Abstract
Inactivation of the breast cancer susceptibility gene BRCA1 plays a significant role in the development of a subset of breast cancers, although the major tumor suppressor function of this gene remains unclear. Previously, we showed that BRCA1 induces antioxidant-response gene expression and protects cells against oxidative stress. We now report that BRCA1 stimulates the base excision repair pathway, a major mechanism for the repair of oxidized DNA, by stimulating the activity of key base excision repair (BER) enzymes, including 8-oxoguanine DNA glycosylase (OGG1), the DNA glycosylase NTH1, and the apurinic endonuclease redox factor 1/apurinic endonuclease 1 (REF1/APE1), in human breast carcinoma cells. The increase in BER enzyme activity appears to be due, primarily, to an increase in enzyme expression. The ability of BRCA1 to stimulate the expression of the three BER enzymes and to enhance NTH1 promoter activity was dependent upon the octamer-binding transcription factor OCT1. Finally, we found that OGG1, NTH1, and REF1/APE1 each contribute to the BRCA1 protection against oxidative stress due to hydrogen peroxide and that hydrogen peroxide stimulates the expression of BRCA1 and the three BER enzymes. These findings identify a novel mechanism through which BRCA1 may regulate the repair of oxidative DNA damage.
Keywords: BRCA BRCB, Breast Cancer, DNA repair, Gene Expression, Oxidative Stress, NTH1, OCT1, OGG1, REF1/APE1, Base Excision Repair
Introduction
The base excision repair (BER)2 pathway is an evolutionarily conserved mechanism for repair of several types of DNA lesions, including oxidative lesions, alkylation, and incorporation of inappropriate bases (1). The primary source of these lesions is reactive oxygen species, whether generated endogenously or due to genotoxic agents. The BER pathway functions to maintain genomic integrity via a high fidelity repair process and is thus anti-mutagenic and anti-carcinogenic (2). This pathway usually has four or five enzymatic steps, involving a DNA glycosylase (e.g. OGG1 or NTH1), an AP endonuclease (REF1/APE1), a DNA polymerase (e.g. polymerase B and D), and a DNA ligase (ligase I or III) (3).
DNA glycosylases recognize specific subsets of damaged bases, excise the damaged base, and may also incise at the site of the excised base due to an intrinsic lyase activity (e.g. OGG1 and NTH1). REF1/APE1 (the only mammalian AP endonuclease) then cleaves the abasic site to form a 3′-OH end and a 5′-deoxyribose phosphate end. The remaining steps may utilize a “long patch” or “short patch” pathway involving repair DNA synthesis and strand ligation by different sets of proteins. The preferred substrates for the DNA glycosylases OGG1 (8-oxoguanine glycosylase 1) and NTH1 (homolog of Escherichia coli endonuclease III) are 8-oxoguanine and thymine glycol (TG) lesions, respectively. REF1/APE1 (redox factor 1/apurinic endonuclease 1) is a multifunctional enzyme with AP endonuclease activity and 3′,5′-exonuclease, 3′-diesterase, and 3′-phosphatase activities. REF1/APE1 also has transcriptional regulatory activity independently of its function in BER (4, 5). Finally, the XRCC1 protein (x-ray repair, cross-complementing defective, in Chinese hamster, 1) associates with several other proteins, polynucleotide kinase, DNA polymerase-β (polymerase B), and DNA ligase III, to form a complex that repairs the single strand DNA breaks generated during the BER process.
The breast cancer susceptibility gene BRCA1 encodes a tumor suppressor protein, mutations of which account for about 2.5–5% of all breast cancers and 40–50% of hereditary breast cancers (6). Even in sporadic (nonhereditary) breast cancers, BRCA1 expression is absent or significantly decreased in about 30–40% of cases (7–9). BRCA1 is a multifunctional protein that has been implicated in regulation of the cell cycle, apoptosis, and chemosensitivity, various transcriptional pathways (including hormone-response pathways), and DNA damage signaling and repair (reviewed in Refs. 10, 11). Studies of its role in cellular responses to DNA damage have identified a function for BRCA1 as a caretaker in the maintenance of genomic integrity. Through interactions with other DNA repair pathway proteins, BRCA1 participates in pathways that: 1) signal DNA damage (i.e. as a target for phosphorylation by upstream kinases, such as ataxia telangiectasia-mutated, ataxia telangiectasia and RAD3-related, and cell cycle checkpoint kinase 1 and 2) (12); 2) mediate the preferential repair of DNA double strand breaks by homology-directed repair (along with Rad51 and BRCA2) (13, 14); and 3) activate several DNA damage-responsive cell cycle checkpoints (15, 16). BRCA1 is also a component of the BRCA-Fanconi anemia pathway, which is essential for the cellular response to DNA cross-linking agents (17). BRCA1, along with another RING domain protein (BARD1), exerts an E3 ubiquitin ligase function that constitutes the only known enzymatic function for BRCA1 (18, 19).
However, it is still unclear which of these functions of BRCA1 is most important for its role in genome maintenance and suppression of breast cancer development. Here, we show that in human breast carcinoma cell lines, BRCA1 stimulates several early steps in the BER pathway through a transcriptional mechanism that involves the octamer-binding transcription factor 1 (OCT1). These findings suggest a role for BRCA1 in protecting cells against oxidative damage to DNA.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture Conditions
MCF-7 and T47D human mammary adenocarcinoma cell lines were obtained from the American Type Culture Collection (Manassas, VA). The cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, nonessential amino acids (100 mm), l-glutamine (5 mm), streptomycin (100 mg/ml), and penicillin (100 units/ml) (all from BioWhittaker, Walkersville, MD). The cells were maintained at 37 °C in a humidified atmosphere of 95% air and 5% CO2 and subcultured weekly, using trypsin (20–22). Oct1−/− (Oct1 null) and Oct1+/+ (wild type) mouse embryonic fibroblasts were generously provided by Dr. Dean Tantin (University of Utah School of Medicine, Salt Lake City, UT) (23).
Expression Vectors and Reagents
The wild-type (WT) BRCA1 expression plasmid (WTBRCA1) was created by cloning the BRCA1 cDNA into the pcDNA3 vector (Invitrogen) using artificially engineered 5′ HindIII and 3′ NotI sites (21). WT-REF1 and dominant negative (DN) REF1 expression plasmids were a kind gift from Dr. Martin L. Smith (Indiana University Cancer Center, Indianapolis). DN-REF1 harbors an alanine substitution for cysteine codon at 65, predicted to block redox signaling to p53 and other REF1-responsive proteins (24). The mutant REF1 is driven by the cytomegalovirus promoter of the pcDNA3.1 vector (25). Dimethyl sulfoxide (DMSO), hydrogen peroxide (H2O2), paraquat, β-mercaptoethanol, 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide (MTT), and all the other chemicals were obtained from the Sigma, unless otherwise stated. T4 DNA ligase was obtained from New England Biolabs (Ipswich, MA). Purified full-length BRCA1 protein was obtained from Protein One (catalog no. 4130-100-EB, Bethesda); and purified OGG1 protein was purchased from R & D Systems (catalog no. P2005-01, Minneapolis, MN).
NTH1 Reporter Assays
NTH1 luciferase vectors driven by a 0.4-kb or 1.2-kb segment of the human NTH1 promoter upstream of the translation start site (within the pGL3-Luc plasmid) were generously provided to us by Dr. Haruhiko Sugimura, Hamamatsu University School of Medicine, Hamamatsu, Shizuoka, Japan (26). To assay NTH1 promoter activity, subconfluent proliferating cells in 24-well dishes were transiently transfected overnight with 0.5 μg of the indicated reporter (0.5 μg) and expression vector (WTBRCA1 or empty pcDNA3 vector) using Lipofectamine 2000 (Invitrogen) as the transfection reagent. The total transfected DNA was kept constant by addition of control vector. The transfected cells were then washed and post-incubated for 24 h in complete Dulbecco's modified Eagle's medium containing 5% fetal bovine serum to allow gene expression. The cells were then harvested and assayed for luciferase activity using the Dual-Luciferase reporter assay system (catalog no. E1910, Promega, Madison, WI), according to the manufacturer instructions. Luciferase values were usually expressed relative to that observed in control (pcDNA3)-transfected cells, as means ± S.E. of three independent experiments. As a negative control, assays were performed using a control reporter missing the NTH1 promoter segment (pGL3-Luc).
Transient Transfections
For ectopic expression, proliferating cells at about 70–80% of confluency were transfected overnight with the indicated expression vector (6 μg of plasmid DNA per well for a 6-well plate or 30 μg per 10-cm dish) using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's protocol, as described before (20, 22, 27). The transfected cells were washed and then incubated for several hours in fresh culture medium to allow them to recover.
Small Interfering (si) RNA Treatments
Asynchronously proliferating cells at about 30–40% of confluency were pretreated with the siRNA of interest (100 nm for 48–72 h) using siPORT amine transfection reagent (Ambion, Foster City, CA). The efficacy of each knockdown was confirmed by Western blotting. The following siRNAs were used in this study: control-siRNA (ON-TARGET plus Nontargeting siRNA (catalog no. D-001810-01, Dharmacon, Chicago)); BRCA1-siRNA (pool of two siRNAs custom-synthesized from Dharmacon (sequences 5′ → 3′ CAGCTACCCTTCCATCATA and CTAGAAATCTGTTGCTATG)); REF1-siRNA (pool of two siRNAs custom synthesized from Dharmacon (5′ → 3′ CAAAGTTTCTTACGGCATA and ACAGCAAGATCCGTTCCAA)); OGG1-siRNA (Santa Cruz Biotechnology, catalog no. SC-43983, Santa Cruz, CA); NTH1-siRNA (Santa Cruz Biotechnology, catalog no. SC-38133)); and OCT1-siRNA (Santa Cruz Biotechnology, catalog no. SC-36119). The OCT1-siRNA is a pool of three target-specific 20–25 nucleotide siRNAs.
Hydrogen Peroxide and Paraquat Treatments
Subconfluent proliferating cells were treated with the indicated concentrations of H2O2 or paraquat in culture medium containing 5% fetal calf serum for 24 h at 37 °C.
MTT Assays of Cell Viability
MTT assays were performed as described before (28). This assay is based on the ability of intact mitochondria to convert MTT, a soluble tetrazolium salt, into an insoluble formazan precipitate, which is quantified by spectrophotometry after solubilization in DMSO. After the indicated cell treatment, cells in 96-well dishes were solubilized and assayed for MTT dye reduction, based on the difference between absorbance at 570 and 630 nm. Cell viability was expressed as the amount of dye reduction relative to untreated control cells. Cell viability values were calculated as means ± S.E. for at least three independent experiments, each of which used eight replicate wells per cell type and assay condition.
Western Blotting
After the indicated treatment, whole cell lysates were prepared. The cells were washed with phosphate-buffered saline and lysed with 200 μl of RIPA buffer (Sigma) supplemented with protease inhibitor mixture (Complete Mini, EDTA-free; 11836170-001, Roche Applied Science) on ice for 30 min. The lysed cells were rocked at 4 °C for 30 min, followed by centrifugation at 12,000 × g for 20 min. The protein content of the supernatant was measured via the Bradford method (Bio-Rad), and the supernatants were stored at −80 °C prior to usage. Aliquots of protein in 4× lithium dodecyl sulfate sample buffer (Invitrogen) were analyzed on 4–20% Tris-glycine gels. Precast gels were obtained from Invitrogen.
The separated proteins were transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA) and blocked for 1 h in blocking buffer (Sigma). The membranes were blotted with primary antibodies, followed by incubation with species-specific horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) at a 1:10,000 dilution. Blotted proteins were visualized using the enhanced chemiluminescence detection system (Santa Cruz Biotechnology). Equal protein loading (40 μg of whole cell protein) was confirmed by blotting with α-actin antibody. Kaleidoscope prestained protein markers (Bio-Rad) were used as molecular size standards.
The primary antibodies were as follows: BRCA1 (C-20, Santa Cruz Biotechnology, 1:200 dilution); REF1/APE1 (E-17, Santa Cruz Biotechnology, 1:200); α-actin (C-11, Santa Cruz Biotechnology, 1:400); OGG1 (ab22766, Abcam (Cambridge, MA), 1:100); NTH1 (AF2675, R & D Systems (Minneapolis, MN), 1:100); XRCC1 (ab47920, Abcam, 1:250); ligase III (ab587, Abcam, 1:200); catalase (F-17, Santa Cruz Biotechnology, 1:150); SOD1 (C-17, Santa Cruz Biotechnology, 1:200); SOD2 (FL-222, Santa Cruz Biotechnology, 1:250); and OCT1 (C-21, SC-232, Santa Cruz Biotechnology, 1:100).
Semi-quantitative RT-PCR Analysis
Semi-quantitative RT-PCR assays were performed as described earlier (27). Briefly, total cell RNA was extracted using the RNase Easy mini kit (Qiagen, Valencia, CA). Aliquots of total cell RNA (200 μg) were reverse-transcribed using 50 units of Superscript III reverse transcriptase (Invitrogen) in a reaction volume of 20 μl. One-μl aliquots of transcribed cDNA were subjected to PCR amplification, using hot-start Taq polymerase (Denville, South Plainfield, NJ). DNA was first denatured for 5 min at 95 °C and then amplified using cycles of 30 s at 95 °C, 30 s at the annealing temperature, and 1 min at 72 °C, with a final 10-min incubation at 72 °C. The cycle number was adjusted so that all reactions fell within the linear range of product amplification. The forward and reverse primers, annealing temperatures, and expected sizes of the PCR products are shown in supplemental Table 1. The size of each PCR product was confirmed by electrophoresis through 2% agarose gels containing 0.1 mg/ml ethidium bromide; and the gels were photographed under ultraviolet light.
Assays of BER Pathway
Preparation of Nuclear Extracts
Nuclear extracts were prepared as described previously (29). Cells (about 1 × 107 per assay condition) were washed once with phosphate-buffered saline and resuspended in 400 μl of hypotonic lysis buffer A (10 mm HEPES, 10 mm KCl, 0.1 mm EGTA, 0.1 mm EDTA, 1 mm dithiothreitol, 2 μg/ml leupeptin, 2 μg/ml pepstatin, and 0.5 mm phenylmethylsulfonyl fluoride, pH 7.9). The cells were allowed to swell on ice for 15 min, after which 25 μl of a 10% solution of Nonidet P-40 (Pierce) was added, and the tube was vortexed vigorously for 10 s. The nuclei were collected by centrifugation at 500 × g at 4 °C for 45 s. The supernatant was considered to be the cytoplasmic fraction. The nuclei were washed three times with buffer A to minimize cytoplasmic contamination. Nuclear proteins were extracted with 200 μl of buffer B (20 mm HEPES, 400 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 2 μg/ml leupeptin, 2 μg/ml pepstatin, and 0.5 mm phenylmethylsulfonyl fluoride, pH 7.9) by vigorous rocking at 4 °C for 30 min, followed by centrifugation at 12,000 × g for 20 min at 4 °C. This supernatant was considered to be the nuclear extract, which was analyzed for protein content by Bradford assay and stored at −80 °C until it was used for the DNA incision or ligation assays.
DNA Incision Assays
The determination of different cleavage activity was performed using a modified oligonucleotide cleavage assay as described previously (30–33). 5′-FITC-labeled oligonucleotides were synthesized containing a single damaged base as follows: 8-oxoguanine, thymine glycol, or a tetrahydrofuranyl artificial AP site at indicated position (see supplemental Table 2). Oligonucleotides containing normal bases in place of the damaged bases were used as controls for specific activities. Complementary oligonucleotides were also synthesized to yield a duplex either with the wild-type or damaged (modified) base containing oligonucleotide. Annealing of the oligonucleotides was performed in freshly prepared annealing buffer (10 mm Tris, 100 mm NaCl, 1 mm EDTA). The sequences of each oligonucleotide used in different assays are provided in supplemental Table 2. Incision assay procedures are described below.
Nuclear extracts were incubated with modified 5′-FITC-labeled duplex oligonucleotides, and in the presence of specific enzyme activity, the modified oligonucleotide was cleaved into smaller oligomers. As a negative control, wild-type (unmodified) 5′-FITC-labeled duplex oligonucleotides were tested at the same time. Reaction mixtures (20 μl) (which contained a fixed quantity of nuclear extract (indicated in the figures), 2.5 pmol of 5′-FITC-labeled duplex oligonucleotide, 20 mm HEPES, 5% glycerol, 100 mm NaCl, 5 mm EDTA, 5 mm dithiothreitol, 100 μg/ml bovine serum albumin, and 25 μg/ml poly(dI)/poly(dC), pH 7.4) were incubated for 45–60 min at 37 °C. All reactions were stopped by incubation with 0.2% SDS and 0.1 mg/ml proteinase K at 68 °C for 10 min. The DNA was extracted using phenol/chloroform/isoamyl alcohol; and the reaction mixture was then admixed with 15 μl of formamide DNA loading dye (85% formamide and 0.03 n NaOH) and heated at 95 °C for 5 min. The products were separated by electrophoresis through 20% polyacrylamide gels containing 7 m urea at 300 V for 70–80 min.
The gels were washed in water and exposed in Ettan DIGE fluorescence imager cassettes (GE Healthcare). The gel images were analyzed using NIH-ImageJ software version 1.38X (rsbweb.nih.gov, National Institutes of Health, Bethesda). The product bands were quantified relative to the sum of the substrate and product bands and expressed as a percentage. At least three independent experiments were performed per assay type per cell line.
Ligase Assay (Ligase III)
A 40-mer oligonucleotide (LigIII-2, see supplemental Table 2) was radiolabeled with [γ-32P]ATP (Amersham Biosciences) at the 5′ end using T4 polynucleotide kinase and subsequently purified using a G-25 mini spin column (Amersham Biosciences). The γ-32P-labeled 40-mer and an unlabeled 30-mer (LigIII-1, supplemental Table 2) were annealed to an unlabeled 70-mer complementary oligonucleotide (see supplemental Table 2). Fifty μg of nuclear extract per assay was incubated with the annealed duplex oligonucleotide and incubated at 37 °C for 60 min in 20 μl of ligation buffer (60 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 50 mg/ml bovine serum albumin, 5 mm dithiothreitol, 1 mm ATP). The reaction was stopped by the addition of 4 volumes of formamide sample loading buffer as above, and the reaction mixture was heated to 85 °C for 10 min and then cooled rapidly on ice. The oligonucleotides were resolved on 12% polyacrylamide gel containing 7 m urea. The gels were washed briefly in water, dried on a gel dryer, and subjected to autoradiography. The appearance of a 70-mer labeled product band is indicative of the ligase activity.
Statistical Methods
Statistical comparisons were made using the two-tailed Student's t test where appropriate. Data are presented as arithmetic means with error bars representing the standard error of mean calculated from at least three independent experiments.
RESULTS
Repair of Thymine Glycol (TG) Lesions
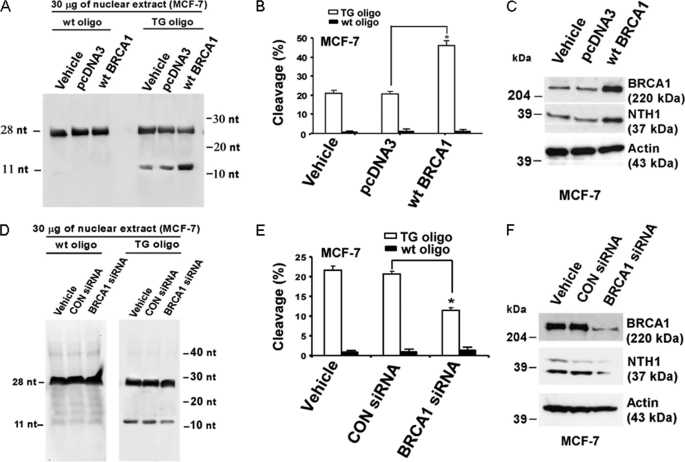
The TG lesion is a common oxidative DNA lesion that is typically repaired starting with an incision by the DNA glycosylase NTH1 (also called NTHL1 or endonuclease III-like 1). We tested the effects of manipulation of BRCA1 levels in MCF-7 and T47D cells on this incision reaction using FITC-conjugated DNA oligonucleotides containing a TG paired with an adenine (see “Experimental Procedures”). Cells were transfected overnight with vehicle only, WTBRCA1, or empty pcDNA3 vector; and nuclear extracts were assayed, as described under “Experimental Procedures.” As illustrated in Fig. 1A, there was very little incision of the wild-type oligonucleotide, whereas WTBRCA1 visibly stimulated incision of TG oligonucleotide in MCF-7 cells. When the percentage of incised DNA was calculated based on densitometry analysis for three independent experiments, it was found that WTBRCA1 increased the amount of incision by about 2.5-fold (p < 0.05) (Fig. 1B). In addition, BRCA1 overexpression also increased the levels of NTH1 protein, as determined by Western blotting (Fig. 1C). Very similar results were observed using T47D cells (see supplemental Fig. S1, A–C).
FIGURE 1.
BRCA1 regulates DNA cleavage at a TG site. A–C, proliferating MCF-7 cells were transiently transfected overnight with vehicle, wild-type BRCA1 (WTBRCA1), or empty pcDNA3 vector, washed, and allowed to recover for several hours prior to assay. Nuclear extracts were prepared, and 30-μg aliquots were tested for their ability to incise duplex oligonucleotides (oligo) containing a TG lesion, using the corresponding wild-type duplex oligonucleotide as a control. A shows a representative DNA gel; B shows the quantitative extents of cleavage, expressed as means ± S.E. of three independent experiments. The protein levels of BRCA1, NTH1, and α-actin for the different transfection conditions are shown in C. D–F, MCF-7 cells were treated with BRCA1-siRNA (100 nm), control (CON)-siRNA, or vehicle only and assayed for incising activity at a TG site as described above. D shows a representative DNA gel; E shows the quantitative extent of cleavage. The BRCA1, NTH1, and actin protein levels for the different treatment conditions are shown in F. B and E, an asterisk represents a statistically significant difference (p < 0.05, two-tailed t test). nt, nucleotide.
Conversely, knockdown of BRCA1 in MCF-7 cells using BRCA1-siRNA caused a significant reduction of incision around TG lesions, as compared with cells treated with control-siRNA or vehicle only (p < 0.05) (Fig. 1, D and E); BECA1 knockdown also caused a reduction of the NTH1 protein levels (Fig. 1F). Again, very similar results were observed using T47D cells (supplemental Fig. S1, D–F). Taken together, these findings suggest that BRCA1 regulates NTH1 incising activity, at least in part, through regulation of NTH1 expression.
Repair of 8-OxoG Lesions
8-Oxoguanine (8-OxoG) is a signature mutagenic DNA lesion caused by oxidative stress that is repaired through BER. We tested whether BRCA1 could modulate the first step in the repair of this lesion, which involves incising of DNA around an 8-OxoG site. This step is mediated by the DNA glycosylase OGG1. This assay tests the ability of nuclear extracts to cause incision of an FITC-conjugated duplex DNA containing an 8-OxoG paired with a cytosine. Cells were transfected overnight with vehicle only, wild-type BRCA1, or empty pcDNA3 vector; and nuclear extracts were assayed as described under “Experimental Procedures.” Here, WTBRCA1 significantly stimulated the cleavage of 8-OxoG containing oligonucleotides (but not a wild-type oligonucleotide) in MCF-7 cells (p < 0.05) (Fig. 2, A and B). WTBRCA1 also caused an increase in the OGG1 protein levels (Fig. 2C). Conversely, BRCA1 knockdown caused a significant reduction of OGG1 incising activity (p < 0.05) and a corresponding reduction of OGG1 protein levels (Fig. 2, D–F); very similar findings were observed using T47D cells (see supplemental Fig. S2). These findings are consistent with the idea that BRCA1 regulates OGG1 incising activity in part through the regulation of OGG1 protein levels.
FIGURE 2.
BRCA1 regulates DNA cleavage at an 8-oxoguanine site. A–C, proliferating MCF-7 cells were transiently transfected overnight with vehicle only, WTBRCA1, or empty pcDNA3 vector, washed, and allowed to recover for several hours prior to assay. Nuclear extracts were prepared, and 100-μg aliquots were tested for their ability to incise duplex oligonucleotides (oligo) containing an 8-OxoG lesion, using the corresponding wild-type duplex oligonucleotide as a negative control. A shows a representative DNA gel; B shows the quantitative extents of cleavage, expressed as means ± S.E. of three independent experiments. Western blots showing the protein levels of BRCA1, OGG1, and α-actin (control for loading and transfer) for the different transfection conditions are provided in C. D–F, MCF-7 cells were treated with BRCA1-siRNA (100 nm), control (CON)-siRNA (100 nm), or vehicle only for 72 h, after which nuclear extracts were prepared and assayed for incising activity as above. D shows a representative DNA gel; E shows the quantified extent of cleavage, expressed as means ± S.E. of three independent experiments. The protein levels of BRCA1, OGG1, and α-actin for the different treatment conditions are shown in F. *, p < 0.05 (two-tailed t test). nt, nucleotide.
As a control, we also tested the ability of a full-length mutant breast cancer-associated BRCA1 protein (T300G, which encodes a point mutation of BRCA1 in the RING domain (C61G)) to alter OGG1 activity. As illustrated in supplemental Fig. S3, expression of T300G had little or no effect on OGG1-related incising activity in MCF-7 cells (supplemental Fig. S3, A and B), although the T300G protein was well expressed but did not stimulate OGG1 expression (supplemental Fig. S3C). These findings suggest that a functional BRCA1 protein is required to stimulate OGG1 activity and expression.
Repair of Abasic Sites
The AP endonuclease (REF1/APE1) mediates the repair of apurinic/apyrimidinic DNA lesions through multiple functionalities, including its ability to incise around AP sites. We tested the effect of BRCA1 on REF1/APE1 incising activity using FITC-labeled DNA duplex oligonucleotides containing an AP site paired with a G nucleotide (see “Experimental Procedures”). Here, we found that overexpression of BRCA1 caused a significant increase in nuclear AP-incising activity in MCF-7 cells (p < 0.01); and it also caused an increase in REF1/APE1 protein levels (Fig. 3, A–C). Conversely, knockdown of BRCA1 caused a significant reduction of AP-incising activity (p < 0.05) that was associated with a reduction of REF1/APE1 protein levels (Fig. 3, D–F). Similar results were observed for T47D cells (supplemental Fig. S4). The effects of BRCA1 overexpression and underexpression on the protein levels of each of the three DNA glycosylases, as quantified by densitometry, corresponding to the experiments shown in Figs. 1–3, are shown in supplemental Fig. S5.
FIGURE 3.
BRCA1 regulates DNA cleavage at an AP site. A–C, MCF-7 cells were transfected with vehicle, WTBRCA1, or empty pcDNA3 vector; and aliquots of nuclear extract (50 μg) aliquots were assayed for incising activity around an AP site in a duplex oligonucleotide (oligo). A shows a representative assay; B shows the quantification of cleavage (based on three independent experiments); C shows the BRCA1, REF1/APE1, and α-actin protein levels for the different transfection conditions. D–F, MCF-7 cells were treated with BRCA1-siRNA (100 nm), control (CON)-siRNA, or vehicle and assayed for incising activity as above. D shows a representative DNA gel; E shows the quantitative extent of cleavage. The BRCA1, REF1/APE1, and actin protein levels for the different treatment conditions are shown in F. *, p < 0.05 (two-tailed t test). nt, nucleotide.
DNA Ligation Activity
As noted earlier, DNA ligase III forms a complex with several other proteins, including XRCC1 that mediates the repair of single strand DNA breaks in the short patch BER pathway. We tested the effect of BRCA1 over/underexpression on the ability of nuclear extracts to ligate two duplex oligonucleotides (a 30- and a 40-mer), one of which (the 40-mer) contains a 5′-FITC label (see “Experimental Procedures” and supplemental Table 2). The product of the ligation is a 70-mer. Representative ligation assays are shown in supplemental Fig. S6, A–D for MCF-7 and T47D cells made to overexpress or underexpress BRCA1. The quantification of the results (based on three independent experiments per cell line and treatment) revealed no consistent and significant changes in ligation activity due to over- or underexpression of BRCA1 (supplemental Fig. S6, E–G).
BRCA1 Stimulates mRNA Expression of BER Enzymes
We tested the effects of BRCA1 overexpression on the mRNA levels of OGG1, NTH1, XRCC1, and REF1/APE1 using semi-quantitative RT-PCR assays. All assays were carried out within the linear range of product amplification. Amplified bands were quantified by densitometry, and mRNA levels were normalized to β-actin and expressed relative to the vehicle control. For both MCF-7 (Fig. 4, A and B) and T47D (supplemental Fig. S7, A and B) cells, WTBRCA1 caused ≥2-fold increases in the mRNA levels of OGG1, NTH1, REF1/APE1, and XRCC1. In addition, in both cell lines, BRCA1 over/underexpression caused a corresponding increase/decrease in XRCC1 protein levels but did not caused any change in ligase III levels (Fig. 4, C and D and supplemental Fig. S7, C and D). These findings suggest that BRCA1 regulates OGG1, NTH1, REF1, and XRCC1 at the mRNA level.
FIGURE 4.
BRCA1 regulates expression of BER enzymes. A and B, proliferating MCF-7 cells were transfected overnight with WTBRCA1, empty pcDNA3 vector, or vehicle only; washed; allowed to recover from the transfection in fresh culture medium, and harvested for semi-quantitative RT-PCR assays, as described under “Experimental Procedures.” Representative gels showing the amplified bands corresponding to BRCA1, OGG1, NTH1, REF1/APE1, XRCC1, and β-actin are provided in A. The PCR bands were quantified by densitometry, and the mRNA levels were normalized to β-actin and expressed relative to that of the vehicle control in B. The fold-change values are means ± S.E. of at least three independent experiments. *, p < 0.05 relative to vehicle control (two-tailed t test). C and D, MCF-7 cells transfected with expression vectors (C) or treated with siRNAs (D) as before were harvested and Western-blotted to detect BRCA1, XRCC1, ligase III, or actin (loading control (CON)).
BRCA1 Does Not Directly Stimulate OGG1 Incision at an 8-OxoG Site
Here, we tested the effects of adding purified BRCA1 or OGG1 protein directly to a nuclear extract on OGG1 incising activity. In control assays, as expected, exogenous BRCA1 both stimulated OGG1 incising activity and increased OGG1 protein levels (supplemental Figs. S8A and S7B, respectively). However, addition of 0.25 or 0.5 μg of BRCA1 protein to an extract prepared from untransfected cells had no effect on incising activity (supplemental Fig. S8C). In contrast, addition of 0.5 μg of OGG1 to the extract caused an obvious increase in incising activity (supplemental Fig. S8D). Taken together with our previous results, these findings suggest that BRCA1 stimulates BER activity (at least OGG1 activity) indirectly, by stimulating gene expression.
Role of BER Enzymes in BRCA1 Protection against Oxidative Stress
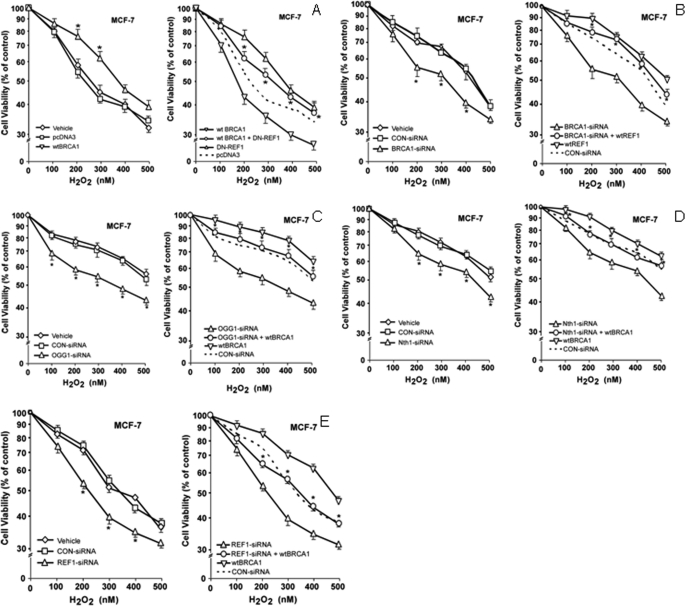
We tested the roles of several of the BER enzymes studied above in BRCA1 protection against oxidative stress due to hydrogen peroxide (H2O2). Briefly, cells were transfected with expression vectors for the protein(s) of interest and/or treated with siRNAs of interest, exposed to different concentrations of H2O2 for 24 h, and tested for cell viability using MTT assays. Here, WTBRCA1 enhanced the survival of H2O2-treated MCF-7 cells (compared with empty vector-transfected cells) at all concentrations of H2O2, and the increases in survival were statistically significant (p < 0.05) at several concentrations (Fig. 5A, left). A dominant negative REF1 vector (DN-REF1) by itself reduced the survival of H2O2-treated cells to below that of empty vector-transfected cells at all H2O2 concentrations. However, survival of cells transfected with WTBRCA1+DN-REF1 was significantly greater than that of cells transfected with DN-REF1 alone at nearly every concentration of H2O2 (Fig. 5A, right). We next tested the ability of WT-REF1 to rescue the reduction in survival of H2O2-treated cells due to BRCA1 knockdown. Compared with control-siRNA, BRCA1-siRNA-treated cells showed significant 10–20% reductions in survival at most doses of H2O2 in MCF-7 cells (Fig. 5B, left). WT-REF1 alone (relative to pcDNA3 vector) increased MCF-7 cell survival to differing degrees, with maximal increases of 15% at some concentrations of H2O2; and WT-REF1 rescued the reduction in survival due to BRCA1-siRNA. In fact, the survival levels of cells treated with BRCA1-siRNA+WT-REF1 were the same as or higher than those of control cells (Fig. 5B, right). Similar results were observed using T47D cells (supplemental Fig. S9, A and B).
FIGURE 5.
Role of BER enzymes in BRCA1 protection against oxidative stress due to H2O2. A, proliferating MCF-7 cells in 6-well dishes were transfected overnight with the indicated expression vectors (using empty pcDNA3 vector to keep the total transfected DNA content constant); washed; post-incubated in fresh medium for 24 h to allow gene expression; harvested, and inoculated into 96-well plates in fresh medium. The cells were allowed to attach and recover for another 24 h; exposed to the indicated concentration of H2O2 for 24 h; and tested for cell viability by MTT assays. Values plotted are means ± S.E. of three independent experiments, with each experiment using 8 replicate wells per assay condition. B, MCF-7 cells in 6-well dishes were treated with BRCA1-siRNA (100 nm), control (CON)-siRNA (100 nm), or vehicle only for 72 h and then inoculated into 96-well plates for H2O2 survival assays as described above. For combination treatment with BRCA1-siRNA+WT-REF1, after incubation of the cells with siRNA (100 nm) for 24 h, was transfected with WT-REF1 overnight and allowed to recover for another 24 h. The cells were then harvested, inoculated into 96-well dishes, and assayed for sensitivity to H2O2 as above. In the left panels, significant increases in survival due to WTBRCA1 (relative to pcDNA3 vector) or reductions in survival due to BRCA1-siRNA (relative to control (CON)-siRNA) are indicated by an asterisk (p < 0.05). In the right panels, the asterisks represent significant comparisons of WTBRCA1+DN-REF1 versus DN-REF alone or BRCA1-siRNA+WT-REF1 versus BRCA1-siRNA alone. C–E, treatments with siRNA alone or combinations of siRNA plus WTBRCA1 vector and subsequent assays of cellular sensitivity to H2O2 were carried out as above. The effects of siRNAs for OGG1 (C), NTH1 (D), and REF1 (E) are shown. Cell viability values are means ± S.E. of three independent experiments and are normalized to the control (vehicle only) treatment. Significant reductions in survival due to OGG1-siRNA, NTH1-siRNA, or REF1-siRNA (left panels) (relative to control (CON)-siRNA) are indicated by an asterisk (p < 0.05). In right panels, increases in survival due to a combination of BER enzyme-siRNA plus WTBRCA1 (relative to BER enzyme-siRNA alone) are shown by an asterisk (p < 0.05).
We next tested the effects of knocking down OGG1, NTH1, or REF1 expression on sensitivity to H2O2 and the ability of WTBRCA1 to rescue these alterations. Here, OGG-siRNA consistently reduced survival of H2O2-treated cells in MCF-7 cells (15–20%) (Fig. 5C, left). The survival levels of WTBRCA1+OGG1-siRNA-transfected cells were higher than those of cells transfected with OGG1-siRNA alone but lower than those of WTBRCA1-transfected cells at each dose of H2O2 tested (Fig. 5C, right). NTH1-siRNA consistently reduced the survival of H2O2-treated MCF-7 (Fig. 5D, left), with reductions of up to 15%, compared with vehicle or control-siRNA-treated cells. As before, WTBRCA1-transfected cells exhibited higher survival than control cells, and the survival of WTBRCA1+NTH1-siRNA-treated cells was similar to that of control-treated cells but higher than that of NTH-siRNA-treated cells. We performed similar experiments using REF1-siRNA to knock down endogenous REF1 rather than using a DN-REF1 expression vector. MCF-7 cells treated with REF1-siRNA alone showed survival reductions of 10–20% (compared with cells treated with control-siRNA) at intermediate doses of H2O2 (Fig. 5E, left). WTBRCA1 alone significantly enhanced the survival of MCF-7 cells at most H2O2 doses. As in experiments using DN-REF1, cells transfected with WTBRCA1+REF1-siRNA showed lower survival than those transfected with WTBRCA1 alone. At most H2O2 doses, survival of WTBRCA1+REF1-siRNA-treated cells was similar to or slightly lower than that of control (vehicle or control-siRNA) cells but higher than that of cells treated with REF1-siRNA alone (Fig. 5E, right). The expression of WT-REF1 and DN-REF1 and the efficacy of the knockdowns of OGG1, NTH1, and REF1 are validated in Fig. 6. Similar results were observed using T47D cells (supplemental Figs. S9, C–E, and S10). These findings suggest that OGG1, NTH1, and REF1 each contribute to cell protection by BRCA1.
FIGURE 6.
Expression of WT-REF1 and DN-REF1 and efficacy of BER enzyme knockdowns. MCF-7 cells were transfected with WT-REF1 (A) or DN-REF (B) or treated with REF1-siRNA (C), OGG1-siRNA (D), or NTH1-siRNA (E), as described above. The cells were then harvested for Western blotting to detect the indicated BER enzyme or actin.
Role of Transcription Factor OCT1 in BRCA1 Stimulation of BER Enzyme Expression and Activity
Because the transcription factors OCT1 and NF-YA have been found to interact directly with BRCA1 and mediate BRCA1 stimulation of the GADD45 promoter (34), we tested whether these transcription factors might also contribute to BRCA1 stimulation of BER enzyme expression. T47D cells were pretreated with control-siRNA or OCT1-siRNA (100 nm for 48 h) and then transfected overnight with WTBRCA1 (+) or empty pcDNA3 vector (−). The cells were then post-incubated for 24 h to allow gene expression and subjected to Western blotting. Knockdown of OCT1 blocked WTBRCA1 stimulation of expression of OGG1, NTH1, and REF1/APE1 (Fig. 7A). Based on densitometric quantification of three independent experiments, the levels of these proteins were 2.5–3-fold higher in the presence of control-siRNA+WTBRCA1 than control-siRNA+pcDNA3 vector (p < 0.05), and in the presence of OCT1-siRNA+WTBRCA1, the protein levels were reduced to nearly control levels (p < 0.05) (Fig. 7B). OCT1-siRNA did not reduce the levels of these proteins to below the base line observed in the absence of WTBRCA1, but it is noted that OCT1-siRNA typically only reduced OCT1 protein levels by 50–60%.
FIGURE 7.
Role of OCT1 in stimulation of BER enzyme expression due to BRCA1 overexpression. A and B, T47D cells were treated with control (CON)-siRNA or OCT1-siRNA (100 nm for 48 h) and then transfected overnight with either a wild-type (wt) BRCA1 expression vector (+) or empty pcDNA3 vector (−). The cells were post-incubated for 24 h to allow gene expression and then subjected to Western blotting to detect the indicated proteins. α-Actin was used as a control for loading and transfer. A shows a typical Western blot. B shows quantification of these experiments by densitometry. Values plotted are means ± S.E. of the fold-change (ratio of protein/actin normalized to control-siRNA and empty vector transfection value) based on three independent experiments. C and D, T47D cells were treated with control (CON)-siRNA or OCT1-siRNA and transfected with WTBRCA1 (+) or empty pcDNA3 vector (−), as above; and 30-μg aliquots of nuclear extracts were tested for incision of duplex oligonucleotides containing a TG lesion, using the corresponding wild-type oligonucleotide as a control. C shows a representative DNA gel and D shows quantification of three separate experiments. *, p < 0.05.
Next, T47D cells were treated with control- versus OCT1-siRNA and then transfected with WTBRCA1 (+) or empty vector (−) as described above, and nuclear extracts were tested for their ability to incise an FITC-conjugated oligonucleotide containing a TG paired with an adenine (a function of NTH1). There was little or no incision of the wild-type oligonucleotide, although OCT1-siRNA blocked the ability of WTBRCA1 to stimulate incision of the TG oligonucleotide (Fig. 7C). Fig. 7D shows quantitative data based on three independent experiments. OCT1-siRNA caused a reduction of the % cleavage due to WTBRCA1 from 44 to 22% (p < 0.05), but cleavage in the presence of OCT1-siRNA+WTBRCA1 was still slightly above base line (17%). OCT1-siRNA by itself did not significantly reduce the basal cleavage level. The ability of OCT1-siRNA to block WTBRCA1-stimulated NTH1 incising activity is consistent with its ability to inhibit WTBRCA1-stimulated NTH1 expression.
Finally, although NF-YA-siRNA reduced NF-YA protein levels to the same extent or greater than OCT1-siRNA-reduced OCT1 protein levels, NF-YA knockdown had little or no effect on the ability of WTBRCA1 to stimulate OGG1, NTH1, and REF1/APE1 protein expression (supplemental Fig. S11). Taken together, these results suggest that OCT1 but not NF-YA contributes to BRCA1 stimulation of the BER pathway.
BRCA1 Regulates NTH1 Promoter Activity in an OCT1-dependent Manner
We next tested the ability of BRCA1 to regulate the promoter activity of the NTH1 gene using two NTH1 luciferase reporters, one of which is driven by a 1.2-kb promoter segment (1.2-kb NTH1-Luc) and the other by a shorter segment (0.4-kb NTH1-Luc) (26). When transfected into T47D cells, the basal activity of the 1.2-kb NTH1-Luc reporter was slightly less than twice as active as the 0.4-kb NTH1-Luc, although the activity of the promoterless reporter plasmid (pGL3-Luc) was negligible (supplemental Fig. S12A). In different experiments, WTBRCA1-transfected cells showed 6–8-fold higher NTH1 promoter activity for the 1.2-kb NTH1-Luc reporter (p < 0.001) (Fig. 8A) and 1.5–2.5-fold higher activity for the 0.4-kb NTH1-Luc reporter (p < 0.05) (supplemental Fig. S12B) than did empty pcDNA3 vector-transfected control cells. BRCA1-siRNA caused significant reductions in the activity of both the 1.2-kb (Fig. 8B) and 0.4-kb (S12C) promoters.
FIGURE 8.
BRCA1 regulates NTH1 promoter activity. A, T47D cells were co-transfected overnight with WTBRCA1 (or empty pcDNA3 vector) and with an NTH1 luciferase (NTH1-Luc) reporter (containing 1.2-kb of NTH1 promoter sequence upstream of the translation start site) or the empty pGL3-Luc reporter plasmid. The cells were then post-incubated for 24 h to allow gene expression and harvested for luciferase assays. Luciferase values are expressed as fold-change relative to control conditions (= 1.0) (pcDNA3+NTH1-Luc). B, T47D cells were pretreated with control (CON)-siRNA or BRCA1-siRNA (100 nm for 48 h), transfected with NTH1-Luc (1.2 kb) or pGL3-Luc, and assayed as above. Luciferase values were expressed as fold-change relative to control conditions (= 1.0) (CON-siRNA+NTH1-Luc). C, T47D cells were pretreated with control-siRNA or OCT1-siRNA, co-transfected overnight with WTBRCA1 (+) or empty vector (−) plus NTH1-Luc (1.2 kb) reporter, post-incubated for 24 h to allow gene expression, and harvested for luciferase assays. Luciferase values are expressed as fold-change relative to control conditions (= 1.0) (CON-siRNA+NTH1-Luc). In all experiments, luciferase values are means ± S.E. of three independent experiments. p values were calculated using two-tailed t tests. *, p < 0.05.
Finally, we tested the effect of OCT1-siRNA on the ability of WTBRCA1 to stimulate NTH1 promoter activity in T47D cells. In pcDNA3-transfected cells, OCT1-siRNA caused modest reductions in the 1.2-kb NTH1-Luc reporter activity that were not statistically significant, compared with control-siRNA-treated cells (Fig. 8C). However, OCT1-siRNA blocked most of the increase in 1.2-kb NTH1-Luc activity due to WTBRCA1 (p < 0.001). Similar results were observed using the 0.4-kb NTH1-Luc reporter, although the increase in activity of this reporter due to WTBRCA1 was smaller than that observed using the 1.2-kb NTH1-Luc reporter (supplemental Fig. S12D). These results suggest that BRCA1 regulates NTH1 promoter activity and that OCT1 is required for stimulation of NTH1 promoter activity due to BRCA1 overexpression, and they are consistent with the finding that OCT1 knockdown blocks the increase in NTH1 protein levels due to WTBRCA1.
WTBRCA1 Fails to Stimulate NTH1 Promoter Activity in Oct1 Null (−/−) Fibroblasts
We compared the NTH1-Luc reporter activity in Oct1−/− mouse embryo fibroblasts (MEFs) (provided by Dr. D. Tantin) (23) with that in wild-type Oct1+/+ MEFs. In Oct1+/+ cells, WTBRCA1 caused a 3.7-fold increase in 1.2-kb promoter activity (p < 0.001), relative to control (pcDNA3)-transfected cells (Fig. 9A). In contrast, WTBRCA1 caused no increase in 1.2-kb promoter activity in Oct1−/− cells (Fig. 9B). Similar results were observed using the 0.4-kb NTH1-Luc reporter (supplemental Fig. S13, A and B). We noted that for both promoter segments, the basal NTH1-Luc activity was significantly greater in Oct1+/+ cells compared with Oct1−/− cells (p < 0.001) (Fig. 9C and supplemental Fig. S13C, respectively). Thus, even though OCT1 knockdown alone caused only a modest reduction, if any, in the basal NTH1 promoter activity in T47D cells, the absence of any functional Oct1 in Oct1−/− is associated with a reduction in the basal promoter activity. These findings suggest that both BRCA1 and OCT1 regulate NTH1 promoter activity.
FIGURE 9.
BRCA1 overexpression does not stimulate NTH1 promoter activity in Oct1 null (Oct1−/−) MEFs. A and B, wild-type (Oct1+/+) (A) or Oct1−/− (B) MEFs were co-transfected with WTBRCA1 (versus empty pcDNA3 vector) and the NTH1-Luc (1.2 kb) or empty pGL3-Luc reporter overnight, post-incubated for 24 h to allow gene expression, and assayed for luciferase activity. Luciferase values are expressed as fold-change relative to control (= 1.0) (pcDNA3+NTH1-Luc). C, compares the activities of the NTH1-Luc (1.2 kb) reporter in Oct1+/+ versus Oct1−/− MEFs. All values are means ± S.E. of three independent experiments. p values were calculated using two-tailed t tests. *, p < 0.05.
Oct1 Is Required for BRCA1 Protection against H2O2
Next, we tested the ability of BRCA1 overexpression to mediate protection against oxidative stress due to H2O2 in Oct1-deficient versus wild-type MEFs. The survival of H2O2-treated Oct1+/+ cells was higher in WTBRCA1-transfected cells than in empty vector-transfected or vehicle-treated cells (p < 0.05 at three different concentrations of H2O2) (Fig. 10A). In contrast, WTBRCA1-transfected Oct1−/− cells showed only very small increases in survival that were not significant at any concentration of H2O2 (Fig. 10B). Fig. 10C compares the survival of control-transfected Oct1+/+ versus Oct1−/− MEFs. Here, the survival of H2O2-treated Oct1+/+ cells was higher than that of H2O2-treated Oct1−/− cells at all except the highest concentration of H2O2. The BRCA1 levels were similarly increased in WTBRCA1-transfected Oct1+/+ versus Oct1−/− cells, as determined by Western blotting (data not shown). These findings suggest that Oct1 contributes to BRCA1 protection against oxidative stress.
FIGURE 10.
Role of Oct1 in BRCA1 protection against H2O2. Oct1+/+ or Oct1−/− fibroblasts were transfected as indicated and then assayed for sensitivity to H2O2, as described in Fig. 5 legend. Significant differences in cell viability (p < 0.05) between WTBRCA1-transfected and pcDNA3 vector-transfected cells are indicated by an asterisk in A and B. C, compares the survival of pcDNA3-transfected Oct1+/+ versus Oct1−/− fibroblasts.
Oxidative Stress Stimulates BRCA1 and BER Enzyme Protein Expression
Here, T47D cells were treated with H2O2 (250 nm) for 0, 2, or 24 h; and the cells were harvested for Western blotting of BRCA1 and the three BER enzymes. As shown in Fig. 11A, H2O2 caused obvious increases in the protein levels of BRCA1, NTH1, OGG1, and REF1/APE1 (but not actin, the loading control) at the 24-h time point but not the 2-h time point. Consistent with these findings, treatment of T47D cells with 250 nm H2O2 for 24 h also caused an increase in NTH1 incising activity (Fig. 11B). Similar results were observed using a concentration of 500 nm H2O2 (supplemental Fig. S14, A and B, respectively). These results are consistent with the previously described findings that knockdown of BRCA1 reduces the survival of H2O2-treated cells, because the BRCA1-siRNA is expected to block the H2O2-induced increases in BRCA1 levels and, consequently, the increases in BER enzyme protein levels.
FIGURE 11.
Oxidative stress due to H2O2 stimulates BRCA1 and BER enzyme expression. A, subconfluent proliferating T47D cells were treated with H2O2 (250 nm) for 0, 2, or 24 h and then harvested for Western blotting to detect BRCA1, NTH1, OGG1, REF1/APE1, and actin (loading control). B, T47D cells were treated with H2O2 (250 nm) for 24 h and then harvested for NTH1 incising activity assays as described in Fig. 1 legend. nt, nucleotide.
DISCUSSION
A major finding of this study was that BRCA1 overexpression increases and underexpression decreases enzymatic activities related to the repair of oxidative DNA lesions by the BER pathway. Thus, BRCA1 stimulated the ability to incise DNA at sites of 8-oxoguanine, thymidine glycol, and an apurinic site. These early steps in the repair of oxidized DNA are related to the activities of the BER proteins OGG1, NTH1, and REF1/APE1, respectively. Consistent with these findings, BRCA1 increased the expression of each of these proteins. It seems likely that BRCA1 would also stimulate other enzymatic activities of the same proteins due to the increase in total protein levels. BRCA1 also increased the expression of XRCC1, a protein that interacts with polymerase B and DNA ligase III and facilitates the repair of the incised DNA (32). However, BRCA1 did not alter the expression or DNA ligation activity of DNA ligase III, suggesting that BRCA1 primarily stimulates the early steps of the BER pathway.
In a recent publication (35), it was reported that breast cancer cells of the triple negative phenotype (i.e. those negative for estrogen receptor expression, progesterone receptor expression, and HER2/Neu overexpression/amplification), including BRCA1-mutant breast cancer cells, show a reduction in the ability to repair oxidative DNA damage by the BER pathway, although no mechanism was described. The assay used to assess BER pathway activity (repair of photodynamically induced damage to an adenovirus plasmid containing a green fluorescent protein coding sequence) was different from those used in our study. Nevertheless, our results are consistent with the findings reported and further indicate that a BRCA1 mutation is not required to confer a reduction in the BER pathway activity, because underexpression of BRCA1 due to RNA interference had the same effect.
The octamer-binding transcription factor OCT1 (so-called because it binds to a variety of specific DNA sequences of eight bases) and the CCAAT-binding transcription factor NF-YA have previously been implicated as mediators of BRCA1 stimulation of GADD45 promoter activity (34). In this study, we found that OCT1, but not NF-YA, is required for stimulation of BER enzyme expression and activity by BRCA1, based on the use of gene-specific siRNAs. These findings suggest that BRCA1 may function as a co-activator for OCT1-dependent transcription factors. Such a role would fit the pattern observed for BRCA1 regulation of various other transcriptional pathways, in which BRCA1, which is not a sequence-specific DNA binding transcription factor, interacts with a transcription factor and functions as a co-regulator to either inhibit or stimulate its transcriptional activity (10). A role for OCT1 in the regulation of BER protein expression is also consistent with previous studies suggesting that OCT1 functions to regulate the response to stress, including oxidative stress (36, 37).
In the previously cited study, BRCA1 was found to interact with OCT1 and to localize to an OCT1-binding site in the GADD45 promoter (34). Interestingly, in our study, although OCT1-siRNA blocks stimulation of BER enzyme expression and activity due to BRCA1 overexpression, OCT1-siRNA did not significantly reduce the basal BER enzyme expression in the absence of exogenous BRCA1. We propose that under basal conditions, BRCA1 but not OCT1 is the limiting regulatory factor. This hypothesis is consistent with the finding that BRCA1 knockdown did significantly reduce BER enzyme expression and activity. We also note that the OCT1-siRNA only reduced OCT1 protein levels by about 60%, which may not be sufficient to reduce basal enzyme expression levels. However, we did note that the basal levels of NTH1 promoter activity were lower in Oct1−/− fibroblasts than in Oct1+/+ fibroblasts, consistent with the idea that OCT1 does become limiting when the levels are sufficiently low.
The MTT cell viability assays revealed that protection against an oxidizing agent (H2O2) by BRCA1 overexpression was attenuated or abrogated by inhibition or knockdown of REF1, NTH1, or OGG1, consistent with roles for these BER enzymes in cell protection by BRCA1. Our previous studies suggest that in the setting of H2O2 treatment, MTT assays correlate well with trypan blue dye exclusion assays of cell survival (38). Exogenous WT-REF1 rescued the increased sensitivity to H2O2 due to BRCA1 knockdown. Interestingly, BRCA1 overexpression was still able to protect against H2O2 to some degree in cells in which REF1, NTH1, or OGG1 were inhibited or underexpressed. This partial protection by BRCA1 in cells in which a single BER enzyme has been inhibited or knocked down may be related to several factors including the following: 1) the ability of BRCA1 to stimulate the activity of other BER enzymes; and 2) the ability of BRCA1 to stimulate cytoprotective pathways other than DNA repair pathways. Thus, in a previous study, we showed that BRCA1 stimulates the activity of the transcription factor NFE2L2 (NRF2), which mediates the transcription of various antioxidant genes, including oxidoreductases and glutathione S-transferases (38). More recently, we found that BRCA1 functions to reduce the intracellular levels of reactive oxygen species (39). Thus, BRCA1 may protect against oxidative stress by multiple mechanisms, including stimulation of expression of some of the key enzymes that repair oxidative DNA lesions.
Studies utilizing Oct1 null fibroblasts further suggest that Oct1 contributes to the survival of H2O2-treated cells and that Oct1 is required for BRCA1-mediated protection against H2O2. Additional studies utilizing human breast carcinoma cells suggest that oxidative stress due to H2O2 stimulates the expression of BRCA1 and three BER enzymes (NTH1, OGG1, and REF1/APE1) and causes an increase in NTH1 incising activity. These findings help to establish the physiological relevance of BRCA1 overexpression to stimulation of the BER pathway, because they suggest that endogenous BRCA1 protein can regulate the pathway in response to oxidative damage. The latter idea is further supported by the findings that knockdown of endogenous BRCA1 causes a reduction of BER enzyme expression, BER enzyme incising activity, and survival of oxidatively stressed cells.
In addition to the above-described experiment, we note that BRCA1 overexpression may be relevant for other reasons. Thus, the range of increases in BRCA1 levels that we typically observe after transfection of cultured cells (e.g. T47D and MCF-7 used in this study) is about 2–5-fold. This fold-change is the same as or smaller than that observed across different cell lines (some of which have endogenous BRCA1 protein levels much higher than T47D or MCF-7) and during different phases of the cell cycle (40–42). Furthermore, Brca1 expression increases dramatically in mammary epithelial cells during puberty and pregnancy in mice and during mammary epithelial cell differentiation in vitro (43–46). These may be key windows of time during which BRCA1 tumor suppressor function is exerted.
We also note that recent studies have demonstrated a cell cycle or proliferation state dependence in the activity of some DNA repair proteins, including several in the BER pathway (47, 48). However, our previous studies have established that although BRCA1 can contribute to stress-activated cell cycle checkpoints, neither overexpression (WTBRCA1) nor underexpression (BRCA1-siRNA) of BRCA1 significantly altered the in vitro growth rate or cell cycle distribution of established cancer cell lines grown under standard cell culture conditions (Dulbecco's modified Eagle's medium plus 5–10% fetal calf serum) (22, 27, 50). Thus, it is unlikely that alterations in the cell cycle or cell growth state due to BRCA1 contributed significantly to the observed changes in BER enzyme activity or expression in this study.
Because oxidative stress is thought to contribute to age-related and degenerative diseases (51), and because BRCA1 is expressed ubiquitously, our findings raise the possibility that BRCA1 could contribute to noncancerous diseases. To our knowledge, BRCA1 mutations have not been clinically linked to noncancer disease processes. However, we are also unaware if this issue has ever been closely investigated, and such studies should be encouraged. Although BRCA1 has not been definitively linked to the aging process, it is interesting to note that mice heterozygous for Brca1 exhibit a shortened life span (52) and that the loss of the full-length Brca1 isoform in mice contributed to both aging and tumorigenesis (53). Finally, centenarians have been found to exhibit certain differences in germ line BRCA1 polymorphisms relative to control populations, suggesting a possible linked of the BRCA1 genotype to longevity (49).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-CA80000, RO1-CA104546, and R01-CA82599 from USPHS (to E. M. R.). This work was also supported by Grant PDF0403044 from the Susan G. Komen Breast Cancer Foundation (to E. M. R. and J. K. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. S1–S14.
- BER
- base excision repair
- AP
- apurinic/apyrimidinic
- DN
- dominant negative
- siRNA
- small interfering RNA
- TG
- thymine glycol
- FITC
- fluorescein isothiocyanate
- RT
- reverse transcription
- MEF
- mouse embryo fibroblast
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide
- 8-OxoG
- 8-oxoguanine.
REFERENCES
- 1.Hegde M. L., Hazra T. K., Mitra S. (2008) Cell Res. 18, 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Errico M., Parlanti E., Dogliotti E. (2008) Mutat. Res. 659, 4–14 [DOI] [PubMed] [Google Scholar]
- 3.Mitra S., Hazra T. K., Roy R., Ikeda S., Biswas T., Lock J., Boldogh I., Izumi T. (1997) Mol. Cells 7, 305–312 [PubMed] [Google Scholar]
- 4.Izumi T., Wiederhold L. R., Roy G., Roy R., Jaiswal A., Bhakat K. K., Mitra S., Hazra T. K. (2003) Toxicology 193, 43–65 [DOI] [PubMed] [Google Scholar]
- 5.Kelley M. R., Parsons S. H. (2001) Antioxid. Redox. Signal. 3, 671–683 [DOI] [PubMed] [Google Scholar]
- 6.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P. A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L. M., Ding W., Bell R., Rosenthal J., Hussey C., Tran T., McClure M., Frye C., Hattier T., Phelps R., Haugen-Strano A., Katcher H., Yakumo H., Gholami Z., Shaffer D., Stone S., Bayer S., Wray C., Bogden R., Dayananth P., Ward J., Tonin P., Narod S., Bristow P. K., Norris F. H., Helvering L., Morrison P., Rosteck P., Lai M., Barrett J. C., Lewis C., Neuhausen S., Cannon-Albright L., Goldgar D., Wiseman R., Kamb A., Skolnick M. H. (1994) Science 266, 66–71 [DOI] [PubMed] [Google Scholar]
- 7.Staff S., Isola J., Tanner M. (2003) Cancer Res. 63, 4978–4983 [PubMed] [Google Scholar]
- 8.Turner N. C., Reis-Filho J. S., Russell A. M., Springall R. J., Ryder K., Steele D., Savage K., Gillett C. E., Schmitt F. C., Ashworth A., Tutt A. N. (2007) Oncogene 26, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 9.Wilson C. A., Ramos L., Villaseñor M. R., Anders K. H., Press M. F., Clarke K., Karlan B., Chen J. J., Scully R., Livingston D., Zuch R. H., Kanter M. H., Cohen S., Calzone F. J., Slamon D. J. (1999) Nat. Genet. 21, 236–240 [DOI] [PubMed] [Google Scholar]
- 10.Rosen E. M., Fan S., Ma Y. (2006) Cancer Lett. 236, 175–185 [DOI] [PubMed] [Google Scholar]
- 11.Weberpals J. I., Clark-Knowles K. V., Vanderhyden B. C. (2008) J. Clin. Oncol. 26, 3259–3267 [DOI] [PubMed] [Google Scholar]
- 12.Ting N. S., Lee W. H. (2004) DNA Repair 3, 935–944 [DOI] [PubMed] [Google Scholar]
- 13.Moynahan M. E., Chiu J. W., Koller B. H., Jasin M. (1999) Mol. Cell 4, 511–518 [DOI] [PubMed] [Google Scholar]
- 14.Stark J. M., Pierce A. J., Oh J., Pastink A., Jasin M. (2004) Mol. Cell. Biol. 24, 9305–9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arlt M. F., Xu B., Durkin S. G., Casper A. M., Kastan M. B., Glover T. W. (2004) Mol. Cell. Biol. 24, 6701–6709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu B., Kim St., Kastan M. B. (2001) Mol. Cell. Biol. 21, 3445–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litman R., Gupta R., Brosh R. M., Jr., Cantor S. B. (2008) Anticancer Agents Med. Chem. 8, 426–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruffner H., Joazeiro C. A., Hemmati D., Hunter T., Verma I. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5134–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starita L. M., Parvin J. D. (2006) Cancer Biol. Ther. 5, 137–141 [DOI] [PubMed] [Google Scholar]
- 20.Fan S., Ma Y. X., Wang C., Yuan R. Q., Meng Q., Wang J. A., Erdos M., Goldberg I. D., Webb P., Kushner P. J., Pestell R. G., Rosen E. M. (2001) Oncogene 20, 77–87 [DOI] [PubMed] [Google Scholar]
- 21.Fan S., Wang J. A., Yuan R. Q., Ma Y. X., Meng Q., Erdos M. R., Brody L. C., Goldberg I. D., Rosen E. M. (1998) Oncogene 16, 3069–3082 [DOI] [PubMed] [Google Scholar]
- 22.Fan S., Yuan R., Ma Y. X., Meng Q., Goldberg I. D., Rosen E. M. (2001) Oncogene 20, 8215–8235 [DOI] [PubMed] [Google Scholar]
- 23.Wang V. E., Schmidt T., Chen J., Sharp P. A., Tantin D. (2004) Mol. Cell. Biol. 24, 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jayaraman L., Murthy K. G., Zhu C., Curran T., Xanthoudakis S., Prives C. (1997) Genes Dev. 11, 558–570 [DOI] [PubMed] [Google Scholar]
- 25.Seo Y. R., Kelley M. R., Smith M. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14548–14553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goto M., Shinmura K., Igarashi H., Kobayashi M., Konno H., Yamada H., Iwaizumi M., Kageyama S., Tsuneyoshi T., Tsugane S., Sugimura H. (2009) Carcinogenesis 30, 1345–1352 [DOI] [PubMed] [Google Scholar]
- 27.Xiong J., Fan S., Meng Q., Schramm L., Wang C., Bouzahza B., Zhou J., Zafonte B., Goldberg I. D., Haddad B. R., Pestell R. G., Rosen E. M. (2003) Mol. Cell. Biol. 23, 8668–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alley M. C., Scudiero D. A., Monks A., Hursey M. L., Czerwinski M. J., Fine D. L., Abbott B. J., Mayo J. G., Shoemaker R. H., Boyd M. R. (1988) Cancer Res. 48, 589–601 [PubMed] [Google Scholar]
- 29.Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adhikari S., Toretsky J. A., Yuan L., Roy R. (2006) J. Biol. Chem. 281, 29525–29532 [DOI] [PubMed] [Google Scholar]
- 31.Nyaga S. G., Lohani A., Jaruga P., Trzeciak A. R., Dizdaroglu M., Evans M. K. (2006) BMC Cancer 6, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trzeciak A. R., Nyaga S. G., Jaruga P., Lohani A., Dizdaroglu M., Evans M. K. (2004) Carcinogenesis 25, 1359–1370 [DOI] [PubMed] [Google Scholar]
- 33.Yacoub A., Kelley M. R., Deutsch W. A. (1997) Cancer Res. 57, 5457–5459 [PubMed] [Google Scholar]
- 34.Fan W., Jin S., Tong T., Zhao H., Fan F., Antinore M. J., Rajasekaran B., Wu M., Zhan Q. (2002) J. Biol. Chem. 277, 8061–8067 [DOI] [PubMed] [Google Scholar]
- 35.Alli E., Sharma V. B., Sunderesakumar P., Ford J. M. (2009) Cancer Res. 69, 3589–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tantin D., Schild-Poulter C., Wang V., Haché R. J., Sharp P. A. (2005) Cancer Res. 65, 10750–10758 [DOI] [PubMed] [Google Scholar]
- 37.Kang J., Gemberling M., Nakamura M., Whitby F. G., Handa H., Fairbrother W. G., Tantin D. (2009) Genes Dev. 23, 208–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bae I., Fan S., Meng Q., Rih J. K., Kim H. J., Kang H. J., Xu J., Goldberg I. D., Jaiswal A. K., Rosen E. M. (2004) Cancer Res. 64, 7893–7909 [DOI] [PubMed] [Google Scholar]
- 39.Saha T., Rih J. K., Rosen E. M. (2009) FEBS Lett. 583, 1535–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaughn J. P., Davis P. L., Jarboe M. D., Huper G., Evans A. C., Wiseman R. W., Berchuck A., Iglehart J. D., Futreal P. A., Marks J. R. (1996) Cell Growth & Differ. 7, 711–715 [PubMed] [Google Scholar]
- 41.Gudas J. M., Li T., Nguyen H., Jensen D., Rauscher F. J., 3rd, Cowan K. H. (1996) Cell Growth & Differ. 7, 717–723 [PubMed] [Google Scholar]
- 42.Chen Y., Farmer A. A., Chen C. F., Jones D. C., Chen P. L., Lee W. H. (1996) Cancer Res. 56, 3168–3172 [PubMed] [Google Scholar]
- 43.Marquis S. T., Rajan J. V., Wynshaw-Boris A., Xu J., Yin G. Y., Abel K. J., Weber B. L., Chodosh L. A. (1995) Nat. Genet. 11, 17–26 [DOI] [PubMed] [Google Scholar]
- 44.Rajan J. V., Marquis S. T., Gardner H. P., Chodosh L. A. (1997) Dev. Biol. 184, 385–401 [DOI] [PubMed] [Google Scholar]
- 45.Rajan J. V., Wang M., Marquis S. T., Chodosh L. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13078–13083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kubista M., Rosner M., Kubista E., Bernaschek G., Hengstschläger M. (2002) Oncogene 21, 4747–4756 [DOI] [PubMed] [Google Scholar]
- 47.Chaudhry M. A. (2007) Cancer Cell Int. 7, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Windhofer F., Wu W., Wang M., Singh S. K., Saha J., Rosidi B., Iliakis G. (2007) Int. J. Radiat. Oncol. Biol. Phys. 68, 1462–1470 [DOI] [PubMed] [Google Scholar]
- 49.Vijg J., Perls T., Franceschi C., van Orsouw N. J. (2001) Ann. N.Y. Acad. Sci. 928, 85–96 [DOI] [PubMed] [Google Scholar]
- 50.Bae I., Rih J. K., Kim H. J., Kang H. J., Haddad B., Kirilyuk A., Fan S., Avantaggiati M. L., Rosen E. M. (2005) Cell Cycle 4, 1641–1666 [DOI] [PubMed] [Google Scholar]
- 51.Galli F., Piroddi M., Annetti C., Aisa C., Floridi E., Floridi A. (2005) Contrib. Nephrol. 149, 240–260 [DOI] [PubMed] [Google Scholar]
- 52.Jeng Y. M., Cai-Ng S., Li A., Furuta S., Chew H., Chen P. L., Lee E. Y., Lee W. H. (2007) Oncogene 26, 6160–6166 [DOI] [PubMed] [Google Scholar]
- 53.Cao L., Li W., Kim S., Brodie S. G., Deng C. X. (2003) Genes Dev. 17, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.