Abstract
Topological properties of DNA influence its mechanical and biochemical interactions. Genomic DNA is maintained in a state of topological homeostasis by topoisomerases and is subjected to mechanical stress arising from replication and segregation. Despite their fundamental roles, the effects of topology and force have been difficult to ascertain. Developments in single-molecule manipulation techniques have enabled precise control and measurement of the topology of individual DNA molecules under tension. This minireview provides an overview of these single-molecule techniques and illustrates their unique capabilities through a number of specific examples of single-molecule measurements of DNA topology and topoisomerase activity.
Keywords: DNA Gyrase, DNA-Protein Interaction, DNA Topoisomerase, DNA Topology, Single-molecule Biophysics, Magnetic Tweezers, Optical Tweezers
Introduction
The topology of DNA, which is defined by how the two complementary single strands are intertwined, has been an important consideration since the double helical structure of DNA was first proposed by Watson and Crick in 1953 (1). The configuration of the complementary strands immediately suggested a replication mechanism in which each antiparallel strand serves as a template for a daughter strand (2). Despite the elegance of this semiconservative replication model, it requires untwisting the double helix, which poses formidable topological problems for long genomic DNA. The semiconservative model of replication was verified by Meselson and Stahl in 1958 (3), but only with the discovery of topoisomerases by James Wang in 1971 (4) was a solution to the topological problem associated with replication identified. By transiently breaking and resealing the DNA backbone, topoisomerases can relieve the excess twist that accumulates during replication (5). Despite the topology implied by the double helical model, the topological properties of DNA were not formally addressed until Vinograd et al. (6) deduced the superhelical twist in closed circular polyoma virus DNA from electron microscopic images. Since these pioneering studies, DNA topology and its regulation by topoisomerases have proved to be a pervasive factor influencing a multitude of DNA processes, including DNA packaging, condensation, transcription, chromosome segregation, and gene expression (7, 8). The fundamental importance of DNA topology is underscored by the strict conservation and necessity of topoisomerases across all cell types and some viruses (7, 9). These enzymes are crucial for maintaining eukaryotic and prokaryotic genomes in well defined topological states. Given the broad impact of topology on DNA processing, fortuitous differences between bacterial and human topoisomerases have led to effective antibiotics (10), whereas human topoisomerase inhibitors are important cancer chemotherapy agents (11).
Topology has been found to play an ever widening role in DNA metabolism within the cell (7, 12), which raises important questions. What are the mechanical properties of DNA under torsional and tensional stress, and what are the consequences of these stresses on the activity of DNA-modifying proteins? Addressing these questions is essential to understanding DNA dynamics and energetics within the cell and to discerning how the myriad of proteins that bind to and modify DNA are influenced by these properties. Despite the pervasive nature of force in vivo, it is a difficult parameter to control or ascertain in most in vitro assays. Over the past 15 years, single-molecule techniques, which complement biochemical and structural approaches, have been developed that can control and measure the topology of individual DNA molecules (13). These techniques enabled the elucidation of DNA mechanics and topology with unprecedented precision, which laid the groundwork for high-resolution measurements of topoisomerases activity. Here, after a review of DNA topology, I survey current single-molecule techniques to manipulate and measure the topology of individual DNA molecules. The power and versatility of these techniques are highlighted by examples demonstrating the contributions they have enabled in the fields of DNA topology and topoisomerase mechanism.
DNA Topology Overview
DNA topology encompasses supercoiling, knots, and catenanes. For the purpose of this minireview, however, I will restrict the discussion to supercoiling. Bates and Maxwell (14) have provided an excellent in-depth treatment of DNA topology. The topology of closed circular DNA is characterized by the linking number (Lk), which, as the name suggests, reflects the number of links between two complementary single strands. This topological quantity is physically manifested in two distinct geometric properties of DNA. Twist (Tw) refers to the number of times the two strands are twisted around each other (formally, around the helical axis), defined as positive for a right-handed helix. Writhe (Wr) refers to the geometric coiling of the double helix, which for DNA usually manifests as an interwound (plectonemic) superhelix. Positive writhe corresponds to a left-handed superhelix. A remarkable mathematical result states that Lk = Tw + Wr (15). Consequently, changes in Lk are partitioned between changes in Tw and Wr: ΔLk = ΔTw + ΔWr. For relaxed B-form DNA, the two strands twist around each other once every ∼10.5 bp; therefore, the relaxed Lk (Lk0) is equal to the relaxed Tw (Tw0) ∼ n/10.5 for n bp. If the ends of the DNA are torsionally constrained, e.g. in a closed circular plasmid, Lk can differ from its relaxed value. An increase in Lk is termed positive supercoiling, whereas a decrease in Lk is termed negative supercoiling. The fractional change in linking number is termed the specific linking difference or supercoiling density (σ): σ = (Lk − Lk0)/Lk0. Supercoiling leads to changes in both Tw and Wr. For example, negative supercoiling resulting from the reduction in Lk is accommodated by reduced Tw and the formation of right-handed plectonemes corresponding to negative Wr.
Supercoiling can be understood by considering a length of flexible tubing. Rotating one end of the tube while holding the other end fixed is akin to changing Lk. Initially, the tube will twist, and the torque (τ) stored in the tube will increase in proportion to the rotation: τ = (C/L)·Δθ, where C is the twist elasticity, L is the length of the tube, and Δθ is the angular rotation of the end. After a certain number of rotations (dependent on the material properties of the tube and the tension), the tube will buckle, forming superhelical plectonemes, which are one possible manifestation of writhe. Further rotations increase the number of plectonemes but do not increase the twist or torque on the tube. The interplay between twist and writhe can be appreciated by stretching the tube. Writhe stored in the plectonemes is converted to twist as the ends of the tube are pulled apart, and the process reverses as the ends are brought back together. This simple model captures the salient features of DNA topology under tension, although it lacks the helicity of DNA.
Single-molecule Manipulation of DNA Topology
Magnetic tweezers and a variant of optical tweezers are the two main techniques used to control and measure the topology of individual DNA molecules (16). Both configurations share a geometry in which one end of double-stranded DNA (1–45 kb) is attached to the surface of a microscope flow cell, and the free end is attached to a micron-sized particle that can be pulled and rotated. If the DNA molecule is devoid of nicks (which act as free swivels) and the attachments at both ends are rotationally constrained, rotating the particle by one full turn changes the Lk of the DNA by 1 unit. By measuring the position of the manipulated particle, the extension of the DNA can be determined with an accuracy of a few nanometers (10−9 m) or better. Extension of the DNA as a function of the linking number difference and the applied force reveals the partitioning between Tw and Wr (17).
Magnetic Tweezers
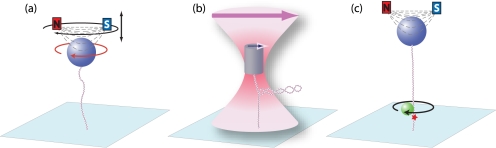
A typical magnetic tweezers instrument consists of a pair of small permanent magnets, arranged with their opposite poles separated by a small gap (∼1 mm), placed above a flow cell on an inverted microscope (Fig. 1) (13). DNA is attached to the anti-digoxigenin-coated surface of the flow cell via multiple digoxigenin moieties incorporated in one end of the DNA. The free end of the DNA is attached to a streptavidin-coated superparamagnetic bead via multiple biotin moieties. The magnets above the flow cell impose an upward force (0.1–100 pN, 10−12 N)2 on the magnetic bead that can be controlled by changing the vertical position of the magnets. The magnetic moment of the bead is entrained by the field of the external magnets so that it rotates in a one-to-one correspondence with magnet rotation. A high-magnification objective images the bead onto a charged-couple device camera, and the three-dimensional position of the bead is obtained with 2–5-nm accuracy in real time (30–200 frames/s) by image processing (18). With this relatively simple setup, Lk of individual DNA molecules, subjected to well controlled pulling forces, can be precisely controlled, and the corresponding changes in DNA extension can be measured with high accuracy.
FIGURE 1.
Single-molecule techniques to control DNA topology (not to scale). a, magnetic tweezers consist of two magnets (red and blue) held above a microscope flow cell in which a magnetic bead (blue) is tethered to the surface (light blue) by a single DNA molecule (red and blue). The magnets impose an upward force on the beads, which depends on the vertical position of the magnets (black arrow). Rotating the magnets (black curved arrow) rotates the bead (red curved arrow), thereby changing the linking number of the DNA. b, optical rotation uses a focused laser (pink) to apply force on a quartz cylinder (blue) that is tethered to the surface of the flow cell by a molecule of DNA. The optical axis of the cylinder (blue arrow) experiences a torque that tends to align it with the polarization direction of the laser (pink arrow), which is focused by a microscope objective below the flow cell. c, rotor bead tracking uses a reporter bead (green) attached near a specific nick in the DNA molecule (red star). A magnetic bead (blue) at the distal end of the DNA allows torque and tension to be applied.
Optical Rotation and Torque
Optical tweezers employ a highly focused laser beam to trap micron-scale objects in solution (13), effectively creating a three-dimensional spring that provides a restoring force on the trapped object. Optical tweezers can apply force (1–100 pN) and measure the position of a trapped object with nanometer precision. Conventional optical tweezers are rotationally isotropic; hence, torque cannot be applied on a trapped particle. However, an optically active particle, i.e. that rotates the polarization of light, can be rotated by manipulating the trapping laser polarization (19). In practice, this has been achieved by fabricating micron-sized quartz cylinders with a strong birefringence perpendicular to their long axis, resulting in their alignment with the polarization of the trapping laser (Fig. 1) (20). Rotating the trapping laser polarization imposes a torque on the cylinder. By tethering the cylinder to the surface of a flow cell in an analogous manner as for magnetic tweezers, Lk and torque can be controlled, and the position of the cylinder can be tracked with high spatial and temporal resolution. Whereas this approach is significantly more complex than magnetic tweezers, it offers a means of directly measuring and controlling torque and affords higher temporal resolution (∼1 ms).
In a third “rotor bead” assay, a small non-magnetic bead is attached adjacent to a nick near the surface-bound end of the DNA in a magnetic tweezers assay (Fig. 1) (21). The nick provides a free swivel about which the DNA can rotate. Tracking the reporter bead provides a direct readout of rotation and torque.
Single-molecule Measurements of DNA Topology
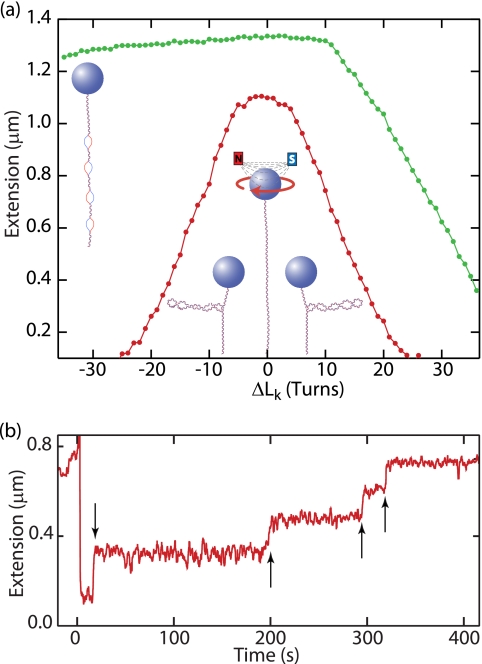
Strick et al. (22) made the first single-molecule measurements of DNA topology using magnetic tweezers to measure the extension as a function of Lk and force for individual molecules of λ-phage DNA. The extension-rotation curves obtained at different forces reveal how twist and writhe are partitioned (Fig. 2). At low force (<0.45 pN), the curves are symmetric, with a small change in extension around Lk0 that reflects a change in only Tw. Past a critical Lk, the extension decreases abruptly, followed by a linear decrease in extension with successive turns. This reflects the buckling of the DNA to form a plectoneme. Further changes in Lk are entirely accommodated by changes in Wr that increase the number of plectonemic crossings and decrease the extension of the DNA by ∼50 nm/turn. At higher forces, the helicity of the DNA duplex becomes apparent as the curves are no longer symmetric. Positive supercoiling is qualitatively similar to the low-force behavior, although the buckling transition occurs at a larger number of turns, and the slope of the linear plectonemic region is smaller. For negative supercoiling, however, it is energetically more favorable to accommodate the decrease in Lk by decreasing Tw rather than Wr, which leads to local melting of ∼10.5 bp of DNA/negative turn. As a result, extension changes very little as Lk is decreased.
FIGURE 2.
Single-molecule measurements of DNA topology and topoisomerase activity. a, extension-rotation (ΔLk) relationship for a 3-kb DNA molecule (Y. Seol and K. C. Neuman, unpublished data). At low force (0.4 pN; red circles), the extension decreases symmetrically as the linking number is changed from Lk0. At ∼5 turns, the DNA buckles, and subsequent turns increase the number of plectoneme crossings (inset schematics) and decrease the extension. At higher force (1.6 pN; green circles), the extension for positive supercoiling is similar, although the buckling transition occurs at ∼11 turns. However, for negative supercoiling, the DNA melts to form locally unwound regions (schematic to the left). b, single-molecule measurement of supercoil relaxation by topoisomerase IV (adapted from Ref. 34). At time 0, the DNA molecule is negatively supercoiled, decreasing Wr by 8 though the introduction of eight right-handed crossings, which decreases the extension from the relaxed length of ∼0.7 μm. Individual relaxation events are observed as abrupt increases in extension (arrows).
In subsequent measurements, Bryant et al. (21) used the rotor bead assay to map out the torque-force phase diagram and directly measure the twist elasticity of DNA. They applied large forces on the DNA to avoid the formation of plectonemes and introduced turns by rotating a bead attached to one end of the DNA while preventing the reporter bead from rotating by introducing flow. Stopping the flow allowed the small reporter bead to rotate, driven by the torque in the DNA. By tracking the rotational velocity of the bead and calibrating it against viscous drag, they were able to directly measure the torque in the DNA. This technique provided a direct measure of the twist elasticity (C = 400 ± 40 pN·nm2), which had previously been measured by several indirect methods that gave conflicting results (17). By recording the torque at varying tensions in the DNA, a torque-force phase diagram of DNA was obtained. Five distinct phases were defined: B-form DNA; unwound or melted DNA past a torque of approximately −10 pN·nm; P-DNA, an overwound helical structure (23) past a torque of ∼10 pN·nm; a supercoiled form of P-DNA at lower forces beyond the critical torque; and overstretched DNA, which is a partially unwound form of DNA obtained at high (>65 pN) stretching forces (21).
More recently, direct optical tweezers measurements of torque have been used to determine the twist elasticity of DNA and the force dependence of the critical buckling torque at which DNA adopts a plectonemic structure (24). The value of C (360 ± 12 pN·nm2) is consistent with previous single-molecule measurements (17). The torque-force relationship provides an important parameter for single-molecule experiments that exploit the constant torque regime past the buckling threshold to maintain a constant torque (see below). These measurements demonstrate that the torque calculated from a simple elastic model of DNA (17) significantly overestimates the torque (24). The data agreed well with a recent model that incorporates torsional elasticity of plectonemes and force-dependent twist elasticity for linear DNA (25).
These micromanipulation techniques have permitted detailed measurements of the mechanical properties of individual DNA molecules subjected to varying degrees of torque and tension. The results of these measurements provide insight into the mechanical properties of genomic DNA subjected to supercoiling and tension and have spurred complementary theoretical and computational approaches (25). Finally, single-molecule topology measurements have paved the way for high-resolution kinetic measurements of topoisomerases (26).
Single-molecule Measurements of Topoisomerase Activity
Once the relationship between Lk and DNA extension has been established, it can be used to measure changes in Lk in real time as changes in extension can be related to changes in linking number (Fig. 2). Single-molecule approaches provide molecular level details of the physical mechanisms of topoisomerase activity. They complement ensemble biochemical assays in several important aspects. Enzymatic rates of individual enzymes are insensitive to the presence of inactive enzyme, which can artificially lower the average rate measured in an ensemble assay. Furthermore, the enzymatic processivity, defined as the number of catalysis cycles per binding event, can be directly measured from single-molecule records (26). This is facilitated by the high degree of positive and negative supercoiling achievable (26). Finally, tension in the DNA can affect enzymatic rates and DNA mechanics.
Strick et al. (27) first measured the relaxation of individual DNA molecules by a type II topoisomerase that passes a segment of duplex DNA (transfer segment) through a transient double-strand break in a second segment (gate segment) in an ATP-dependent reaction, thereby changing Lk by 2 units (5, 9). Using a magnetic tweezers, the DNA linking number was increased or decreased to introduce positive or negative plectonemes that were relaxed by topoisomerase II. Relaxation was measured as the stepwise increase in the height of the magnetic bead accompanying the change in Lk (Fig. 2). At low ATP concentrations, Lk changed in steps of 2, consistent with the removal of one plectonemic crossing (28) and with the two-gate mechanism of topoisomerase II (29).
Unlike most other type II topoisomerases, topoisomerase IV, a bacterial type II topoisomerase, relaxes positively supercoiled DNA at least 20-fold faster than negatively supercoiled DNA (30). Possible mechanisms explaining this behavior include preferential binding of positively versus negatively supercoiled DNA and activity that is sensitive to the differences in the twist or writhe of the supercoiled DNA (31). Two elegant experiments distinguished between these possibilities by attaching two torsionally unconstrained (nicked) DNA molecules between a rotatable bead and a fixed surface (31, 32). The two DNA molecules were twisted around one another, or “braided,” by rotating the bead. Monte Carlo simulations and measurements of braided DNA indicated that it adopts a conformation similar to that of a superhelical plectoneme (33). The braided molecules could therefore adopt positive or negative writhe in the absence of twist. Relaxation of braided molecules by topoisomerase IV, but not the homologous yeast topoisomerase, displayed an almost absolute preference for positive versus negative writhe (32). This result, in conjunction with ensemble measurements demonstrating that topoisomerase IV has no significant binding preference for positively supercoiled DNA (31), is consistent with the proposal that chiral discrimination is based on differences in writhe (31). Further experiments demonstrated that the sign of the writhe has little effect on the rate of strand passage, but it strongly influences the processivity (34).
DNA gyrase, the second bacterial type II topoisomerase, is the only topoisomerase that uses the energy of ATP hydrolysis to negatively supercoil DNA (5). In vivo, gyrase maintains the bacterial genome in a negatively supercoiled state (12). On binding to DNA, gyrase wraps a segment of DNA into a left-handed crossing, which is converted to a right-handed (negative writhe) crossing by the strand passage reaction. Gore et al. (35) used the rotor bead assay to investigate the details of this reaction. Tracking rotation of the reporter bead revealed that gyrase imposes ∼1.3 positive turns upon binding to the DNA substrate in the absence of ATP. Binding kinetics were extremely sensitive to the tension in the DNA, from which the amount of DNA wrapped was estimated to be ∼110 bp (35). In the presence of ATP, successive steps of two complete negative turns were observed, consistent with the decrease in Lk by 2 per reaction cycle (28). Interestingly, the negative supercoiling rate was insensitive to the force on the DNA, but the processivity and initiation rate were exquisitely sensitive to force. These results led to a mechanochemical model for gyrase in which the tension-dependent wrapping is in kinetic competition with dissociation (35). In follow-up experiments, Nöllmann et al. (36) further explored the mechanochemistry of gyrase by measuring its activity as a function of force and DNA topology. They described three modes of activity that arise under different regimes of force and topology. At low forces (less than ∼0.5 pN), gyrase introduces negative supercoils into DNA in the presence of ATP through a DNA-wrapping mechanism. At higher forces, gyrase can remove positive writhe and decatenate DNA in a wrapping-independent mode akin to topoisomerase IV. Finally, in the absence of ATP, high concentrations of gyrase will slowly remove negative writhe.
The mechanisms of supercoil relaxation by type I topoisomerases, which transiently cut one strand of DNA in an isoenergetic process that does not require an external energy source, have also been explored with single-molecule manipulation techniques. Koster et al. (37) used magnetic tweezers to measure supercoil relaxation by topoisomerase IB, which introduces a transient nick in the DNA backbone that allows a variable number of supercoils to be relaxed (9). By tracking individual topoisomerase IB relaxation events, the authors determined that the number of supercoils relaxed is exponentially distributed. Moreover, by taking advantage of the constant torque regime of the DNA force-linking number relation, they were able to show that the rate and extent of relaxation depended on the torque. These results suggested a model in which rotation of the DNA at the transiently generated nick is hindered by rotational friction within the enzyme (37). In follow-up experiments, they determined how topotecan, a potent chemotherapy agent, affects supercoil relaxation by topoisomerase IB (38).
In addition to the study of topoisomerases, single-molecule approaches for controlling and measuring topology have been applied to a number of protein-DNA interaction measurements. For example, Revyakin et al. (39) measured promoter opening and transcription initiation by bacterial RNA polymerase on supercoiled DNA molecules. Promoter opening unwinds the DNA, and the extent of unwinding can be measured from the introduction of writhe that compensates for the decrease in twist. By comparing the change in extension on promoter opening for both positively and negatively supercoiled DNA, 1.2 turns of helix melting could be distinguished from 15 nm of compaction associated with promoter opening. Moreover, promoter opening was found to be highly sensitive to DNA twist. More recently, DNA “scrunching” during abortive initiation and promoter escape by RNA polymerase was directly observed (40). These experiments relied in part on the large amplification of small changes in twist due to the generation of compensating writhe. Similar approaches have been used to estimate the helical tracking of proteins as they translocate on DNA (41, 42) and to estimate the amount of twist that is trapped in a protein-mediated DNA loop (43).
Conclusions and Outlook
Single-molecule manipulation techniques have opened up new and exciting avenues of DNA topology research by permitting detailed measurements of DNA mechanics as a function of topology and tension. These techniques have enabled fundamental contributions to our understanding of DNA topology and topoisomerase mechanochemistry. Expanding and extending these results will shed light on the influence of topology on DNA interactions and bring us closer to understanding cellular DNA dynamics and interactions. Recent work investigating the topological properties of chromatin fibers (44) is a particularly intriguing direction, as is the prospect of directly measuring the torque of rotary motors such as RNA polymerases. Addressing these questions will undoubtedly drive continuing improvements in single-molecule techniques.
Supplementary Material
Acknowledgments
I thank Amanda Gentry, Richard Neuman, and members of the Neuman laboratory for critical reading of the manuscript.
This work was supported by the NHLBI Intramural Research Program of the National Institutes of Health. This is the second article in the Thematic Minireview Series on Single-molecule Measurements in Biochemistry and Molecular Biology. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- N
- newtons.
REFERENCES
- 1.Watson J. D., Crick F. H. (1953) Nature 171, 737–738 [DOI] [PubMed] [Google Scholar]
- 2.Watson J. D., Crick F. H. (1953) Nature 171, 964–967 [DOI] [PubMed] [Google Scholar]
- 3.Meselson M., Stahl F. W. (1958) Proc. Natl. Acad. Sci. U.S.A. 44, 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J. C. (1971) J. Mol. Biol. 55, 523–533 [DOI] [PubMed] [Google Scholar]
- 5.Wang J. C. (1998) Q. Rev. Biophys. 31, 107–144 [DOI] [PubMed] [Google Scholar]
- 6.Vinograd J., Lebowitz J., Radloff R., Watson R., Laipis P. (1965) Proc. Natl. Acad. Sci. U.S.A. 53, 1104–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J. C. (2002) Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 8.Travers A., Muskhelishvili G. (2005) Nat. Rev. Microbiol. 3, 157–169 [DOI] [PubMed] [Google Scholar]
- 9.Champoux J. J. (2001) Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 10.Drlica K., Malik M. (2003) Curr. Top. Med. Chem. 3, 249–282 [DOI] [PubMed] [Google Scholar]
- 11.Li T. K., Liu L. F. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 53–77 [DOI] [PubMed] [Google Scholar]
- 12.Espeli O., Marians K. J. (2004) Mol. Microbiol. 52, 925–931 [DOI] [PubMed] [Google Scholar]
- 13.Neuman K. C., Lionnet T., Allemand J. F. (2007) Annu. Rev. Mater. Res. 37, 33–67 [Google Scholar]
- 14.Bates A. D., Maxwell A. (2005) DNA Topology, 2nd Ed., Oxford University Press, New York [Google Scholar]
- 15.White J. H. (1969) Am. J. Math. 91, 693–728 [Google Scholar]
- 16.Neuman K. C., Nagy A. (2008) Nat. Methods 5, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charvin G., Allemand J. F., Strick T. R., Bensimon D., Croquette V. (2004) Contemp. Phys. 45, 383–403 [Google Scholar]
- 18.Lionnet T., Allemand J.-F., Revyakin A., Strick T. R., Saleh O. A., Bensimon D., Croquette V. (2008) in Single-Molecule Techniques: A Laboratory Manual (Selvin P. R., Ha T. eds) pp. 347–369, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19.La Porta A., Wang M. D. (2004) Phys. Rev. Lett 92, 190801-1–190801-4 [DOI] [PubMed] [Google Scholar]
- 20.Deufel C., Forth S., Simmons C. R., Dejgosha S., Wang M. D. (2007) Nat. Methods 4, 223–225 [DOI] [PubMed] [Google Scholar]
- 21.Bryant Z., Stone M. D., Gore J., Smith S. B., Cozzarelli N. R., Bustamante C. (2003) Nature 424, 338–341 [DOI] [PubMed] [Google Scholar]
- 22.Strick T. R., Allemand J. F., Bensimon D., Bensimon A., Croquette V. (1996) Science 271, 1835–1837 [DOI] [PubMed] [Google Scholar]
- 23.Allemand J. F., Bensimon D., Lavery R., Croquette V. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14152–14157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forth S., Deufel C., Sheinin M. Y., Daniels B., Sethna J. P., Wang M. D. (2008) Phys. Rev. Lett. 100, 148301-1–148301-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marko J. F. (2007) Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 76, 021926-1–021926-4 [Google Scholar]
- 26.Charvin G., Strick T. R., Bensimon D., Croquette V. (2005) Annu. Rev. Biophys. Biomol. Struct. 34, 201–219 [DOI] [PubMed] [Google Scholar]
- 27.Strick T. R., Croquette V., Bensimon D. (2000) Nature 404, 901–904 [DOI] [PubMed] [Google Scholar]
- 28.Brown P. O., Cozzarelli N. R. (1979) Science 206, 1081–1083 [DOI] [PubMed] [Google Scholar]
- 29.Roca J., Berger J. M., Harrison S. C., Wang J. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4057–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crisona N. J., Strick T. R., Bensimon D., Croquette V., Cozzarelli N. R. (2000) Genes Dev. 14, 2881–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone M. D., Bryant Z., Crisona N. J., Smith S. B., Vologodskii A., Bustamante C., Cozzarelli N. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8654–8659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charvin G., Bensimon D., Croquette V. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9820–9825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charvin G., Vologodskii A., Bensimon D., Croquette V. (2005) Biophys. J. 88, 4124–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuman K. C., Charvin G., Bensimon D., Croquette V. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gore J., Bryant Z., Stone M. D., Nöllmann M., Cozzarelli N. R., Bustamante C. (2006) Nature 439, 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nöllmann M., Stone M. D., Bryant Z., Gore J., Crisona N. J., Hong S. C., Mitelheiser S., Maxwell A., Bustamante C., Cozzarelli N. R. (2007) Nat. Struct. Mol. Biol. 14, 264–271 [DOI] [PubMed] [Google Scholar]
- 37.Koster D. A., Croquette V., Dekker C., Shuman S., Dekker N. H. (2005) Nature 434, 671–674 [DOI] [PubMed] [Google Scholar]
- 38.Koster D. A., Palle K., Bot E. S., Bjornsti M. A., Dekker N. H. (2007) Nature 448, 213–217 [DOI] [PubMed] [Google Scholar]
- 39.Revyakin A., Ebright R. H., Strick T. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4776–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Revyakin A., Liu C., Ebright R. H., Strick T. R. (2006) Science 314, 1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seidel R., van Noort J., van der Scheer C., Bloom J. G., Dekker N. H., Dutta C. F., Blundell A., Robinson T., Firman K., Dekker C. (2004) Nat. Struct. Mol. Biol. 11, 838–843 [DOI] [PubMed] [Google Scholar]
- 42.Saleh O. A., Pérals C., Barre F. X., Allemand J. F. (2004) EMBO J. 23, 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lia G., Praly E., Ferreira H., Stockdale C., Tse-Dinh Y. C., Dunlap D., Croquette V., Bensimon D., Owen-Hughes T. (2006) Mol. Cell 21, 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bancaud A., Conde e Silva N., Barbi M., Wagner G., Allemand J. F., Mozziconacci J., Lavelle C., Croquette V., Victor J. M., Prunell A., Viovy J. L. (2006) Nat. Struct. Mol. Biol. 13, 444–450 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.