Abstract
In DNA replication, the antiparallel nature of the parental duplex imposes certain constraints on the activity of the DNA polymerases that synthesize new DNA. The leading-strand polymerase advances in a continuous fashion, but the lagging-strand polymerase is forced to restart at short intervals. In several prokaryotic systems studied so far, this problem is solved by the formation of a loop in the lagging strand of the replication fork to reorient the lagging-strand DNA polymerase so that it advances in parallel with the leading-strand polymerase. The replication loop grows and shrinks during each cycle of Okazaki fragment synthesis. The timing of Okazaki fragment synthesis and loop formation is determined by a subtle interplay of enzymatic activities at the fork. Recent developments in single-molecule techniques have enabled the direct observation of these processes and have greatly contributed to a better understanding of the dynamic nature of the replication fork. Here, we will review recent experimental advances, present the current models, and discuss some of the exciting developments in the field.
Keywords: DNA/Polymerase, DNA/Primase, DNA-Protein Interaction, DNA/Replication, Nucleic Acid/Enzymology, Protein-Nucleic Acid Interaction, Electron Microscopy, Single-molecule Biophysics
The Replisome
The replisome operates according to a set of highly conserved principles (reviewed in Refs. 1–3). A helicase unwinds the parental double-stranded DNA (dsDNA),3 allowing two DNA polymerases, complexed with processivity factors, to each synthesize DNA on the resulting single-stranded templates. The 5′ to 3′ direction of polymerase-dependent nucleic acid synthesis permits one of these enzymes to synthesize DNA in a continuous fashion on the leading strand but requires the polymerase on the lagging strand to periodically restart using short RNA primers made by a DNA primase. The discontinuous synthesis of DNA on the lagging strand gives rise to a succession of Okazaki fragments that are later processed and ligated into one continuous strand (4). Single-stranded DNA (ssDNA)-binding proteins remove any secondary structure that may inhibit synthesis by the DNA polymerase. These basic activities have been fully reconstituted in vitro in three prokaryotic replication systems, Escherichia coli and its bacteriophages T7 and T4 (reviewed in Refs. 1–3). In this minireview, we will use the minimal T7 replication model system to illustrate the key mechanisms underlying coordination at the replication fork and comment on the other two systems as needed.
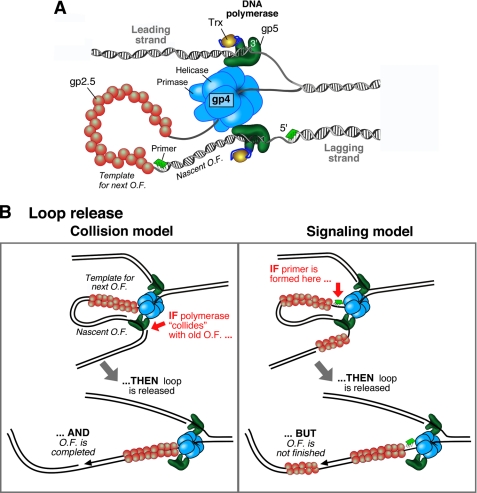
In bacteriophage T7, a fully functional replisome can be reconstituted with only four proteins (Fig. 1A) (3). The T7 DNA polymerase consists of a 1:1 complex of the T7 gene 5 protein (gp5) and the E. coli thioredoxin (Trx) processivity factor; this complex will be referred to as gp5-Trx. The T7 gene 4 protein (gp4) provides both helicase and primase activities. The helicase activity is located in the C-terminal half, and the primase activity resides in the N-terminal half. Finally, the T7 gene 2.5 protein (gp2.5) codes for the ssDNA-binding activity. The replisomes of E. coli and T4 are more complex and require several extra proteins such as processivity clamps and clamp loaders (1–3).
FIGURE 1.
A, organization of the bacteriophage T7 replication fork. gp4 encircles the lagging strand and mediates both the unwinding of dsDNA via its helicase domain and the synthesis of RNA primers via the primase domain. The T7 DNA polymerases are stably bound to gp4 and incorporate nucleotides on the leading and lagging strands. The DNA polymerase is a 1:1 complex of T7 gp5 and E. coli Trx. The ssDNA extruded behind the helicase is coated by the ssDNA-binding protein gp2.5. A replication loop is formed in the lagging strand to align it with the leading strand. The lagging-strand DNA polymerase initiates Okazaki fragment (O.F.) synthesis using RNA primers (green segments). B, schematic depiction of the two models that describe replication loop release. In the collision model (left panel), the replication loop is released when the lagging-strand polymerase collides with the 5′ terminus of the previous Okazaki fragment. In the signaling model (right panel), the synthesis of a new primer triggers the release of the replication loop prior to the completion of the nascent Okazaki fragment.
Protein-protein interactions coordinate the various activities of the replisome. In T7, the interaction of Trx with gp5 increases processivity of polymerization from 1–15 nucleotides to 700–1000 nucleotides (5–7). The interaction of gp4 with gp5-Trx coordinates DNA unwinding with nucleotide polymerization on duplex DNA (8–10) and increases the processivity of leading-strand synthesis to tens of kb (6). The presence of the helicase and primase within a single polypeptide allows the primase immediate access to the ssDNA generated behind the translocating helicase (Fig. 1A). The primase interacts with the lagging-strand polymerase to place the primer in the active site of the polymerase (11). gp2.5 interacts with both gp5-Trx and gp4 to stimulate polymerization and primer synthesis, respectively (12, 13).
Coordination of Leading- and Lagging-strand Synthesis
Highly processive synthesis of DNA on the leading and lagging strands relies on a stable association of polymerases with the replication fork (14–17). To allow for the opposite directions of synthesis, Alberts et al. (17) proposed the “trombone model,” in which a loop reorients the lagging-strand DNA so that its polymerase can replicate DNA in parallel with the leading-strand polymerase while bound to the replisome (Fig. 1A). This replication loop grows and shrinks, like the slide of a trombone, during each cycle of Okazaki fragment synthesis. The replication loop contains both the nascent Okazaki fragment produced by the lagging-strand DNA polymerase and the single-stranded product of the helicase. The replication loop is held in place by the physical interaction of the lagging-strand gp5-Trx with gp4. In E. coli and T4, the homodimerization of the DNA polymerase provides a structure to hold the replication loop. These homodimeric DNA polymerases are anchored to the fork through their interactions with the helicase as in the case of the T7 system (1–3).
Formation of the replication loop is initiated by primer synthesis. Primer synthesis is a template-directed process that requires the primase to first recognize a specific sequence on the single-stranded helicase product. Using only ATP and CTP, the T7 primase catalyzes the synthesis of the tetraribonucleotides pppACCC, pppACCA, and pppACAC (reviewed in Ref. 18). The primase remains bound to the primer to prevent the dissociation of the short tetraribonucleotide from the template until transferred to the lagging-strand polymerase (11). The subsequent extension of the primer into an Okazaki fragment results in the formation of the replication loop.
The observation that synthesis on the leading and lagging strands proceeds at identical rates (19, 20) suggests a coordination between the polymerization activities on both strands. In T7, at least three steps are required to reset the replication loop at the end of synthesis of an Okazaki fragment: release of the Okazaki fragment, the synthesis of a primer, and its handoff to the lagging-strand polymerase. With the complexity of the multiple steps involved in lagging-strand synthesis and the relatively straightforward continuous incorporation of nucleotides on the leading strand, it is not clear how this coordination is maintained.
Efforts of a large number of researchers have resulted in a progressively refined picture of the coordination of events at the replication fork. In particular, two models have been proposed that describe how and when the replication loop is released. In one mechanism called the signaling model, the synthesis of a primer triggers the immediate release of the replication loop whether or not the nascent Okazaki fragment is completed (Fig. 1B, right panel). Supporting this model, an analysis of Okazaki fragment lengths in the T4 system demonstrated the existence of gaps of ssDNA between Okazaki fragments, suggesting the release of replication loops before completion of the nascent Okazaki fragments (21). Also, the size of Okazaki fragments produced by E. coli, T7, and T4 replisomes is dependent on primase activity (21–23). Furthermore, Okazaki fragment size is sensitive to helicase-primase interactions in E. coli (25) and dependent on the concentration of two important components for the lagging-strand synthesis in T4, the clamp and clamp loader (21).
In an alternative model of replication loop release called the collision mechanism, the lagging-strand DNA polymerase dissociates from the fork after encountering the 5′ terminus of the previously synthesized Okazaki fragment (Fig. 1B, left panel). In support of this model, the T4 DNA polymerase and E. coli DNA polymerase III holoenzyme have been shown to dissociate rapidly when they encounter a 5′ terminus while extending a primer on ssDNA (23, 26–29). Even though a large amount of suggestive evidence has been reported to support both the signaling and collision mechanisms, it is unclear which method is operative within the replisome during coordinated DNA replication.
Characterization of Replication Loop Length Distributions by Electron Microscopy
Okazaki fragments as visualized on denaturing agarose gels display a wide range of lengths (19). This distribution could result from individual replisomes producing differently sized Okazaki fragments or individual replisomes producing a constant length of Okazaki fragment, with different lengths from one replisome to another. The exact nature of the length distributions within individual replisomes is important to understand the timing mechanisms underlying Okazaki fragment synthesis. In the collision model, the lagging-strand DNA polymerase will complete the nascent Okazaki fragment before a new primer is utilized and the next loop is formed. If a primer is readily available upon completion of the nascent fragment, the next Okazaki fragment is expected to be of equal length. If a primer is not available upon collision, leading-strand synthesis needs to continue to allow the primase to search for its recognition sequence to initiate primer synthesis. In this case, the size of the subsequent Okazaki fragment would increase by the extra amount of ssDNA generated during the primase search. Therefore, the collision model predicts a gradual increase in Okazaki fragment size as the replisome progresses. In the signaling model, primer synthesis takes place before the lagging-strand polymerase encounters the previous Okazaki fragment and signals the premature release of the replication loop. As a result, the amount of ssDNA accumulated in the loop that is available for the next Okazaki fragment will decrease, which results in a gradual reduction of the Okazaki fragment size as the replisome progresses.
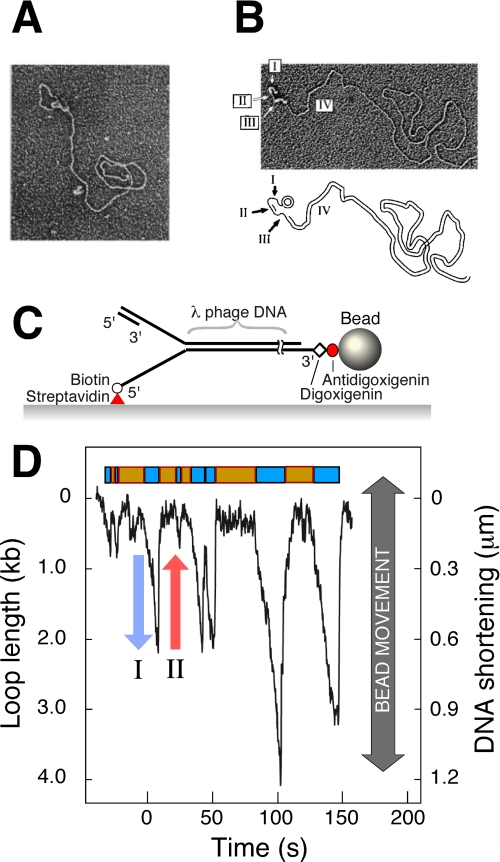
The ability of electron microscopy (EM) to visualize structural properties of replication intermediates and products has played an important role in understanding the origin of the heterogeneity in Okazaki fragment length. The imaging of rolling-circle replication intermediates revealed the presence of a dsDNA replication loop extending from the protein mass at the replication fork (Fig. 2A). Not only did these experiments provide direct visual confirmation of the trombone model, they also allowed a quantitative characterization of loop lengths (22, 30, 31). Interestingly, only half of the replication intermediates showed a replication loop. To visualize the ssDNA, the reaction product was deproteinized and then stretched by incubation with E. coli SSB (single-stranded DNA-binding protein). Nearly half of the DNA molecules contained two stretches of ssDNA flanking the nascent Okazaki fragment, consistent with the configuration of a replication loop (Fig. 2B). The remaining half showed dsDNA flanked by only one stretch of ssDNA and likely corresponds to those intermediate states that do not involve a replication loop. In these experiments, the majority of the Okazaki fragments were completed, which makes it difficult to obtain precise information on their length distribution. To circumvent this problem, the T4 replication system was used with an exonuclease-deficient T4 DNA polymerase capable of strand displacement activity. The short ssDNA flaps that it generates at the junction between Okazaki fragments can be used to precisely measure the length of each fragment (32). These experiments revealed that the majority of replisomes produced Okazaki fragments with random length. The absence of a clear trend in Okazaki fragment length within individual replisomes prevented discrimination between the various models of loop release.
FIGURE 2.
A, observation of replication loops by EM. Coordinated DNA synthesis is carried out on a 70-bp minicircle substrate. The replication proteins appear as a globular object. The dsDNA loop extending from the replisome represents the nascent Okazaki fragment. Both the small minicircle and the condensed gp2.5-ssDNA complex are obscured by the protein mass containing the replisome. The figure is from Ref. 22. B, observation of Okazaki fragments by EM. The replication reaction was carried out as described for A, deproteinated, and treated with SSB. SSB extends ssDNA and enables its visualization as a DNA segment with increased diameter. Segment I indicates the template for the next Okazaki fragment produced by the helicase, segment II is the nascent Okazaki fragment, segment III is the template of the nascent Okazaki fragment, and segment IV is the long dsDNA of previously synthesized Okazaki fragments. The figure is from Ref. 22. C, flow-stretching individual DNA molecules. Duplex λ-DNA (48.5 kb) is modified to contain a replication fork. The lagging strand of the forked end is coupled to the surface. The other end of the DNA is attached to a bead. A constant laminar flow applies a well controlled drag force to the bead and stretches the DNA molecule. The length of the individual DNA molecules is measured by imaging the beads and tracking their positions. The figure is from Ref. 38. D, dynamic single-molecule observation of replication loops produced by individual replisomes. The trajectory shows a time course of the length of a single DNA molecule during replication. The DNA shortening corresponds to loop growth during leading- and lagging-strand synthesis (blue arrow) and is followed by a rapid length increase when a loop is released (red arrow). The lag phases between looping events and the loop growth phases are shown as orange and cyan boxes, respectively. The figure is from Ref. 38.
The fixed proteins in the EM images appear as a globular mass at the replication fork (Fig. 2A). To obtain information on the architecture of the replisomes, various components of the T4 replication proteins were biotin-tagged, and their location was mapped using nanoscale DNA biopointers (33). The combination of observing both proteins and DNA provided detailed information on several key intermediary steps at the replication fork. Replication intermediates containing the replication loop and two markers for leading- and lagging-strand polymerases were imaged, providing direct evidence for their association with the replisome. Some of these molecules contain a third polymerase that is not associated with the replisome, extending previously synthesized Okazaki fragments that remained unfinished. This observation is of particular interest for the signaling model. In the signaling model, gaps of ssDNA will be generated as a result of the premature release of the replication loop. Some of these gaps might be large and will need a processive polymerase to be filled. The EM images provide a model in which the lagging-strand polymerase departs the replisome with the incomplete nascent Okazaki fragment, and the new primer is extended by another polymerase bound at the replication fork. This picture is in agreement with biochemical studies that showed the presence of three polymerases in the E. coli replisome (34) and the observation that DNA polymerases can be recruited from solution to exchange with polymerases at the fork (8, 35, 36).
Observation of Replication Loop Formation and Release by Dynamic Single-molecule Methods
The static nature of EM studies makes it challenging to infer information on the dynamics of replication loop formation and release. Understanding the molecular mechanisms underlying replication loop dynamics and the timeline controlling the various enzymatic activities at the fork requires the direct observation of loops while being formed and released. Recent advances in imaging and molecular manipulation techniques have made it possible to observe individual molecules and record “molecular movies” that provide insight into their dynamics and reaction mechanisms (37).
In the T7 replication system, the replisome has been reconstituted and dynamically visualized at the single-molecule level (6, 38). Individual DNA molecules were stretched by laminar flow, and their lengths were monitored by tracking the positions of small beads attached to the ends of the DNA molecules (Fig. 2C). Conversion from dsDNA to ssDNA was monitored through a decrease in total DNA length at the low force used. By using a forked DNA template, a complex of one gp5-Trx and gp4 could be assembled on one end of the DNA molecule. Leading-strand synthesis catalyzed by gp5-Trx converts one DNA strand arising from gp4 helicase activity into dsDNA. In the absence of the lagging-strand gp5-Trx, the lagging strand will remain in the single-stranded form. By attaching the DNA to the surface of the flow cell by the 5′-end of the lagging strand, leading-strand synthesis could be detected by an effective shortening of the DNA (6).
The ability to monitor leading-strand synthesis in real time provides a powerful assay to monitor fork kinetics during the synthesis of a primer on the lagging strand. Upon activation of the primase activity by the addition of the required ribonucleotides, the leading-strand synthesis displayed transient pauses (6) with an average duration in agreement with the known kinetics of primer synthesis (39, 40). Lowering ATP and CTP concentrations resulted in longer pauses,4 confirming that the observed pauses were caused by the synthesis of a primer on the lagging strand. These observations suggest how the slow enzymatic priming on the lagging strand takes place without leading-strand synthesis progressing too far ahead of lagging-strand synthesis. Recent single-molecule experiments on the T7 leading-strand synthesis reaction (41) and the T4 helicase-primase complex (42) suggest an alternative mechanism to coordinate leading- and lagging-strand synthesis. These studies demonstrated the formation of a small ssDNA loop as a result of the continuation of leading-strand synthesis (in the T7 study) and helicase activity (in the T4 study) during primer synthesis. Further work is needed to reconcile these two models.
Using similar flow-stretching techniques as described above, Hamdan et al. (38) recently visualized the growth and release dynamics of replication loops produced by single replisomes. During replication, individual DNA molecules show repeated cycles of DNA shortening (Fig. 2D, blue arrow) and lengthening (red arrow) as a result of the formation and release, respectively, of a replication loop in the surface-tethered lagging strand. In the presence of gp2.5, ssDNA has roughly the same length per nucleotide as dsDNA. As a result, the assay reports exclusively on loop formation and is insensitive to length differences between ssDNA and dsDNA. The average rate of DNA shortening observed during loop growth is nearly twice the rate observed for leading-strand synthesis alone (6), consistent with the notion that loop growth is supported by both leading- and lagging-strand synthesis. This rate of fork movement has been confirmed at the single-molecule level using fluorescently stained M13 rolling circles (43).
The single-molecule DNA length measurements reveal that the release of a replication loop and the initiation of a new one are separated by a lag phase during which the overall length of DNA does not change. Because there is no contrast between ssDNA and dsDNA in the presence of gp2.5, it cannot be directly determined whether the replisome is continuing DNA synthesis without a replication loop or whether the complex has halted synthesis to allow other enzymatic steps, such as primer synthesis and handoff, to take place in preparation for the next replication loop. From these single-molecule experiments, it appeared that half of the time, the replisome was not engaged in the production of a replication loop (Fig. 2D). This observation is consistent with EM, which revealed that only half of the active replisomes contained a loop (22, 30, 31, 33).
The observation of replication loop release and a lag preceding the formation of the next loop enables the characterization of the molecular mechanisms by which the replication loops are released and the replication cycle is reset. T7 primer synthesis takes place in two distinct steps (24, 39, 44). First, the primase condenses ATP and CTP to form pppAC. Subsequently, pppAC is extended in a much slower step to a full-length tetraribonucleotide primer in a sequence-dependent manner (24, 39, 44). T7 DNA primase can utilize preformed pAC and extend it efficiently using only ATP and CTP (24). Therefore, the requirement of the condensation step can be bypassed. In the coordinated replication reaction, reducing the concentrations of ATP and CTP results in significant increase in both loop length and lag time. The replication loop length can be restored under low ATP and CTP concentration upon providing the reaction with a high concentration of preformed pAC. The lag time remains unaffected, however. These results demonstrate that the formation of pppAC is sufficient to trigger loop release. As a consequence, the lag time has to include the slow extension step of pppAC to form a full tetraribonucleotide.
The observation that primer synthesis can trigger replication loop release supports the signaling mechanism. As discussed above, this mechanism will result in a gradual reduction of the loop lengths produced by a single replisome as time progresses. Conversely, the collision mechanism will results in a gradual increase in loop length. However, comparison between subsequent replication loops formed by individual replisomes suggests no apparent trend in loop size. Consistent with the random distribution of Okazaki fragments detected by EM (32), this observation raises the possibility that both signaling and collision models are operative, preventing a net change in loop length. The additional presence of the collision mechanism is confirmed by the observation that an increase in length from one loop to the next is correlated with the lag time separating the loops. Loop pairs that showed a decrease in length displayed no correlation between length change and lag time, consistent with the absence of additional leading-strand synthesis during the lag time as predicted by the signaling mechanism.
The primase-induced leading-strand pausing studies demonstrated that priming is a stochastic process, with a limited probability of the primase recognizing and utilizing a priming sequence during its scanning of the lagging strand (6). The utilization of both signaling and collision mechanisms provides an elegant way to allow the replisome to deal with the stochastic nature of the primase activity. The signaling mechanism will release the replication loop if the primase locates one of its sequences before the nascent Okazaki fragment is finished. On the other hand, if the nascent Okazaki fragment is finished and the primase did not engage a recognition sequence, then the collision mechanism acts as a fail-safe mechanism to trigger loop release and ensure a proper reset of the cycle of enzymatic events at the replication fork. The molecular mechanisms of the primase-induce pausing and loop release are thus far not understood. The extension of the initial pppAC to a full-length tetraribonucleotide is mediated by an interaction between two adjacent primases in a hexameric gp4 ring (44). This transient interaction might result in conformational changes that inhibit the gp4 helicase activity and/or release the loop. Correlating such structural models with replisome function will require the development of even more direct single-molecule techniques, such as the simultaneous visualization of DNA length changes with fluorescently labeled replication proteins at the fork.
Supplementary Material
Acknowledgment
We are grateful to Jack Griffith for critical reading of the manuscript and helpful comments.
This is the fourth article in the Thematic Minireview Series on Single-molecule Measurements in Biochemistry and Molecular Biology. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
S. M. Hamdan, J.-B. Lee, C. C. Richardson, and A. M. van Oijen, unpublished data.
- dsDNA
- double-stranded DNA
- ssDNA
- single-stranded DNA
- Trx
- thioredoxin
- EM
- electron microscopy.
REFERENCES
- 1.Benkovic S. J., Valentine A. M., Salinas F. (2001) Annu. Rev. Biochem. 70, 181–208 [DOI] [PubMed] [Google Scholar]
- 2.Johnson A., O'Donnell M. (2005) Annu. Rev. Biochem. 74, 283–315 [DOI] [PubMed] [Google Scholar]
- 3.Hamdan S. M., Richardson C. C. (2009) Annu. Rev. Biochem. 78, 205–243 [DOI] [PubMed] [Google Scholar]
- 4.Ogawa T., Okazaki T. (1980) Annu. Rev. Biochem. 49, 421–457 [DOI] [PubMed] [Google Scholar]
- 5.Tabor S., Huber H. E., Richardson C. C. (1987) J. Biol. Chem. 262, 16212–16223 [PubMed] [Google Scholar]
- 6.Lee J. B., Hite R. K., Hamdan S. M., Xie X. S., Richardson C. C., van Oijen A. M. (2006) Nature 439, 621–624 [DOI] [PubMed] [Google Scholar]
- 7.Wuite G. J., Smith S. B., Young M., Keller D., Bustamante C. (2000) Nature 404, 103–106 [DOI] [PubMed] [Google Scholar]
- 8.Hamdan S. M., Johnson D. E., Tanner N. A., Lee J. B., Qimron U., Tabor S., van Oijen A. M., Richardson C. C. (2007) Mol. Cell 27, 539–549 [DOI] [PubMed] [Google Scholar]
- 9.Stano N. M., Jeong Y. J., Donmez I., Tummalapalli P., Levin M. K., Patel S. S. (2005) Nature 435, 370–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamdan S. M., Marintcheva B., Cook T., Lee S. J., Tabor S., Richardson C. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5096–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato M., Ito T., Wagner G., Richardson C. C., Ellenberger T. (2003) Mol. Cell 11, 1349–1360 [DOI] [PubMed] [Google Scholar]
- 12.He Z. G., Rezende L. F., Willcox S., Griffith J. D., Richardson C. C. (2003) J. Biol. Chem. 278, 29538–29545 [DOI] [PubMed] [Google Scholar]
- 13.He Z. G., Richardson C. C. (2004) J. Biol. Chem. 279, 22190–22197 [DOI] [PubMed] [Google Scholar]
- 14.Kadyrov F. A., Drake J. W. (2001) J. Biol. Chem. 276, 29559–29566 [DOI] [PubMed] [Google Scholar]
- 15.Debyser Z., Tabor S., Richardson C. C. (1994) Cell 77, 157–166 [DOI] [PubMed] [Google Scholar]
- 16.Kim S., Dallmann H. G., McHenry C. S., Marians K. J. (1996) J. Biol. Chem. 271, 21406–21412 [DOI] [PubMed] [Google Scholar]
- 17.Alberts B. M., Barry J., Bedinger P., Formosa T., Jongeneel C. V., Kreuzer K. N. (1983) Cold Spring Harbor Symp. Quant. Biol. 47, 655–668 [DOI] [PubMed] [Google Scholar]
- 18.Frick D. N., Richardson C. C. (2001) Annu. Rev. Biochem. 70, 39–80 [DOI] [PubMed] [Google Scholar]
- 19.Lee J., Chastain P. D., 2nd, Kusakabe T., Griffith J. D., Richardson C. C. (1998) Mol. Cell 1, 1001–1010 [DOI] [PubMed] [Google Scholar]
- 20.Salinas F., Benkovic S. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7196–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J., Nelson S. W., Benkovic S. J. (2006) Mol. Cell 21, 153–164 [DOI] [PubMed] [Google Scholar]
- 22.Lee J., Chastain P. D., 2nd, Griffith J. D., Richardson C. C. (2002) J. Mol. Biol. 316, 19–34 [DOI] [PubMed] [Google Scholar]
- 23.Li X., Marians K. J. (2000) J. Biol. Chem. 275, 34757–34765 [DOI] [PubMed] [Google Scholar]
- 24.Kusakabe T., Richardson C. C. (1997) J. Biol. Chem. 272, 12446–12453 [DOI] [PubMed] [Google Scholar]
- 25.Tougu K., Marians K. J. (1996) J. Biol. Chem. 271, 21398–21405 [DOI] [PubMed] [Google Scholar]
- 26.López de Saro F. J., Georgescu R. E., O'Donnell M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14689–14694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carver T. E., Jr., Sexton D. J., Benkovic S. J. (1997) Biochemistry 36, 14409–14417 [DOI] [PubMed] [Google Scholar]
- 28.Georgescu R. E., Kurth I., Yao N. Y., Stewart J., Yurieva O., O'Donnell M. (2009) EMBO J. 28, 2981–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hacker K. J., Alberts B. M. (1994) J. Biol. Chem. 269, 24221–24228 [PubMed] [Google Scholar]
- 30.Chastain P. D., 2nd, Makhov A. M., Nossal N. G., Griffith J. (2003) J. Biol. Chem. 278, 21276–21285 [DOI] [PubMed] [Google Scholar]
- 31.Park K., Debyser Z., Tabor S., Richardson C. C., Griffith J. D. (1998) J. Biol. Chem. 273, 5260–5270 [DOI] [PubMed] [Google Scholar]
- 32.Chastain P. D., 2nd, Makhov A. M., Nossal N. G., Griffith J. D. (2000) Mol. Cell 6, 803–814 [DOI] [PubMed] [Google Scholar]
- 33.Nossal N. G., Makhov A. M., Chastain P. D., 2nd, Jones C. E., Griffith J. D. (2007) J. Biol. Chem. 282, 1098–1108 [DOI] [PubMed] [Google Scholar]
- 34.McInerney P., Johnson A., Katz F., O'Donnell M. (2007) Mol. Cell 27, 527–538 [DOI] [PubMed] [Google Scholar]
- 35.Johnson D. E., Takahashi M., Hamdan S. M., Lee S. J., Richardson C. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5312–5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J., Zhuang Z., Roccasecca R. M., Trakselis M. A., Benkovic S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8289–8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Single-molecule Themed Volume (2008) Annu. Rev. Biochem. 77, 45–22818518817 [Google Scholar]
- 38.Hamdan S. M., Loparo J. J., Takahashi M., Richardson C. C., van Oijen A. M. (2009) Nature 457, 336–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frick D. N., Kumar S., Richardson C. C. (1999) J. Biol. Chem. 274, 35899–35907 [DOI] [PubMed] [Google Scholar]
- 40.Kusakabe T., Richardson C. C. (1997) J. Biol. Chem. 272, 5943–5951 [DOI] [PubMed] [Google Scholar]
- 41.Pandey M., Syed S., Donmez I., Patel G., Ha T., Patel S. S. (2009) Nature 462, 940–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manosas M., Spiering M. M., Zhuang Z., Benkovic S. J., Croquette V. (2009) Nat. Chem. Biol. 5, 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanner N. A., Loparo J. J., Hamdan S. M., Jergic S., Dixon N. E., van Oijen A. M. (2009) Nucleic Acids Res. 37, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qimron U., Lee S. J., Hamdan S. M., Richardson C. C. (2006) EMBO J. 25, 2199–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.