Abstract
Mitomycin C (MMC) is a commonly used and extensively studied chemotherapeutic agent requiring biological reduction for activity. Damage to nuclear DNA is thought to be its primary mechanism of cell death. Due to a lack of evidence for significant MMC activation in the nucleus and for in vivo studies demonstrating the formation of MMC-DNA adducts, we chose to investigate alternative nucleic acid targets. Real-time reverse transcription-PCR was used to determine changes in mitochondrial gene expression induced by MMC treatment. Although no consistent effects on mitochondrial mRNA expression were observed, complementary results from reverse transcription-PCR experiments and gel-shift and binding assays demonstrated that MMC rapidly decreased the transcript levels of 18S ribosomal RNA in a concentration-dependent manner. Under hypoxic conditions, transcript levels of 18S rRNA decreased by 1.5-fold compared with untreated controls within 30 min. Recovery to base line required several hours, indicating that de novo synthesis of 18S was necessary. Addition of MMC to an in vitro translation reaction significantly decreased protein production in the cell-free system. Functional assays performed using a luciferase reporter construct in vivo determined that protein translation was inhibited, further confirming this mechanism of toxicity. The interaction of MMC with ribosomal RNA and subsequent inhibition of protein translation is consistent with mechanisms proposed for other natural compounds.
Keywords: Cancer, RNA, RNA/Ribosomal RNA, Translation, Anticancer Drug, Bioreductive Drugs, Cancer Chemotherapy, Mitomycin C
Introduction
Nuclear DNA has long been considered the primary target for bioreductive anticancer drugs. These agents, which include the mitomycins, anthracyclines, nitroimidazoles, quinones, and nitrogen mustards, are used by oncologists primarily because of their toxicity toward radiation-insensitive hypoxic fractions in solid tumors (1–4). The historical mechanism for bioreductive drugs, based on studies with the prototype mitomycin C (MMC),3 involves intracellular activation, binding G/C-rich regions of nuclear DNA, and subsequent cell death (5–7). Many aspects of this hypothesis, however remain problematic and unsubstantiated.
The initial evidence for an MMC interaction with DNA was presented in the early 1960s and demonstrated cross-linking of purified bacterial DNA in the presence of cell lysates after exposure to MMC in vitro (5, 6, 8, 9). Subsequent studies using labeled MMC or the MMC analogue, porfiromycin, in various cell-free or in situ conditions have described the metabolic activation, chemical intermediates, and drug-DNA adducts formed from these exposures (9–11). MMC-DNA adducts have also been detected in various studies using cultured cell lines, but these DNA-drug interactions have only been demonstrated following exposure to extremely high concentrations of MMC (12, 13). Other studies using very high concentrations of MMC have also shown MMC-DNA adducts when tested in cultured cell lines transfected with MMC-activating enzymes modified to be targeted to the nucleus (21, 22). Thus, all direct evidence for a MMC-DNA adduct comes from in vitro or in situ studies or under nonbiologically relevant conditions.
MMC is a naturally occurring antibiotic originally isolated from the Gram-negative bacteria Streptomyces caespitosus. It is routinely used as a chemotherapeutic agent in the treatment of several types of cancer, such as bladder, colon, and breast cancers (14). Within the cell, several of the enzymes capable of activating MMC have been extensively characterized. NAD(P)H dehydrogenase, quinone 1 (NQO1; DT-diaphorase) (15, 16), and xanthine dehydrogenase (17) are capable of activating MMC by a two-electron reduction, while NADPH-cytochrome P450 reductase, xanthine oxidase (18), and cytochrome b5 reductase (19) are responsible for one-electron reduction. Differential toxicity can be obtained depending on whether MMC undergoes one- or two-electron reduction. Additionally, the possibility of other as yet unidentified enzymes capable of activating MMC remains.
Activity levels of several enzymes responsible for bioactivation of MMC are often increased in cancer cells compared with the normal surrounding tissue (20). Additionally, in both human tumors and cell lines, decreases in reductive enzymes lead to an increased resistance to MMC. In human breast and gastric tumors, a poor patient prognosis correlates with lower xanthine oxidase levels (21, 22). The enzyme NQO1 has been shown to be overexpressed in a number of cancerous tissues compared with normal tissues (23). Reduced activity of NQO1 due to a genetic polymorphism may increase the risk of developing bladder cancer (23, 24). Pretreatment with dicumarol, an inhibitor of NQO1, decreases toxicity of MMC under aerobic conditions in vitro (25).
Most enzymes that reduce MMC are located in the cytosol, suggesting that drug activation in vivo is also cytosolic. Therefore, to bind DNA, reactive species must translocate to the nucleus. Translocation is highly speculative, as the existence of translocator proteins is unproven and the feasibility of reactive redox compounds binding transiently to such proteins or diffusing across biological membranes is improbable. The alternative, that the prodrug enters the nucleus and then becomes reduced, also is unlikely because levels of nuclear reductive enzymes are prohibitively low (26). Finally, the only in vivo MMC-DNA adducts ever detected occurred in cell lines engineered to overexpress nuclear enzymes and exposed to considerable MMC concentrations of 10 μm or more (27, 28). Because nuclear DNA appears an unlikely target for bioreductive drugs in vivo, we speculated that other nucleic acids may be more important physiologically (29).
Mitochondria are a plausible target for bioreductive drug interactions because they contain enzymes capable of reducing MMC (30), whereas mtDNA has no true introns and constitutively lacks nucleotide excision repair, which removes MMC-DNA adducts (31, 32). Accordingly, almost all adducts would be likely to affect gene expression. The other possible nucleic acid target of MMC, RNA, comprises several species located largely in the cytosol. The most abundant RNA species in eukaryotes is rRNA, which is required to form the ribosomal complex and produce all cellular proteins (33). rRNA is also G/C-rich (34), exists in close proximity to the enzymes that activate MMC, and lacks some of the protection afforded to nuclear DNA by membranes and extensive repair mechanisms. It has been noted that there is an increase in ribosome number in tumor cells versus their normal counterparts, further increasing the probability of MMC interacting with rRNA (35). If nuclear DNA is the primary target of MMC, there is greater opportunity for MMC-DNA adducts to affect rRNA transcription than most other genes because rRNAs are encoded on several different chromosomes. Decreases in 18S rRNA of this magnitude would be expected to cause deficient ribosomal assembly and/or function, genome-wide translational inhibition, and cell death (36). The concept that bioreductive drugs bind and inhibit rRNA as their cytotoxic mechanism of action in human cells is novel but consistent with the mechanism of action for other antibacterial compounds, such as erythromycin, azithromycin, and cyclohexamide (37).
MATERIALS AND METHODS
Reagents
Mitomycin C was obtained from MP Biomedicals, Inc. (Solon, OH). Cell culture media and plastic ware were from Fisher Scientific (Hampton, NH). All other chemicals and reagents of analytical grade or better were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
Cell Culture
MCF-7 human breast cancer cells (American Type Culture Collection, Rockville, MD) were grown and maintained in Eagle's minimal essential medium supplemented with 0.1 mm nonessential amino acids, 2 mm glutamine, 10% fetal bovine serum (BioWhittakerTM, Cambrex, Walkersville, MD), 0.01 mg/ml insulin, and 1 mm sodium pyruvate in 95% air and 5% CO2. Prior to treatment with MMC or vehicle control, cells were plated in media at an appropriate density and allowed to adhere overnight. Fresh media was added to the cells prior to treatment.
RNA Isolation and Purification
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions following treatment with MMC at the concentrations and times indicated for each subsequent procedure. Residual DNA was removed by treatment with DNase I using the on-column digestion protocol used in combination with RNeasy mini-columns (Qiagen, Valencia, CA). RNA was quantified spectrophotometrically (Beckman) at 260 nm, with a 260/280 nm ratio of >1.7 used to assess purity.
Real-time RT-PCR
All primers were designed using Primer Express software (Applied Biosystems, Foster City, CA) and checked against the NCBI database using the BLASTn algorithm to ensure species and gene specificity. Oligonucleotides were custom synthesized (MWG Biotech, High Point, NC).
Primer optimization to determine the minimum primer concentrations generating fluorescence with nonspecific amplification was performed using three different concentrations of each primer (600, 300, and 150 nm) with PCR Master Mix and thermal cycling parameters identical to that performed during real-time PCR (detailed below). Products were electrophoresed on a 3% agarose gel, visualized with ethidium bromide, and documented with a Gel Doc imaging system (Bio-Rad, Hercules, CA). 300 nm was selected for all future primer concentrations, and efficiency for each transcript was determined using standard curves from calibrator cDNAs made with 10-fold serial dilutions of total RNA.
cDNA was synthesized using the Stratascript First-Strand synthesis system (Stratagene, La Jolla, CA) and a GeneAmp 9700 Thermal Cycler (Applied Biosystems) with a separate mix prepared for each set of primers. Reactions were prepared in 25 μl as follows: 1.5 μl of each forward and reverse primer (300 nm), 7.5 μl RNase-free water, 2 μl of cDNA, and 12.5 μl of 2× SYBR Green Master Mix. Real-time PCR for all genes was carried out on a 7800HT sequence detection system (Applied Biosystems) as follows: heating for 2 min at 50 °C, denaturation for 10 min at 95 °C, amplification and quantification, 40 cycles of 15 s at 95 °C and 1 min at 60 °C with a single fluorescence measurement; dissociation, 55 °C to 85 °C with continuous fluorescence measurements and cooling to 25 °C. PCR data were collected using ABI Prism 7000 SDS software (Applied Biosystems). After PCR, products were electrophoretically separated on 3% agarose gels, and bands were visualized with ethidium bromide and documented using a Gel Doc imaging system (Bio-Rad) to confirm primer specificity.
Electrophoretic Mobility Assays
Total RNA was extracted and purified from MCF-7 cells treated with 0–1 μm MMC for 1.5 h using the method outlined above. A 1.5% denaturing agarose gel containing 20 mm MOPS, 2 mm sodium acetate, 1 mm EDTA, and 6.6% formaldehyde was used to separate 2 μg of purified RNA. Gels were visualized by ethidium bromide staining and documented with a Gel Doc imaging system (Bio-Rad). Bands corresponding to 18S, 28S, and 5S ribosomal RNA were clearly visible, and the presence of a band migrating more slowly that would indicate that MMC was bound to the RNA was also evaluated. Three separate gels were prepared from each of three experiments.
Competitive Binding Assays
The ability of MMC to displace or inhibit binding of another nucleic acid interacting agent was evaluated. Ethidium bromide intercalation into DNA and RNA has been characterized previously and used to evaluate the binding ability of other agents (38, 39). RNA was isolated and purified as described previously from cells treated with 0–1 μm MMC for 1.5 h under normal oxygen conditions. RNA (2 μg) was diluted to a volume of 90 μl in 15 mm Tris-HCl buffer and transferred to one well of a UV-transparent 96-well microtiter plate. An ethidium bromide solution (ranging from 0.1 μg/ml to 5 μg/ml final concentration) was added to the wells to bring the total volume to 100 μl. Fluorescence was monitored every 30 s for 1 h using λ = 530 nm excitation and λ = 590 nm emission filters. Experiments were performed in triplicate.
MTT Cytotoxicity Assays
Cell death over time was measured by using 3-(4,5-dimethylthiaziazol-2-yl)2,5-diphenyl tetrazolium bromide (MTT). Cells were cultured under normal conditions and plated with an n = 6 for each treatment group at densities of 500, 1000, 5000, and 10,000 cells per well in a 96-well plate. After allowing the cells to adhere, the cells were exposed to 0–1 μm MMC for 0–96 h under both normoxic and hypoxic conditions. At the end of the treatment, media was removed, the cells were rinsed, and fresh medium containing 0.5 mg/ml of MTT was added to each well. After incubation for 4 h in normal cell culture conditions, the medium containing MTT was removed, and 200 μl of dimethyl sulfoxide was added to each well to dissolve the metabolized MTT formazan product. The OD was measured at 550 nm, with a greater absorbance indicating more live cells. The values obtained were plotted versus the original number of cells plated for each treatment group. Linear regression analysis was performed, and the percentage of cell death versus the untreated control was calculated by dividing the slope of the treated by the slope of the control.
In Vitro Translation
The rate of protein translation following treatment with MMC was measured in vitro using a cell-free system (Promega). A reaction mixture consisting of 35 μl rabbit reticulocyte lysate, 0.5 μl of amino acid mixture minus leucine, 0.5 μl of amino acid mixture minus methionine, and 1 μl RNasin (40 units/μl, Promega) was prepared in a microcentrifuge tube. Cycloheximide (0.1–1 μg/ml final concentration) was added as a positive control for inhibition of protein translation. Mitomycin C (0–10 μm) or vehicle control was added in a 1-μl volume to the reaction tube immediately prior to the addition of 1 μl messenger RNA-encoding firefly luciferase, which served as the translation template (luciferase control mRNA, 1 μg/μl, Promega). The reaction was allowed to proceed for 90 min at 30 degrees, as per the manufacturer's instructions. At the end of the incubation, 1-μl aliquots of the reaction mixture were added to 49 μl of GloLysis buffer and transferred to an opaque white 96-well plate, and 50 μl of BrightGlo luciferase substrate was added. Luminescence was measured in triplicate, and relative decreases in luminescence were compared with a standard curve. The model fit used in Fig. 5 is a one-phase exponential association with the equation: Y = Y0 + (Plateau − Y0) × (1 − exp(−K× x)). The fit used least-squares regression and iterated until the sum of squares changed by <0.001% before assigning the best fit line. All MMC lines have very good to excellent fits, whereas cycloheximide lines fit less well, with only moderate associations because the inhibition was so strong.
FIGURE 5.
Time-course for MMC and cycloheximide inhibition of protein translation in a cell-free system. Rabbit reticulocyte lysates were incubated with 0–10 μm MMC or 0.1–1 μg/μl cycloheximide (CX) along with mRNA-encoding firefly luciferase, and protein translation was allowed to proceed for various amounts of time under normoxic conditions. Luciferase activity was measured, with the luminescence intensity correlating with the amount of luciferase protein present.
Protein Translation in Vivo
A luciferase reporter assay was used to determine a relative decrease in protein translation following MMC exposure in MCF-7 breast cancer cells transfected with the construct. A plasmid (pGL3 control, Promega) containing a firefly luciferase construct constitutively expressed under the control of a cytomegalovirus promoter, was propagated in chemically competent Escherichia coli cells (DH5α, Invitrogen). Plasmid DNA was prepared using a commercially available kit (Promega Maxi-Prep). MCF-7 cells plated at a density of 500,000 cells into each well of a 24-well plate were transfected with 2 μg of pGL3 control plasmid using FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's instructions. After a 24-h recovery period, the cells were treated with 0–1 μm of MMC continuously for up to 6 h in normal cell culture conditions. Alternatively, cells were treated with 0–1 μm of MMC for 1 h, followed by removal of the media and incubation of 0–72 h in fresh drug-free media. At the conclusion of each time point, cell lysate was prepared by removing the media and adding 200 μl of GloLysis buffer (Promega) to each well and incubated for ∼5 min at room temperature with gentle agitation to ensure thorough lysis. Firefly luciferase activity was measured in a 100-μl aliquot of the sample using the Bright-Glo assay system (Promega) according to the manufacturer's instructions. Quantitations were calculated relative to the appropriate untreated (vehicle) controls.
Statistical Analysis and Modeling
Relative quantification of target genes in comparison to the reference gene (Cytb) were established as described previously (40), and results were tested for significance using the Relative Expression Software Tool (REST) (41). Normalization of mRNA expressions to the 18S rRNA copy number (100%) was performed using the equation: (½(Gene Ct − 18S Ct)) × 100, where Ct represents cycle threshold.
RESULTS
Abundance of Intracellular RNA Species following Exposure to MMC
Using the MCF-7 human breast cancer cell line, we confirmed by real-time RT-PCR that 18S rRNA was a highly abundant intracellular species (Fig. 1a) when compared with the levels of other RNAs. Mitochondrial mRNAs were 15.0 ± 11.3% as abundant as 18S rRNA, and the least abundant species that was investigated, nuclear-encoded glyceraldehyde-3-phosphate dehydrogenase mRNA, was only 1.7 ± 0.8% of 18S rRNA.
FIGURE 1.
Real-time RT-PCR investigation of the effects of MMC on the relative levels of mitochondrial and nuclear mRNA and 18S rRNA. a, base-line relative RNA expression of several mitochondrial mRNAs, 18S rRNA, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA in MCF-7 cells under normal culture conditions (normalized to 18S rRNA, 100%). Bars are means ± S.D. b and c, mitochondrial and nuclear gene expression changes caused by MMC effects. RNA levels in normoxic (b) and hypoxic (c) MMC-treated MCF-7 cells normalized to the base line. Values <−1 imply a decrease in RNA expression (down-regulation) and values >+1 indicate an increase in RNA expression (up-regulation). Values between −1 and +1 do not, by definition, exist.
To investigate MMC interactions with mitochondrial- and nuclear-encoded RNA, the effects of treatment of MCF-7 cells with MMC (LD50, 1 μm) were assessed with real-time RT-PCR. MMC exposures of up to 60 min were designed to observe immediate MMC-RNA interactions. Changes in gene transcription that could indicate a MMC-DNA interaction were investigated by exposing cells to MMC for 1 h followed by a 5.5-h drug-free period to allow for the degradation of mRNA that existed prior to treatment (42, 43). Marginal decreases in a few of the mitochondrial mRNAs were observed within 30–60 min of exposure under normal oxygen conditions, none of which were consistently observed or statistically significant (Fig. 1b). Under hypoxic conditions, treatment with MMC did not significantly decrease any of the mitochondrial genes being investigated (Fig. 1c). Levels of 18S rRNA, typically considered to be a “housekeeping” gene and commonly used for normalizing real-time RT-PCR experiments, decreased marginally after 60 min of continuous MMC exposure. Hypoxia in combination with MMC was able to diminish levels of 18S rRNA to ∼75% of control at both 30- and 60-min time points. Rapid loss of 18S rRNA and the several hours needed for recovery indicate that this most likely occurs through direct MMC-rRNA interaction and transcript degradation and that de novo synthesis is required to restore expression.
Mitomycin C Treatment Slows RNA Mobility through a Gel following Treatment in Vivo
Electrophoresis on a denaturing gel was performed to determine whether a decrease in ribosomal RNA or difference in mobility could be observed in samples treated with MMC compared with untreated controls (Fig. 2). In the absence of a degradation product, a band shift representing retarded migration of the MMC-RNA adducts was observed with both hypoxic and normoxic treatments. This band shift corresponded to an increase in size of the rRNA by ∼150 nucleotides. Density analysis of the bands determined that there were no significant changes in the ratio of 18S to 28S rRNA in the treated versus control samples, and there was no significant change in the overall amount of rRNA present. These data thus suggest that MMC is interacting with rRNA to slow its mobility through the gel. The diffusion pattern of the bands was different between normoxic and hypoxic treatments; however, the patterns within treatment groups did not appear to change with or without MMC. Thus, it appears that MMC interactions with rRNA result in a band shift but not band broadening. The exact nature of these MMC-RNA interactions remains to be elucidated.
FIGURE 2.
The effects of MMC on the binding and degradation of 18S rRNA in vivo. Total RNA was prepared as noted and electrophoresed on a denaturing 1.5% agarose gel. The effects on 18S and 28S rRNA were observed and noted. a, total RNA isolated from MCF-7 cells treated with MMC (0–1 μm) under normoxic conditions for 1.5 h in vivo. b, total RNA isolated from MCF-7 cells treated with MMC (0–1 μm) under hypoxic conditions for 1.5 h in vivo. The 18S and 28S rRNA bands appear to “shift” and correspond to a calculated increase in mass of ∼150 bases.
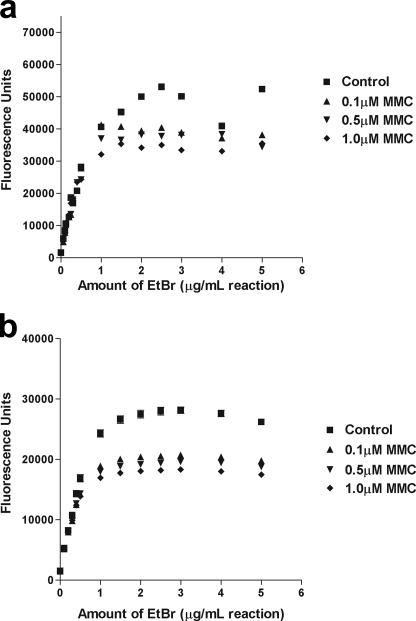
Treatment with MMC in Vivo Displaces Ethidium Bromide Binding to RNA
Another method of determining the presence of RNA adducts was implemented to further verify the effects observed with the gel shift assay. Ethidium bromide, like MMC, binds preferentially to GC-rich regions in DNA and RNA although ethidium bromide intercalates, whereas MMC binds covalently (38). When cells were treated with MMC for 1.5 h in vivo, the RNA isolated from the treated cells showed decreased binding and fluorescence of ethidium bromide in a concentration-dependent manner (Fig. 3). Increasing amounts of ethidium bromide were added to equal amounts of control and treated RNA. The fluorescence of the treated RNA became saturated at a lower concentration of ethidium bromide under both normoxic and hypoxic conditions. Additionally, kinetic measurements of ethidium bromide incorporation showed a more rapid incorporation and increase in fluorescence in untreated samples compared with treated (data not shown). There was little difference between samples that were exposed to MMC under normoxic conditions compared with those treated under hypoxic conditions. These results demonstrate a competitive inhibition of ethidium bromide, implying that MMC binds to RNA with greater affinity than ethidium bromide.
FIGURE 3.
Inhibition of ethidium bromide binding to RNA treated with MMC. Increasing concentrations of ethidium bromide (0–5 μg/ml) were added to a solution containing total RNA extracted from MCF-7 cells treated in vivo with MMC (0–1 μm) for 1.5 h under either normoxic (a) or hypoxic (b) conditions. In untreated control cells, maximum ethidium bromide binding and fluorescence was reached with a concentration of 2 μg/ml ethidium bromide per 1 μg RNA. In contrast, RNA from cells treated with MMC had a maximum ethidium bromide binding and fluorescence at 1.25 μg/ml ethidium bromide per 1 μg RNA.
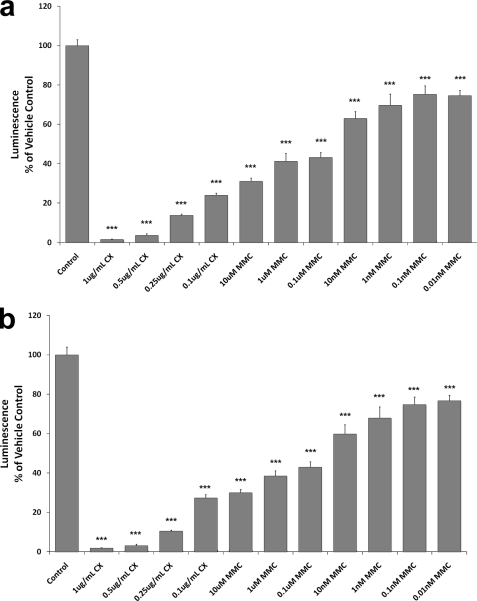
Mitomycin C Inhibits Protein Translation in a Dose-dependent Manner in a Cell-free System
Protein translation of firefly luciferase was significantly decreased dose-dependently when the lysates were incubated in the presence of MMC under both normoxic and hypoxic conditions (Fig. 4). Luciferase protein, quantitated using a standard luminescence assay, was only 33% of that observed in untreated vehicle control samples at 10 μm MMC under both aerobic and hypoxic conditions. Concentrations as low as 0.01 nm MMC were sufficient to cause protein inhibition under either condition. While this similar degree of inhibition under both aerobic and hypoxic conditions initially appears counterintuitive based upon preferential cytotoxicity of MMC under hypoxia, one must remember that this preferential cytotoxicity is largely due to an induction of MMC activating enzymes under hypoxia, in vivo. The protein translation system used in these studies is a cell-free or static system unresponsive to changing levels of oxygenation. Cycloheximide, a known inhibitor of translation, was used as a positive control in these studies. Fig. 4 shows that cycloheximide inhibits protein translation in a dose-dependent manner similarly to MMC. These experiments are useful in determining the actual levels of protein synthesized over time, without the interfering effect of existing protein, as is the case in cells transfected with a luciferase reporter.
FIGURE 4.
MMC decreases protein translation in a cell-free system. Rabbit reticulocyte lysates were incubated with 0–10 μm MMC along with mRNA encoding firefly luciferase and protein translation allowed to proceed for 90 min. Additionally, positive control reactions were run using 0.1–1 μg/μl cycloheximide (CX). Reactions were carried out under normoxic (a) or hypoxic (b) conditions. Luciferase activity was measured, with the luminescence intensity correlating with amount of luciferase protein present. Values are expressed as percent luminescence compared with an untreated vehicle control. Bars are means ± S.E. ***, p < 0.01 versus untreated control.
Mitomycin C Inhibition of Protein Translation in a Cell-free System Is Time-dependent
Protein translation of firefly luciferase was measured at various time points at several MMC and cycloheximide concentrations (Fig. 5). The data suggest that the reaction is completed between 30 and 60 min under control conditions. The data show that although cycloheximide reacts quickly to inhibit in vitro translation, MMC inhibition is much slower. This slower onset of inhibition may be due to several factors, including the rate of MMC activation in the cell-free system. It is important to note, however, that even at very low MMC concentrations, the reaction appears to be completely or nearly completely inhibited by 60 min. Thus, it appears that MMC may, in fact, be a powerful inhibitor of translation in this system, but the somewhat slow onset of inhibition by MMC allows for an initial burst of translation, which masks its inhibitory effects in an in vitro assay under standard assay conditions.
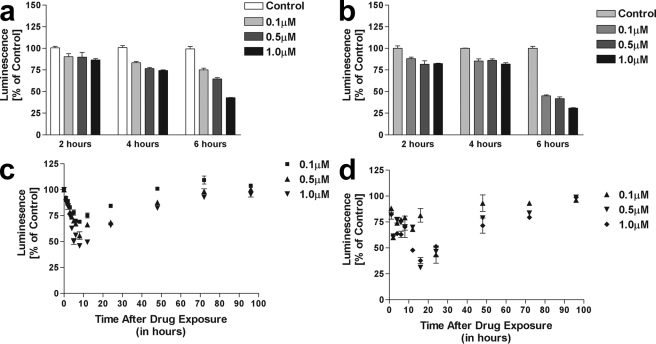
Cells Transfected with a Luciferase Reporter Construct Show Decreased Protein Levels following MMC Treatment in a Time- and Concentration-dependent Manner
The use of a luciferase reporter assay was used to determine whether protein translation was affected by MMC and oxygen concentration. Decreased protein levels of a luciferase reporter were observed following treatment with MMC in MCF-7 cells transiently transfected with a luciferase reporter construct. Concentrations as low as 0.1 μm were sufficient to cause an inhibition in protein translation, although further decreases were dose-dependent. The decrease in luminescence, correlating with amount of luciferase protein was also time-dependent, because the half-life of luciferase is 3.5 h, and any luciferase protein translated prior to MMC treatment would still be present. Under normoxic conditions (Fig. 5a), the decrease in luciferase was observed at ∼4 h, and persisted for up to 24 h (data not shown). Cells exposed to MMC under hypoxia (Fig. 6b) showed a delayed decrease in luciferase protein starting at ∼6 h, but levels were significantly lower than those observed at the same time point under normoxia. Luminescence was calculated as percent of the untreated control and adjusting for the cell death observed over the time course, as determined by the MTT assay.
FIGURE 6.
Treatment with MMC decreases protein translation in cells transfected with a luciferase reporter in vivo. MCF-7 cells were transiently transfected with pGL-3 control vector prior to treatment with MMC. a, cells were exposed to 0–1 μm MMC constantly for the times indicated under normal oxygen conditions. b, cells were exposed to 0–1 μm MMC constantly for the times indicated under hypoxic conditions. c, cells were exposed to 0–1 μm MMC for 2 h under normal oxygen conditions, followed by removal of the drug and addition of drug-free medium for the times indicated. d, cells were exposed to 0–1 μm MMC for 2 h under hypoxia, followed by removal of the drug and addition of drug-free medium and incubation under normal oxygen conditions for the times indicated. Luciferase activity was measured at the end of treatment and compared with untreated controls at the same time point.
To determine the duration of the MMC 18S binding effect, a series of experiments were performed comparing cells exposed to MMC throughout the duration of the experiment to those exposed for only 1.5 h and allowed to continue the rest of the incubation time in drug-free media. Under normoxia, a similar decrease in luciferase activity was observed 2 h after drug removal, persisting for at least 24 h (Fig. 6c). Luciferase protein levels gradually returned to base line by 72 h. Cells treated with MMC under hypoxia were allowed to recover in drug-free media and under normal oxygen conditions for the times indicated. At higher concentrations of MMC, the decrease in luciferase activity was more pronounced, and recovery of protein translation was observed at ∼24 h post-treatment (Fig. 6d). However, it is noteworthy to mention that the luciferase readings are adjusted for the number of living cells. Therefore, luciferase activity in the living cells is not a good indicator of whether the drug is working more efficiently at causing cell death under normoxic or hypoxic conditions.
DISCUSSION
Although most studies with real-time PCR report changes in gene expression upwards of 10-fold, we observed significant 2-fold decreases in 18S rRNA levels. Although these differences approach the level of sensitivity of real-time RT-PCR, because 18S rRNA exists in the cell in far greater amounts than other RNA species, 2-fold (75%) decreases in expression correspond to a tremendous reduction in absolute 18S rRNA quantities. The results are further strengthened by the use of REST software for data analysis, which corrects for primer efficiency and is more accurate than models that assume 100% efficiency and uses a repeated randomization test employed that is superior to parametric methods and nonparametric methods based on ranks in generating lower type I errors.
It is indisputable that MMC binds DNA in vitro. However, the present study suggests that in vivo, DNA may not be the primary target of the drug. The chemical similarities to DNA and the relative abundance of RNA in the cytosol, the primary cellular site for MMC activation, make RNA a likely target. That rRNA constitutes ∼71% of total cellular RNA in eukaryotes and that it contains G/C-rich regions for preferred MMC binding (34) suggest that rRNA is likely the primary cellular RNA target. We propose that MMC rapidly binds 18S rRNA in the cytosol causing degradation and profound decreases in cytosolic levels. Decreases of this magnitude likely prevent the formation and/or function of ribosomal complexes causing cell death through genome-wide translational silencing.
The ribosome content of cells can be regulated by control of rRNA transcription, by degradation of rRNA during nuclear processing, or by turnover of cytoplasmic ribosomes (44). Normally, there are conditions in which ribosomal concentrations change in response to different stimuli. Contact inhibition of cells growing in culture has been shown to alter rRNA levels (45). In eukaryotic cells undergoing growth arrest, decreases in rRNA synthesis and altered accumulation of mature 28S and 18S rRNA have been described (46, 47). Conversely, an increase in rRNA synthesis has been observed when cells transition from a resting to a proliferating state. It has been theorized that the regulation of ribosome formation may play a role in the pathogenesis of various diseases, including cancer (35, 48). In the case of exposure to MMC, it is unlikely that control of rRNA synthesis at the transcriptional level is responsible for the decrease in 18S, as it would require several hours, instead of minutes, for such a dramatic effect to be observed.
Although novel in human cells, degradation of rRNA by MMC was previously reported in bacteria as early as 1967 (49, 50) and is common for drugs obtained from natural sources such as the antibiotics erythromycin, azithromycin, and cyclohexamide that have similar structures as MMC (37). Based on the evidence presented and empirical reasoning, MMC-rRNA interactions are more functionally plausible with respect to drug distribution and metabolism than the current hypothesis asserting MMC-DNA interactions. As attention increasingly turns to RNA as a therapeutic target for small molecules, RNA interference, and genomics (33, 37), this paradigm shift for the mechanism of action of bioreductive drugs provides great potential for the development of more specific and effective cytotoxic agents.
This work was supported in part by the Nevada Agricultural Experiment Station.
- MMC
- mitomycin C
- MTT
- 3-(4,5-dimethylthiaziazol-2-yl)2,5-diphenyl tetrazolium bromide
- RT
- reverse-transcriptase
- MOPS
- 4-morpholinepropanesulfonic acid.
REFERENCES
- 1.Rauth A. M., Mohindra J. K., Tannock I. F. (1983) Cancer Res. 43, 4154–4158 [PubMed] [Google Scholar]
- 2.Keohane A., Godden J., Stratford I. J., Adams G. E. (1990) Br. J. Cancer 61, 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKeown S., Cowen R., Williams K. (2007) Clin. Oncol. (R Coll. Radiol.) 19, 427–442 [DOI] [PubMed] [Google Scholar]
- 4.Teicher B. A., Lazo J. S., Sartorelli A. C. (1981) Cancer Res. 41, 73–81 [PubMed] [Google Scholar]
- 5.Iyer V. N., Szybalski W. (1963) Proc. Natl. Acad. Sci. U.S.A. 50, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer V. N., Szybalski W. (1964) Science 145, 55–58 [DOI] [PubMed] [Google Scholar]
- 7.Teng S. P., Woodson S. A., Crothers D. M. (1989) Biochemistry 28, 3901–3907 [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto I., Lark K. G. (1963) Exp. Cell Res. 32, 192–196 [DOI] [PubMed] [Google Scholar]
- 9.Szybalski W., Iyer V. (1964) Microbiol. Genet. Bull. 21, 16–17 [Google Scholar]
- 10.Weissbach A., Lisio A. (1965) Biochemistry 4, 196–200 [Google Scholar]
- 11.Tomasz M., Lipman R., Chowdary D., Pawlak J., Verdine G. L., Nakanishi K. (1987) Science 235, 1204–1208 [DOI] [PubMed] [Google Scholar]
- 12.Tomasz M., Hughes C. S., Chowdary D., Keyes S. R., Lipman R., Sartorelli A. C., Rockwell S. (1991) Cancer Commun. 3, 213–223 [DOI] [PubMed] [Google Scholar]
- 13.Bizanek R., Chowdary D., Arai H., Kasai M., Hughes C. S., Sartorelli A. C., Rockwell S., Tomasz M. (1993) Cancer Res. 53, 5127–5134 [PubMed] [Google Scholar]
- 14.Verweij J., Pinedo H. M. (1990) Anticancer Drugs 1, 5–13 [PubMed] [Google Scholar]
- 15.Siegel D., Gibson N. W., Preusch P. C., Ross D. (1990) Cancer Res. 50, 7483–7489 [PubMed] [Google Scholar]
- 16.Siegel D., Beall H., Senekowitsch C., Kasai M., Arai H., Gibson N. W., Ross D. (1992) Biochemistry 31, 7879–7885 [DOI] [PubMed] [Google Scholar]
- 17.Gustafson D. L., Pritsos C. A. (1992) J. Natl. Cancer Inst. 84, 1180–1185 [DOI] [PubMed] [Google Scholar]
- 18.Pan S. S., Andrews P. A., Glover C. J., Bachur N. R. (1984) J. Biol. Chem. 259, 959–966 [PubMed] [Google Scholar]
- 19.Hodnick W. F., Sartorelli A. C. (1993) Cancer Res. 53, 4907–4912 [PubMed] [Google Scholar]
- 20.Fitzsimmons S. A., Workman P., Grever M., Paull K., Camalier R., Lewis A. D. (1996) J. Natl. Cancer Inst. 88, 259–269 [DOI] [PubMed] [Google Scholar]
- 21.Linder N., Lundin J., Isola J., Lundin M., Raivio K. O., Joensuu H. (2005) Clin Cancer Res. 11, 4372–4381 [DOI] [PubMed] [Google Scholar]
- 22.Linder N., Haglund C., Lundin M., Nordling S., Ristimäki A., Kokkola A., Mrena J., Wiksten J. P., Lundin J. (2006) J. Clin. Pathol. 59, 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danson S., Ward T. H., Butler J., Ranson M. (2004) Cancer Treat. Rev. 30, 437–449 [DOI] [PubMed] [Google Scholar]
- 24.Park S. J., Zhao H., Spitz M. R., Grossman H. B., Wu X. (2003) Mutat. Res. 536, 131–137 [DOI] [PubMed] [Google Scholar]
- 25.Marshall R. S., Paterson M. C., Rauth A. M. (1991) Biochem. Pharmacol. 41, 1351–1360 [DOI] [PubMed] [Google Scholar]
- 26.Briggs L. A., Pritsos C. A. (1999) Biochem. Pharmacol. 58, 1609–1614 [DOI] [PubMed] [Google Scholar]
- 27.Seow H. A., Penketh P. G., Belcourt M. F., Tomasz M., Rockwell S., Sartorelli A. C. (2004) J. Biol. Chem. 279, 31606–31612 [DOI] [PubMed] [Google Scholar]
- 28.Seow H. A., Belcourt M. F., Penketh P. G., Hodnick W. F., Tomasz M., Rockwell S., Sartorelli A. C. (2005) Mol. Pharmacol. 67, 417–423 [DOI] [PubMed] [Google Scholar]
- 29.Pritsos C. A., Briggs L. A., Gustafson D. L. (1997) Oncol Res. 9, 333–337 [PubMed] [Google Scholar]
- 30.Spanswick V. J., Cummings J., Smyth J. F. (1996) Biochem. Pharmacol. 51, 1623–1630 [DOI] [PubMed] [Google Scholar]
- 31.Bianchi N. O., Bianchi M. S., Richard S. M. (2001) Mutat. Res. 488, 9–23 [DOI] [PubMed] [Google Scholar]
- 32.Cullinane C., Bohr V. A. (1998) Cancer Res. 58, 1400–1404 [PubMed] [Google Scholar]
- 33.Doudna J. A., Rath V. L. (2002) Cell 109, 153–156 [DOI] [PubMed] [Google Scholar]
- 34.Schmid M., Nanda I., Steinlein C., Epplen J. T. (1994) Hum. Genet. 93, 375–382 [DOI] [PubMed] [Google Scholar]
- 35.Montanaro L., Treré D., Derenzini M. (2008) Am. J. Pathol. 173, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruggero D., Pandolfi P. P. (2003) Nat. Rev. Cancer 3, 179–192 [DOI] [PubMed] [Google Scholar]
- 37.Ecker D. J., Griffey R. H. (1999) Drug Discov. Today 4, 420–429 [DOI] [PubMed] [Google Scholar]
- 38.Feigon J., Leupin W., Denny W. A., Kearns D. R. (1982) Nucleic Acids Res. 10, 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatti C., Houssier C., Fredericq E. (1975) Biochim. Biophys. Acta 407, 308–319 [PubMed] [Google Scholar]
- 40.Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfaffl M. W., Horgan G. W., Dempfle L. (2002) Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cantatore P., Flagella Z., Fracasso F., Lezza A. M., Gadaleta M. N., de Montalvo A. (1987) FEBS Lett. 213, 144–148 [DOI] [PubMed] [Google Scholar]
- 43.Perry R. P., Greenberg J. R., Kelley D. E., LaTorre J., Schochetman G. (1973) Basic Life Sci. 1, 149–168 [DOI] [PubMed] [Google Scholar]
- 44.Perry R. P. (1973) Biochem. Soc. Symp. 37, 105–116 [PubMed] [Google Scholar]
- 45.Emerson C. P. (1971) Nat. New Biol. 232, 101–106 [DOI] [PubMed] [Google Scholar]
- 46.Cooper H. L. (1973) J. Cell Biol. 59, 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mauck J. C., Green H. (1973) Proc. Natl. Acad. Sci. U.S.A. 70, 2819–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holland E. C., Sonenberg N., Pandolfi P. P., Thomas G. (2004) Oncogene 23, 3138–3144 [DOI] [PubMed] [Google Scholar]
- 49.Kato N., Okabayashi K., Mizuno D. (1970) J. Biochem. 67, 175–184 [DOI] [PubMed] [Google Scholar]
- 50.Suzuki H., Kilgore W. W. (1967) J. Bacteriol. 94, 666–676 [DOI] [PMC free article] [PubMed] [Google Scholar]