Abstract
In this study, we aim to determine cellular mechanisms linking nutrient metabolism to the regulation of inflammation and insulin resistance. The nutrient sensors AMP-activated protein kinase (AMPK) and SIRT1 show striking similarities in nutrient sensing and regulation of metabolic pathways. We find that the expression, activity, and signaling of the major isoform α1AMPK in adipose tissue and macrophages are substantially down-regulated by inflammatory stimuli and in nutrient-rich conditions, such as exposure to lipopolysaccharide (LPS), free fatty acids (FFAs), and diet-induced obesity. Activating AMPK signaling in macrophages by 5-aminoimidazole-4-carboxamide-1-β4-ribofuranoside or constitutively active α1AMPK (CA-α1) significantly inhibits; although inhibiting α1AMPK by short hairpin RNA knock-down or dominant-negative α1AMPK (DN-α1) increases LPS- and FFA-induced tumor necrosis factor α expression. Chromatin immunoprecipitation and luciferase reporter assays show that activation of AMPK by CA-α1 in macrophages significantly inhibits LPS- or FFA-induced NF-κB signaling. More importantly, in a macrophage-adipocyte co-culture system, we find that inactivation of macrophage AMPK signaling inhibits adipocyte insulin signaling and glucose uptake. Activation of AMPK by CA-α1 increases the SIRT1 activator NAD+ content and SIRT1 expression in macrophages. Furthermore, α1AMPK activation mimics the effect of SIRT1 on deacetylating NF-κB, and the full capacity of AMPK to deacetylate NF-κB and inhibit its signaling requires SIRT1. In conclusion, AMPK negatively regulates lipid-induced inflammation, which acts through SIRT1, thereby contributing to the protection against obesity, inflammation, and insulin resistance. Our study defines a novel role for AMPK in bridging the signaling between nutrient metabolism and inflammation.
Keywords: AMP-activated Kinase (AMPK), Fatty Acid, Inflammation, Insulin Resistance, Macrophage, SIRT1
Introduction
Obesity is causally associated with insulin resistance and type 2 diabetes (1, 2). The underlying mechanisms linking obesity to insulin resistance/type 2 diabetes have received intense investigation during the past decade. Obesity is characterized by a state of chronic inflammation featuring increased production of pro-inflammatory cytokines and chemokines, such as tumor necrosis factor-α (TNF-α),3 interleukin-6 (IL-6), and monocyte chemoattractant protein 1, and activation of the inflammatory signaling networks, including inhibitor of NF-κB (IκB) kinase β (IKK-β)/NF-κB and the c-Jun NH2-terminal kinase (JNK) pathways in key metabolic tissues as well as macrophages (1–4). Loss of function studies have demonstrated pivotal roles of these inflammatory pathways in the development of obesity-induced insulin resistance (3, 4).
Adipose tissue plays a key role in the generation of inflammatory responses and mediators in obesity (1, 2). Obese adipose tissue exhibits increased infiltration of macrophages, and acrophages may be a significant source of inflammation (5, 6). The importance of macrophages in obesity-induced inflammation and insulin resistance has been well documented in genetic studies (7–10). Genetic disruption of IKK-β in myeloid lineage-protected mice from high fat (HF)-induced inflammation and insulin resistance (7). Likewise, disruption of JNK1 in hematopoietically derived cells including macrophages via bone marrow transplantation also ameliorated obesity-induced inflammation and insulin resistance in mice (8). In contrast, targeted deletion of peroxisome proliferator-activated receptor-γ in macrophages increased systemic inflammation and impaired insulin signaling and sensitivity in muscle, fat, and liver (9, 10). These studies support the notion that macrophage inflammation is a key component of obesity-induced inflammation and insulin resistance.
Chronic nutrient overload leads to obesity and an array of associated metabolic disorders, including insulin resistance and type 2 diabetes (11, 12). Inflammatory signaling pathways in obesity can be activated in nutrient-rich conditions (e.g. exposure to lipids and glucose) (11, 12), suggesting a clear cross-talk between metabolic and inflammatory pathways. However, how excess nutrients are sensed within the cells (e.g. macrophages and adipocytes) and alter inflammatory programs in these cells is not fully understood. Two signaling proteins that stand out as candidates linking nutrient metabolism and inflammation are the two nutrient sensors: AMP-activated protein kinase (AMPK) and SIRT1.
AMPK, an evolutionarily conserved sensor of cellular energy status, is emerging as a chief regulator of whole body energy homeostasis (13). AMPK functions as a cellular energy gauge that regulates metabolic pathways in lipid and glucose metabolism by integrating nutritional and hormonal signals in the periphery and hypothalamus (13). SIRT1, which also functions as an energy sensor, is an NAD+-dependent deacetylase that mediates the effects of caloric restriction to extend longevity (14). AMPK and SIRT1 show striking similarities in sensing nutrient supply and regulating metabolic pathways and are likely to interact to perform these functions. Emerging evidence suggests that both AMPK and SIRT1 also regulate inflammatory signaling in various cells (15–17). We therefore hypothesize that the nutrient sensors AMPK and SIRT1 also serve as key determinants of lipid-induced inflammatory signaling events and cooperate to regulate inflammatory responses in macrophages; disruption of these pathways contributes to abnormal inflammatory signaling and may contribute to insulin resistance in obesity. To test this hypothesis, we examined macrophage AMPK signaling and SIRT1 expression in response to the inflammatory stimulus lipopolysaccharide (LPS), lipids or a high fat diet. We also examined the inflammatory pathways in free fatty acid (FFA)-treated macrophages with gain or loss functions of AMPK or SIRT1, and explored the relationship of AMPK and SIRT1 in the regulation of macrophage inflammation. We further investigated the effects of inactivation of macrophage AMPK signaling on adipocyte insulin signaling and sensitivity in a macrophage-adipocyte co-culture system.
EXPERIMENTAL PROCEDURES
Animals
For diet-induced obesity studies, 6-week-old male C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME), and were fed chow (D12450B, 10% kcal from fat, Research Diets Inc., New Brunswick, NJ) or HF diets (D12492, fat content 60% by calorie, Research Diets Inc.) for 24 weeks. For lipid infusion studies, 12-week-old male C57BL/6J mice were implanted with indwelling catheters (Micro-Renathane tubing, MRE-025; Braintree Scientific Inc.). After a 5-day recovery, mice were infused with lipids (5 ml/kg·h; Liposyn II; Abbott) and heparin (6 units/h) or saline for 8 h with a microdialysis pump (CMA/102; CMA Microdialysis). For LPS injection experiments, 8-week-old mice were injected with LPS (2 mg/kg body weight).
Cell Culture
Raw264.7 macrophages were cultured in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum. Bone marrow was flushed from the femur and tibia, dispersed, and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 30% L929 conditional medium for 8 days. Peritoneal macrophages were isolated by lavage 4 days after intraperitoneal injection of 3% thioglycollate (2 ml; Difco, BD Biosciences, San Jose, CA). Fatty acids (stearate or palmitate/oleate/stearate (equal molars) mixture, Sigma) were conjugated with BSA at a 4:1 molar ratio before treatment. TNF-α protein secreted into medium was measured using a TNF-α ELISA kit (R&D Systems, Minneapolis, MN).
For the macrophage-adipocyte co-culture experiment, macrophages were plated in a transwell insert (0.4 μm porous membrane) and placed into wells containing 3T3-L1 adipocytes at day 8 after the induction of differentiation (18). Four days after co-culture, adipocytes were used for insulin signaling and glucose transport assays as described below.
AMPK Activity and NAD+ and NADH Content Assays
Cell lysates (50 μg) were immunoprecipitated with specific antibodies (Upstate, Lake Placid, NY) against the α1 subunit bound to protein G-Sepharose beads. The kinase activity of the immunoprecipitates was measured using “SAMS” peptide and [γ-32P]ATP (19). NAD+ and NADH contents were measured using a NAD+/NADH quantification kit (BioVision, Mountain View, CA).
Plasmid Constructs and Transfection
Construction of constitutively active (CA) and dominant-negative (DN) α1AMPK expression vectors were described previously (20). pCruz-SIRT1, pAd-Track SIRT1, pAd-Track-SIRT1 H355A, SIRT2-SIRT7, p300, and p65 expression vectors were all purchased from Addgene (Cambridge, MA). The expression constructs that were not carried on the pcDNA3.1 backbones were further subcloned into the pcDNA3.1 expression vectors. Raw264.7 macrophages were transfected with expression vectors using a SuperFect Transfection Reagent kit (Qiagen, Valencia, CA). For some AMPK studies, cells transfected with CA- or DN-α1AMPK were selected with G418 and positively transfected cells were expanded for further experiments.
Luciferase Reporter Assay
pNFκB-Luc and pRL-SV40 vectors were purchased from Clontech. Luciferase activity was measured using a Dual Luciferase Reporter Assay kit (Promega, Madison, WI), according to the manufacturer's instructions.
Lentiviral Short Hairpin RNA (shRNA) Knockdown
The SIRT1 lentiviral shRNA vector constructs were purchased from Open Biosystems (Huntsville, AL). The SIRT1 shRNA lentivirus was generated according to the instructions. Briefly, the SIRT1 or control lentiviral vectors were co-transfected with the packaging plasmid (pCMV-dR8.91, the Broad Institute, Cambridge, MA) and the envelope plasmid (vesicular stomatitis virus-G, the Broad Institute) into 293T cells. Medium containing the lentivirus was harvested and filtered, and used to infect Raw264.7 cells. For α1AMPK knockdown, the infected cells were selected with puromycin (8 μg/ml) for 8 days, and the surviving cells were pooled and evaluated for α1AMPK mRNA and protein expression by real time RT-PCR and immunoblotting, respectively. For SIRT1 knockdown, the infected cells were selected with puromycin (8 μg/ml) for 2 weeks, and the surviving colonies were picked, expanded, and evaluated for SIRT1 mRNA and protein expression by real time RT-PCR and immunoblotting, respectively. The SIRT1 knockdown cell line was established and used to transfect CA-α1AMPK expression vectors and other constructs in acetylation and luciferase assays.
Total RNA Extraction and Quantitative RT-PCR
Adipose tissue or macrophage total RNA was extracted using the Tri-Reagent kit (Molecular Research Center, Cincinnati, OH), according to the manufacturer's protocol. The expression of genes of interest was assessed by quantitative RT-PCR (ABI Universal PCR Master Mix, Applied Biosystems, Foster City, CA) using a Stratagene Mx3000p thermocycler (Stratagene, La Jolla, CA), as previously described (18). The primer and probe pairs used in the assays were purchased from Applied Biosystems.
Immunoblotting
Immunoblotting was conducted as previously described (18) with modifications. Briefly, the transferred membranes were blocked, washed, incubated with various primary antibodies overnight at 4 °C, and followed by Alexa Fluor 680-conjugated secondary antibodies (Invitrogen) at room temperature for 2 h. The blots were developed with a Li-COR Odyssey Infrared Imager system (Li-COR Biosciences, Lincoln, NE). Rabbit polyclonal antibodies against phospho-JNK, phospho-AMPK (Thr172), and acetyl-p65 (lysine 310) were purchased from Cell Signaling (Beverly, MA). Rabbit polyclonal antibodies against IRS-1, SIRT1, α1AMPK, and phospho-acetyl-CoA carboxylase (ACC) (Ser79) were obtained from Upstate (Lake Placid, NY). Rabbit polyclonal antibodies against IR, p65, mouse monoclonal antibody against LKB1, and goat polyclonal antibodies against actin were from Santa Cruz (Santa Cruz, CA). Rabbit polyclonal antibodies against IR (Tyr1162/Tyr1163) and IRS-1 (Tyr612) were from BIOSOURCE International (Camarillo, CA). Total ACC was detected using streptavidin-conjugated horseradish peroxidase (Amersham Biosciences) (19).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP was conducted using a ChIP assay kit (Upstate) as previously described (18).
Insulin Signaling and Glucose Uptake Assay
Adipocytes were stimulated with 100 nm insulin for 5 min. Cells were harvested and homogenized in modified RIPA buffer. IR (Tyr1162/Tyr1163) and IRS-1 (Tyr612) phosphorylation was measured by immunoblotting. Glucose transport assay was conducted as previously described (21).
Statistics
All data are expressed as mean ± S.E. Data were evaluated for statistical significance by one-way analysis of variance, and statistical significance for comparison of means of different groups was calculated by the least significant difference test using the SPSS software package version 11.5. p < 0.05 was considered significant.
RESULTS
Inflammatory Stimuli and Lipids Down-regulate AMPK in Macrophages and Adipose Tissue
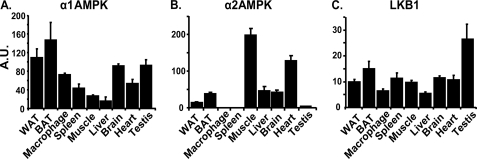
Using real time RT-PCR, we found that α1AMPK is more abundantly expressed in adipose tissue, peritoneal macrophages, and spleen than in muscle, whereas the expression of α2AMPK is undetectable in peritoneal macrophages and spleen and low in adipose tissue compared with muscle (Fig. 1, A and B). These data indicate that α1AMPK is the predominant isoform in immune-effector cells, which likely serves as a target for immunoregulation. The expression of LKB1, one of the upstream kinases for AMPK, is ubiquitously distributed (Fig. 1C).
FIGURE 1.
Tissue distribution of α1AMPK (A), α2AMPK (B), and LKB1 (C). Total RNA was isolated from individual tissues and the expression of α1AMPK, α2AMPK, and LKB1 was measured by real time RT-PCR and normalized to cyclophilin. BAT, brown adipose tissue; Macrophage, thioglycollate-elicited peritoneal macrophage. Data are expressed as mean ± S.E., n = 3. A.U., arbitrary units.
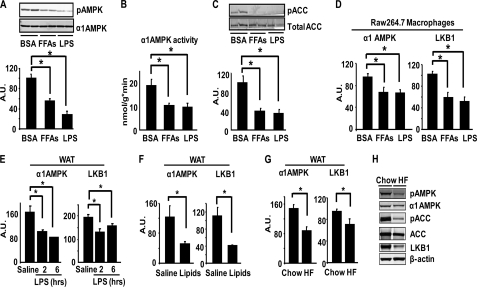
More importantly, we found that LPS treatment significantly inhibited AMPK phosphorylation and α1AMPK activity by approximately 60 and 40%, respectively, in bone marrow macrophages (Fig. 2, A and B). Similar results were observed in macrophages treated with FFAs (mixture of palmitate, oleate, and stearate) (Fig. 2, A and B). Consistent with this, the phosphorylation of acetyl-CoA carboxylase (ACC), a direct downstream target of AMPK, was also reduced by LPS and FFA treatment (Fig. 2C). LPS and FFAs not only inhibit AMPK signaling but also suppress the expression of the signaling molecules. LPS treatment for 24 h inhibited α1AMPK and LKB1 mRNA by 30 and 50%, respectively, with a similar result observed with FFA treatment (Fig. 2D). These data suggest that LKB1/AMPK signaling and expression are down-regulated in response to FFAs and inflammatory stimuli in macrophages and may play an important role in maintaining immune homeostasis. Consistent with the data from macrophages, LPS administration inhibited α1AMPK and LKB1 mRNA in the epididymal adipose tissue of mice (Fig. 2E). We then examined AMPK signaling and expression in fat in the physiological context of obesity. Increasing circulating FFAs by lipid infusion for 8 h reduced α1AMPK and LKB1 mRNA by 50% (Fig. 2F). Similar results were observed in the fat of mice challenged with HF diets, although to a lesser extent (Fig. 2G). The decreased expression of AMPK signaling molecules by HF diets is associated with the reduced AMPK signaling in fat, evident by the decreased AMPK and ACC phosphorylation levels (Fig. 2H). This is also accompanied by reduced LKB1 protein levels (Fig. 2H), indicating that down-regulated upstream kinase LKB1 may partly explain the suppressed AMPK signaling in adipose tissue of obese animals. These data strongly suggest that the AMPK signaling pathway, primarily via the α1AMPK isoform, is down-regulated in conditions that are characterized by over-nutrition and/or inflammation, such as exposure to LPS, lipids, and diet-induced obesity.
FIGURE 2.
AMPK signaling and expression in macrophages and adipose tissue are down-regulated by LPS, FFAs, HF diets, and lipid infusion. A–C, LPS and FFAs inhibit AMPK phosphorylation (A), activity (B), and ACC phosphorylation (C) in macrophages. Bone marrow macrophages were treated with LPS (100 ng/ml) or FFAs (palmitate, oleate, and stearate mixture, 500 μm) for 2 h. D, LPS and FFAs inhibit the expression of α1AMPK and LKB1 in macrophages. RAW264.7 macrophages were treated with LPS (100 ng/ml) or FFAs (250 μm) overnight. n = 6/group. E, LPS inhibits the mRNA expression of α1AMPK and LKB1 in epididymal WAT of mice. C57/BL6J mice (male, n = 4/group) were intraperitoneally injected with LPS (2 mg/kg body weight) and WAT were dissected 2 or 6 h after injection. F, increasing circulating FFAs inhibits the mRNA expression of α1AMPK and LKB1 in epididymal WAT in mice (n = 5/group). Mice were infused with lipids (5 ml/kg·h, liposyn II; coupled with 6 units/h of heparin) for 8 h. G and H, HF feeding inhibits the mRNA expression of α1AMPK and LKB1 (G) and AMPK signaling (H) in epididymal WAT of mice (n = 8/group). For A, C, and H, AMPK signaling was measured by immunoblotting. For B, α1AMPK activity was measured using an immunocomplex assay with SAMS peptide. For D–G, mRNA levels of target genes were measured by real time RT-PCR and normalized to cyclophilin. All data are expressed as mean ± S.E. *, p < 0.05. A.U., arbitrary units.
Activation of the AMPK Signaling Pathway Suppresses LPS- and FFA-induced Inflammation in Macrophages
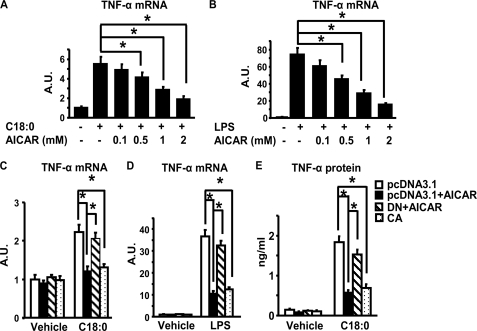
FFAs are important mediators of obesity-induced inflammation and insulin resistance (18). We showed previously that inflammatory signaling in macrophages can be activated by FFAs, among which stearate (C18:0) is the most potent stimulator of cytokine expression (18). Here we found that activation of AMPK by 5-aminoimidazole-4-carboxamide-1-β4-ribofuranoside (AICAR), a specific agonist of AMPK, significantly suppressed both LPS- and FFA-induced TNF-α expression in a dose-dependent manner in macrophages (Fig. 3, A and B). The effect of AICAR was likely mediated by activation of AMPK signaling, as it can be mimicked by overexpression of the CA-α1AMPK (causing sustained activation of AMPK signaling), and was blocked by overexpression of the DN-α1AMPK (causing sustained inactivation of AMPK signaling) (Fig. 3, C and D). Moreover, activating AMPK signaling by AICAR or CA-α1AMPK also markedly attenuated stearate-induced TNF-α protein secretion (Fig. 3E).
FIGURE 3.
A and B, activation of AMPK signaling by AICAR suppresses stearate (C18:0) (A)- or LPS (B)- induced TNF-α mRNA expression in macrophages. RAW264.7 macrophages were pre-treated with various concentrations of AICAR as indicated, and then stimulated with LPS (100 ng/ml) or stearate (C18:0) (500 μm) for 4 h. TNF-α mRNA was measured by real time RT-PCR. C and D, activation of AMPK by CA-α1AMPK or AICAR inhibits stearate (C)- or LPS (D)-induced TNF-α expression. RAW264.7 macrophages were transfected with pcDNA3.1, DN-α1AMPK, or CA-α1AMPK, and then treated with vehicle (BSA for stearate and H2O for LPS), LPS (100 ng/ml), stearate (500 μm), with or without AICAR (2 mm) as indicated for 4 h. E, activation of AMPK by AICAR or CA-α1AMPK attenuates TNF-α secretion by macrophages. RAW264.7 macrophages transfected with pcDNA3.1, DN-α1AMPK, or CA-α1AMPK or treated with AICAR (2 mm) were stimulated with stearate (250 μm) for 24 h. All data are expressed as mean ± S.E., n = 6–8, *, p < 0.05. A.U., arbitrary units.
Inactivation of α1AMPK Promotes Basal and LPS-/FFA-induced Inflammation in Macrophages
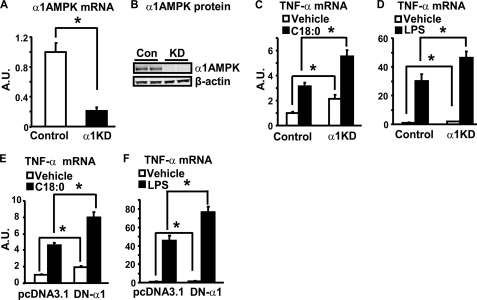
To investigate the role of endogenous α1AMPK expression in the regulation of inflammation, we established macrophages with reduced α1AMPK expression by lentiviral shRNA knockdown. The α1AMPK mRNA in the knockdown cells was suppressed by ∼80%, compared with that of macrophages infected with control lentivirus (Fig. 4A), as assessed by real time RT-PCR, whereas the protein expression was also significantly decreased, as assessed by immunoblotting (Fig. 4B). α1AMPK knockdown increased basal TNF-α mRNA expression by 1-fold, and further promoted stearate- or LPS-stimulated TNF-α expression (Fig. 4, C and D). Similar results were observed in macrophages with inactivation of α1AMPK by overexpression of DN-α1AMPK (Fig. 4, E and F). These data suggest that endogenous α1AMPK is a key determinant of basal inflammatory signaling and an important suppressor of LPS- and FFA-induced inflammation in macrophages.
FIGURE 4.
Inactivation of α1AMPK increases basal and stearate-stimulated TNF-α mRNA. A and B, establishment of a macrophage cell line with α1AMPK knockdown (α1KD). RAW264.7 macrophages were infected with the α1AMPK shRNA lentivirus or control lentivirus, and selected with puromycin for 8 days. α1AMPK RNA (A) and protein (B) levels were evaluated by real time RT-PCR and immunoblotting, respectively. C and D, α1AMPK knockdown increases basal and stearate- or LPS-stimulated TNF-α mRNA. The α1AMPK-knockdown and control macrophages were treated with vehicle (BSA for stearate and H2O for LPS), stearate (500 μm), or LPS (100 ng/ml) for 4 h. E and F, inactivation of α1AMPK by DN-α1AMPK overexpression increases basal and stearate- or LPS-stimulated TNF-α mRNA. RAW264.7 macrophages were transfected with pcDNA3.1 or DN-α1AMPK, and then treated with vehicle (BSA for stearate and H2O for LPS), LPS (100 ng/ml), or stearate (500 μm) as indicated for 4 h. TNF-α mRNA was measured by real time RT-PCR. Data are expressed as mean ± S.E., n = 6. *, p < 0.05. A.U., arbitrary units.
Activation of α1AMPK Antagonizes NF-κB in Macrophages
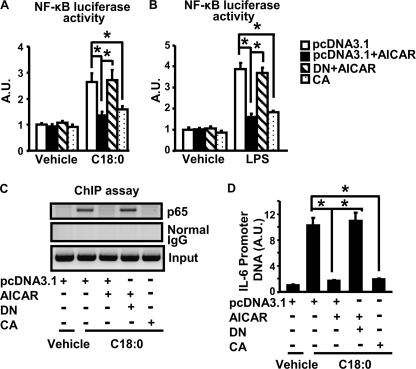
We further explored whether AMPK acts on the NF-κB pathway to antagonize cytokine expression. Activation of the AMPK signaling pathway by AICAR or CA-α1AMPK significantly suppressed NF-κB luciferase reporter activity induced by stearate (Fig. 5A) or LPS (Fig. 5B) in macrophages. To further confirm the inhibitory effect of AMPK on NF-κB signaling, we combined ChIP assays with SYBR Green quantitative PCR to examine the NF-κB subunit p65 binding to the consensus sequence of the IL-6 promoter in vivo. Activating AMPK by AICAR prevented stearate-induced p65 DNA binding to the IL-6 promoter, which can be mimicked by overexpression of CA-α1AMPK and blocked by DN-α1AMPK (Fig. 5C). Quantitation of immunoprecipitated DNA by the SYBR Green PCR further confirmed these findings (Fig. 5D). These data suggest that the anti-inflammatory effect of the AMPK signaling pathway is mainly mediated via inactivation of NF-κB signaling.
FIGURE 5.
Activation of AMPK suppresses NF-κB signaling in macrophages. A and B, activation of AMPK by AICAR or CA-α1AMPK inhibits NF-κB luciferase reporter activity. RAW264.7 macrophages expressing pcDNA3.1, CA-α1AMPK, or DN-α1AMPK were co-transfected with pNFκB-Luc vectors. After 5 h, cells were then treated overnight with vehicle (BSA for stearate and H2O for LPS), LPS (100 ng/ml), and stearate (250 μm) with or without AICAR (2 mm). n = 6. C, activation of AMPK blocks NF-κB (p65) binding to the IL-6 promoter in ChIP assay. D, SYBR Green quantitative PCR was used to measure the promoter DNA immunoprecipitated by the anti-p65 antibody; n = 3. All data are expressed as mean ± S.E. *, p < 0.05. A.U., arbitrary units.
Inactivation of Macrophage α1AMPK Inhibits Adipocyte Insulin Signaling
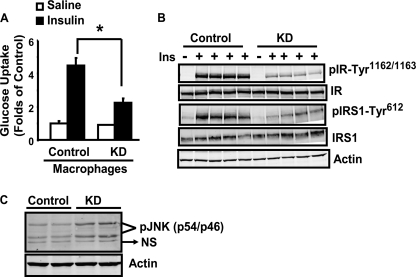
To mimic the physiological milieu of adipose tissue where infiltrated macrophages affect adipocyte insulin sensitivity in a paracrine fashion, we employed a co-culture system where control or α1AMPK-knockdown macrophages were grown in transwell inserts, which was then placed into the lower well chambers containing differentiated 3T3-L1 adipocytes. After a 4-day co-culture, insulin-stimulated glucose uptake was decreased by 50% in adipocytes co-cultured with α1AMPK-knockdown macrophages, compared with those with control macrophages (Fig. 6A). Consistent with this, adipocytes co-cultured with knockdown macrophages exhibited decreased insulin-stimulated phosphorylation of insulin receptor (IR) and insulin receptor substrate (IRS-1) (Fig. 6B). We further examined the JNK signal, a key inflammatory signal causing insulin resistance. JNK phosphorylation was up-regulated in adipocytes co-cultured with α1AMPK-knockdown macrophages, indicating activation of the JNK pathway (Fig. 6C). These data suggest that inactivation of macrophage α1AMPK likely increases the production of inflammatory cytokines, which activate adipocyte inflammatory signals such as JNK and lead to impaired insulin signaling and sensitivity in a paracrine fashion.
FIGURE 6.
Inactivation of macrophage α1AMPK inhibits adipocyte insulin signaling in a co-culture system. A and B, 3T3-L1 adipocytes co-cultured with α1AMPK-knockdown macrophages exhibit reduced insulin (Ins)-stimulated glucose uptake (A) and signaling (B). Insulin-stimulated glucose uptake and signaling were conducted in 3T3-L1 adipocytes co-cultured with either the knockdown (KD) or control macrophages. C, inactivation of macrophage α1AMPK stimulates JNK phosphorylation in adipocytes in the co-culture system. NS, non-specific. Data are expressed as mean ± S.E., n = 4; *, p < 0.05.
SIRT1 Is a Negative Regulator of Macrophage Inflammation
We found that SIRT1 is ubiquitously expressed in various tissues, with relatively high expression in immune-effector cells such as macrophages, WAT, and spleen (supplemental Fig. S1A). More importantly, both LPS and FFAs significantly inhibited SIRT1 expression in RAW264.7 macrophages (supplemental Fig. S1B), suggesting that SIRT1, like α1AMPK, also responds to inflammatory stimuli, such as LPS and lipids, and may thereby play a role in maintaining immune homeostasis. Consistent with this, LPS injection suppressed SIRT1 mRNA in white adipose tissue (WAT) of mice (supplemental Fig. S1C). In addition, both a high fat diet treatment and lipid infusion in mice suppressed SIRT1 mRNA in epididymal WAT (supplemental Fig. S1, D and E). The seemingly parallel response of α1AMPK and SIRT1 to inflammatory and metabolic stimuli suggests a close cross-talk between these two proteins in regulating inflammation in obesity and insulin resistance.
Several studies have shown that SIRT1 regulates inflammatory signaling pathways in various cell types (16, 17, 22). We evaluated the abilities of all SIRT family members (SIRT1–7) to antagonize inflammation. Raw264.7 macrophages were transiently transfected with expression vectors for SIRT1–7 and an NF-κB luciferase reporter construct, and were then stimulated with LPS. We found that only SIRT1 significantly inhibited LPS-induced luciferase activity. A slight inhibition by SIRT6 was also observed (supplemental Fig. S2). However, SIRT6 is not detectable in macrophages (data not shown) and SIRT3–5 are mainly located in mitochondria (23), SIRT1 is therefore the only SIRT family member that potentially plays a major role in regulating macrophage inflammation. To determine whether SIRT1 antagonizes FFA-induced inflammation via inhibiting the inflammatory networks, we examined the inflammatory signaling pathways of NF-κB. Forced expression of the wild-type SIRT1 in macrophages substantially blocked FFA- or LPS-stimulated NF-κB luciferase activity, but the mutant SIRT1(H355A) that lacks the deacetylase activity did not, suggesting that the deacetylation function is required for SIRT1 to antagonize NF-κB signaling (supplemental Fig. S3A).
We further performed a study of loss of SIRT1 function to investigate the role of endogenous SIRT1 expression in the regulation of inflammation. We established a macrophage cell line with reduced SIRT1 expression by lentiviral shRNA knockdown. The SIRT1 mRNA level in the knockdown cells were suppressed by more than 80%, compared with that of control macrophages (supplemental Fig. S3B), as assessed by real time RT-PCR, whereas the protein expression was not detectable (supplemental Fig. S3C). Real time RT-PCR analysis showed that SIRT1 knockdown in macrophages increased basal expression of inflammatory genes including TNF-α, IL-6, IL-1β, and iNOS (supplemental Fig. S3, D–G). These data suggest that SIRT1 is required for suppressing the basal inflammatory state.
Activation of AMPK Increases NAD+ Levels and SIRT1 Expression in Macrophages
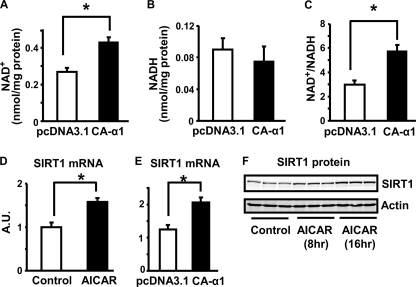
We further explored the downstream signals that mediate the anti-inflammatory effects of AMPK. A recent study showed that AMPK enhances SIRT1 activity via increasing cellular NAD+ levels, resulting in deacetylation of peroxisome profiferator-activated receptor γ coactivator 1α in muscle (24). We investigated whether AMPK increases the content of the SIRT1 activator, NAD+, in macrophages. Activation of AMPK signaling by overexpression of CA-α1AMPK enhanced the cellular NAD+/NADH ratio by 60%, mainly due to the increase of NAD+ levels (Fig. 7, A–C). We also found that activation of AMPK by AICAR or CA-α1AMPK overexpression in macrophages increased SIRT1 mRNA by 50% (Fig. 7, D and E). In parallel, AICAR treatment also increased SIRT1 protein levels in a time course study (Fig. 7F). These data suggest that activation of AMPK increases NAD+ content and SIRT1 expression levels, which would enhance SIRT1 deacetylase activity in cells. Therefore, SIRT1 is likely to be a downstream signal that mediates the anti-inflammatory actions of AMPK in macrophages.
FIGURE 7.
A–C, activation of AMPK by CA-α1AMPK expression increases NAD+ content and NAD+/NADH ratio in macrophages. RAW264.7 macrophages were transfected with CA-α1AMPK expression vectors. NAD+ and NADH nucleotides were measured with a NAD+/NADH quantification kit. D and E, activation of AMPK increases SIRT1 mRNA expression in macrophages. RAW264.7 macrophages were treated with 2 mm AICAR (D) or transfected with CA-α1AMPK overnight (E). F, AICAR increases SIRT1 protein expression in macrophages. Peritoneal macrophages were treated with AICAR (2 mm) at the indicated time points. SIRT1 mRNA was measured by real time RT-PCR. SIRT1 protein expression was measured by immunoblotting. All data are expressed as mean ± S.E., n = 4; *, p < 0.05.
SIRT1 Is Required for AMPK to Deacetylate NF-κB and Antagonize Its Signaling in Macrophages
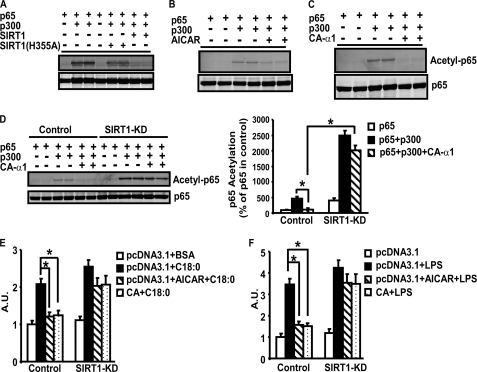
Previous studies have shown that SIRT1 antagonizes NF-κB (p65) activity by deacetylating its lysine 310 in several cell lines (17). We tested whether AMPK is also capable of deacetylating NF-κB in macrophages. Because NF-κB acetylation levels remain low in the basal state, we expressed p300, an acetyltransferase widely used to acetylate the subunit p65 of NF-κB (17), to stimulate NF-κB acetylation in macrophages. Transfection of the p300 expression vector effectively led to p65 acetylation, which was largely inhibited by the wild type SIRT1 but not by the mutant SIRT1(H355A) lacking the full strength of deacetylase activity (Fig. 8A). Similarly, activation of AMPK by AICAR also decreased p300-induced acetylation of p65, mimicking the ability of SIRT1 to deacetylate p65 (Fig. 8B). In parallel, genetic activation of AMPK by CA-α1AMPK overexpression also led to decreases in p300-induced p65 acetylation (Fig. 8C).
FIGURE 8.
A, SIRT1 deacetylates p65 in macrophages. RAW264.7 macrophages were transfected with pcDNA3.1 or expression vectors for p65, p300, SIRT1, or SIRT1(H355A). Cell lysates were used to examine p65 acetylation (lysine 310) by immunoblotting. B and C, activation of AMPK by AICAR (B) or CA-α1AMPK (C) deacetylates p65 in macrophages. RAW264.7 macrophages were transfected with expression vectors for p65, p300, and CA-α1AMPK, or treated with AICAR (2 mm). D, SIRT1 is required for full capacity of AMPK to deacetylate NF-κB. The SIRT1-knockdown or control cells were transfected with expression vectors for p65, p300, and CA-α1AMPK. A representative blot was shown in the left panel. The blots were quantitated with a Li-COR Odyssey Infrared Imager system (right panel). *, p < 0.05. E and F, SIRT1 is required for full capacity of AMPK to inhibit NF-κB transcriptional activity. The control or SIRT1-knockdown macrophages transfected with pNFκB-Luc vectors were treated with 2 mm AICAR or co-transfected with CA-α1AMPK expression vectors, and then stimulated with stearate (250 μm) (E) or LPS (100 ng/ml) (F) for 24 h. All data are expressed as mean ± S.E., n = 4; *, p < 0.05.
We tested whether the ability of AMPK to deacetylate NF-κB requires SIRT1. As shown above, we have established a macrophage cell line with SIRT1 knockdown by lentiviral shRNA infection (supplemental Fig. 3, B and C). In control cells, activation of AMPK by CA-α1AMPK overexpression largely blocked acetylation of p65 induced by p300 (Fig. 8D, left panel). Quantitation of the blot showed that overexpression of p300 increased p65 acetylation by 450% in control cells, which was largely prevented by CA-α1AMPK (reduced to 110%, p < 0.05, Fig. 8D, right panel). In contrast, CA-α1AMPK overexpression was unable to fully deacetylate p65 in SIRT1-knockdown cells (Fig. 8D, left panel). Quantitation of the blot showed that p300 overexpression significantly increased p65 acetylation in SIRT1-knockdown cells compared with control cells (2480% in SIRT1 knockdown cells versus 450% in control cells, p < 0.05, Fig. 8D, right panel). Although there was a trend of a slight decrease of p65 acetylation by CA-α1AMPK in the knockdown cells (2480 versus 2010%, Fig. 8D, right panel), it did not reach statistical significance (p = 0.12). We further determined whether SIRT1 is required for AMPK to antagonize NF-κB signaling. Activation of AMPK by AICAR or CA-α1AMPK largely prevented LPS- or stearate-induced NF-κB reporter activity in control macrophages, whereas AICAR or CA-α1AMPK failed to exert the same actions in SIRT1-knockdown cells (Fig. 8, E and F). Expression of the other SIRT2–7 was not affected by SIRT1 knockdown in these macrophages (data not shown), excluding their roles in mediating the inhibitory effects of AMPK in NF-κB acetylation and activity. Therefore, these data demonstrate that SIRT1 mediates the anti-inflammatory effects of AMPK in macrophages.
DISCUSSION
This study was designed to address the hypothesis that two energy sensors AMPK and SIRT1 “sense” lipids or the state of HF diet-induced obesity and serve as negative regulators of FFA-induced inflammation in macrophages. The plausibility of this hypothesis derives from several prior findings targeting obesity-induced insulin resistance. First, inflammation is a key link between obesity and insulin resistance/type 2 diabetes (1–4), and macrophage inflammation is a key component of obesity-induced inflammation and plays a key role in obesity-induced insulin resistance (5–10). Second, excess nutrients, such as lipids and glucose, can lead to obesity, and hyperlipidemia and hyperglycemia are commonly seen with obesity (1). Excessive lipids are highly detrimental in the pathological development of obesity-induced inflammation and insulin resistance (1). FFAs, levels of which are elevated in obesity due to increased release from the expanded fat tissue or due to dietary sources, may impair insulin sensitivity by several actions (25), one of which is FFA-induced inflammation in metabolic cells and macrophages (18, 26–28). FFAs are a bona fide fuel source for energy metabolism. We believe that a signaling protein(s) may regulate both FFA metabolism and FFA-induced inflammatory response. This signaling protein(s) might be expected to be (a) a nutrient and energy sensor; (b) abundantly expressed in both macrophages and adipocytes, two critical cell types involved in obesity-induced inflammation/insulin resistance and exhibiting inflammatory properties when overloaded with lipids; (c) regulated by both FFAs and inflammatory stimuli; and (d) a regulator of both FFA metabolism and inflammatory response. AMPK and SIRT1 are ideal candidates to meet these criteria and fulfill the role as coordinators for FFA metabolism and inflammation.
Our data show that the AMPK signaling pathway in macrophages, possibly via the α1AMPK isoform, is down-regulated by FFAs or HF diets. Our studies with loss or gain of function show that inactivation of AMPK signaling promotes a FFA-induced inflammatory response, whereas activation of AMPK signaling prevents FFA activation of inflammatory pathways in macrophages. Therefore, AMPK may serve as an important signaling protein regulating both FFA metabolism and FFA-induced inflammatory signaling pathways in response to changes of cellular nutrient and energy status. Consistent with our findings, a few studies have also demonstrated that activation of AMPK prevents LPS-induced expression of pro-inflammatory cytokines and NF-κB signaling in several cell types (15, 29). Our studies further link the anti-inflammatory functions of AMPK to nutrient- and obesity-induced inflammation and provide a mechanism in which SIRT1 is a downstream signal that mediates the anti-inflammatory actions of AMPK. Our data strongly indicate that AMPK is a negative regulator of innate immunity, and serves as an important energy sensor-linking nutrient metabolism to the regulation of inflammatory signaling. Disruption of AMPK signaling during metabolic disturbance, such as excessive FFA exposure and HF diets, may be responsible for the abnormal activation of inflammatory signaling networks in obesity.
The anti-inflammatory function of AMPK may have a physiological consequence of preventing inflammation-induced insulin resistance. Emerging evidence shows that AMPK plays an important role in regulating insulin sensitivity. The insulin sensitizers metformin and rosiglitazone can activate AMPK (29–31), suggesting a possible role of AMPK in regulating insulin sensitivity. Indeed, activation of AMPK increases basal and insulin-stimulated glucose uptake in skeletal muscle and inhibits gluconeogenesis in the liver, leading to improved systemic insulin sensitivity and glucose homeostasis (32–34). These beneficial effects are largely attributed to AMPK actions on glucose and lipid metabolism and mitochondrial biogenesis in skeletal muscle, liver, and hypothalamus (13). However, our data derived from macrophage-adipocyte co-culture studies indicate that AMPK may also have impact on insulin sensitivity through its ability to antagonize FFA-induced inflammation. This effect is exerted primarily through the α1AMPK isoform in macrophages. Down-regulation of AMPK by excess nutrients (e.g. FFAs) in macrophages may likely contribute to obesity-induced inflammation and insulin resistance; whereas activation of macrophage AMPK may serve as a therapeutic target in preventing obesity-induced inflammation and insulin resistance. Genetic models with gain or loss of function of α1AMPK in specific tissues (e.g. macrophages) will be required to determine the role of α1AMPK in regulating obesity-induced inflammation and insulin resistance in vivo.
Like AMPK, SIRT1 expression was suppressed by lipids and HF diets. Moreover, overexpression of SIRT1 prevented FFA- or LPS-stimulated NF-κB signaling in macrophages, whereas SIRT1 knockdown increased basal expression of inflammatory genes. This is consistent with previous studies revealing that SIRT1 regulates inflammatory signaling pathways (16, 17, 22). SIRT1 physically binds to the p65/RelA subunit of NF-κB and deacetylates p65 at lysine 310, which down-regulates the transcriptional activity of NF-κB and leads to reduced inflammatory response (17). SIRT1 has therefore emerged as a novel anti-inflammatory target. Indeed, genetic studies show that global overexpression of SIRT1 in transgenic mice improves glucose tolerance, which is associated with down-regulation of inflammatory cytokine expression and NF-κB signaling (35). In contrast, specific deletion of SIRT1 in the liver increases inflammation and decreases insulin sensitivity in liver (36). Recent studies from Olefsky's group (16, 22) directly linked the anti-inflammatory effects of SIRT1 in adipocytes and macrophages to improved insulin sensitivity. Additional studies, involving deletion or overexpression of SIRT1 in specific tissues (e.g. macrophages) are required to separate the systemic effects on energy metabolism from the direct effect of inflammation.
It is intriguing that SIRT1 mimics many of the effects of AMPK in energy sensing (e.g. NAD+/NADH and AMP/ATP) and metabolism (e.g. mitochondrial biogenesis and FFA oxidation). The striking similarities of these two signaling proteins in nutrient sensing, and their parallel functions in regulating metabolic pathways, suggest a close relationship between them. Indeed, several recent studies suggested that the SIRT1 agonist resveratrol stimulates AMPK (37, 38). Several studies have been conducted to determine whether and how SIRT1 and AMPK cooperate to regulate metabolic pathways. A recent study reported that AMPK activates SIRT1 by increasing the expression of nicotinamide phosphoribosyltransferase (Nampt), an enzyme that catalyzes NAD+ synthesis from nicotinamide (39). More convincingly, a further study showed that AMPK enhances SIRT1 activity via increasing cellular NAD+ levels, resulting in the deacetylation of peroxisome profiferator-activated receptor γ coactivator 1α in muscle (24). However, other studies also indicated that SIRT1 activates AMPK via deacetylating and activating LKB1, the upstream kinase of AMPK (40, 41). No matter what the hierarchy of signals between the two proteins, AMPK and SIRT1 seem to be coordinately regulated and work in concert to regulate metabolic pathways. It is possible that AMPK and SIRT1 can activate and feed off each other depending on different cells or physiological scenarios. We found that in macrophages, activation of AMPK increases the SIRT1 activator NAD+ content and SIRT1 expression in macrophages. We further demonstrate that α1AMPK mimics the effect of SIRT1 on deacetylating NF-κB, and the full capacity of AMPK to deacetylate NF-κB and inhibit its signaling requires SIRT1, suggesting that SIRT1 is the downstream target for AMPK in the regulation of macrophage inflammation.
In sum, our data demonstrate that AMPK signaling and expression in adipose tissue and macrophages are down-regulated by inflammatory stimuli, lipids, and HF diets. Activation of macrophage AMPK signaling by pharmacological or genetic approaches significantly inhibits LPS- and FFA-induced TNFα expression, whereas inactivation of AMPK does the opposite. In addition, the anti-inflammatory effect of AMPK is likely mediated through antagonism of NF-κB signaling. More importantly, inactivation of macrophage AMPK signaling inhibits adipocyte insulin sensitivity and signaling in a macrophage-adipocyte co-culture system. We further demonstrate that SIRT1 is a downstream signal that mediates the anti-inflammatory functions of AMPK. AMPK activates SIRT1 by increasing its expression and production of its activator NAD+ in macrophages. Furthermore, α1AMPK activation mimics the effect of SIRT1 on deacetylation of NF-κB, and SIRT1 is required for the ability of AMPK to deacetylate NF-κB and inhibit its signaling. In conclusion, our data strongly indicate that AMPK is a negative regulator of innate immunity, and serves as an important energy sensor linking nutrient metabolism to the regulation of inflammatory signaling. AMPK and SIRT1, two classic energy sensors that play key roles in regulating lipid and glucose metabolism and systemic energy homeostasis, may also be negative regulators for lipid-induced inflammation in macrophages.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK084172 (to H. S.) and P01DK56116 (to B. B. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- TNF-α
- tumor necrosis factor-α
- IL
- interleukin
- JNK
- c-Jun NH2-terminal kinase
- IKK-β
- IκB kinase β
- HF
- high fat
- AMPK
- AMP-activated protein kinase
- CA
- constitutively active
- DN
- dominant-negative
- ChIP
- chromatin immunoprecipitation
- AICAR
- 5-aminoimidazole-4-carboxamide-1-β4-ribofuranoside
- IRS-1
- insulin receptor substrate 1
- IR
- insulin receptor
- WAT
- white adipose tissue
- RT
- reverse transcription
- LPS
- lipopolysaccharide
- FFA
- free fatty acid
- BSA
- bovine serum albumin
- shRNA
- short hairpin RNA
- ACC
- acetyl-CoA carboxylase.
REFERENCES
- 1.Hotamisligil G. S. (2006) Nature 444, 860–867 [DOI] [PubMed] [Google Scholar]
- 2.Shoelson S. E., Lee J., Goldfine A. B. (2006) J. Clin. Invest. 116, 1793–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan M., Konstantopoulos N., Lee J., Hansen L., Li Z. W., Karin M., Shoelson S. E. (2001) Science 293, 1673–1677 [DOI] [PubMed] [Google Scholar]
- 4.Hirosumi J., Tuncman G., Chang L., Görgün C. Z., Uysal K. T., Maeda K., Karin M., Hotamisligil G. S. (2002) Nature 420, 333–336 [DOI] [PubMed] [Google Scholar]
- 5.Xu H., Barnes G. T., Yang Q., Tan G., Yang D., Chou C. J., Sole J., Nichols A., Ross J. S., Tartaglia L. A., Chen H. (2003) J. Clin. Invest. 112, 1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisberg S. P., McCann D., Desai M., Rosenbaum M., Leibel R. L., Ferrante A. W., Jr. (2003) J. Clin. Invest. 112, 1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arkan M. C., Hevener A. L., Greten F. R., Maeda S., Li Z. W., Long J. M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M. (2005) Nat. Med. 11, 191–198 [DOI] [PubMed] [Google Scholar]
- 8.Solinas G., Vilcu C., Neels J. G., Bandyopadhyay G. K., Luo J. L., Naugler W., Grivennikov S., Wynshaw-Boris A., Scadeng M., Olefsky J. M., Karin M. (2007) Cell Metab. 6, 386–397 [DOI] [PubMed] [Google Scholar]
- 9.Hevener A. L., Olefsky J. M., Reichart D., Nguyen M. T., Bandyopadyhay G., Leung H. Y., Watt M. J., Benner C., Febbraio M. A., Nguyen A. K., Folian B., Subramaniam S., Gonzalez F. J., Glass C. K., Ricote M. (2007) J. Clin. Invest. 117, 1658–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odegaard J. I., Ricardo-Gonzalez R. R., Goforth M. H., Morel C. R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A. W., Chawla A. (2007) Nature 447, 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hotamisligil G. S., Erbay E. (2008) Nat. Rev. Immunol. 8, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schenk S., Saberi M., Olefsky J. M. (2008) J. Clin. Invest. 118, 2992–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue B., Kahn B. B. (2006) J. Physiol. 574, 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haigis M. C., Guarente L. P. (2006) Genes Dev. 20, 2913–2921 [DOI] [PubMed] [Google Scholar]
- 15.Sag D., Carling D., Stout R. D., Suttles J. (2008) J. Immunol. 181, 8633–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshizaki T., Milne J. C., Imamura T., Schenk S., Sonoda N., Babendure J. L., Lu J. C., Smith J. J., Jirousek M. R., Olefsky J. M. (2009) Mol. Cell. Biol. 29, 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. (2004) EMBO J. 23, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H., Kokoeva M. V., Inouye K., Tzameli I., Yin H., Flier J. S. (2006) J. Clin. Invest. 116, 3015–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue B., Pulinilkunnil T., Murano I., Bence K. K., He H., Minokoshi Y., Asakura K., Lee A., Haj F., Furukawa N., Catalano K. J., Delibegovic M., Balschi J. A., Cinti S., Neel B. G., Kahn B. B. (2009) Mol. Cell. Biol. 29, 4563–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein S. C., Woods A., Jones N. A., Davison M. D., Carling D. (2000) Biochem. J. 345, 437–443 [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H., Tzameli I., Bjørbaek C., Flier J. S. (2004) J. Biol. Chem. 279, 34733–34740 [DOI] [PubMed] [Google Scholar]
- 22.Yoshizaki T., Schenk S., Imamura T., Babendure J. L., Sonoda N., Bae E. J., Oh da Y., Lu M., Milne J. C., Westphal C., Bandyopadhyay G., Olefsky J. M. (2010) Am. J. Physiol. Endocrinol. Metab. 298, E419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto H., Schoonjans K., Auwerx J. (2007) Mol. Endocrinol. 21, 1745–1755 [DOI] [PubMed] [Google Scholar]
- 24.Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosen E. D., Spiegelman B. M. (2006) Nature 444, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen M. T., Satoh H., Favelyukis S., Babendure J. L., Imamura T., Sbodio J. I., Zalevsky J., Dahiyat B. I., Chi N. W., Olefsky J. M. (2005) J. Biol. Chem. 280, 35361–35371 [DOI] [PubMed] [Google Scholar]
- 27.Gao Z., Zhang X., Zuberi A., Hwang D., Quon M. J., Lefevre M., Ye J. (2004) Mol. Endocrinol. 18, 2024–2034 [DOI] [PubMed] [Google Scholar]
- 28.Kim J. K., Kim Y. J., Fillmore J. J., Chen Y., Moore I., Lee J., Yuan M., Li Z. W., Karin M., Perret P., Shoelson S. E., Shulman G. I. (2001) J. Clin. Invest. 108, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilon G., Dallaire P., Marette A. (2004) J. Biol. Chem. 279, 20767–20774 [DOI] [PubMed] [Google Scholar]
- 30.Zou M. H., Kirkpatrick S. S., Davis B. J., Nelson J. S., Wiles W. G., 4th, Schlattner U., Neumann D., Brownlee M., Freeman M. B., Goldman M. H. (2004) J. Biol. Chem. 279, 43940–43951 [DOI] [PubMed] [Google Scholar]
- 31.Saha A. K., Avilucea P. R., Ye J. M., Assifi M. M., Kraegen E. W., Ruderman N. B. (2004) Biochem. Biophys. Res. Commun. 314, 580–585 [DOI] [PubMed] [Google Scholar]
- 32.Fisher J. S., Gao J., Han D. H., Holloszy J. O., Nolte L. A. (2002) Am. J. Physiol. Endocrinol. Metab. 282, E18–23 [DOI] [PubMed] [Google Scholar]
- 33.Pold R., Jensen L. S., Jessen N., Buhl E. S., Schmitz O., Flyvbjerg A., Fujii N., Goodyear L. J., Gotfredsen C. F., Brand C. L., Lund S. (2005) Diabetes 54, 928–934 [DOI] [PubMed] [Google Scholar]
- 34.Foretz M., Ancellin N., Andreelli F., Saintillan Y., Grondin P., Kahn A., Thorens B., Vaulont S., Viollet B. (2005) Diabetes 54, 1331–1339 [DOI] [PubMed] [Google Scholar]
- 35.Pfluger P. T., Herranz D., Velasco-Miguel S., Serrano M., Tschöp M. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9793–9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purushotham A., Schug T. T., Xu Q., Surapureddi S., Guo X., Li X. (2009) Cell Metab. 9, 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., Prabhu V. V., Allard J. S., Lopez-Lluch G., Lewis K., Pistell P. J., Poosala S., Becker K. G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K. W., Spencer R. G., Lakatta E. G., Le Couteur D., Shaw R. J., Navas P., Puigserver P., Ingram D. K., de Cabo R., Sinclair D. A. (2006) Nature 444, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dasgupta B., Milbrandt J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7217–7222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fulco M., Cen Y., Zhao P., Hoffman E. P., McBurney M. W., Sauve A. A., Sartorelli V. (2008) Dev. Cell 14, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lan F., Cacicedo J. M., Ruderman N., Ido Y. (2008) J. Biol. Chem. 283, 27628–27635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou X., Xu S., Maitland-Toolan K. A., Sato K., Jiang B., Ido Y., Lan F., Walsh K., Wierzbicki M., Verbeuren T. J., Cohen R. A., Zang M. (2008) J. Biol. Chem. 283, 20015–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.