Abstract
Oxidative phosphorylation in mitochondria requires the synthesis of proteins encoded in the mitochondrial DNA. The mitochondrial translation machinery differs significantly from that of the bacterial ancestor of the organelle. This is especially evident from many mitochondria-specific ribosomal proteins. An important site of the ribosome is the polypeptide tunnel exit. Here, nascent chains are exposed to an aqueous environment for the first time. Many biogenesis factors interact with the tunnel exit of pro- and eukaryotic ribosomes to help the newly synthesized proteins to mature. To date, nothing is known about the organization of the tunnel exit of mitochondrial ribosomes. We therefore undertook a comprehensive approach to determine the composition of the yeast mitochondrial ribosomal tunnel exit. Mitochondria contain homologues of the ribosomal proteins located at this site in bacterial ribosomes. Here, we identified proteins located in their proximity by chemical cross-linking and mass spectrometry. Our analysis revealed a complex network of interacting proteins including proteins and protein domains specific to mitochondrial ribosomes. This network includes Mba1, the membrane-bound ribosome receptor of the inner membrane, as well as Mrpl3, Mrpl13, and Mrpl27, which constitute ribosomal proteins exclusively found in mitochondria. This unique architecture of the tunnel exit is presumably an adaptation of the translation system to the specific requirements of the organelle.
Keywords: Mitochondria, Protein Translocation, Ribosome Structure, Ribosomes, Translation
Introduction
The membrane-embedded complexes driving oxidative phosphorylation in mitochondria consist of subunits that are encoded in either the nuclear or the organellar DNA. In yeast mitochondria, the genetic system expresses eight proteins, of which seven are hydrophobic membrane proteins that represent the catalytic core of the enzymes (1). Membrane insertion of the hydrophobic translation products is achieved in a presumably co-translational fashion (2). This insertion is mediated by Oxa1 and Mba1. To allow a tight spatial coupling between protein synthesis and membrane insertion, Oxa1 and Mba1 bind to mitochondrial ribosomes to either mediate or coordinate membrane insertion (3–7).
Mitochondrial ribosomes evolved from those of the bacterial ancestor of mitochondria. This ancestry is reflected by sensitivity to a similar set of antibiotics, by a set of interchangeable translation factors (8), and by many conserved proteins and RNA moieties. At the same time, mitochondrial ribosomes have a strikingly different composition. In contrast to the eubacterial ribosome that contains 55 proteins and three rRNAs, mitochondrial ribosomes typically contain between 70 (yeast) and 80 (mammals) proteins and two rRNAs (9). In addition to numerous mitochondria-specific proteins, mitochondrial ribosomes have many conserved proteins that contain extensions at their N and C termini. The significance of these additional proteins and domains is unclear; they might compensate for the sequence losses of the mitochondrial rRNAs and thus help to stabilize the structure of the ribosome (10, 11), or they might introduce novel features for organization or regulation (12, 13).
One region of the ribosome that is of special importance for the fate of newly synthesized polypeptides is the ribosomal tunnel exit where nascent chains are exposed to a hydrophilic milieu for the first time. To help the nascent polypeptide to mature, a variety of biogenesis factors interact with the ribosome in close proximity to the tunnel exit (14, 15). The rim of the exit tunnel of the eubacterial ribosome is formed by four proteins (L22, L23, L24, and L29) that are highly conserved and found in all ribosomes.
The exact composition and structure of the mitochondrial ribosomal tunnel exit are not known. Cryo-electron microscopy reconstruction of mitochondrial ribosomes indicated that their tunnel exits differ substantially from that of the bacterial counterpart (16, 17). Specifically, numerous protuberances and additional mass are present around this site. The resolution of these structures, however, did not allow the identification of the proteins located at this region. In this study, we undertook a comprehensive approach to identify the composition of the mitochondrial ribosomal tunnel exit. These analyses led to the identification of mitochondria-specific ribosomal proteins that are located in the vicinity of the tunnel exit. Moreover, we demonstrate that Mba1 binds in proximity to this site, substantiating the important role Mba1 plays for the coordination of synthesis and insertion of mitochondrially encoded proteins.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Media
All strains used in this study were isogenic to the wild type strain YPH499. His7-tagged variants of Mrpl40, Mrp20, Mrpl4, Mrpl22, Mrpl3, Mrpl13, Mrpl27, and Mba1 were generated by replacing the stop codons of the endogenous open reading frames by a sequence encoding a heptahistidine tag followed by a HIS3 selection cassette (18). MRPL3, MRPL13, and MRPL27 were disrupted using a kanamycin resistance cassette. Yeast cultures were grown at 30 °C in lactate medium and YP (1% yeast extract, 2% peptone) medium supplemented with 2% dextrose, 2% galactose, or 2% glycerol. Mitochondria were isolated as described previously (19).
Cross-linking and Denaturing Purification
Mitochondria from yeast strains expressing His7-tagged proteins were incubated in isotonic buffer (0.6 m sorbitol, 20 mm Hepes, pH 7.4). The membrane-permeable, non-cleavable cross-linkers DFDNB,2 bismaleimidohexane, disuccinimidyl glutarate, and 3-maleimidobenzoyl-N-hydroxy-succinimide ester were dissolved in DMSO and used at a final concentration of 200 μm. DMSO without cross-linker served as vehicle control. Cross-linking was performed at 25 °C for 45 min, at 30 °C for 30 min, and at 37 °C for 15 min. Cross-linking was stopped by the addition of 100 mm Tris/HCl, pH 8.0, or 100 mm β-mercaptoethanol and incubating the samples for 10 min at the respective temperature. Next, mitochondria were reisolated by centrifugation at 25,000 × g for 10 min at 4 °C.
For small scale approaches, 50 μg of mitochondria were incubated with cross-linking reagents, directly lysed in SDS loading buffer, subjected to SDS-PAGE, and analyzed by Western blotting. To purify the cross-linking products for mass spectrometry, 40 mg of mitochondria were treated with the indicated cross-linker. After quenching excessive cross-linker, mitochondria were reisolated. The mitochondrial pellet was lysed for 30 min in 30 ml of buffer containing 1% Triton X-100, 50 mm KCl, 0.5 mm MgCl2, 20 mm Hepes/KOH, pH 7.4, 1 mm phenylmethylsulfonyl fluoride, and 1× Complete protease inhibitor mix (Roche Applied Science). The lysate was transferred to Ti70 tubes (Beckman Coulter) containing a 2-ml sucrose cushion (1.2 m sucrose, 50 mm KCl, 0.5 mm MgCl2, 20 mm Hepes, pH 7.4). The samples were centrifuged in the Ti70 rotor at 30,000 rpm for 18 h at 4 °C. After discarding the supernatant, the ribosomal pellet was lysed in a total volume of 1.4 ml of 1% SDS and shortly heated at 98 °C. Next, the sample was adjusted to 10 ml with binding buffer (0.1% dodecyl maltoside, 300 mm NaCl, 20 mm imidazole, pH 7.4, 20 mm KPi, pH 7.4), and the His-tagged proteins and their cross-linking partners were purified on Ni-NTA beads.
Ni-NTA purification was performed by incubating the respective samples with 10 μl of Ni-NTA beads tumbling for 2 h at 4 °C for small approaches and 150 μl of beads for 5 h at 4 °C for large approaches. The beads were then washed three times with binding buffer and eluted with sample buffer containing 500 mm imidazole, pH 7.4. Small scale samples were separated via SDS-PAGE and analyzed by Western blotting. Large scale samples were separated via SDS-PAGE following silver staining and controlled by Western blots with anti-His7 antibody. Bands were excised from the silver gel and analyzed by mass spectrometry.
Trypsin Digest and Mass Spectrometry
In-gel digests were performed as described in standard protocols. Briefly, following the SDS-PAGE and washing of the excised gel slices, proteins were reduced by adding 10 mm dithiothreitol (Sigma-Aldrich) prior to alkylation with 55 mm iodoacetamide (Sigma-Aldrich). After washing and shrinking of the gel pieces with 100% acetonitrile, trypsin (sequencing grade modified, Promega) was added, and proteins were digested overnight in 40 mm ammonium bicarbonate at 37 °C. For protein identification, probes were directly used for nano-electrospray mass ionization-LC-tandem MS. Each sample was first separated on a C18 reversed phase column via a linear acetonitrile gradient (UltiMate 3000 system (Dionex)) and column (75-μm inner diameter × 15 cm, packed with C18 PepMapTM, 3 μm, 100 Å; LC Packings) before MS, and tandem MS spectra were recorded on an Orbitrap mass spectrometer (Thermo Electron). The resulting spectra where then analyzed via the MascotTM Software (Matrix Science) using the National Center for Biotechnology Information (NCBI) nr Protein Database.
Miscellaneous
Mitochondrial translation products were labeled with [35S]methionine and cross-linked with DFDNB as described (6). The antibody against the His7 tag was purchased from Qiagen. Antibodies against Mrp20, Mrpl40, and Mrpl4 were obtained by immunizing rabbits with purified mature Mrpl40, Mrpl4, and MBP-Mrp20, respectively.
RESULTS
Strategy to Identify Proteins Located at the Mitochondrial Ribosomal Tunnel Exit
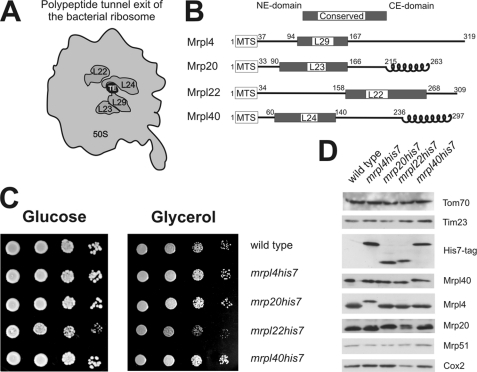
The rim of the tunnel exit of bacterial ribosomes is composed of the conserved proteins L22, L23, L24, and L29 (Fig. 1A). We identified by BLAST searches the homologues of these bacterial proteins that are present in yeast mitochondrial ribosomes. Thereby, the proteins Mrpl22, Mrp20, Mrpl40, and Mrpl4 were identified as homologous of L22, L23, L24, and L29, respectively. All four mitochondrial proteins have a conserved domain and additionally N-terminal mitochondrial targeting signals (MTS) and mitochondria-specific domains at the N and C termini (Fig. 1B). For Mrp20 and Mrpl40, these C-terminal extensions show a high probability to form coiled-coil structures.
FIGURE 1.
Proteins located around the mitochondrial ribosomal tunnel exit. A, bottom view of the large ribosomal subunit of bacteria. The localization of the conserved proteins located around the tunnel exit (TE) of the 50 S subunit is depicted. B, mitochondrial proteins that are homologous to the conserved tunnel exit proteins in bacteria. These mitochondrial proteins contain, in addition to a conserved region N-terminal mitochondrial targeting signals (MTS) as well as N- and C-terminal extension domains (NE-domain and CE-domain, respectively). Furthermore, Mrp20 and Mrpl40 have C-terminal domains that show a high propensity to form a coiled-coil structure. Numbers indicate amino acid positions. C, C-terminal His7 tags on Mrpl4, Mrp20, and Mrpl40 do not impair respiratory growth. Yeast cells of the indicated strains were plated in serial 10-fold dilutions onto media requiring (Glycerol) or not requiring respiration (Glucose) and incubated for 2 days. D, C-terminal His7 tags on Mrpl4, Mrp20, and Mrpl40 do not destabilize the proteins. Equal amounts of cell lysates of the indicated strains were separated on SDS-PAGE and analyzed by Western blotting.
Ribosomal Proteins Are Functional with an Engineered C-terminal His7 Tag
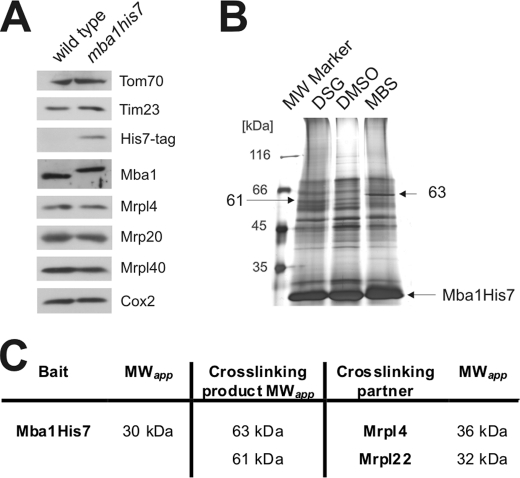
Although the high resolution structures of bacterial ribosomes (20) revealed the molecular composition of the bacterial ribosomal tunnel exit, no such information exists for mitochondrial ribosomes. To identify proteins located in proximity to the mitochondrial ribosomal tunnel exit, we mapped the vicinity of the tunnel exit proteins using cross-linking and characterization of the purified cross-linking products via mass spectrometry. To allow efficient purification of the proteins of tunnel exit, we introduced tags consisting of seven histidines (His7) at the C termini of Mrpl4, Mrp20, Mrpl22, and Mrpl40 by directed homologous recombination. First, we tested whether the tagging compromises the growth of the yeast cells on media containing either fermentable (Glucose) or non-fermentable (Glycerol) carbon sources (Fig. 1C). All strains with C-terminal His7 tags were able to grow on a non-fermentable carbon source similar to the wild type with the exception of mrpl22his7, that showed a decreased growth rate.
Next, we checked whether the His7-tagged variants of the proteins accumulated to similar levels. When proteins of whole cell lysates were analyzed by Western blotting, no changes in the steady state levels of Mrpl4, Mrp20, and Mrpl40 were detected when the proteins contained a C-terminal His7 tag (Fig. 1D). Similarly, the amounts of Mrp51, a constituent of the small ribosomal subunit, or of the mitochondrially encoded Cox2 were not affected in these strains, indicating that the variants with an engineered C-terminal His7 tag are fully functional. In contrast, mrpl22his7 showed reduced levels of Cox2. From this, we concluded that the addition of the C-terminal His7 tag impaired the function of Mrpl22 and therefore continued our analysis only with the fully functional proteins Mrpl4His7, Mrp20His7, and Mrpl40His7.
Identification of Proteins Located around the Mitochondrial Tunnel Exit
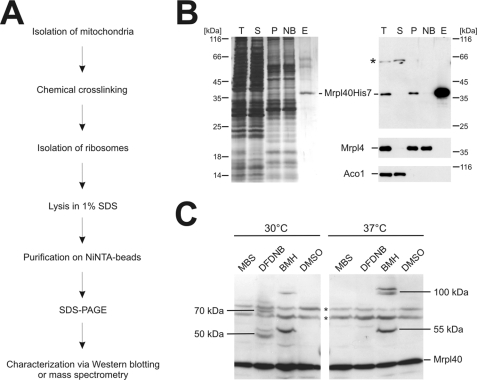
To purify the C-terminally His7-tagged proteins from isolated mitochondria, we established a protocol consisting of a two-step strategy (Fig. 2A). First, mitochondria were solubilized, and the mitochondrial ribosomes were sedimented through a high density sucrose cushion. Next, the ribosomes were disintegrated by boiling in 1% SDS, and the liberated His7-tagged proteins were purified using immobilized metal affinity chromatography. This procedure led to a strong enrichment of the His7-tagged proteins (here Mrpl40 as an example), whereas other mitochondrial proteins such as aconitase and Mrpl4 were efficiently removed (Fig. 2B).
FIGURE 2.
Strategy to identify proteins located at the mitochondrial ribosomal tunnel exit. A, flow chart of the strategy to identify interaction partners of the tunnel exit proteins. B, purification of Mrpl40His7. Mitochondria from a strain expressing His7-tagged Mrpl40 were lysed in Triton X-100. Ribosomes of this lysate were centrifuged through a high density sucrose cushion. The resulting ribosomal pellet was resuspended in 1% SDS, and Mrpl40His7 was purified. Left panel, silver-stained gel; right panel, Western blotting using the indicated antibodies. T, total; S, supernatant after centrifugation; P, ribosomal pellet after centrifugation; NB, not bound material of the Ni-NTA purification; E, elution fraction. C, cross-linking of Mrpl40 in isolated mitochondria. Wild type mitochondria were incubated with a variety of cross-linking agents at different temperatures. DMSO was used as control. The mitochondria were reisolated, subjected to SDS-PAGE, and analyzed by Western blotting. A number of specific cross-linking products are indicated with their apparent molecular mass. * indicates nonspecific cross reactions. MBS, 3-maleimidobenzoyl-N-hydroxysuccinimide ester; BMH, bismaleimidohexane.
Next, we established conditions under which cross-linking products to the His7-tagged proteins were obtained efficiently using isolated mitochondria. Specifically, we used membrane-permeable cross-linking reagents that are amine-specific (DFDNB, disuccinimidyl glutarate), sulfhydryl-specific (bismaleimidohexane), or hetero-bifunctional (3-maleimidobenzoyl-N-hydroxy-succinimide ester). First, we tested under which conditions and with which cross-linking reagent robust cross-linking products of Mrpl40, Mrpl4, and Mrp20 could be generated. Depending on the cross-linking conditions, additional bands were repeatedly obtained, indicating successful formation of cross-linking products of Mrpl40 (Fig. 2C) as well as of Mrpl4 and Mrp20 (not shown).
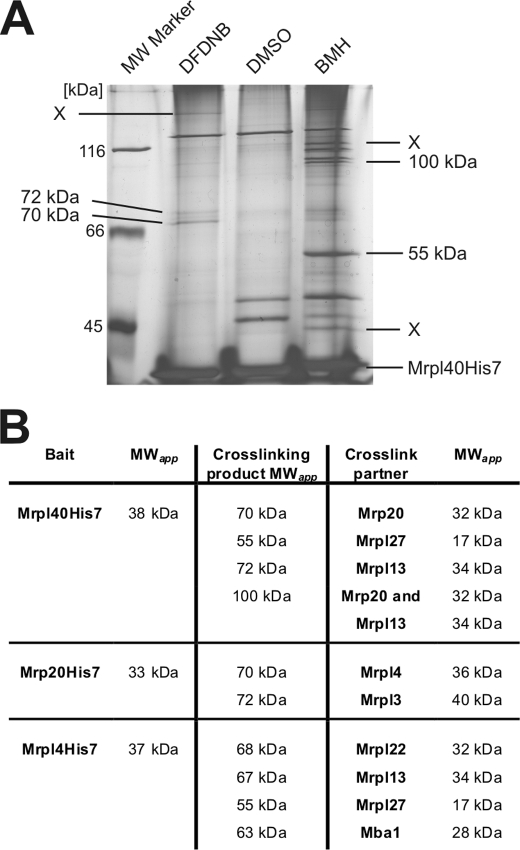
Next, we repeated this cross-linking in preparative scale using isolated mitochondria containing His7-tagged variants of Mrpl40 (Fig. 3A), Mrpl4, and Mrp20 (supplemental Fig. S1). Next, the His7-tagged proteins and their cross-linking products were purified. This procedure resulted in the purification of Mrpl40 and, depending on the cross-linker, a variety of additional bands (Fig. 3A). These bands were excised and analyzed by liquid chromatography-mass spectrometry (LC-MS). To qualify as an unambiguously identified cross-linking product, it had to fulfill three criteria. 1) At least four different peptides are identified from the His7-tagged ribosomal protein and from the cross-linking partner. 2) It has an apparent molecular weight that represents the sum of the apparent molecular weights of the proteins. 3) The peptides of both proteins are absent from a gel slice that was excised from the vehicle control purification at the identical position.
FIGURE 3.
Identification of proteins located around the mitochondrial ribosomal tunnel exit. A, silver-stained gel of the elution fractions after a large scale experiment with cross-linking and purification using mitochondria that contain Mrpl40His7. Isolated mitochondria (40 mg each) were incubated with the indicated cross-linkers or a control (DMSO). Mrpl40His7 was purified from isolated ribosomes. In comparison with the control, the additional bands represent cross-linking products of Mrpl40His7 (indicated by the apparent molecular mass; X, not identified) and were excised from the gel and analyzed by LC-MS. B, identified cross-linking partners of Mrpl4, Mrp20, and Mrpl40. MWapp, apparent molecular weight.
This approach resulted in the identification of proteins that could be cross-linked to Mrpl40 (Fig. 3B; for a summary of the MS analysis, see supplemental Table 1). These proteins include Mrp20, Mrpl13, and Mrpl27. Hence, our work supports earlier findings that Mrpl40 is in close proximity to Mrp20 (21). Moreover, we identified the subribosomal localization of Mrpl13 and Mrpl27 that represent ribosomal proteins found exclusively in mitochondria.
These experiments were repeated with the His7-tagged variants of Mrpl4 and Mrp20 (Fig. 3B, supplemental Fig. S1). Specifically, we found that Mrp20 is located in proximity to Mrpl4 and Mrpl3, the latter again representing a mitochondria-specific ribosomal protein. Likewise, our analysis of cross-linking partners of Mrpl4 revealed the localization of Mrpl22, Mrpl13, Mrpl27, and Mba1 in close proximity to Mrpl4. Taken together, our analysis suggests that Mrpl3, Mrpl13, and Mrpl27 as well as Mba1 are located in proximity to the mitochondrial ribosomal tunnel exit.
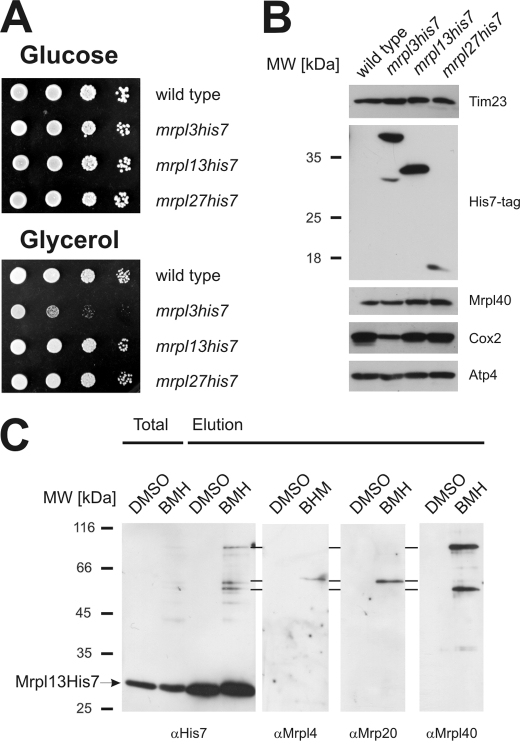
To confirm the subribosomal localization of Mrpl3, Mrpl13, and Mrpl27, we prepared strains expressing His7-tagged variants. Although the His7 tag on the C terminus of Mrpl13 or Mrpl27 did not disturb respiratory growth, cells containing Mrpl3His7 showed a marked growth defect on a non-fermentable carbon source (Fig. 4A). When mitochondria isolated from these strains were analyzed, the His7-tagged proteins migrated at the expected position as judged from the calculated molecular weight (Fig. 4B). mrpl13his and mrpl27his accumulated Cox2 to wild type level, whereas mrpl3his7 did not. Hence, the C-terminal His7 tag compromised the function of Mrpl3. We therefore did not use mrpl3his7 for further analyses. Next, we repeated the cross-linking experiments with mitochondria isolated from mrpl13his7 (Fig. 4C) and mrpl27his7 (not shown) and tested by Western blotting for the formation of cross-links to Mrpl4, Mrp20, and Mrpl40. These analyses confirmed that Mrpl13 is located in proximity to Mrpl4, Mrp20, and Mrpl40 (Fig. 4C) and that Mrpl27 can be cross-linked to Mrpl4 and Mrpl40 (not shown).
FIGURE 4.
The identified components are located at the ribosomal tunnel exit of mitochondria. A, growth test of yeast strains harboring C-terminal His7-tagged variants of Mrpl3, Mrpl13, and Mrpl27. Yeast cells of the indicated strains were plated in serial 10-fold dilutions onto media requiring (Glycerol) or not requiring respiration (Glucose) and incubated for 2 days. B, C-terminal His7 tags on Mrpl13 and Mrpl27, but not on Mrpl3, allow the accumulation of mitochondrially encoded Cox2 to wild type level. Equal amounts of mitochondria isolated from the indicated strains were separated on SDS-PAGE and analyzed by Western blotting. C, Mrpl13 can be cross-linked to Mrpl4, Mrp20, and Mrpl40. Mitochondria containing Mrpl13His7 were incubated with the cross-linker bismaleimidohexane (BMH) or a control (DMSO). Next, Mrpl13His7 was purified following the scheme presented in Fig. 2A. The elution fraction was analyzed by Western blotting using antibodies against the His7 tag, Mrpl4, Mrp20, or Mrpl40, respectively.
Mba1 Is Located in Proximity to the Exit Tunnel
Mba1 acts as a ribosome receptor that aligns the ribosome with the insertion site of the inner membrane to facilitate co-translational protein insertion (7). The binding partner of Mba1 on the ribosome remained unknown. It therefore was exciting to identify the L29 homologue Mrpl4 as a cross-linking partner of Mba1. To characterize the binding site of Mba1 in more detail, we generated a C-terminally His7-tagged variant of Mba1 that accumulated to similar levels as the untagged protein and did not change the steady state level of Cox2 (Fig. 5A). Hence, we concluded that the protein was functional. Next, we isolated mitochondria from this strain and generated cross-linking products of Mba1 in preparative scale. Mba1 and its cross-linking products were enriched (Fig. 5B) and separated on SDS-PAGE. These cross-linking products were excised and analyzed by LC-MS. These experiments confirmed Mrpl4 as a cross-linking partner of Mba1. Moreover, we found that Mrpl22 is located in close proximity to Mba1 (Fig. 5C). We therefore concluded that Mba1 is positioned at the ribosome in proximity to the ribosomal proteins Mrpl4 and Mrpl22. This subribosomal localization of Mba1 indicates that the protein has a binding site on the ribosome that is distinct from that of Oxa1, which has been shown to be located near Mrpl40 and Mrp20 (3, 21).
FIGURE 5.
Mba1 is located in proximity to the tunnel exit. A, a C-terminal His7 tag on Mba1 does not destabilize the protein. Lysates of the indicated strains were separated on SDS-PAGE and analyzed by Western blotting. B, large scale purification of cross-linking products to Mba1His7. The experiment was performed as depicted in Fig. 3A. MBS, 3-maleimidobenzoyl-N-hydroxysuccinimide ester. C, identified cross-linking partners of Mba1. MWapp, apparent molecular weight; DSG, disuccinimidyl glutarate.
Contacts of Proteins Located at the Tunnel Exit with Nascent Chains
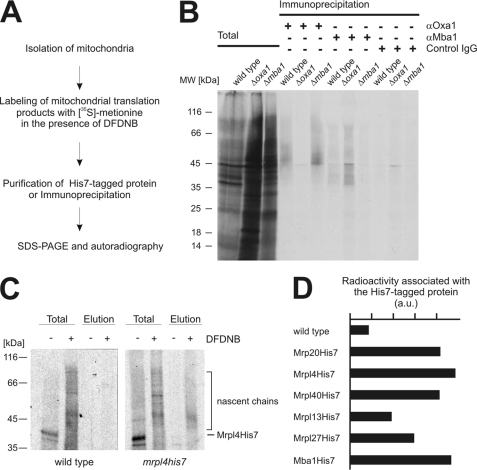
Mba1 and Oxa1 can be cross-linked to short nascent chains (6), indicating that they are in contact with newly synthesized proteins. It is currently not known whether both components cooperate in establishing these contacts or whether they can act independently from each other. To address this question, we isolated mitochondria from wild type, Δoxa1, and Δmba1 cells and labeled their translation products with [35S]methionine in the presence or absence of the cross-linking reagent DFDNB (Fig. 6A). DFDNB is characterized by a short (3 Å) spacer between the reactive groups, hence allowing cross-linking of polypeptides that are in very close proximity. After cross-linking, Oxa1 and Mba1 were immunoprecipitated, and nascent chains cross-linked to both proteins were detected by autoradiography as diffuse smears that are migrating about 3 kDa above the molecular mass of Oxa1 and Mba1 (Fig. 6B). In mitochondria isolated from wild type cells, nascent chains could be detected after immunoprecipitation with antibodies against Oxa1 and Mba1 but not with control IgGs (Fig. 6B). In the absence of either Oxa1 or Mba1, the other component was still able to contact nascent chains (Fig. 6B), indicating that both proteins interact independently of each other with nascent chains.
FIGURE 6.
Contacts of proteins located at the tunnel exit with nascent chains. A, strategy to cross-link nascent chains to proteins. B, Oxa1 and Mba1 can contact nascent chains independently of each other. Translation products of mitochondria isolated from wild type, Δmba1 or Δoxa1 cells were labeled with [35S]methionine in the presence of the cross-linking reagent DFDNB. Oxa1 and Mba1 with cross-linked nascent chains were precipitated from SDS-denatured lysates using specific antibodies or control IgGs. The samples were separated on SDS-PAGE, blotted to nitrocellulose, and analyzed by autoradiography. Nascent chains migrate as a diffuse smear above the position of Oxa1 (39 kDa) and Mba1 (28 kDa) in the gel. Total, 10% of total. C, contact of ribosomal proteins to nascent chains. The experiments were performed as in B with the exception that mitochondria containing His7-tagged proteins were used. The His7-tagged proteins were purified and eluted from the resin. Total, 1% of total. D, the experiment in C was repeated with mitochondria containing the indicated His7-tagged proteins. The radioactive signals of a region starting at the position of the His7-tagged protein plus an additional 10 kDa were densitometrically quantified in the elution fractions. The background signal of the control was subtracted from the signal of the DFDNB-cross-linked nascent chains. a.u., arbitrary units.
These results inspired us to test whether nascent chains can be cross-linked to the ribosomal proteins that are located around the tunnel exit. We therefore repeated the experiments as described above with the exception that we used mitochondria from the strains expressing His7-tagged versions of these proteins (Fig. 6A) to allow purification. Although no radioactive signals were obtained after mock purification using wild type mitochondria (Fig. 6C, left panel), a clear signal of nascent chains was recovered when the mitochondria contained His7-tagged Mrpl4 (Fig. 6C, right panel). Next, we analyzed densitometrically the autoradiographic signals of nascent chains associated with the His7-tagged versions of Mrp20, Mrpl4, Mrpl40, Mrpl13, Mrpl27, and Mba1 (Fig. 6D). This revealed that nascent chains can be cross-linked to Mrpl4, Mrp20, and Mrpl40 as well as Mba1 and presumably with a lower efficiency to Mrpl13 and Mrpl27 (Fig. 6D).
Because the bacterial homologues of Mrpl4, Mrp20, and Mrpl40 are building the rim of the bacterial ribosomal tunnel exit, we conclude that these three mitochondrial proteins indeed have a subribosomal localization similar to their bacterial counterparts. In contrast, Mrpl13 and Mrpl27, which appear not to be similarly exposed to nascent chains, are possibly not directly contributing to the formation of the polypeptide exit tunnel.
DISCUSSION
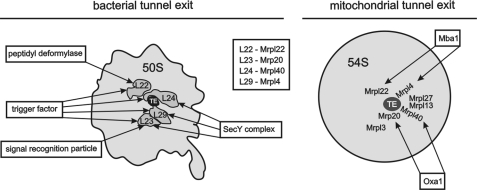
The tunnel exit of ribosomes is the site where nascent chains are exposed to a hydrophilic environment for the first time. Hence, many biogenesis factors interact with the tunnel exit to accept the newly synthesized proteins and guide them through their early biogenesis. In the bacterial system, these factors include molecular chaperones and translocases mediating protein insertion as well as certain processing enzymes (Fig. 7) (14). To accept the newly synthesized proteins, these factors interact with the ribosome by the help of a conserved set of ribosomal proteins, namely L22, L23, L24, and L29. Although the architecture and composition of the tunnel exit of bacterial ribosomes are well known due to high resolution structures (20), no such information exists for the mitochondrial ribosome. In this study, we followed a comprehensive approach to identify proteins located in proximity to the tunnel exit of yeast mitochondrial ribosomes. This analysis revealed a previously uncharacterized network of proteins with a composition unique to these ribosomes (Fig. 7).
FIGURE 7.
A comparison of interactions at the bacterial and mitochondrial ribosomal tunnel exit (TE). Left, the tunnel exit of bacterial ribosomes and the mapped binding sites of biogenesis factors are depicted. Right, a hypothetical model of the tunnel exit of mitochondrial ribosomes and the mitochondria-specific proteins as revealed in this study. Mba1 is positioned in proximity to Mrpl22 and Mrpl4. Oxa1 interacts with the mitochondrial ribosome close to Mrp20 and Mrpl40. The hypothetical distribution of the ribosomal proteins as suggested by the cross-linking analysis is delineated.
Mba1 is part of this network. Specifically, we found that Mba1 is in proximity to Mrpl4 and Mrpl22. Such localization of Mba1 to the ribosomal tunnel exit supports its function in co-translational protein insertion where Mba1 acts as a ribosome receptor. This function is required to coordinate the ribosome with the insertion site to allow a tight coupling between protein synthesis and membrane insertion (7). Mba1 cooperates in membrane insertion of mitochondrially encoded proteins with Oxa1, a protein translocase of the inner mitochondrial membrane. Overexpressed Oxa1 can be cross-linked to Mrp20 (3) and to Mrpl40 (21). We also observed cross-links of overexpressed Oxa1 to Mrpl40 but were unable to detect them in wild type mitochondria (not shown). A cryo-electron microscopy reconstruction of a heterologous system composed of Oxa1 and a bacterial ribosome exposing a hydrophobic nascent chain revealed a close contact of Oxa1 with L23 and L24, the bacterial homologues of Mrp20 and Mrpl40 (22), thus supporting the published cross-linking data (3, 20). Hence, the localization of Oxa1 (to Mrp20 and Mrpl40) and of Mba1 (to Mrpl4 and Mrpl22) would be compatible with a simultaneous binding of both proteins to the mitochondrial ribosome (Fig. 7) (7).
Mitochondrial ribosomes evolved from those of the bacterial ancestor of the organelle and still share many features with prokaryotic ribosomes. However, mitochondrial ribosomes underwent a major remodeling during evolution and contain a variety of additional proteins. The function and precise ribosomal localization of these mitochondria-specific proteins are currently not known. We revealed with this work that the mitochondria-specific ribosomal proteins Mrpl13, Mrpl3, and Mrpl27 are located in proximity to the tunnel exit. Deletion of these proteins provokes the loss of mitochondrial DNA (not shown). A likely explanation for this is that the loss of ribosomal proteins directly impairs the stability of the mitochondrial ribosome. A decreased stability of the translation machinery will eventually lead to the loss of the ability to synthesize mitochondrially encoded proteins. Importantly, it is well documented that a block in mitochondrial translation destabilizes mitochondrial DNA (23). Hence, it is likely that the mitochondrial ribosomal proteins that were discovered in this study are critical for stability of the ribosomes.
However, it is possible that these mitochondria-specific ribosomal proteins were recruited to the tunnel exit to support an additional, organelle-specific mechanism. We are therefore currently aiming to identify interaction partners of Mrpl3, Mrpl13, and Mrpl27 to address whether these proteins are involved in recruiting biogenesis factors to the mitochondrial ribosome.
Supplementary Material
Acknowledgments
We are grateful to Andrea Trinkaus and Sabine Knaus, Technische Universität Kaiserslautern, for expert technical assistance and to Eli van der Sluis, Ludwig-Maximilians-Universität München, and Jan Riemer, Technische Universität Kaiserslautern, for stimulating discussions.
This work was supported by a grant from the Stiftung Rheinland-Pfalz für Innovation and by Deutsche Forschungsgemeinschaft (Research Unit FOR967).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
- DFDNB
- 1,5-difluoro-2,4-dinitrobenzene
- DMSO
- dimethyl sulfoxide
- LC-MS
- liquid chromatography-mass spectrometry
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Borst P., Grivell L. A. (1978) Cell 15, 705–723 [DOI] [PubMed] [Google Scholar]
- 2.Ott M., Herrmann J. M. (December2, 2009) Biochim. Biophys. Acta 10.1016/j.bbamcr.2009.11.010 [DOI] [Google Scholar]
- 3.Jia L., Dienhart M., Schramp M., McCauley M., Hell K., Stuart R. A. (2003) EMBO J. 22, 6438–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preuss M., Ott M., Funes S., Luirink J., Herrmann J. M. (2005) J. Biol. Chem. 280, 13004–13011 [DOI] [PubMed] [Google Scholar]
- 5.Szyrach G., Ott M., Bonnefoy N., Neupert W., Herrmann J. M. (2003) EMBO J. 22, 6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preuss M., Leonhard K., Hell K., Stuart R. A., Neupert W., Herrmann J. M. (2001) J. Cell Biol. 153, 1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ott M., Prestele M., Bauerschmitt H., Funes S., Bonnefoy N., Herrmann J. M. (2006) EMBO J. 25, 1603–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spremulli L. L., Coursey A., Navratil T., Hunter S. E. (2004) Prog. Nucleic Acid Res. Mol. Biol. 77, 211–261 [DOI] [PubMed] [Google Scholar]
- 9.Smits P., Smeitink J. A., van den Heuvel L. P., Huynen M. A., Ettema T. J. (2007) Nucleic Acids Res. 35, 4686–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki T., Terasaki M., Takemoto-Hori C., Hanada T., Ueda T., Wada A., Watanabe K. (2001) J. Biol. Chem. 276, 33181–33195 [DOI] [PubMed] [Google Scholar]
- 11.Koc E. C., Burkhart W., Blackburn K., Moyer M. B., Schlatzer D. M., Moseley A., Spremulli L. L. (2001) J. Biol. Chem. 276, 43958–43969 [DOI] [PubMed] [Google Scholar]
- 12.Williams E. H., Bsat N., Bonnefoy N., Butler C. A., Fox T. D. (2005) Eukaryot. Cell 4, 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prestele M., Vogel F., Reichert A. S., Herrmann J. M., Ott M. (2009) Mol. Biol. Cell 20, 2615–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer G., Boehringer D., Ban N., Bukau B. (2009) Nat. Struct. Mol. Biol. 16, 589–597 [DOI] [PubMed] [Google Scholar]
- 15.Houben E. N., Zarivach R., Oudega B., Luirink J. (2005) J. Cell Biol. 170, 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma M. R., Koc E. C., Datta P. P., Booth T. M., Spremulli L. L., Agrawal R. K. (2003) Cell 115, 97–108 [DOI] [PubMed] [Google Scholar]
- 17.Sharma M. R., Booth T. M., Simpson L., Maslov D. A., Agrawal R. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9637–9642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafontaine D., Tollervey D. (1996) Nucleic Acids Res. 24, 3469–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daum G., Böhni P. C., Schatz G. (1982) J. Biol. Chem. 257, 13028–13033 [PubMed] [Google Scholar]
- 20.Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. (2000) Science 289, 905–920 [DOI] [PubMed] [Google Scholar]
- 21.Jia L., Kaur J., Stuart R. A. (2009) Eukaryot. Cell 8, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohler R., Boehringer D., Greber B., Bingel-Erlenmeyer R., Collinson I., Schaffitzel C., Ban N. (2009) Mol. Cell 34, 344–353 [DOI] [PubMed] [Google Scholar]
- 23.Myers A. M., Pape L. K., Tzagoloff A. (1985) EMBO J. 4, 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.