Abstract
SNM1A is a member of the SNM1 family of nucleases required for cellular processing of interstrand DNA crosslinks (ICLs). Little is known about the molecular function of SNM1A, in terms of its recruitment to ICL lesions or its DNA damage processing activity. Here we show that SNM1A contains a functional PIP box (PCNA-interacting protein box) and a UBZ (ubiquitin binding zinc finger), required for assembly of SNM1A into nuclear focus. Moreover, RAD18-dependent monoubiquitination of PCNA is required for Mitomycin C and Ultraviolet Light inducible SNM1A nuclear focus assembly. Taken together, our results identify a novel RAD18-PCNA(Ub)-SNM1A pathway required for nuclear focus formation and ICL resistance.
Keywords: DNA Damage, DNA Repair, Protein-Protein Interactions, Ubiquitination, Zinc Finger
Introduction
The repair of DNA interstrand crosslinks (ICLs)4 in mammalian cells requires the coordinate activity of three discrete pathways, including nucleotide excision repair (NER), homologous recombination (HR), and translesion DNA synthesis (TLS) (1). In addition, yeast SNM1 (also known as Pso2) is required for ICL repair and appears to function independently of these three pathways. Pso2 is a member of the β-CASP metallo-β-lactamase family of nucleases (2). Human cells have three homologs of SNM1 (referred to as SNM1A, SNM1B/Apollo, and SNM1C/Artemis), and their relative contribution to ICL repair remains unknown.
Recent studies have elucidated some of the functions of the SNM1A protein. First, human SNM1A, unlike SNM1B and SNM1C, can rescue the crosslinker hypersensitivity of a yeast Pso2 mutant, suggesting that SNM1A is the homologue of the yeast protein (3). Second, a murine knock-out of SNM1A is hypersensitive to MMC, but not to ionizing radiation (IR) (4–6). Third, in response to DNA damage, SNM1A assembles in subnuclear foci (7), further supporting its role in the DNA damage response. Fourth, SNM1A can function as a DNA exonuclease in vitro, suggesting that it may have a direct role in excising DNA ICLs (3, 8).
DNA repair proteins are often recruited to specific sites of DNA damage through protein-protein interaction domains. For example, the FHA domain of NBS1 and the BRCT domains of BRCA1 are required for recruitment of these DNA repair proteins to complexes containing phosphopeptide motifs (9). Ubiquitin binding motifs (UBMs) and ubiquitin-binding zinc fingers (UBZs) are found in some DNA repair proteins and can target these proteins to monoubiquitinated complexes (10–12). The UBZ motif of Polη for example, can recruit this protein to monoubiquitinated PCNA. However, the molecular mechanism by which SNM1A is recruited to sites of ICL repair is currently unknown (7).
In this study, we propose a mechanism for the damage-inducible recruitment of SNM1A to sites of DNA damage. We identified a conserved UBZ domain in SNM1A, which is functional for monoubiquitin binding. DNA damage, by MMC or ultraviolet light, activated the UBZ-dependent recruitment of SNM1A into nuclear foci. Interestingly, this recruitment was RAD18 dependent, suggesting that RAD18 monoubiquitinates PCNA, leading to the direct interaction of UBZ-SNM1A with PCNA-Ub at ICL-stalled replication forks.
EXPERIMENTAL PROCEDURES
Cell Culture and Plasmids
HeLa and HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Sigma) and 1% Pen-Strep glutamine (Invitrogen). Wild-type and RAD18-deficient HCT116 cells were grown as previously described (13). Human SNM1A cDNA has been described previously (8) and was subcloned into pEGFP-C1 vector (BglII and SalI). Mutants of the PIP box and UBZ domain were further generated based on the wild-type vector. Plasmid transfection was performed using Lipofectamine 2000 (Invitrogen), according to the product manual.
Antibodies
Anti-GFP antibody (JL-8) was from Clontech. Anti-FancD2 antibody (FI-17) and anti-PCNA antibody (PC-10) were both from Santa Cruz Biotechnology. Anti-actin monoclonal antibody (AC-40) was from Sigma. Rabbit anti-UAF1 antibody was described previously (14).
Induction of DNA Damage and GFP Fluorescence
For UV damage, cells on imaging chambers were washed once with phosphate-buffered saline and UV-irradiated at various doses using UV Stratalinker 2400 (Stratagene). Afterward, fresh medium was added, and cells were incubated for indicated time before being processed for microscopy.
For MMC (Sigma) treatment, a stock solution of 1 mg/ml was prepared and added onto cells to final concentrations as indicated. Cells were further incubated for indicated time before being processed for microscopy.
For GFP fluorescence, cells were fixed with 4% formaldehyde (Sigma), and then treated with 0.15% Triton X-100 (Sigma), followed by staining with DAPI. Photography was performed using a Zeiss AX10 fluorescence microscope. For focus quantification, GFP-positive cells were first identified, and among these cells, those containing more than 10 subnuclear foci were scored as positive for focus formation.
GST Pull-down Assay
GST fusion proteins were expressed in Escherichia coli BL21(DE3) Strain, and purified using glutathione-Sepharose beads. GFP-tagged SNM1Awt or mutants were transfected into HEK293T cells, and cell lysates were prepared using lysis buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40, and protease mixture inhibitor (Roche). The total protein concentration of the lysates was normalized using Bradford Assay. Beads with equal amount of GST fusion protein were incubated with indicated cell lysates for 3 h at 4 °C. Beads were then washed with lysis buffer for three times and boiled in 1× SDS loading buffer.
Western Blot
Cells were harvested, and total protein extract was prepared using radioimmune precipitation assay buffer (50 mm Tris-HCl pH 7.3, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.05% SDS). Protein extracts were normalized using Bradford Assay, loaded onto polyacrylamide gels, and transferred onto nitrocellulose membranes. Immunoblotting was performed using the indicated antibodies.
RESULTS
The UBZ Domain of SNM1A Can Interact with Monoubiquitin
The ubiquitin binding zinc finger, UBZ, has been identified in the Y-family of translesion DNA polymerases (10). For instance, the UBZ domain of polymerase η binds to the monoubiquitin moiety of ubiquitinated PCNA, thereby allowing Polη to participate in translesion DNA synthesis (TLS).
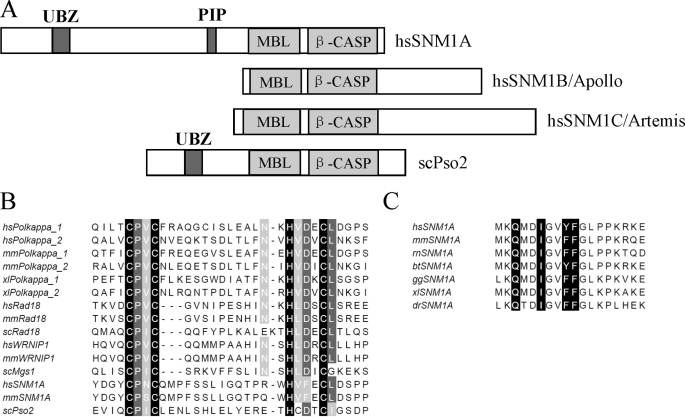
Previous studies have indicated that the proteins Polκ (10, 15), RAD18 (10, 13, 16), and WRNIP1 (17, 18) contain a similar UBZ domain (C2HC). We performed a search with the Hidden Markov Model (HMM) (19, 20), using this conserved UBZ domain as bait, and found that SNM1A shares critical amino acid residues in a UBZ domain in its N-terminal region (Fig. 1, A and B). This putative UBZ domain was not observed in SNM1B or SNM1C. Because RAD18 accumulates in a UBZ-dependent manner at sites of DNA damage (13), we reasoned that the UBZ of SNM1A might be important for recruitment in subnuclear foci.
FIGURE 1.
SNM1A has conserved ubiquitin binding zinc finger and PIP box. A, schematic representation of the domain structures of SNM1 family from human and budding yeast. The UBZ domain and PIP box are labeled in dark gray. B, sequence alignment of the UBZ domain from Polκ, Rad18, WRNIP1, and SNM1A from multiple species. The critical cysteines and histidines of the zinc fingers are labeled in black, and other highly conserved amino acids are labeled in gray. C, sequence alignment of the PIP box in SNM1A from different species. The consensus amino acids QXXIXXY(F)F are labeled in black. hs, Homo sapiens; mm, Mus musculus; rn: Rattus norvegicus; bt, Bos taurus; gg, Gallus gallus; dr, Danio rerio; xl, Xenopus laevis; dm, Drosophila melanogaster; sc, Saccharomyces cerevisiae.
Some UBZ-containing proteins, such as Polη (21) or Polκ (22), also contain a PIP box (PCNA interaction motif). Sequence analysis of SNM1A revealed a conserved PIP box at amino acid sequence 556–563 (Fig. 1C). A PIP box was not observed in SNM1B or SNM1C, though scPso2 does contain a degenerative PIP box at a different location, compared with human SNM1A. The presence of a PIP box and a UBZ domain in the human SNM1A primary amino acid sequence suggested a model in which SNM1A is recruited to monoubiquitinated PCNA (PCNA-Ub). We reasoned that SNM1A might function downstream of PCNA-Ub in a DNA repair pathway.
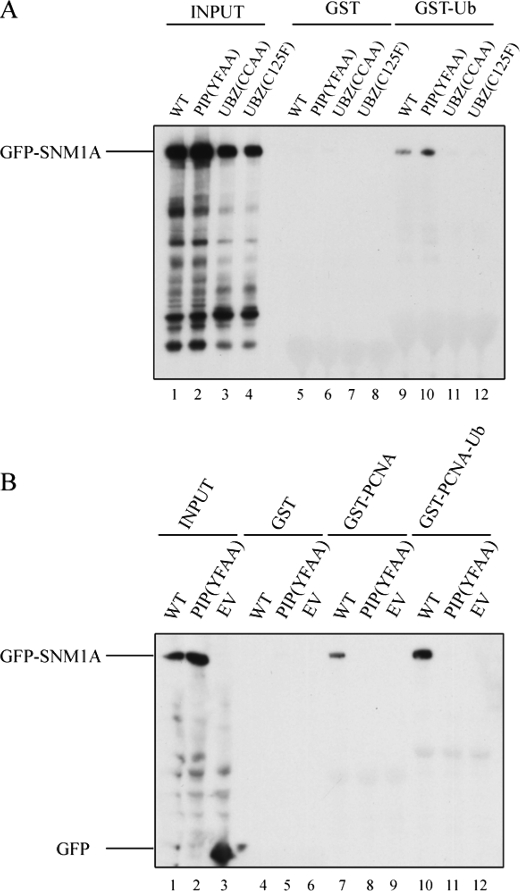
Initially, we determined whether the putative UBZ domain of SNM1A can bind ubiquitin. For this purpose, we generated the cDNA-encoding GFP-tagged full-length wild-type SNM1A protein or mutant SNM1A protein. Specifically, we generated two mutant forms of SNM1A, each containing amino acid changes in the UBZ domain (C125F and CC(122/125)AA mutants). Fig. 2A demonstrates that a GST-Ub fusion protein can selectively pull down GFP- SNM1Awt but cannot pull down SNM1A (CCAA) or SNM1A (C125F) mutant proteins. These results indicate that the UBZ domain of SNM1A is required for monoubiquitin binding.
FIGURE 2.
SNM1A physically interacts with Ub and PCNA through its UBZ domain and PIP box. A, GST and GST-Ub were used to pull down human wild-type and mutant GFP-SNM1A overexpressed in HEK 293T cells. PIP(YFAA) is the PIP Box mutant with mutation of YF (562/563) to AA. UBZ(CCAA) is the UBZ mutant with the mutation CC (122/125) to AA. UBZ(C125F) is another UBZ mutant with the mutation C125F. B, GST, GST-PCNA, and GST-PCNA-Ub agarose were used to pull down human wild-type and mutant GFP-SNM1A overexpressed in HEK 293T cells. EV is the empty vector.
We next examined the PCNA binding activity of the PIP box of SNM1A (Fig. 2B). A GST-PCNA fusion protein precipitated GFP-SNM1Awt (lane 7). Consistently, a GST-PCNA-Ub fusion protein precipitated GST-SNM1Awt even more strongly (lane 10), suggesting that the combination of a PIP box and a UBZ domain results in enhanced binding of SNM1A to monoubiquitinated PCNA. A mutant version of SNM1A (GFP-SNM1A-YFAA), with a disrupted PIP box sequence, failed to bind to either GST-PCNA or GST-PCNA-Ub (lanes 8 and 11, respectively). Taken together, these results confirm that PCNA-Ub and SNM1A have multiple sites of interaction.
DNA Damage Activates the UBZ-dependent Assembly of SNM1A into Nuclear Foci
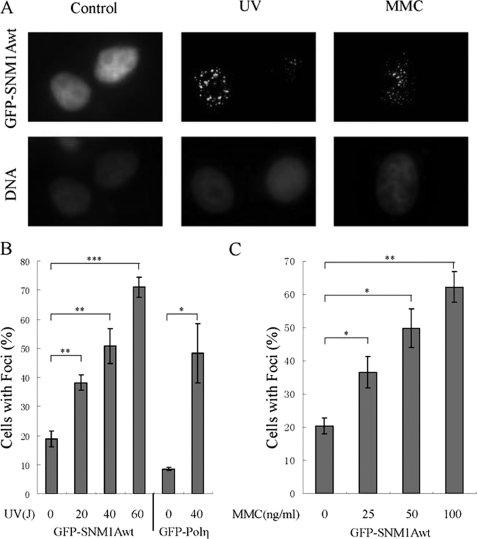
The presence of a functional UBZ domain and PIP box in the SNM1A protein suggested that this domain may be required for DNA damage-inducible foci assembly. We therefore transfected HeLa cells with the cDNA encoding GFP-SNM1Awt, and examined spontaneous and damage-inducible focus formation (Fig. 3). MMC and UV activated the assembly of GFP-SNM1Awt nuclear foci (Fig. 3A), and this focus formation demonstrated dose dependence (Fig. 3, B and C). Specifically, there was a dose-dependent increase in SNM1Awt foci after cellular exposure to UV light in the 0–60 J/m2 range and a dose-dependent increase following MMC exposure in the 0–100 ng/ml range.
FIGURE 3.
SNM1A forms damage-inducible nuclear foci after UV and MMC treatment. A, representative images of GFP-SNM1A foci without cellular damage or after cellular treatment with UV or MMC. GFP-SNM1A was transfected into HeLa cells, and the cells were treated with UV (40J/m2, 6h) or MMC (25 ng/ml, 12 h). B, quantification of SNM1A foci after different doses of UV treatment (0, 20, 40, 60J/m2, 6 h). Measurement indicates the percentage of cells with more than 10 subnuclear foci among all the cells expressing GFP-SNM1A. GFP-Polη was used as a positive control for focus formation. The data shown were an average of results from four independently repeated experiments. Student's t test was performed on the original data sets, * indicates that p < 0.05, ** indicates that p < 0.01, and *** indicates that p < 0.001. C, quantification of SNM1A foci after different doses of MMC treatment (0, 25, 50, 100 ng/ml, 12 h). The data shown was an average of results from three independently repeated experiments. Student's t test was performed on the original data sets; * indicates that p < 0.05; ** indicates that p < 0.01.
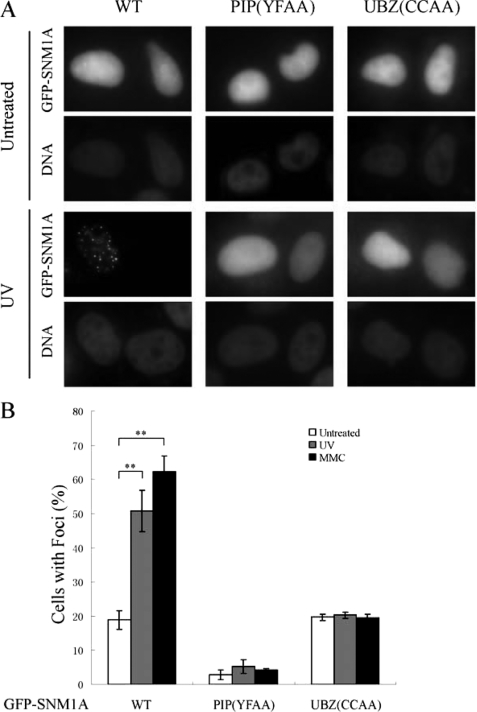
We next examined the requirement of the UBZ domain and the PIP box in DNA damage-inducible nuclear focus formation (Fig. 4). Approximately 20% of transfected cells expressing GFP-SNM1Awt exhibited nuclear foci, even in the absence of exogenous DNA damage. UV activated up to 50% of cells with nuclear foci, and MMC activated up to ∼60% of cells with nuclear foci. Disruption of the PIP box (YFAA mutant) resulted in nearly complete loss of nuclear foci. These results indicate that the PIP box, and likely intracellular PCNA binding, is required for both spontaneous and damage inducible foci assembly. Interestingly, disruption of the UBZ domain did not affect spontaneous SNM1A focus formation but instead blocked the DNA damage-inducible SNM1A focus formation.
FIGURE 4.
The PIP box and UBZ domain are required for SNM1A focus formation. A, representative images of focus formation of wild-type and mutant SNM1A in HeLa cells without damage and after treated with UV. cDNA encoding wild-type, PIP box mutant (YF(562/563)AA), UBZ mutant (CC(122/125)AA) were transfected into HeLa cells, and the cells were treated with UV (40J/m2, 6 h). B, quantification of the wild-type and mutant SNM1A foci after UV (40J/m2, 6 h) and MMC (100 ng/ml, 12 h) was measured by counting the percentage of cells with more than 10 subnuclear foci among all the cells that were GFP-positive. The data shown was an average of results from four independently repeated experiments. Student's t test was performed on the original data sets; ** indicates that p < 0.01. For PIP(YFAA) and UBZ(CCAA), the p values between the data sets of before and after DNA damage were much higher than 0.05, indicating that there were no significant increases in the percentage of cells with foci after UV or MMC treatment of these two mutants.
RAD18 Is Required for DNA Damage-inducible SNM1A Recruitment to Nuclear Foci
Like SNM1A, the E3 ubiquitin ligase, RAD18, is required for cellular resistance to MMC (6). RAD18 is also required for UV resistance. In response to certain kinds of DNA damage, RAD18 is known to monoubiquitinate the DNA replication processing factor, PCNA. We reasoned that monoubiquitinated PCNA may be the site of SNM1A recruitment, for a number of reasons. First, RAD18-deficient murine cells are hypersensitive to crosslinking agents, such as MMC, though the molecular basis for this hypersensitivity has remained unknown (23, 24). Second, a DT40 knock-in cell line containing PCNA (K164R) (i.e. a point mutation at the site of RAD18-dependent monoubiquitination) is known to be hypersensitive to DNA crosslinking agents (23, 25).
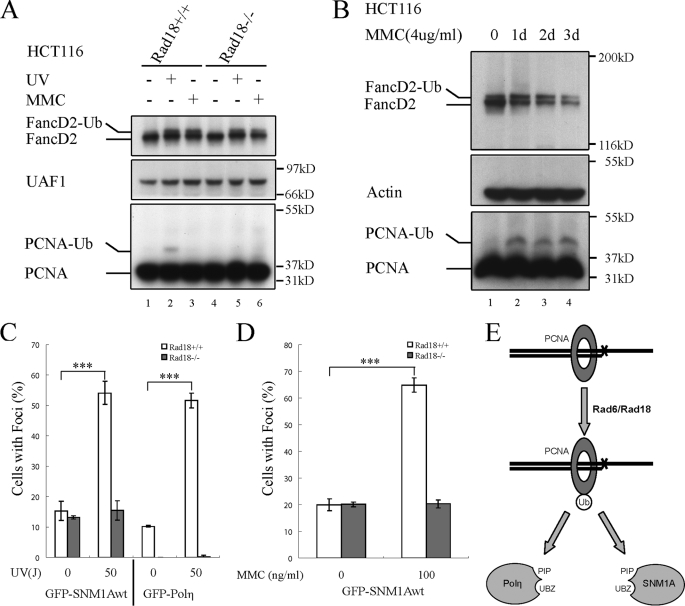
We therefore tested the hypothesis that RAD18 mediates SNM1A recruitment through PCNA monoubiquitination (Fig. 5). For this purpose, we compared two human colon cancer cell lines, one which is wild-type for RAD18 and one which has a biallelic disruption of RAD18. Stimulation of the RAD18(+/+) cells with UV light resulted in the formation of a low level of PCNA-Ub (Fig. 5A, lane 2), consistent with previous studies (26). The RAD18(−/−) mutant cells failed to activate PCNA-Ub after UV exposure (lane 5). MMC failed to activate the monoubiquitination of PCNA in either the RAD18(+/+) cells (lane 3) or the RAD18(−/−) cells (lane 6) under the conditions used in this experiment (MMC, 100 ng/ml, 12 h).
FIGURE 5.
SNM1A foci assembly is dependent on RAD18. A, characterization of the HCT116 RAD18(+/+) and RAD18(−/−) cell lines. HCT116 RAD18(+/+) and RAD18(−/−) cells were either untreated or treated with UV (50J/m2, for 6 h) or with MMC (100 ng/ml, 12 h). Cells were harvested and Western blot was performed to determine the ubiquitination level of PCNA. FANCD2 was used as the positive control for damage caused by UV or MMC, and UAF1 (14) was used as the loading control. B, high dose of MMC treatment could induce PCNA monoubiquitination. HCT116 wild-type cells (parental) were exposed continuously to the indicated MMC concentration for 0–3 days. Whole cell lysates were prepared at the indicated time points and immunoblotted with antibodies to FANCD2, actin, and PCNA. C, UV-inducible SNM1A foci are dependent on RAD18. The cDNA encoding GFP-SNM1Awt was transfected into HCT116 RAD18(+/+) and RAD18(−/−) cells. Cells were either untreated or treated with UV (50J/m2, 6 h). Foci were measured by counting the percentage of cells with more than 10 subnuclear foci among all the cells that express GFP-SNM1A. GFP-Polη was used as a positive control for the focus formation. Student's t test was performed on the original data sets, *** indicates that p < 0.001. In RAD18(−/−) cells for both SNM1A and Polη, the p values between the datasets of before and after UV treatment were much higher than 0.05, indicating that there were no significant increases in the percentage of cell with foci after UV treatment. D, MMC-inducible SNM1A foci are dependent on RAD18. HCT116 RAD18(+/+) and RAD18(−/−) cells were transfected with the cDNA encoding GFP-SNM1Awt, and the cells were either untreated or treated with MMC (100 ng/ml, 12 h), as indicated. Foci were quantified using the same methods described in C. Student's t test was performed on the original data sets, *** indicates that p < 0.001. In RAD18(−/−) cells, the p value between the datasets of before and after MMC treatment was much higher than 0.05, indicating that there were no significant increases in the percentage of cell with foci after MMC treatment. E, proposed model of RAD18-dependent PCNA monoubiquitination and SNM1A recruitment onto PCNA-Ub through its PIP box and UBZ domain.
More recent studies have indicated that crosslinking agents can activate PCNA monoubiquitination (27, 28). To further examine MMC-inducible monoubiquitination of PCNA in the RAD18(+/+) cells, we next tested a higher concentration of MMC (4 μg/ml) over a three-day time course (Fig. 5B). Under these conditions, MMC activated PCNA monoubiquitination (Fig. 5B, lanes 2–4), consistent with these earlier studies.
We next transfected the RAD18(+/+) and RAD18(−/−) cell lines with the cDNA encoding GFP-SNM1Awt. Both cell lines assembled SNM1A foci spontaneously (i.e. in the absence of external DNA damage). The RAD18(+/+) cells exhibited an increase in UV-damage inducible SNM1A foci, whereas the RAD18(−/−) cells were unable to mount a damage-inducible response (Fig. 5C). Interestingly, the RAD18(+/+) cells, but not the RAD18(−/−) cells, also exhibited a MMC-inducible increase in GFP-SNM1A focus formation (Fig. 5D).
DISCUSSION
In the current study, we examined the primary amino acid sequence of SNM1A and identified a PIP box and a UBZ. We demonstrated that these domains have direct (in vitro) binding activity for PCNA and ubiquitin, respectively, suggesting that they may participate in recruitment of SNM1A to the replication fork. We therefore examined the DNA damage-inducible assembly of the SNM1A protein into nuclear foci. Efficient focus formation was dependent on the PIP box and UBZ domain of SNM1A, as well as on the cellular function of the RAD18 protein. Our results support a model (Fig. 5E) in which DNA damage activates RAD18, leading to monoubiquitination of PCNA at the sites of stalled replication forks. This ultimately leads to PIP box and UBZ-mediated SNM1A recruitment.
SNM1A can also assemble in nuclear foci in the absence of PCNA monoubiquitination. For instance, cells expressing a UBZ mutant form of SNM1A still form SNM1A foci (20% of cells). Also, cells with a disruption of RAD18 can form SNM1A foci (20% of cells). But in these situations there was no enhancement of foci number or size after DNA damage. Consistent with these observations, a mutant form of SNM1A, in which the PIP box (located from residues 556 to 563 on SNM1A) is disrupted, cannot form foci, either spontaneously or after cellular exposure to DNA-damaging agents. These results suggest that SNM1A can associate directly with unubiquitinated PCNA at the replication fork. The results are also consistent with a previous study that identified a region from residues 394 to 615 on SNM1A, which is required for spontaneous focus formation (7).
Our results are consistent with previous studies indicating that RAD18 is required for DNA crosslink repair. A murine knock-out of RAD18 results in both MMC (crosslinker) and UV hypersensitivity (4, 8). How crosslinking agents activate the ubiquitin ligase activity of RAD18 remains unknown. Recent studies suggest that RAD18 recruitment to damaged chromatin and its subsequent activation may require RAD51C (29). Also, blockage of a replication fork by an ICL may activate local ssDNA formation and RPA binding, leading to the recruitment and activation of RAD18 (30).
Previous work suggested that RAD18 is not essential for SNM1A activity. In chicken DT40 cells, for example, SNM1A and Rad18 are not epistatic for crosslink repair (31). Moreover, Kannouche et al. (21) originally did not detect MMC-inducible PCNA ubiquitination, suggesting that crosslinks do not activate this RAD18-inducible PCNA modification. More recent studies, however, demonstrate that MMC or other interstrand crosslinking agents, such as cisplatin, can activate PCNA-Ub, although this activation appears to occur at higher doses of MMC and only after a delay of 24–48 h (27, 28). Indeed, we observed that SNM1A does assemble in RAD18-dependent DNA damage-induced foci after MMC treatment and that MMC can activate PCNA monoubiquitination after 24 h in our cell system. We also considered the possibility that RAD18 may directly interact with SNM1A after crosslink damage and may recruit SNM1A to these sites, similar to the mechanism described for polymerase η activation and recruitment (32). However, based on repeated attempts, we found no evidence of a direct interaction between RAD18 and SNM1A proteins (data not shown). Finally, we cannot exclude the possibility that other monoubiquitinated targets of RAD18 are required for MMC-induced SNM1A focus formation. Overall, these results support a model in which RAD18-dependent PCNA monoubiquitination enhances, but is not essential for, SNM1A recruitment.
SNM1A therefore appears to be recruited to either unmodified PCNA or to monoubiquitinated PCNA at the stalled replication fork. The molecular events which follow recruitment remain unknown. It is likely that SNM1A then provides an exonuclease function required for crosslink repair. Crosslink repair requires initial endonuclease incisions, on one DNA strand, on each side of the crosslink. These incisions result in the looping out of the crosslink, bound to a short stretch of single strand DNA. It is possible that the exonuclease activity of SNM1A will be required to trim this ssDNA, before translesion synthesis can proceed. Future studies will address this model.
Finally, the RAD18-PCNA(Ub)-SNM1A activation pathway appears to be independent of another DNA crosslink repair pathway, the FA pathway (33). MMC inducible assembly of SNM1A occurs normally in FA pathway-deficient cells (data not shown). Also, FA pathway-deficient cells appear to be hyperdependent on a functional RAD18-PCNA(Ub)-SNM1A pathway. Knockdown of RAD18 (34) or SNM1A (5) in an FA pathway-deficient cell results in a further increase in crosslinker-inducible chromosome breakage. The crosstalk between these two independent DNA crosslink repair pathways will be the subject of future experiments.
Acknowledgments
We thank Dr. Cyrus Vaziri for providing HCT116 RAD18(−/−) cells, Dr. Ivan Dikic for providing GST-PCNA and GST-PCNA-Ub constructs, and Dr. Robb Moses and Dr. James Heija for providing the human SNM1A cDNA. We would also like to express our thanks to Dr. Kay Hofmann, Dr. David Pellman, Dr. Daniel Finley, Dr. Lee Zou, and members of the D'Andrea laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK43889, R01HL52725, and P01CA092584 (to A. D. D.).
- ICL
- interstrand crosslinks
- PCNA
- proliferating cell nuclear antigen
- PCNA-Ub
- monoubiquitinated PCNA
- FancD2-Ub
- monoubiquitinated FancD2
- Ub
- ubiquitin
- UBZ
- ubiquitin binding zinc finger
- PIP Box
- PCNA-interacting protein box
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- MMC
- mitomycin C.
REFERENCES
- 1.Kennedy R. D., D'Andrea A. D. (2006) J. Clin. Oncol. 24, 3799–3808 [DOI] [PubMed] [Google Scholar]
- 2.Callebaut I., Moshous D., Mornon J. P., de Villartay J. P. (2002) Nucleic Acids Res. 30, 3592–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazrati A., Ramis-Castelltort M., Sarkar S., Barber L. J., Schofield C. J., Hartley J. A., McHugh P. J. (2008) DNA Repair 7, 230–238 [DOI] [PubMed] [Google Scholar]
- 4.Dronkert M. L., de Wit J., Boeve M., Vasconcelos M. L., van Steeg H., Tan T. L., Hoeijmakers J. H., Kanaar R. (2000) Mol. Cell. Biol. 20, 4553–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemphill A. W., Bruun D., Thrun L., Akkari Y., Torimaru Y., Hejna K., Jakobs P. M., Hejna J., Jones S., Olson S. B., Moses R. E. (2008) Mol. Genet. Metab. 94, 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tateishi S., Niwa H., Miyazaki J., Fujimoto S., Inoue H., Yamaizumi M. (2003) Mol. Cell. Biol. 23, 474–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richie C. T., Peterson C., Lu T., Hittelman W. N., Carpenter P. B., Legerski R. J. (2002) Mol. Cell. Biol. 22, 8635–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hejna J., Philip S., Ott J., Faulkner C., Moses R. (2007) Nucleic Acids Res. 35, 6115–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., Hofmann K., Dikic I. (2005) Science 310, 1821–1824 [DOI] [PubMed] [Google Scholar]
- 11.Hofmann K. (2009) DNA Repair 8, 544–556 [DOI] [PubMed] [Google Scholar]
- 12.Moldovan G. L., Pfander B., Jentsch S. (2007) Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 13.Nakajima S., Lan L., Kanno S., Usami N., Kobayashi K., Mori M., Shiomi T., Yasui A. (2006) J. Biol. Chem. 281, 34687–34695 [DOI] [PubMed] [Google Scholar]
- 14.Cohn M. A., Kowal P., Yang K., Haas W., Huang T. T., Gygi S. P., D'Andrea A. D. (2007) Mol. Cell 28, 786–797 [DOI] [PubMed] [Google Scholar]
- 15.Guo C., Tang T. S., Bienko M., Dikic I., Friedberg E. C. (2008) J. Biol. Chem. 283, 4658–4664 [DOI] [PubMed] [Google Scholar]
- 16.Notenboom V., Hibbert R. G., van Rossum-Fikkert S. E., Olsen J. V., Mann M., Sixma T. K. (2007) Nucleic Acids Res. 35, 5819–5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bish R. A., Myers M. P. (2007) J. Biol. Chem. 282, 23184–23193 [DOI] [PubMed] [Google Scholar]
- 18.Crosetto N., Bienko M., Hibbert R. G., Perica T., Ambrogio C., Kensche T., Hofmann K., Sixma T. K., Dikic I. (2008) J. Biol. Chem. 283, 35173–35185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann K., Bucher P. (1996) Trends Biochem. Sci. 21, 172–173 [PubMed] [Google Scholar]
- 20.Hofmann K., Falquet L. (2001) Trends Biochem. Sci. 26, 347–350 [DOI] [PubMed] [Google Scholar]
- 21.Kannouche P., Broughton B. C., Volker M., Hanaoka F., Mullenders L. H., Lehmann A. R. (2001) Genes Dev. 15, 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogi T., Kannouche P., Lehmann A. R. (2005) J. Cell Sci. 118, 129–136 [DOI] [PubMed] [Google Scholar]
- 23.Arakawa H., Moldovan G. L., Saribasak H., Saribasak N. N., Jentsch S., Buerstedde J. M. (2006) PLoS Biol. 4, e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szüts D., Simpson L. J., Kabani S., Yamazoe M., Sale J. E. (2006) Mol. Cell. Biol. 26, 8032–8041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edmunds C. E., Simpson L. J., Sale J. E. (2008) Mol. Cell 30, 519–529 [DOI] [PubMed] [Google Scholar]
- 26.Kannouche P. L., Wing J., Lehmann A. R. (2004) Mol. Cell 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 27.Niimi A., Brown S., Sabbioneda S., Kannouche P. L., Scott A., Yasui A., Green C. M., Lehmann A. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terai K., Abbas T., Jazaeri A. A., Dutta A. (2010) Mol. Cell 37, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J., Huen M. S., Kim H., Leung C. C., Glover J. N., Yu X., Chen J. (2009) Nat. Cell Biol. 11, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies A. A., Huttner D., Daigaku Y., Chen S., Ulrich H. D. (2008) Mol. Cell 29, 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishiai M., Kimura M., Namikoshi K., Yamazoe M., Yamamoto K., Arakawa H., Agematsu K., Matsushita N., Takeda S., Buerstedde J. M., Takata M. (2004) Mol. Cell. Biol. 24, 10733–10741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. (2004) EMBO J. 23, 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moldovan G. L., D'Andrea A. D. (2009) Annu. Rev. Genet. 43, 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirchandani K. D., McCaffrey R. M., D'Andrea A. D. (2008) DNA Repair 7, 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]