavage site of SpoIIQ proteins that are sufficiently closely related to allow a reliable alignment of their N-terminal domain.
We more precisely followed degradation of three cleavage-defective mutants and two permissive mutants using larger scale sporulations (Fig. 3B). Each of the cleavage-defective mutants showed very slow and inefficient degradation, with little accumulation of the C-terminal product and no notable decrease in the levels of full-length SpoIIQ even after 5 h of sporulation. In contrast the permissive mutants showed normal or slightly slower (V72C) degradation (Fig. 3B), accumulating a stable C-terminal breakdown product. To determine if these proteins were equally resistant or sensitive to in vitro proteolysis by purified SpoIVB, we introduced the cleavage-defective substitution V72M and the permissive substitution V72A into GST-SpoIIQ. As expected, V72M was resistant to SpoIVB cleavage at the usual cleavage site, although it generated a larger proteolysis product that might correspond to cleavage at a more C-terminal SpoIVB cleavage (G2 in Fig. 2C). We could find no evidence that SpoIVB cleaved V72M at this site in vivo, because no intermediate size product was observed in whole cell extracts (Fig. 3B). Interestingly, V72A, which is permissive for proteolysis in intact cells, was degraded in vitro more slowly than wild-type SpoIIQ (Fig. 2C).
Effects of SpoIIQ Cleavage Mutants on σG and σK Activity
Both SpoIIQ proteolysis and activation of σG and σK depend on engulfment, raising the possibility that SpoIIQ proteolysis might mediate extracellular signal transduction via an RIP-like mechanism (post-proteolytic model in Fig. 1E). However, full-length SpoIIQ also acts during engulfment to recruit mother cell membrane proteins required for σG and σK activation to the septum (17, 23, 24), raising the possibility that an event that occurs before proteolysis might mediate signal transduction (pre-proteolytic model in Fig. 1D). To distinguish between these models, we compared the levels of σG and σK activity (using sspB-lacZ and cotD-lacZ, respectively) in strains carrying mutations that block (V72M and G73E) or allow (V72C and G73V) SpoIIQ proteolysis. These mutants showed σG and σK activities nearly identical to wild type (Fig. 3, D and E). Thus, the proteolysis-defective mutants have no effect on σG or σK activity, suggesting that SpoIIQ proteolysis is not essential for engulfment-dependent gene expression, in keeping with the pre-proteolytic model.
The activation of σK is governed by two checkpoints, the prior activation of σG (the forespore checkpoint) (11) and a separate checkpoint governed by the completion of engulfment (the engulfment checkpoint). The forespore checkpoint can be bypassed by a bofA mutation, which allows σK activity in the absence of σG (37), but the bofA mutation does not bypass either engulfment or the requirement for SpoIIQ and SpoIIIAH for σK activity (17). We tested whether cleavage-defective SpoIIQ supported σK activation in a bofA mutant lacking σG, by introducing the cleavage-defective or -permissive mutants into a cell that has null mutations in spoIIIG (encoding σG), bofA and spoIIQ. Again, both cleavage-defective and cleavage-permissive mutations supported σK activation in these assays, with a slight reduction in activity seen in both classes of mutations at late times (Fig. 3F).
Thus, SpoIVB-mediated proteolysis of SpoIIQ is dispensable for both σG and σK activity, as well as for the production of heat-resistant spores (Table 2). These results suggest that either SpoIIQ proteolysis is normally dispensable for sporulation or that cytoplasmic proteolysis does not depend on SpoIVB-mediated extracellular proteolysis.
TABLE 2.
Sporulation efficiency of spoIIQ mutants
| Strain | Genotype | Cleavage | Spore titer |
|---|---|---|---|
| PY79 | spoIIQ+ | + | 2.7 × 108 |
| SCB3 | ΔspoIIQ::spc, amyE::cat | 0 | |
| SCB4 | ΔspoIIQ::spc, amyE::pspoIIQspoIIQΩcat | + | 2.5 × 108 |
| SCB50 | ΔspoIIQ::spc, amyE::pspoIIQspoIIQV72CΩcat | + | 2.6 × 108 |
| SCB71 | ΔspoIIQ::spc, amyE::pspoIIQspoIIQV72MΩcat | − | 2.0 × 108 |
| SCB72 | ΔspoIIQ::spc, amyE::pspoIIQspoIIQG73VΩcat | + | 1.7 × 108 |
| SCB73 | ΔspoIIQ::spc, amyE::pspoIIQspoIIQG73EΩcat | − | 2.0 × 108 |
| SCB83 | ΔspoIIQ::spc, amyE::pspoIIQspoIIQK74RΩcat | + | 1.8 × 108 |
| SCB80 | ΔspoIIQ::spc, amyE::pspoIIQspoIIQK74EΩcat | − | 1.9 × 108 |
Cytoplasmic Cleavage of SpoIIQ Depends on Extracytoplasmic Cleavage
The absence of smaller breakdown products in SpoIIQ proteins resistant to SpoIVB-mediated extracellular proteolysis (Fig. 3) and in the absence of SpoIVB (17) suggests that, in intact cells, cytoplasmic proteolysis occurs after extracellular proteolysis. To further test this hypothesis, we introduced the cleavage-resistant mutations V72Y and V72E into GFP-SpoIIQ and tested whether the cytoplasmic cleavage occurred to release the N-terminal GFP tag into the cytoplasm. Western blot analysis demonstrated that GFP-SpoIIQ was partially stabilized by these mutations, with a decreased amount of the N-terminal GFP-containing breakdown product relative to wild type at t3 (Fig. 4A). Prior studies have demonstrated that GFP-SpoIIQ is more permissive for proteolysis than native SpoIIQ. Unlike the wild-type protein, GFP-SpoIIQ is not completely stabilized by the absence of either SpoIVB or SpoIIIAH (17). This proteolysis might occur during sample preparation, because fluorescence microscopy demonstrated that, similar to GFP-SpoIIQ in the spoIVB and spoIIIAH mutants (17), the proteolysis-resistant mutant V72Y remained membrane-bound after engulfment, with little cytoplasmic GFP fluorescence (Fig. 4B). Together with the ability of mutations at the site of extracellular cleavage to completely stabilize native SpoIIQ (with no slightly smaller breakdown products, Fig. 3), these results suggest that cytoplasmic cleavage of SpoIIQ normally depends on its extracellular cleavage by SpoIVB (Fig. 1C).
This mechanism of SpoIIQ proteolysis shows similarity to that of RIP, in which degradation of a signal transduction protein is typically initiated by an extracellular cleavage that allows a second intracellular or intramembrane cleavage by a second protease. However, despite the observation that SpoIIQ proteolysis is governed by the same morphological checkpoint (engulfment) and protease (SpoIVB) as late mother cell gene expression, we found no evidence that it participates in intracellular signal transduction.
Release of SpoIIQ from an Immobile Complex after Engulfment
We reasoned that the completion of engulfment might mediate a rearrangement in the complex between SpoIIQ and its mother cell ligand SpoIIIAH or its unidentified tether (19, 23) and that this rearrangement might allow both SpoIIQ proteolysis and signal transduction. If this is the case, then SpoIIQ might show different diffusion kinetics before and after the membrane fusion event that is the final step of engulfment. Indeed, a FRAP analysis of the diffusion kinetics of wild-type GFP-SpoIIQ during and after engulfment demonstrated that SpoIIQ is relatively immobile during engulfment with somewhat increased mobility after engulfment (22). However, wild-type SpoIIQ is degraded after engulfment, so this could reflect loss of the extracellular domain rather than release from a complex. We therefore performed a FRAP analysis of the protease-resistant mutant V72Y. During engulfment, V72Y showed the same restricted mobility as wild-type SpoIIQ (supplemental Fig. S2) (22), with equilibration between the bleached and unbleached regions requiring at least 200 s (supplemental Fig. S2) (22). After engulfment, V72Y showed both the punctate localization seen in wild type before proteolysis, and a smooth localization pattern not observed in wild type (Fig. 4B), unless it is expressed in the absence of SpoIVB (17). These two patterns each had distinct FRAP results (Fig. 4C): sporangia with punctate localization showed low mobility (equilibration times or teq ≥ 240 s), whereas those with smooth localization showed a high mobility (teq ≤ 10 s) similar that of forespore-expressed MalFTM1–2-GFP (teq ≤ 6 s) (22). We obtained essentially the same results with another cleavage-defective SpoIIQ protein (V72E), and wild-type SpoIIQ in the proteolysis-defective spoIVB strain (supplemental Fig. S2).
Thus after membrane fusion, protease-resistant SpoIIQ is released from a punctate or helical structure in which it is essentially immobile, attaining a rapid diffusion rate similar to a non-localized protein. This event could allow both RIP and intracellular signal transduction necessary for engulfment-dependent gene expression in the forespore and the mother cell.
Reduced Interaction between the Extracellular Proteolysis Product of SpoIIQ and SpoIIIAH
We were interested in determining if the proteolytic products of SpoIIQ interacted with its mother cell ligand SpoIIIAH. We therefore used non-denaturing sucrose density gradient analysis (31) to compare the apparent molecular weight of full-length SpoIIQ, its C-terminal degradation product, and SpoIIIAH. Full-length SpoIIQ and SpoIIIAH were present in the same fractions (Fig. 5A), with apparent molecular masses of ~100 kDa, significantly higher than their predicted molecular masses of 31 kDa and 24 kDa, respectively. The high molecular weight was likely a consequence of the interaction between SpoIIQ and SpoIIIAH, because both proteins had a reduced apparent molecular mass in strains lacking the other protein (~40 kDa). The level of SpoIIIAH was reduced in the absence of SpoIIQ (Fig. 5A, supplemental Fig. S3D), suggesting that SpoIIQ protects SpoIIIAH from proteolysis.
Samples harvested after SpoIIQ proteolysis commenced demonstrated that, although the differences in molecular mass between full-length SpoIIQ and its C-terminal product is only ~8 kDa (31 kDa versus 23 kDa), these two proteins showed strikingly different apparent sizes, with full-length SpoIIQ behaving as a ~100 kDa protein, and the C-terminal product as a ~30 kDa protein (Fig. 5B). This suggests a reduced interaction between the C-terminal fragment of SpoIIQ and SpoIIIAH. We confirmed this hypothesis by immunoprecipitating SpoIIIAH-FLAG and probing the eluate with anti-SpoIIQ antibody. While full-length SpoIIQ was efficiently co-precipitated with SpoIIIAH-FLAG, the C-terminal fragment of SpoIIQ was not precipitated (Fig. 5C). Together these results indicate that if there is any interaction between the C-terminal degradation product of SpoIIQ and SpoIIIAH, the affinity is too low to be detected by either sucrose density gradient analysis or co-immunoprecipitation.
SpoIVB-mediated SpoIIQ proteolysis occurs after V72, leaving the N-terminal transmembrane segment and almost 30 amino acids outside the cell that might interact with SpoIIIAH. In an attempt to determine if this region interacted with SpoIIIAH, we expressed the N-terminal fragment of SpoIIQ1–72 and used immunofluorescence and Western blotting to determine if it was able to recruit SpoIIIAH to the sporulation septum and protect it from proteolysis. SpoIIIAH localization and stability was reduced in the SpoIIQ1–72 strain. This might be due to the rapid degradation of the N-terminal fragment, because GFP fusions to the first 60 or 72 amino acids of SpoIIQ were quickly degraded to soluble GFP (data not shown), as is the N-terminal cleavage product of full-length GFP-SpoIIQ (which fails to accumulate). This suggests that the N-terminal product of SpoIVB cleavage is too unstable to sustain an interaction with SpoIIIAH. Thus, SpoIIQ appears to make a high affinity interaction with SpoIIIAH only before proteolysis, raising the possibility that premature degradation of SpoIIQ might compromise its ability to recruit SpoIIIAH and other proteins to the sporulation septum.
SpoIIQ Proteolysis Occurs in a Membrane Fusion-dependent Manner
Although SpoIVB is synthesized during engulfment (18), three lines of evidence indicate that SpoIVB-mediated proteolysis of SpoIIQ occurs only after engulfment. First, cell biological studies of GFP-SpoIIQ degradation demonstrate that the N-terminal GFP moiety is released from the membrane only in sporangia that have completed membrane fusion, demonstrating that cytoplasmic proteolysis occurs only after fusion (19). Second, Western blot analysis of native SpoIIQ (with anti-SpoIIQ antiserum) demonstrates that low levels of SpoIIQ proteolysis is first observed at 2.5 h of sporulation (Fig. 5C), consistent with observations that membrane fusion is first completed in a few sporangia just before 2 h of sporulation (at 105 min (32)). Finally, proteolysis of native SpoIIQ does not occur in spoIID or spoIIP mutants, which block engulfment prior to the onset of membrane migration (17).
We were interested in determining if SpoIIQ proteolysis was regulated by membrane migration or by the membrane fusion event that is the final step of engulfment. To address this question, we made use of a spoIIIE mutant that completes membrane migration but not membrane fusion (and which translocates DNA (33)). The membrane fusion-defective mutant abolished degradation of native SpoIIQ (Fig. 6B) and accumulated full-length SpoIIQ. These results indicate that SpoIVB-mediated degradation of SpoIIQ depends on the final step of engulfment, membrane fusion, in keeping with prior cell biological studies.
FIGURE 6. SpoIIQ proteolysis depends on engulfment membrane fusion.
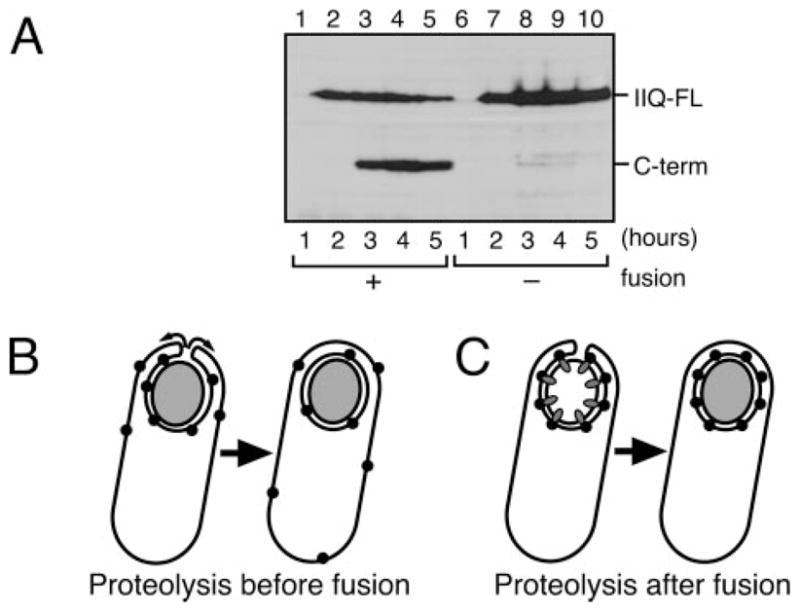
A, SpoIIQ proteolysis assessed by Western blot analysis of strains KP6012 (gfp-spoIIIE+; lanes 1–5) and the membrane fusion-defective KP6111 (gfp-spoIIIE121; lanes 6 –10). B and C, schematics depicting the potential importance of coupling SpoIIQ proteolysis to membrane fusion. B, if SpoIIQ proteolysis occurs before fusion, mother cell proteins that interact with SpoIIQ (such as SpoIIIAH) would be released from the septum and therefore be distributed throughout the mother cell membrane. C, if SpoIIQ proteolysis occurs after fusion, binding proteins cannot escape from the outer forespore membrane. Coupling SpoIIQ proteolysis to membrane fusion might therefore be necessary for the efficient localization of mother cell membrane proteins to the outer forespore membrane.
DISCUSSION
Our results demonstrate that SpoIIQ is directly cleaved by the SpoIVB protease that is also required for activation of the late mother cell transcription factor σK. Interestingly, both σK activation and SpoIIQ proteolysis depend on the phagocytosis-like process of engulfment (17, 38). We here provide evidence that SpoIIQ proteolysis more specifically depends on the final step of engulfment, membrane fusion, which releases the forespore into the mother cell cytoplasm. Although SpoIIQ proteolysis is not essential for sporulation, it in some ways provides a better model for SpoIVB-mediated proteolysis than the other identified SpoIVB substrate, SpoIVFA. Specifically, unlike SpoIVFA, SpoIIQ proteolysis releases stable proteolysis products that can readily be detected by both immunoblot analysis and cell biological methods. SpoIIQ thereby provides a tractable system to investigate the mechanism by which SpoIVB activity is governed by the morphological checkpoint of engulfment.
The Checkpoint for SpoIIQ Proteolysis Might Retain the Active Signal Transduction Complex at the Septum
It remains unclear why SpoIIQ is subject to engulfment-dependent proteolysis, because we have failed to identify any phenotypic consequence of blocking SpoIIQ proteolysis. However, our results allow us to propose a reason for why it is important to delay SpoIIQ proteolysis until after the completion of membrane fusion. The interaction between SpoIIQ and SpoIIIAH is required to retain SpoIIIAH and SpoIVFB in the outer forespore membrane (17, 23, 24), where they are involved in intracellular signal transduction cascades that result in activation of late forespore and mother cell transcription factors. We have been unable to detect an interaction between the SpoIIQ proteolysis products and SpoIIIAH in living cells. Thus, if SpoIIQ were degraded prior to membrane fusion the decreased affinity of the interaction between SpoIIQ and SpoIIIAH might result in the release of SpoIIIAH and SpoIVFB from the outer forespore membrane (Fig. 6B), thereby compromising intracellular signal transduction. Thus, using membrane fusion as a checkpoint for SpoIIQ proteolysis might serve to maintain the interaction between SpoIIQ and SpoIIIAH and SpoIVFB until after engulfment, ensuring that proteins required for σG and σK activation localize exclusively to the outer forespore membrane where cell-to-cell communication occurs (Fig. 6C).
A Membrane Fusion-dependent Reorganization in the SpoIIQ-SpoIIIAH Complex
Our studies also suggest a membrane fusion-dependent rearrangement in the complex between SpoIIQ, SpoIIIAH, and other proteins involved in late forespore and mother cell gene expression. Specifically, our FRAP studies demonstrate that, although SpoIIQ is essentially immobile during engulfment (22), after membrane fusion it diffuses through the forespore membrane at rates nearly identical to a non-localized MalF-GFP protein (Fig. 4). This result is most easily interpreted as reflecting a remodeling of the interaction between SpoIIQ and its binding partner, SpoIIIAH (or another unidentified SpoIIQ-interacting protein), an event that could easily initiate intracellular signal transduction. It is therefore tempting to speculate that remodeling of the SpoIIQ complex provides a signal both for σG and σK activation and for SpoIIQ proteolysis, because this would explain both the coordinate regulation of these events by membrane fusion and the dispensable nature of SpoIIQ proteolysis.
Supplementary Material
Acknowledgments
We thank Ruanbao Zhou, Lee Kroos, and Charles Moran for providing strains and Dan Broder and Amber Dance for providing helpful comments on the manuscript.
Footnotes
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
The abbreviations used are: RIP, regulated intramembrane proteolysis; FRAP, fluorescence recovery after photobleaching; PBS, phosphate buffered saline; GFP, green fluorescent protein; AMS, 4-acetamido-4′-maleimidylstilbene-2;2′-disulfonic acid; KLH, Keyhole limpet hemocyanin; GST, glutathione S-transferase; BSA, bovine serum albumin; t0, t1, t2 …, time of sporulation, with the number indicating hours after the initiation of sporulation by resuspension; FM, forespore membrane; OFM, outer forespore membrane.
This work was supported in part by National Institutes of Health Grant GM 57045.
References
- 1.Errington J. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 2.Piggot PJ, Hilbert DW. Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Rudner DZ, Losick R. Dev Cell. 2001;1:733–742. doi: 10.1016/s1534-5807(01)00094-6. [DOI] [PubMed] [Google Scholar]
- 4.Kroos L, Yu YT. Curr Opin Microbiol. 2000;3:553–560. doi: 10.1016/s1369-5274(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B, Hofmeister A, Kroos L. J Bacteriol. 1998;180:92434–92441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu YT, Kroos L. J Bacteriol. 2000;182:3305–3309. doi: 10.1128/jb.182.11.3305-3309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou R, Kroos L. Proc Natl Acad Sci U S A. 2004;101:6385–6390. doi: 10.1073/pnas.0307709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudner DZ, Fawcett P, Losick R. Proc Natl Acad Sci U S A. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrmann M, Clausen T. Annu Rev Genet. 2004;38:709–724. doi: 10.1146/annurev.genet.38.072902.093416. [DOI] [PubMed] [Google Scholar]
- 10.Brown MS, Ye J, Rawson RB, Goldstein JL. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 11.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 12.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 13.Rudner DZ, Losick R. Genes Dev. 2002;16:1007–1018. doi: 10.1101/gad.977702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong TC, Cutting SM. Mol Microbiol. 2003;49:1425–1434. doi: 10.1046/j.1365-2958.2003.03651.x. [DOI] [PubMed] [Google Scholar]
- 15.Campo N, Rudner DZ. Mol Cell. 2006;7:25–35. doi: 10.1016/j.molcel.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 16.Zhou R, Kroos L. Mol Microbiol. 2005;58:835–846. doi: 10.1111/j.1365-2958.2005.04870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X, Rubio A, Chiba S, Pogliano K. Mol Microbiol. 2005;58:102–115. doi: 10.1111/j.1365-2958.2005.04811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez M, Cutting SM. Microbiology. 1996;142:3453–3457. doi: 10.1099/13500872-142-12-3453. [DOI] [PubMed] [Google Scholar]
- 19.Rubio A, Pogliano K. EMBO J. 2004;23:1636–1646. doi: 10.1038/sj.emboj.7600171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun YL, Sharp MD, Pogliano K. J Bacteriol. 2000;182:2919–2927. doi: 10.1128/jb.182.10.2919-2927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Londono-Vallejo JA, Frehel C, Stragier P. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- 22.Broder DH, Pogliano K. Cell. 2006;126:917–928. doi: 10.1016/j.cell.2006.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blaylock B, Jiang X, Rubio A, Moran CP, Jr, Pogliano K. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doan T, Marquis KA, Rudner DZ. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- 25.Dubnau D, Davidoff-Abelson R. J Mol Biol. 1971;56:209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- 26.Sawano A, Miyawaki A. Nucleic Acids Res. 2000;28:78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sterlini JM, Mandelstam J. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abanes-De Mello A, Sun YL, Aung S, Pogliano K. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 30.Pogliano K, Hofmeister AE, Losick R. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saikawa N, Akiyama Y, Ito K. J Struct Biol. 2004;146:123–129. doi: 10.1016/j.jsb.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 32.Sharp MD, Pogliano K. Proc Natl Acad Sci U S A. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu NJ, Dutton RJ, Pogliano K. Mol Microbiol. 2006;59:1097–1113. doi: 10.1111/j.1365-2958.2005.05004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez AR, Abanes-De Mello A, Pogliano K. J Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K. Proc Natl Acad Sci U S A. 1997;94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wakeley PR, Dorazi R, Hoa NT, Bowyer JR, Cutting SM. Mol Microbiol. 2000;36:1336–1348. doi: 10.1046/j.1365-2958.2000.01946.x. [DOI] [PubMed] [Google Scholar]
- 37.Ricca E, Cutting S, Losick R. J Bacteriol. 1992;174:3177–3184. doi: 10.1128/jb.174.10.3177-3184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubio A, Jiang X, Pogliano K. J Bacteriol. 2005;187:5000–5002. doi: 10.1128/JB.187.14.5000-5002.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Youngman P, Perkins JB, Losick R. Mol Gen Genet. 1984;195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.