Abstract
Homologous recombination (HR) plays a critical role in facilitating replication fork progression when the polymerase complex encounters a blocking DNA lesion, and it also serves as the primary mechanism for error-free repair of DNA double strand breaks. Rad51 is the central catalyst of HR in all eukaryotes, and to this point studies of human Rad51 have focused exclusively on events occurring within the nucleus. However, substantial amounts of HR proteins exist in the cytoplasm, yet the function of these protein pools has not been addressed. Here, we provide the first demonstration that Rad51 and the related HR proteins Rad51C and Xrcc3 exist in human mitochondria. We show stress-induced increases in both the mitochondrial levels of each protein and, importantly, the physical interaction between Rad51 and mitochondrial DNA (mtDNA). Depletion of Rad51, Rad51C, or Xrcc3 results in a dramatic decrease in mtDNA copy number as well as the complete suppression of a characteristic oxidative stress-induced copy number increase. Our results identify human mtDNA as a novel Rad51 substrate and reveal an important role for HR proteins in the maintenance of the human mitochondrial genome.
Keywords: DNA-Protein Interaction, DNA Recombination, DNA Repair, Mitochondria, Mitochondrial DNA, Mitochondrial Rad51 Paralogs
Introduction
In mammalian cells, mitochondria contain 2–10 copies of a 16.5-kbp genome (mtDNA) that encodes rRNAs and tRNAs involved in mitochondrial translation, as well as several proteins that contribute to ATP generation via the oxidative phosphorylation pathway (1). mtDNA is located within the mitochondrial matrix in close proximity to the inner membrane and, therefore, suffers significant damage resulting from the production of reactive oxygen species (ROS)2 via the oxidative phosphorylation pathway (2). Although there have been reports of DNA end-joining activity in mitochondrial extracts (3), base excision repair is currently the only mitochondrial DNA repair pathway for which all required enzymes have been identified in mammalian cells (1, 2).
The occurrence of genetic recombination in the mitochondrial genomes of fungi and plants has been clearly established (4, 5), but evidence of mtDNA recombination in animal cells has long been elusive as inheritance is almost exclusively maternal (4). An earlier study suggested a DNA recombination activity in mammalian mitochondrial extracts (6), and several studies indicated the rare occurrence of genetic recombination events in human mitochondria (7–9). However, it is not clear whether these activities contribute in any way to mitochondrial genome integrity, nor have the proteins responsible been identified. The significant level of cytoplasmic Rad51 observed in numerous studies (10–17) has recently been shown to contribute to a DNA damage-induced increase in nuclear Rad51 levels (18), but given previous suggestions of a recombination activity in vertebrate mitochondria, the question remains as to whether some amount of cytoplasmic Rad51 may be present and function in mitochondria.
In this study, we report for the first time the presence of the Rad51 recombinase as well as the Rad51C and Xrcc3 paralog proteins in mitochondria of cultured human cells. We observe a DNA damage-induced increase in mitochondrial levels of each protein as well as an oxidative stress-induced increase in the physical association of Rad51 with mtDNA. Strikingly, depletion of Rad51, Rad51C, or Xrcc3 from cells results in a significant decrease in mtDNA copy number following oxidative stress, supporting a novel role for Rad51-mediated recombination processes in the maintenance of mtDNA genome stability.
EXPERIMENTAL PROCEDURES
Cell Culture, Exposure of Cells to DNA Damage, and siRNA Knockdowns
HCT116 cells (ATCC no. CCL-247) were grown at 37 °C in McCoy's 5A medium (Invitrogen) with 5% CO2, and HeLa (ATCC no. CCL-2) and U20S (ATCC no. HTB-96) cells in DMEM (Invitrogen) both supplemented with 10% fetal bovine serum and antibiotics. Cells were treated with IR using a Gammacell 40 137Cs irradiator (Atomic Energy of Canada, Ottawa, ON). For oxidative stress, glucose oxidase (GO; Sigma) stock solutions were prepared fresh prior to each experiment and added at the indicated concentrations. Cells were washed once with nonsupplemented medium that was then replaced with nonsupplemented medium containing the indicated concentration of GO. The resulting concentrations of hydrogen peroxide produced were determined using a Hydrogen Peroxide Assay kit (National Diagnostics). Protein expression was knocked down by treating cells with pools of siRNAs targeting Rad51, Rad51C, Xrcc3, or CDK9 (10 nm final: Dharmacon L-003530-00, M-010534-01, M-012067-00, or M-003243-03, respectively) delivered using Dharmafect1. A pool of nonsilencing siRNAs was used as an additional negative control.
Subcellular Fractionation, Isolation of Mitochondria, and Proteinase K Protection Assay
Cells at 75–85% confluence were harvested and resuspended in mitochondria isolation buffer (10 mm Tris-HCl, pH 7.8, 0.2 mm EDTA, 250 mm sucrose, 150 mm KCl, 0.15% digitonin (Sigma), protease inhibitors (Roche)), incubated on ice for 20 min, then centrifuged at 1,000 × g at 4 °C for 10 min. The supernatant was removed to a fresh tube and centrifuged at 12,000 × g at 4 °C for 15 min to pellet a crude mitochondrial fraction. The new supernatant was removed and saved as the cytosolic fraction. Pellets containing mitochondria were combined and washed multiple times with isolation buffer containing 1 m KCl to yield the enriched mitochondrial fraction. For the proteinase K protection assay, equal aliquots of the resuspended mitochondrial fraction were treated with 0.8 mg/ml proteinase K (Qiagen), in the presence or absence of digitonin (0.2 mg/ml), or SDS (1%) for 20 min at room temperature. Proteinase K activity was halted with the addition of 2 volumes of 20% trichloroacetic acid and incubation on ice for 20 min. Precipitated protein pellets were washed once with ice-cold acetone, resuspended in 2 × Laemmli sample buffer, and evaluated by Western blotting as described below.
Immunoblotting
Total protein in each mitochondrial fraction was determined using the BCA Protein Assay kit (Pierce). Samples were prepared by adding Laemmli sample buffer (Sigma) to 1 × and heating to 95 °C for 5 min. These samples were run on 4–12% Bis-Tris acrylamide gels (Invitrogen) and transferred to polyvinylidene difluoride membranes using a semidry transfer system (Bio-Rad). Membranes were incubated with blocking buffer (10 mm Tris-HCl, pH 8.0, 300 mm NaCl, 0.25% Tween 20, 15% nonfat dry milk) and then with blocking buffer containing 2% nonfat dry milk and primary antibodies (mouse anti-Rad51 (clone 14B4), mouse anti-Rad51C (clone 2H11/6), mouse anti-Xrcc3 (clone 10F1/6) (Novus Biologicals), mouse anti-ATP synthase (BD Biosciences), goat anti-lamin A/C (Santa Cruz Biotechnologies), mouse anti-GAPDH (Millipore), rabbit anti-TFAM (Abcam), rabbit anti-PCNA (Abcam), and rabbit anti-OPA1 (Abcam)). Blots were then washed with blocking buffer (without milk) followed by incubation with horseradish peroxidase-conjugated secondary antibody (goat anti-mouse (Millipore) or rabbit anti-goat (Jackson Laboratories)). Following additional washing, blots were incubated with chemiluminescent visualizer (Denville) and developed using an LAS-4000 imaging instrument (Fuji).
DNA Isolation and Determination of mtDNA Copy Number
Total cellular DNA was isolated using the QIAamp DNA Mini Kit (Qiagen), and DNA concentrations were determined spectrophotometrically. Equal amounts of total DNA were assayed by quantitative PCR (MJR Research) using QuantiFast SYBR Green mix (Qiagen) with primers designed to amplify a 100-bp segment of the mtDNA genome. Amplification of a 100-bp segment of the 18 S ribosomal RNA gene was used as a normalization factor for the determination of changes in mtDNA copy number. Primer pair sequences for qPCR (quantitative PCR) experiments are as follows: 18 S rRNA gene (5′-AGCCATGCATGTCTAAGTACGCACG-3′ and 5′-CAAGTAGGAGAGGAGCGAGCGACCA-3′) and mtDNA (5′-CAGGAGTAGGAGAGAGGGAGGTAAG-3′ and 5′-TACCCATCATAATCGGAGGCTTTGG-3′).
mtDNA Immunoprecipitation
Cells were cross-linked by adding formaldehyde to 1% and incubating at 37 °C for 15 min. The reaction was terminated by adding glycine to 125 mm and incubating at 37 °C for a further 15 min. Cells were harvested by scraping and pelleted at 1,000 × g for 5 min. Pellets were resuspended in lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1, protease inhibitors) and incubated on ice for 20 min. Following another centrifugation at 1,000 × g for 10 min, the soluble fraction was removed to a fresh tube, and protein concentrations were determined as described above. For each immunoprecipitation reaction, 100 μg of chromatin was diluted 10-fold with dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, 167 mm NaCl) and precleared with 10 μl of protein G Dynabeads (Invitrogen) for 1 h at 4 °C. One microgram of anti-Rad51, anti-TFAM, anti-PCNA, or anti-mouse IgG (mock) antibody was added, and tubes were incubated overnight at 4 °C with rocking. The following day, immune complexes were precipitated by adding 10 μl of protein G Dynabeads with rocking at 4 °C for 1 h. Beads were washed once with buffer 1 (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 150 mm NaCl), once with buffer 2 (0.1% SDS, 1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, 500 mm NaCl), once with buffer 3 (250 mm LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, 10 mm Tris-HCl, pH 8.1), and finally twice with TE (10 mm Tris-HCl, pH 7.5, 1 mm EDTA). Cross-linking was reversed by adding TE buffer + 1% SDS and rocking overnight at 65 °C. DNA was purified and analyzed by qPCR as described above. Fold enrichment of mtDNA signal was determined relative to mock immunoprecipitation and normalized to the input signal.
RESULTS AND DISCUSSION
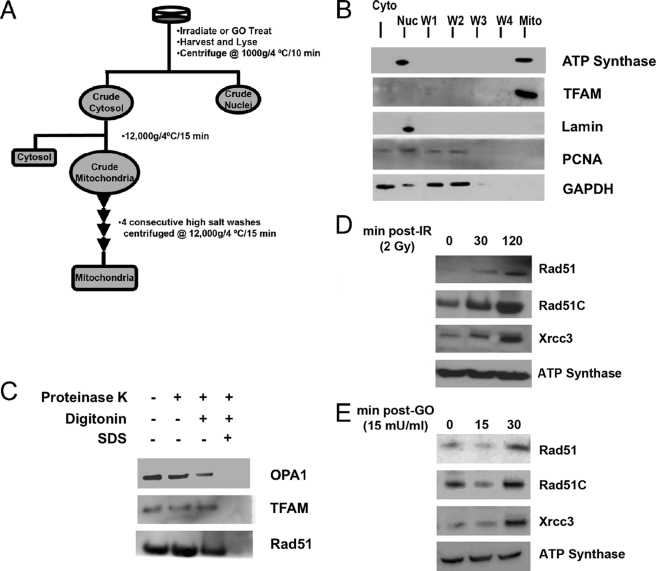
High levels of cytosolic Rad51 have been observed in previous studies from our group (15, 18, 19) and others (10–14, 16, 17). Therefore, we explored the possibility that part of the cytoplasmic Rad51 pool may be present in mitochondria. We developed a stringent fractionation protocol to ensure that mitochondria are free of contaminating nuclear and cytosolic Rad51 (Fig. 1A). The resulting mitochondrial fraction shows clear separation of the mitochondrial markers ATP synthase and TFAM from cytosolic (GAPDH) and nuclear markers (lamin and PCNA) (Fig. 1B, Mito lane). After verification that detectable contaminants had been removed, Western blot analyses revealed the presence of Rad51, Rad51C, and Xrcc3 in mitochondria isolated from normal cycling cells that had not been exposed to exogenous DNA damage (Fig. 1, D and E). To investigate the mitochondrial localization of Rad51 in greater depth, isolated mitochondria were subjected to proteinase K treatment in the presence or absence of digitonin to remove the outer mitochondrial membrane, or SDS to solubilize both membranes. As shown in Fig. 1C, the inner membrane space protein OPA1 is sensitive to proteinase K digestion when mitochondria are treated with digitonin, whereas Rad51 in the mitochondrial fraction is largely resistant to proteinase K treatment unless treated with SDS. This indicates that mitochondrial Rad51 is not present at the external surface of the mitochondria and resides predominantly in the inner matrix compartment.
FIGURE 1.
DNA damage-induced increases in mitochondrial Rad51, Rad51C, and Xrcc3. A, following either IR or GO treatment, cells were subjected to a gentle lysis procedure followed by a series of high salt washes and centrifugation steps (for a complete description, see “Experimental Procedures”). B, Western blot analysis showing separation of mitochondria from cytosolic and nuclear contaminants. Markers are ATP synthase and TFAM (mitochondria (Mito)), GAPDH (cytosolic (Cyto)), and lamin A/C and PCNA (nuclear (Nuc)). C, isolated mitochondria were treated with proteinase K in the presence or absence of digitonin or SDS, and blots were developed using antibodies against OPA1, an inner membrane space marker, TFAM, an inner matrix marker, and Rad51. D, HCT116 cells were treated with 2 Gy IR, grown for 30 min or 2 h, and fractionated as described in A. Equal amounts of total mitochondrial protein were immunoblotted for Rad51, Rad51C, and Xrcc3, as well as the loading control, ATP synthase. E, HCT116 cells were treated with 15 milliunits/ml GO and fractionated at 15 and 30 min. Equal amounts of total mitochondrial protein were immunoblotted for Rad51, Rad51C, and Xrcc3, as well as the loading control, ATP synthase. All blots are representative of at least three independent experiments.
We also note that of these proteins, only Rad51C is predicted to localize to the mitochondria with a probability of 30% or 20% when analyzed using either MitoProt II 1.0a4 (20) or Predotar 1.03, respectively. These programs evaluate protein N-terminal domains for the presence of mitochondrial targeting signals without defining a specific peptide sequence. Thus, further work will define the possible presence and function of such a signal in the Rad51C protein.
Given that exposure of cells to exogenous DNA damage induces a response that includes an increase in the nuclear levels of Rad51 (16, 18), we asked whether a similar increase occurs in mitochondria following DNA damage. In fact, cells treated with low levels of ionizing radiation (2 Gy IR) show increases in the level of each protein at 30 and 120 min post-IR (Fig. 1D). We next asked what effect oxidative stress, the major endogenous source of DNA damage encountered by mtDNA, would have on HR protein localization. To generate this stress, GO was added to the culture medium using amounts that result in chronic exposure of cells to a low level of H2O2 (23) (supplemental Fig. S1). HCT116 cells treated with 15 milliunits/ml GO consistently showed an increase in the mitochondrial levels of Rad51, Rad51C, and Xrcc3 over a 30-min time course (Fig. 1E). Similar results were obtained using HeLa cells (data not shown). Interestingly, increases in mitochondrial levels of the human apurinic/apyrimidinic endonuclease APE/Ref-1, a component of the base excision repair pathway, have been observed in response to DNA damage (24). Together with this and other studies (18, 25), our results support the idea that cellular redistribution of HR and other signaling and repair proteins is part of a general cellular response to DNA damage.
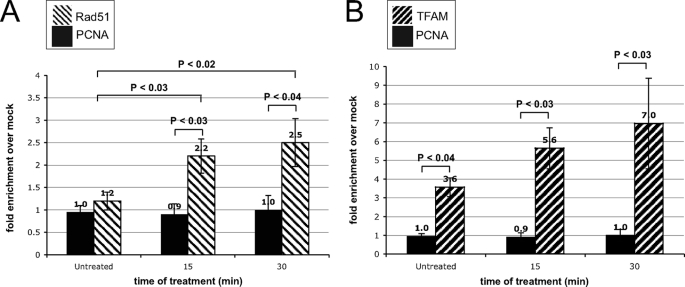
To support a role for HR in the oxidative stress response further, we evaluated the physical interaction between Rad51 and mtDNA using mtDNA immunoprecipitation. We observed no significant interaction between Rad51 and mtDNA in unstressed U20S cells relative to a mock mtDNA immunoprecipitation (Fig. 2A). However, following exposure of cells to oxidative stress there is a >2-fold increase in the association of Rad51 with mtDNA at 15 min of GO treatment, with a further increase at 30 min (Fig. 2A). As controls for the immunoprecipitation conditions, we used TFAM, a known mtDNA-binding protein, and PCNA, a nuclear DNA-binding protein not present in the mitochondria. As expected, there was no enrichment of mtDNA observed when precipitating with PCNA antibody (Fig. 2), and we observe a ∼3.5-fold enrichment with TFAM (Fig. 2B). We also find that oxidative stress increases the association of TFAM with mtDNA up to 7-fold over mock after 30 min of GO treatment, a result consistent with previous studies by Yoshida et al. (26) in which TFAM was shown to bind oxidatively damaged DNA preferentially. These results are consistent with Rad51 playing a significant role in regulating mtDNA copy number in response to stress relative to nonstress conditions.
FIGURE 2.
Rad51 physically interacts with mtDNA in an oxidative stress-dependent manner. A, at 0, 15, and 30 min following treatment with 15 milliunits/ml GO, U20S cells were incubated with formaldehyde, and mtDNA was immunoprecipitated using anti-Rad51 or anti-PCNA antibodies. Resulting DNA samples were evaluated by SYBR Green qPCR, and changes in fold enrichment of mtDNA over mock IP were calculated and normalized to input DNA (n = 12). Similar results were obtained using HeLa and HCT116 cells (not shown). B, experiments similar to those in A were performed, but immunoprecipitations were done using anti-TFAM and anti PCNA antibodies.
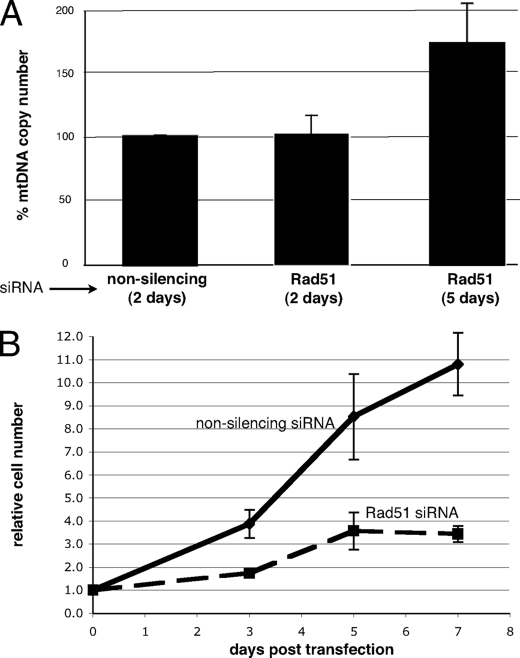
What is the mitochondrial function of human Rad51 and other HR proteins? Hundreds of mitochondria can exist in a given cell with each individual organelle containing multiple copies of the 16.5-kbp genome. In fact, the mtDNA copy number of a given cell type has been shown to be tissue-specific and linked to cell-specific metabolic requirements, as perturbations in copy number are associated with a variety of human diseases (1). In light of its mitochondrial localization (Fig. 1) and its interaction with mtDNA (Fig. 2), we sought to evaluate the role that Rad51 may play in the maintenance of mtDNA copy number under both nonstress and stress conditions. Rad51 protein was depleted from cycling cells, and mtDNA copy number was monitored as a function of time by qPCR. At 48 h after transfection with Rad51-specific siRNAs, no significant change in copy number was observed relative to cells transfected with a pool of nonsilencing siRNAs (Fig. 3A). However, mtDNA copy number increased by ∼70% following 5 days of Rad51 depletion. This undoubtedly results from prolonged cell cycle arrest induced by Rad51 depletion (Fig. 3B), as similar arrest-induced increases in mtDNA copy number have been reported previously (27). Therefore, changes in mtDNA copy number under these conditions appear to result from a general inhibition of cell cycle progression rather than being due to loss of a specific Rad51 function. However, given the important role of Rad51 in the nuclear DNA damage response and the fact that we observed increases in the mitochondrial levels of Rad51, Rad51C, and Xrcc3 in response to DNA damage, we next assessed the effect of Rad51 depletion on mtDNA copy number in cells exposed to low levels of oxidative stress.
FIGURE 3.
Depleting cells of Rad51 leads to cell cycle arrest and a characteristic arrest-induced increase in mtDNA copy number. A, U20S cells were transfected with either nonsilencing or Rad51-specific siRNAs (10 nm final concentration). Total cellular DNA was isolated at 2 and 5 days, and mtDNA copy number was evaluated by SYBR Green qPCR relative to the 18 S rRNA gene internal control. Quantification incorporates three independent experiments (error bars indicate S.E.). B, representative cell count of control or Rad51 siRNA-treated cells harvested prior to processing for total DNA. Error bars indicate the S.E. of five independent experiments.
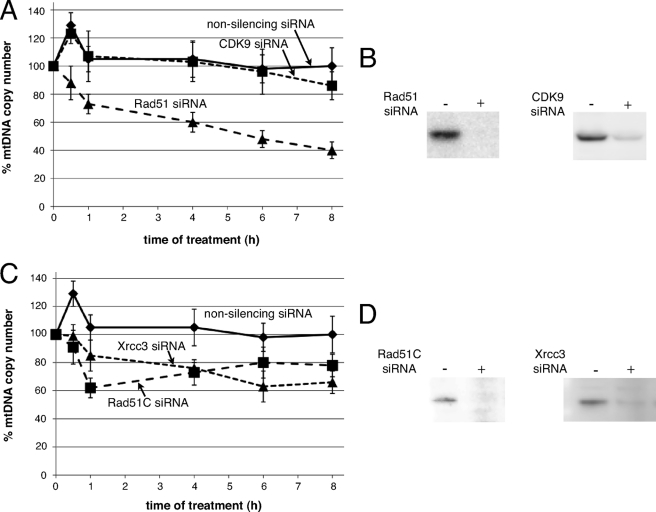
The production of ATP by enzymes of the electron transport chain located in the inner mitochondrial membrane is the primary source of ROS in the cell. Because of its close proximity to this membrane, the mtDNA is routinely subjected to oxidative stress leading to a variety of DNA lesions, including oxidized bases, protein-DNA cross-links, abasic sites, and strand breaks. Although base excision repair is the predominant DNA repair pathway in mammalian mitochondria (1, 2, 28), these lesions also represent replication blocks that can lead to fork stalling or collapse. Cells transfected with nonsilencing siRNAs, followed by exposure to 5 milliunits/ml GO, showed a characteristic, transient increase in mtDNA copy number at 30 min (Fig. 4A), consistent with previous reports of stress-induced increases observed in various cell types (27). This stress-induced increase was also observed when cells were treated with a control siRNA targeting CDK9 (Fig. 4, A and B), a protein not associated with Rad51-mediated recombination. In both cases, copy number returned to pre-stress levels at 1 h. In contrast, cells depleted of Rad51 failed to display this characteristic initial rise in copy number (Fig. 4, A and B). Strikingly, 4 h of treatment with GO led to an approximate 40% decrease in copy number in cells treated with Rad51 siRNAs relative to the nonsilencing and CDK9 controls (Fig. 4A). Analysis of mtDNA copy number at 8 h of GO treatment showed an overall 60% decrease in cells depleted of Rad51 relative to an approximate 10% decrease in control knockdown cells (Fig. 4, A and B; cells analyzed at 6 and 8 h were exposed to 2.5 milliunits/ml continuous GO rather than 5 milliunits/ml for all earlier time points, to ensure that H2O2 concentration remained at a level that imposed damage specifically on mitochondrial and not nuclear DNA (supplemental Fig. S1)) (23). These data show that depletion of Rad51 leads to a biphasic decrease in mtDNA copy number following treatment of cells with low levels of GO: (i) an initial rapid decrease in response to ROS instead of an early ROS-induced increase (0.5 h) and (ii) a continuous decrease over extended periods of exposure to GO. We observe a related biphasic decrease in mtDNA copy number following GO exposure with depletion of Rad51C or Xrcc3 (Fig. 4, C and D). Together, these results strongly suggest that a Rad51-mediated activity is critical for regulating mtDNA copy number under conditions of oxidative stress and that this activity requires the functions of Rad51C and Xrcc3.
FIGURE 4.
Depleting cells of Rad51, Rad51C, or Xrcc3 leads to a stress-induced reduction in mtDNA copy number. A and B, U20S cells were transfected with either nonsilencing, CDK9-specific siRNAs or Rad51-specific siRNAs (10 nm final concentration). 48 h following transfection, cells were treated with 5 milliunits/ml GO, and DNA was isolated at 0, 0.5, 1, and 4 h, or cells were treated with 2.5 milliunits/ml GO and isolated at 6 and 8 h. mtDNA copy number was determined by qPCR, with changes calculated relative to unstressed controls and normalized to changes in the 18 S rRNA gene. Error bars indicate S.E. (nonsilencing n = 6, CDK9 n = 4, Rad51 n = 4). C and D, U20S cells were transfected as above with siRNAs targeting Rad51C or Xrcc3 transcripts and mtDNA copy number determined by qPCR (Rad51C n = 3, Xrcc3 n = 3).
It is well established that DNA replication and recombination processes are intimately linked and that recombination proteins carry out functions essential to the restoration of the replication fork following its encounter with a blocking lesion (29–32). Particularly relevant to a role in mtDNA replication, human Rad51 and Xrcc3 have been shown to cooperate in regulating replication fork progression on damaged chromosomes (33). In a related finding, Shedge et al. (5) found that the mitochondrially targeted plant RECA3 and MSH1 proteins appear to be components of a recombination-based genome surveillance mechanism that directs recombination-dependent replication to long DNA repeats. Therefore, a likely role for mitochondrial Rad51, Rad51C, and Xrcc3 is to ensure faithful completion of mtDNA replication as the fork encounters blocking lesions. Mitochondria, like their bacterial ancestors, also have the unique problem of resolving circular chromosomes following DNA replication, a process known to involve recombination in bacteria (30).
Studies in yeast have led to the finding that a homologous DNA-pairing protein, Mhr1, associates with an oxidative stress-induced double stranded DNA break in the control region of the mtDNA (34). The authors present a model in which this recombinagenic DNA end is used to initiate rolling circle replication, presumably using an undamaged homologous mtDNA molecule as an error-free template (34). This ROS-induced use of a rapid, amplification mode of DNA replication could explain the transient increase in mtDNA copy number observed in this (Fig. 4, A and C) and other studies (27). Therefore, it is possible that an additional function of human Rad51 in regulating mtDNA copy number involves facilitating the initiation of an alternative replication mechanism when the cell senses an increase in the oxidative stress load. In fact, all potential components required for oxidative stress-induced initiation and catalysis of rolling circle replication are present in human mitochondria. The human Nth1 endonuclease, the homolog of yeast Ntg1, partitions to both the nucleus and mitochondria of human cells (35). Its endonuclease activity targets oxidized bases and, therefore, is appropriate for a role in initiating this replication mechanism (36). Potential mitochondrial candidates for double strand break end processing include Dna2 (37), Xrn2 (38), and Exo1 (38), with Exo1 being the most likely due to its processive 5′–3′ exonclease activity (38). Rad51 would be expected to function as the required Mhr1-like homologous pairing activity, and lastly, the complex containing mitochondrial polymerase γ, the helicase TWINKLE, and mitochondrial SSB protein (mtSSB) has been shown to carry out efficient rolling circle replication in vitro (39). In further studies we will investigate whether a rolling circle mechanism contributes to mtDNA replication and if any of these components play roles in this process.
This work highlights an important new role for Rad51 in the oxidative stress response of human cells. Specifically, the copy number and mtDNA immunoprecipitation data identify mtDNA as a novel Rad51 substrate in a pathway that likely facilitates the completion of mtDNA replication in the presence of DNA lesions. This conclusion is in keeping with what is currently known about mitochondrial DNA depletion syndrome, a clinically heterogeneous group of disorders in which all known disease-associated mutations are found in proteins involved in DNA replication (1). Our studies demonstrate a specific requirement for Rad51, Rad51C, and Xrcc3 function following an increased oxidative load, and given the excessive amount of oxidative damage occurring during periods of metabolic stress, there is a clear rationale for the need for recombinase function. Future work will focus on clarifying the mechanisms by which these HR proteins facilitate mtDNA replication in a stress-dependent manner.
Supplementary Material
Acknowledgments
We thank Dr. Elisabet Mandon for assistance with the mitochondrial isolation procedure and Siobhan O'Brien for advice regarding development of the mtDNA immunoprecipitation protocol and for providing CDK9 siRNAs and antibody.
This work was supported by National Institutes of Health Grants GM65851 and GM44772 (to K. L. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ROS
- reactive oxygen species
- siRNA
- small interfering RNA
- IR
- ionizing radiation
- GO
- glucose oxidase
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TFAM
- mitochondrial transcription factor A
- PCNA
- proliferating cell nuclear antigen
- qPCR
- quantitative PCR
- Gy
- gray
- HR
- homologous recombination.
REFERENCES
- 1.Graziewicz M. A., Longley M. J., Copeland W. C. (2006) Chem. Rev. 106, 383–405 [DOI] [PubMed] [Google Scholar]
- 2.Stuart J. A., Brown M. F. (2006) Biochim. Biophys. Acta 1757, 79–89 [DOI] [PubMed] [Google Scholar]
- 3.Lakshmipathy U., Campbell C. (1999) Nucleic Acids Res. 27, 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr C. M., Neiman M., Taylor D. R. (2005) New Phytol. 168, 39–50 [DOI] [PubMed] [Google Scholar]
- 5.Shedge V., Arrieta-Montiel M., Christensen A. C., Mackenzie S. A. (2007) Plant Cell 19, 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thyagarajan B., Padua R. A., Campbell C. (1996) J. Biol. Chem. 271, 27536–27543 [DOI] [PubMed] [Google Scholar]
- 7.Kajander O. A., Karhunen P. J., Holt I. J., Jacobs H. T. (2001) EMBO Rep. 2, 1007–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Aurelio M., Gajewski C. D., Lin M. T., Mauck W. M., Shao L. Z., Lenaz G., Moraes C. T., Manfredi G. (2004) Hum. Mol. Genet. 13, 3171–3179 [DOI] [PubMed] [Google Scholar]
- 9.Kraytsberg Y., Schwartz M., Brown T. A., Ebralidse K., Kunz W. S., Clayton D. A., Vissing J., Khrapko K. (2004) Science 304, 981. [DOI] [PubMed] [Google Scholar]
- 10.Davies A. A., Masson J. Y., McIlwraith M. J., Stasiak A. Z., Stasiak A., Venkitaraman A. R., West S. C. (2001) Mol. Cell 7, 273–282 [DOI] [PubMed] [Google Scholar]
- 11.Essers J., Houtsmuller A. B., van Veelen L., Paulusma C., Nigg A. L., Pastink A., Vermeulen W., Hoeijmakers J. H., Kanaar R. (2002) EMBO J. 21, 2030–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraakman-van der Zwet M., Overkamp W. J., van Lange R. E., Essers J., van Duijn-Goedhart A., Wiggers I., Swaminathan S., van Buul P. P., Errami A., Tan R. T., Jaspers N. G., Sharan S. K., Kanaar R., Zdzienicka M. Z. (2002) Mol. Cell. Biol. 22, 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarsounas M., Davies D., West S. C. (2003) Oncogene 22, 1115–1123 [DOI] [PubMed] [Google Scholar]
- 14.Liu N., Lim C. S. (2005) J. Cell. Biochem. 95, 942–954 [DOI] [PubMed] [Google Scholar]
- 15.Bennett B. T., Knight K. L. (2005) J. Cell. Biochem. 96, 1095–1109 [DOI] [PubMed] [Google Scholar]
- 16.Mladenov E., Anachkova B., Tsaneva I. (2006) Genes Cells 11, 513–524 [DOI] [PubMed] [Google Scholar]
- 17.Henson S. E., Tsai S. C., Malone C. S., Soghomonian S. V., Ouyang Y., Wall R., Marahrens Y., Teitell M. A. (2006) Mutat. Res. 601, 113–124 [DOI] [PubMed] [Google Scholar]
- 18.Gildemeister O. S., Sage J. M., Knight K. L. (2009) J. Biol. Chem. 284, 31945–31952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forget A. L., Bennett B. T., Knight K. L. (2004) J. Cell. Biochem. 93, 429–436 [DOI] [PubMed] [Google Scholar]
- 20.Claros M. G., Vincens P. (1996) Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 21.Deleted in proof
- 22.Deleted in proof
- 23.Salazar J. J., Van Houten B. (1997) Mutat. Res. 385, 139–149 [DOI] [PubMed] [Google Scholar]
- 24.Frossi B., Tell G., Spessotto P., Colombatti A., Vitale G., Pucillo C. (2002) J. Cell. Physiol. 193, 180–186 [DOI] [PubMed] [Google Scholar]
- 25.Tembe V., Henderson B. R. (2007) Cell. Signal. 19, 1113–1120 [DOI] [PubMed] [Google Scholar]
- 26.Yoshida Y., Izumi H., Ise T., Uramoto H., Torigoe T., Ishiguchi H., Murakami T., Tanabe M., Nakayama Y., Itoh H., Kasai H., Kohno K. (2002) Biochem. Biophys. Res. Commun. 295, 945–951 [DOI] [PubMed] [Google Scholar]
- 27.Lee H. C., Wei Y. H. (2005) Int. J. Biochem. Cell Biol. 37, 822–834 [DOI] [PubMed] [Google Scholar]
- 28.Mandavilli B. S., Santos J. H., Van Houten B. (2002) Mutat. Res. 509, 127–151 [DOI] [PubMed] [Google Scholar]
- 29.Flores-Rozas H., Kolodner R. D. (2000) Trends Biochem. Sci. 25, 196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreuzer K. N. (2005) Annu. Rev. Microbiol. 59, 43–67 [DOI] [PubMed] [Google Scholar]
- 31.Heller R. C., Marians K. J. (2006) Nat. Rev. Mol. Cell Biol. 7, 932–943 [DOI] [PubMed] [Google Scholar]
- 32.Li X., Heyer W. D. (2008) Cell Res. 18, 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry-Mowatt J., Jackson D., Masson J. Y., Johnson P. A., Clements P. M., Benson F. E., Thompson L. H., Takeda S., West S. C., Caldecott K. W. (2003) Mol. Cell 11, 1109–1117 [DOI] [PubMed] [Google Scholar]
- 34.Hori A., Yoshida M., Shibata T., Ling F. (2009) Nucleic Acids Res. 37, 749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trzeciak A. R., Nyaga S. G., Jaruga P., Lohani A., Dizdaroglu M., Evans M. K. (2004) Carcinogenesis 25, 1359–1370 [DOI] [PubMed] [Google Scholar]
- 36.Ikeda S., Biswas T., Roy R., Izumi T., Boldogh I., Kurosky A., Sarker A. H., Seki S., Mitra S. (1998) J. Biol. Chem. 273, 21585–21593 [DOI] [PubMed] [Google Scholar]
- 37.Zheng L., Zhou M., Guo Z., Lu H., Qian L., Dai H., Qiu J., Yakubovskaya E., Bogenhagen D. F., Demple B., Shen B. (2008) Mol. Cell 32, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., Mootha V. K. (2008) Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korhonen J. A., Pham X. H., Pellegrini M., Falkenberg M. (2004) EMBO J. 23, 2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.