Abstract
Microtubule-associated protein Tau is known to bind to and stabilize microtubules, thereby regulating microtubule dynamics. However, recent evidence has indicated that Tau can also interact with various components of intracellular signaling pathways, leading to the possibility that Tau might have a role in signal transduction. Here we provide evidence that during growth factor stimulation of neuronal cells, Tau has functions in advance of the neurite elongation stage. Using Tau-depleted neuronal cell lines, we demonstrate that Tau is required for neurite initiation in a manner that does not involve its microtubule binding function. In addition, we demonstrate that Tau potentiates AP-1 transcription factor activation in response to nerve growth factor (NGF). The effect of Tau on AP-1 activation is mediated through its ability to potentiate the activation of mitogen-activated protein kinase (MAPK), which occurs in response to both NGF and epidermal growth factor. Phosphorylation of Tau at Thr-231 also occurs in response to NGF and is required for Tau to impact on MAPK signaling, whereas the ability of Tau to bind to microtubules is not required. Together, these findings indicate a new functional role for Tau in early neuronal development independent of its established role in microtubule stabilization.
Keywords: AP-1 Transcription Factor, MAP Kinases (MAPKs), Neurodifferentiation, Protein Phosphorylation, Tau, Epidermal Growth Factor, Nerve Growth Factor, Neurite Outgrowth
Introduction
Microtubule-associated proteins are a class of proteins that includes Tau, MAP2, and MAP4. These proteins are so named for their ability to bind to and stabilize microtubules, thereby contributing to the regulation of microtubule dynamics. Tau in particular has been extensively studied due to its prominent role in the pathogenesis of neurodegenerative diseases such as Alzheimer disease and the fronto-temporal dementias (reviewed in Refs. 1–4). In general, research into the physiological functions of Tau has focused on its interactions with microtubules. However, additional roles for Tau beyond those associated with microtubules have been suggested by numerous studies. We have previously demonstrated that the amino terminus of Tau, a domain not involved in microtubule binding, is associated with the plasma membrane and can affect nerve growth factor (NGF)2-induced neurite outgrowth (5). Also, neurite outgrowth was shown to require phosphorylation of Tau at Ser-262, a modification that reduces the ability of Tau to bind to microtubules (6). In addition, interactions between Tau and various signaling proteins such as Src, Fyn, Abl, the p85 subunit of phosphatidylinositol 3-kinase, phospholipase Cγ, 14-3-3, and Grb2 have been described (7–12). Such findings suggest that non-microtubule-associated Tau species may be associated with signaling components at the plasma membrane during early neurogenesis and differentiation.
Recent reports have also indicated that Tau can be linked to the increased expression of cell cycle proteins. Mice expressing human Tau in the absence of mouse Tau and mice expressing FTDP-17 mutant Tau were found to have abnormal expression of cell cycle proteins (13–15) and a cell culture model expressing P301L mutant Tau was found to up-regulate genes associated with cell cycle re-entry (16). These findings, combined with the growing body of data showing that Tau interacts with signaling proteins, led us to further investigate the potential role of Tau in neuronal signal transduction.
Here we have used the well studied PC12 system to investigate the role of Tau in early events following NGF stimulation. In undifferentiated PC12 cells, Tau is expressed at low levels, whereas stimulation with NGF leads to increases in Tau expression starting after ∼3 days of treatment (17, 18). The subcellular distribution of Tau in PC12 has been investigated by its detergent solubility (19). This study showed that before NGF treatment, the cells contained approximately half of the Tau in cytoskeletal, detergent-insoluble pellets, whereas the other half was detergent soluble. This indicated that in undifferentiated PC12, half of the endogenous Tau was not associated with microtubules. Tau is known to be required for microtubule stabilization at later stages of NGF-induced neurite elongation (5, 18, 19), but the functional role of microtubule-free Tau present before NGF and at the early stages of NGF stimulation remains unclear.
In this report, we demonstrate that after NGF stimulation of PC12-derived cell lines, Tau is required for neurite initiation. Moreover, Tau acts to potentiate activation of AP-1 transcription factors through the mitogen-activated protein kinase (MAPK) pathway. The ability of Tau to potentiate MAPK activation occurs independently of its ability to bind to microtubules and our data indicates that Tau acts through a mechanism that is common to both NGF and epidermal growth factor (EGF) activated pathways and likely to be upstream of Ras GTPase. Furthermore, we show that phosphorylation of Tau at Thr-231, a phospho-site found in both fetal brain development and neurodegenerative disease, occurs in response to NGF and is required for this activity. These findings indicate a new role for Tau in the context of neuronal differentiation, independent of its established role as a microtubule stabilizing protein.
EXPERIMENTAL PROCEDURES
Cell Culture
COS7 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. PC6-3 cells (20) were cultured on collagen (BD Biosciences)-coated dishes using RPMI 1640 medium with 10% horse serum and 5% fetal bovine serum. Medium for stable cell lines was supplemented with 200 μg/ml of G418.
Generation of Stable Cell Lines
Clonal PC6-3 cell lines stably expressing human Tau (352 residue, 0N3R isoform) were generated by transfecting PC6-3 cells with pRc/CMVn123c (21), followed by selection in 500 μg/ml of G418. Stable colonies were screened for human Tau expression by Western blotting with Tau13, a human Tau-specific antibody (22). Clonality was confirmed by immunofluorescent analysis. Relative Tau expression level was quantified by densitometry (NIH ImageJ analysis software) on Western blots probed with Tau5, which reacts to both human and rat Tau (23). To develop PC6-3 cell lines with stable depletion of endogenous Tau, we first identified candidate RNA interference sequences in rat Tau that had several mismatches with the corresponding human Tau sequence, aiming to specifically target rat Tau. A panel of candidate rat dsRNA oligonucleotides was generated with the Ampliscribe T7 polymerase kit as previously described (24) and screened in COS7 cells by transient co-transfection of rat or human Tau expression plasmids (1 μg) and increasing quantities (0–5 μg) of dsRNA oligonucleotides. Rat Tau plasmid encoded the 2N4R rat Tau isoform (5) and, as described above, pRc/CMVn123c (also referred to as “hTau”) encoded 0N3R human Tau. Based on the results from our screening, the sequence (5′-gatccccGTGTCCGCCTCTTTGGCCAttcaagagaTGGCCAAAGAGGCGGACACtttttggaaa-3′) was synthesized and subcloned into the pNTO vector (25), generously provided by Dr. Stefan Strack (University of Iowa). The short hairpin RNA (shRNA) expressed from this plasmid (pNTO-Rtau) targets a sequence in rat Tau exon 13 (5′-GTGTCCGCCTCTTTGGCCA-3′). pNTO-Rtau was transfected into PC6-3 and stable cell lines were selected as described above. Empty vector control cell lines (EV) were also generated by transfection with the pNTO vector. Extent of Tau depletion was determined by Western blotting with Tau5, using lysates harvested from cells that had been induced with 100 ng/ml of NGF (Research Diagnostics, Flanders, NJ) for 3 days. Table 1 summarizes the characteristics of all PC6-3 cell lines, with and without altered Tau expression, used in this study.
TABLE 1.
Characteristics of PC6-3 cell lines with altered Tau expression
| Cell line | Parental line | Stably expressed plasmid | Tau expression levels |
|---|---|---|---|
| PC6-3 | None | Control | |
| D5 | PC6-3 | pRc/CMVn123c | 0N3R human Tau was expressed 4-fold higher than endogenous Tau in undifferentiated cells and 2-fold higher after differentiation |
| rTau4 | PC6-3 | pNTO-Rtau | No Tau was detected in undifferentiated or differentiated cells |
| rTau7 | PC6-3 | pNTO-Rtau | No Tau was detected in undifferentiated cells and Tau expression was 15% of control cell level after differentiation |
| EV | PC6-3 | pNTO Vector | Tau levels were equivalent to parental PC6-3 cells, with or without NGF |
Tau expression levels were determined by quantitative Western blotting, with or without NGF (3 days at 100 ng/ml).
Neurite Initiation Assays
PC6-3 cells and stable cell lines were differentiated in 100 ng/ml of NGF (Research Diagnostics) for 36 h prior to glutaraldehyde fixation and permeabilization with Nonidet P-40 (26). In some experiments, 0.1 μm taxol (Calbiochem) was added simultaneously with NGF. After fixation, cells were stained with anti-tubulin (DM1A, Sigma), rhodamine-coupled anti-mouse secondary antibody, and viewed by epifluorescence. Neurite initiation for each cell was scored as the number of processes with a length greater than one cell diameter extending from the cell body. Random fields of cells were evaluated and a minimum of 200 cells per cell line was assessed in each experiment. For each experiment, neurite initiation was calculated as the average number of neurites per cell and from three independent experiments, neurite initiation was expressed as the mean ± S.E. For neurite initiation assays performed on transfected cells, Tau-depleted cells were transfected with Lipofectamine 2000 (Invitrogen) and hTau or S262D/S356D mutant Tau for 24 h prior to NGF or NGF-taxol addition. Thirty-six hours after NGF addition, cells were fixed and double-labeled with anti-tubulin (DM1A) and polyclonal anti-Tau (CR (27)). A minimum of 200 CR-positive cells was scored for neurite initiation in each experiment. Confocal microscopy was performed on a Bio-Rad MRC-1024 system with a Nikon E600 microscope. Statistical analysis of variance and post-hoc testing for neurite initiation assays was performed as with luciferase assays below (see “Statistical Analysis”).
AP-1 and MAPK Reporter Assays
The level of endogenous AP-1 transcription factor activity was measured by an AP-1 reporter plasmid 3X-AP-1-Luc (generously provided by Dr. Paul Rothman, University of Iowa), which contains three AP-1 consensus binding sites (TGACTAA), in tandem, upstream of a minimal murine Fos promoter regulating the expression of firefly luciferase. The internal transfection control plasmid, pRL-SV40 (Promega), expresses Renilla luciferase under the control of the SV40 early promoter. Used together, these two plasmids will be referred to as the “AP-1 reporter system plasmids.”
For MAPK activation assays, the PathDetect Trans-Reporting System (Stratagene), comprising pFR-Luc and pFA2-ELK1, was used according to the manufacturer's protocol with the addition of the internal transfection control pRL-SV40 Renilla luciferase plasmid. Used together, these three plasmids will be referred to as the “MAPK reporter system plasmids.”
Cells were grown on 24-well collagen-coated plates to ∼50% confluency and transfections were performed in triplicate with Lipofectamine 2000 (Invitrogen). For AP-1 assays, cells were transfected with 1.1 μg of DNA (500 ng of 3X-AP-1-Luc, 100 ng of pRL-SV40, and 500 ng of either pRc/CMV control vector or hTau). For MAPK assays, cells were transfected with 1.1 μg of DNA (500 ng of pFR-Luc, 50 ng of pFA2-ELK1, 50 ng of pRL-SV40, and 500 ng of either pRc/CMV control vector or Tau plasmid (hTau, S262D/S356D, T231D/S235D, T231A/S235A, T231D, S235D, or T231A)). The pRc/CMV control vector was used as a control for human Tau plasmids and to maintain equivalent amounts of total DNA in each transfection.
NGF (2.5S, Sigma) and EGF (Sigma) treatments were carried out 36–48 h after transfection at 50 and 25 ng/ml, respectively. For both AP-1 and MAPK reporter assays, a time course of growth factor treatment of up to 24 h was carried out in preliminary experiments to determine the point of maximum reporter activation. In both assays, a 3-h growth factor induction proved to have the highest amount of reporter activity and therefore, this time point was used in all subsequent experiments. Cells were harvested and AP-1 (or MAPK) activation was assayed using the Dual Luciferase Assay Kit (Promega) according to the manufacturer's protocol, measuring Firefly and Renilla luciferase activities with a tube luminometer. For data analysis, firefly luciferase values were first normalized to Renilla luciferase values from the same sample to control for transfection efficiency. To calculate the fold-increase in reporter activity after growth factor treatment, the normalized firefly luciferase activity from the growth factor-stimulated sample was divided by the normalized firefly luciferase activity from the non-stimulated control cells.
For experiments with the MEK1 inhibitor, 50 μm U0126 (Promega) or dimethyl sulfoxide vehicle control was added to the cells 15 min prior to NGF treatment. For experiments with oncogenic Ras (G12V mutant (28)), cells were co-transfected with MAPK reporter system plasmids and FLAG-RasV12 (generously provided by Dr. Stefan Strack), and harvested after 36 h in the absence of growth factors. The amount of FLAG-RasV12 DNA used was determined by preliminary experiments indicating the amount of plasmid required to yield reporter activation levels similar to those present after a 3-h NGF treatment. Fold-increase in MAPK reporter activity was calculated by dividing the normalized firefly luciferase reading from the RasV12 containing condition with the normalized firefly luciferase reading from the control vector containing condition.
Statistical Analysis
For AP-1 and MAPK luciferase assays, the results for each condition were reported as mean ± S.E. from three independent assays. In addition, each assay used transfections that were performed in triplicate. Statistical significance was determined by analysis of variance (linear mixed model) with the Statistical Analysis System software package. Reporter activity from all assays (n = 3) was analyzed as the random effect, with each cell line/Tau transfection/treatment as the fixed effect. In all figures, the data for each condition are shown as the mean from all assays ± S.E. However, for statistical analysis, the data were log transformed to account for proportional differences between groups. All p values <0.05 calculated from post-hoc Tukey comparisons between groups were considered to be statistically significant.
Plasmids
Plasmids expressing mutant Tau (T231D, S235D, T231A/S235A, and T231A, numbered according to 2N4R hTau40 human Tau isoform) were synthesized by site-directed mutagenesis (QuikChange Mutagenesis Kit, Stratagene) using the 0N3R human Tau plasmid, pRc/CMVn123c, as template (21). Plasmids expressing the 0N3R human Tau mutants S262D/S356D and T231D/S235D have been previously described (29).
Western Blotting
Parental PC6-3 cells and stable cell lines were treated with NGF or EGF as above for various time points and lysed in 2× Laemmli buffer containing 10% β-mercaptoethanol. Samples were resolved in 8% SDS-PAGE gels, transferred to polyvinylidene difluoride (Millipore), and probed with one of the following antibodies: Tau5, Tau13 (generous gifts from Dr. Lester Binder), phospho-ERK1/2 (Thr202/Tyr204, Cell Signaling 9101), ERK1/2 (Santa Cruz K-23), or glyceraldehyde-3-phosphate dehydrogenase (Chemicon). PerkinElmer Western Lightning Plus-ECL was used for signal detection. Quantification of phospho-ERK1/2 and total ERK1/2 signals was performed by densitometry with ImageJ software. The phospho-ERK1/2 level at each time point was normalized to the total ERK1/2 level from the same sample and each normalized value was expressed as a percentage relative to the highest value, which was assigned 100%. Statistical significance was determined by Student's t test. Expression levels of the Tau mutants used in the MAPK reporter assays were determined by probing lysates of rTau4 cells transfected under identical conditions used for the MAPK reporter assays. Lysates were harvested from 24-well plates and probed with Tau13 and glyceraldehyde-3-phosphate dehydrogenase.
Immunoprecipitation
PC6-3 cells stably expressing 0N3R human Tau (D5) were plated in 150-mm dishes and serum starved overnight prior to growth factor treatment (50 ng/ml of NGF, Sigma). Cells were lysed in 1 ml of ice-cold RIPA buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS) containing protease and phosphatase inhibitors. After centrifugation at 6,200 × g for 20 min at 4 °C, Nonidet P-40 was increased to 1.5% and supernatants were precleared with protein G-Sepharose beads for 20 min. The resulting supernatants were then incubated with either 3.5 μg of nonspecific mouse IgG or CP17 (anti-phospho-Thr-231 Tau, generously provided by Dr. Peter Davies (30)) for 1 h. Protein G-Sepharose beads were added to the supernatants for an additional 1 h. Immunoprecipitates were resolved by SDS-PAGE and transferred. Total Tau (human and rat) was detected with Tau5-horseradish peroxidase (1:4000), prepared using EZ-Link Plus Activated Peroxidase Kit (Pierce).
RESULTS
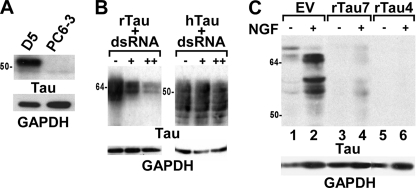
Stable cell lines that either overexpress Tau or have Tau depletion were developed in PC6-3 cells, a subclone of the well characterized PC12 rat pheochromocytoma cell line (20). Four clonal cell lines that stably express the fetal isoform of human Tau (352 residues, 0N3R) were generated and one representative cell line, D5, was used for further studies (Fig. 1A). Quantification of human Tau expression in D5 showed that in undifferentiated cells, the human Tau level was 4-fold higher than the endogenous rat Tau level (Table 1). To generate cell lines with Tau depletion, we first identified a rat Tau dsRNA oligonucleotide that would effectively suppress rat Tau expression without affecting human Tau expression using a COS7 co-transfection assay (Fig. 1B). A vector expressing the rat Tau dsRNA sequence as a shRNA was then used to generate stably transfected PC6-3 lines. Control cell lines were generated using the empty vector. To determine the extent of Tau depletion, cells were first differentiated with NGF for 3 days to up-regulate Tau expression. Like PC12 cells that are known to increase Tau expression after NGF treatment (17), the control EV cell line also up-regulated Tau after NGF differentiation (Fig. 1C, lanes 1 and 2). Other control cell lines acted similarly. In contrast, the Tau-depleted cell line rTau4 showed almost complete depletion of endogenous rat Tau after NGF-induced differentiation (Table 1 and Fig. 1C, lane 6), whereas the cell line rTau7 showed an intermediate level of Tau depletion (Table 1 and Fig. 1C, lane 4). In the undifferentiated state, both rTau4 and rTau7 had less Tau than EV (Table 1 and Fig. 1C, lanes 1, 3, and 5).
FIGURE 1.
Generation of stable cell lines with Tau overexpression or depletion. A, cell lysates from D5, a representative cell line with stable expression of human Tau, and from control PC6-3 cells were probed with a human Tau specific antibody, Tau13. B, cell lysates from COS7 cells co-transfected with increasing amounts of rat Tau-specific dsRNA oligonucleotides and either rat Tau (rTau, left panel) or human Tau (hTau, right panel) plasmids, were probed with Tau5. “−, +, and ++” indicate the addition of 0, 3, or 5 μg, respectively, dsRNA. C, cell lysates from cell lines stably expressing shRNA against endogenous rat Tau (rTau4, rTau7), or from an empty pNTO vector control cell line (EV) were harvested before or after NGF treatment for 3 days as indicated. Lysates were probed with Tau5. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control on all panels. 1 μg of lysate was loaded in each lane in A–C.
Tau Depletion Impairs Early Neurite Initiation during the NGF Response
The differentiation of PC12 cells in response to NGF involves an early stage of neurite initiation in which a nascent process buds from the cell body, followed by a period of process elongation and stabilization. The rate of neurite elongation and neurite stability depend on the NGF-induced up-regulation of Tau that occurs 3 days after NGF addition (17–19). However, Tau is present at low levels before NGF addition as well as during the earliest phase of the NGF response. To determine whether these low levels of Tau have a role in the early NGF response, we first investigated neurite initiation. Neurite initiation takes place before the up-regulation of Tau because process budding and nascent neurites are already visible in many cells within 36 h after NGF addition.
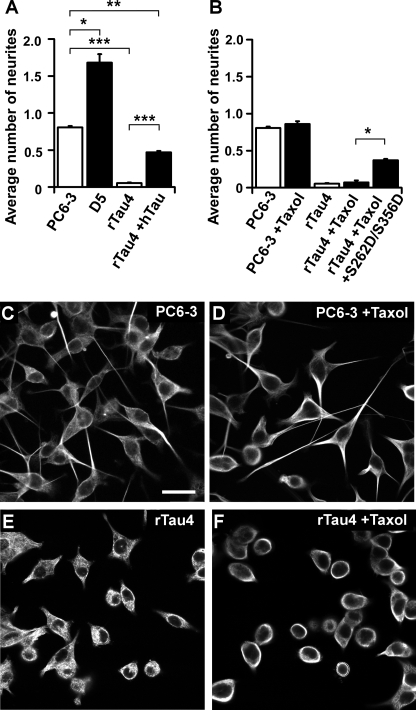
To determine the effect of Tau on neurite initiation, the morphologies of Tau-depleted and Tau overexpressing cell lines were evaluated after a 36-h NGF induction, and processes greater than one cell diameter were scored as neurites. Neurite initiation, represented by the average number of neurites per cell, was decreased in Tau-depleted rTau4 cells compared with control PC6-3 cells (Fig. 2, A, C, and E), whereas in Tau overexpressing D5 cells, the average number of neurites per cell was increased (Fig. 2A). Similar results were obtained with the Tau-depleted rTau7 cell line and with cells evaluated at an earlier time point (20 h, data not shown). Neurite initiation could also be restored in rTau4 after expressing human Tau by transient transfection (Fig. 2A), although rTau4 rescued with human Tau exhibited 42% less neurite initiation than PC6-3 (**, p < 0.01, Fig. 2A). These results suggest a role for Tau in events leading to neurite initiation.
FIGURE 2.
Tau expression level affects early neurite initiation induced by NGF. A, average number of neurites per cell in PC6-3 or in stable cell lines D5 or rTau4 was determined after a 36-h NGF differentiation. rTau4 +hTau denotes rTau4 transfected with hTau and scored for neurite initiation as described under “Experimental Procedures.” Results are shown as the average ± S.E. from three independent experiments (*, p < 0.05; **, p < 0.01; ***, p < 0.005). B, average number of neurites per cell was determined as in A for PC6-3 or rTau4 cells differentiated in the presence of NGF, with or without the addition of 0.1 μm taxol. rTau4+taxol+S262D/S356D denotes rTau4 transfected with the S262D/S356D mutant Tau plasmid 24 h prior to simultaneous taxol-NGF treatment. Neurite initiation was scored and results are shown as described in A (*, p < 0.05). C–F, PC6-3 or rTau4, differentiated with NGF for 36 h in the presence or absence of taxol as indicated, were fixed, labeled with anti-tubulin, and viewed by confocal microscopy as described under “Experimental Procedures.” Scale bar = 10 μm.
One possible explanation for the observed effect of Tau depletion on neurite initiation is a requirement for Tau in promoting microtubule assembly during process initiation. To address this possibility, we tested the ability of the microtubule stabilizing drug taxol to restore process initiation during NGF differentiation in rTau4. We have previously shown that a low concentration of taxol (0.1 μm), which allows microtubules to retain dynamicity (31), mimics the effects of Tau expression on process outgrowth in PC12 cells treated with cytochalasin (26). Therefore, taxol treatment would be expected to restore neurite initiation in Tau-depleted cells if Tau was required solely for microtubule stabilization during the early phase of process formation. Subsequently, we found that although taxol treatment did not affect neurite initiation in control PC6-3 cells, it did not increase the average number of neurites per cell in rTau4 after NGF differentiation (Fig. 2, D and F, quantification in Fig. 2B). Because process initiation in Tau-depleted cell lines could not be restored by providing a microtubule stabilizing agent, these findings suggested that a Tau function other than microtubule stabilization was involved in neurite initiation. This was further investigated by testing a phosphomimetic Tau mutant, S262D/S356D, which has a significantly reduced affinity for microtubules both in vitro and in cells (29, 32–34) (supplemental Fig. S1). By expressing Tau mutant S262D/S356D in Tau-depleted cells and then scoring for its ability to restore neurite initiation, we found that the mutant restored neurite initiation in taxol-treated rTau4 cells 7-fold (Fig. 2B). This was similar to the 8.8-fold increase obtained when neurite initiation was restored to rTau4 cells by expressing wild-type Tau (Fig. 2A) and indicated that the requirement for Tau during neurite initiation was not likely to involve its ability to interact with microtubules.
Tau Levels Affect the DNA Binding Activity of AP-1 Transcription Factors
In PC12 cells, during the first 36 h of NGF treatment, a sustained activation of ERKs (MAPK) and a consequent up-regulation of the AP-1 transcription factor c-Fos accompanies neurite outgrowth (35). Because gene expression profiling of Tau-deficient mice had indicated that Tau depletion affected the expression of AP-1 transcription factors (36), we questioned whether Tau would have a role in NGF-induced AP-1 activity during the early phase of NGF treatment. To investigate this possibility, we assayed AP-1 activity with a luciferase reporter system; activity was measured after 3 h of NGF treatment as described under “Experimental Procedures.”
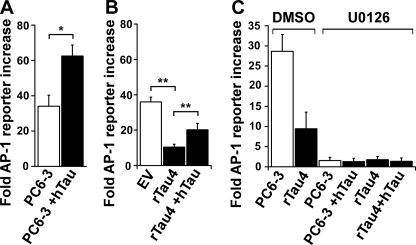
As expected, control PC6-3 cells showed a 34-fold increase in AP-1 activity over non-stimulated cells following NGF treatment (Fig. 3A). Interestingly, PC6-3 cells transiently overexpressing wild-type human Tau (PC6-3 + hTau) were found to have a significant increase in NGF-induced AP-1 activity (1.8-fold) relative to control cells, whereas the Tau-depleted line rTau4 had a significant decrease in AP-1 activity (3.5-fold) when compared with control EV cells (Fig. 3B). Moreover, transient expression of human Tau in rTau4 was sufficient to significantly increase AP-1 activity (2.0-fold) relative to control rTau4 (Fig. 3B; note that rTau4 + hTau was 44% less than EV). Similar results were obtained in rTau7 (2.7-fold decrease relative to EV), as well as in assays performed at later time points of NGF treatment (12 and 24 h, data not shown). Together, these data indicated that Tau potentiates the NGF-mediated activation of AP-1 transcription factor DNA binding activity.
FIGURE 3.
Tau expression level affects DNA binding activity of AP-1 transcription factors. A, PC6-3 cells were transfected with AP-1 reporter system plasmids and either control vector (PC6-3) or human Tau (PC6-3 +hTau). 36 h after transfection, NGF was added for 3 h prior to harvesting. Fold-increase in reporter activity relative to minus NGF (naive) values was calculated for each cell line or condition as described under “Experimental Procedures” (*, p < 0.01). B, control (EV) and Tau-depleted rTau4 cells were transfected with AP-1 reporter system plasmids and hTau as indicated (EV control and rTau4 received control vector DNA). NGF addition and reporter activity was assayed and expressed as in A (**, p < 0.005). C, PC6-3 and rTau4 were transfected as in A, adding hTau as indicated (controls received control vector DNA). Where indicated, MEK1 inhibitor (U0126, 50 μm) or vehicle control (DMSO, dimethyl sulfoxide) was added to the cells 15 min before induction with NGF as above. Reporter activity was assayed and expressed as in A. For A–C, data shown are the mean ± S.E. from three independent experiments; for each experiment, transfections were performed in triplicate for each condition.
Tau Affects AP-1 Activation through the MAPK Pathway
AP-1 activity in response to NGF can be regulated through the Ras/MAPK pathway (35). To determine whether Tau affected the MAPK pathway, cells were treated with 50 μm U0126, which inhibits the dual specificity kinase MEK1 (MAPKK) that is responsible for activating ERK1/2 (MAPK). Pre-treatment of cells with U0126 almost completely abrogated AP-1 reporter activity in both control PC6-3 and rTau4 (Fig. 3C). Furthermore, the presence of human Tau in either cell line, which had previously increased AP-1 activity (1.8–2.0-fold), failed to increase AP-1 activity when the cells were pre-treated with U0126. These data implicate the Ras/MAPK pathway in mediating the effect of Tau on AP-1 activity.
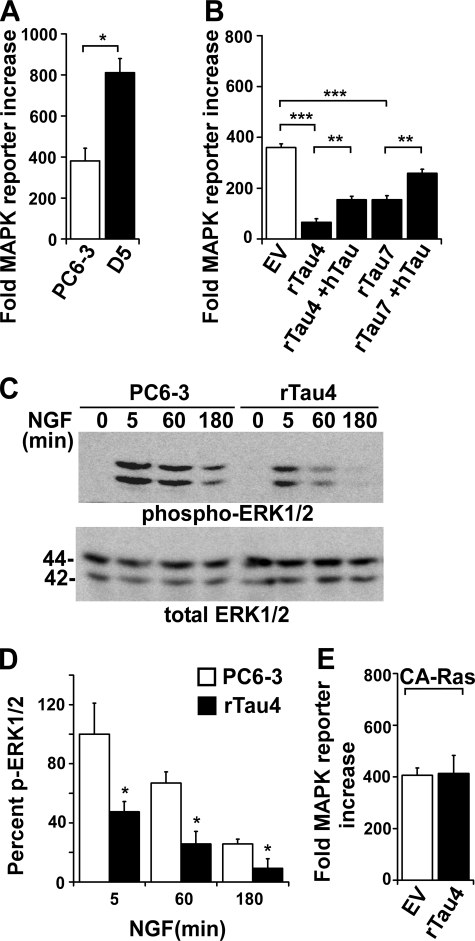
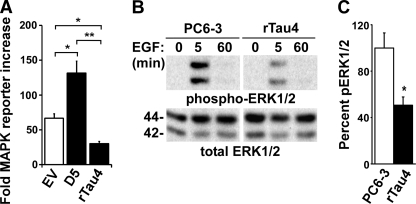
Accordingly, we tested the effects of Tau on the activation of MAPK using a MAPK luciferase reporter system to measure changes in ERK1/2 activation. We first compared control PC6-3 cells to PC6-3 cells overexpressing wild-type human Tau and found that overexpression of human Tau resulted in a significant increase in MAPK reporter activity (2.1-fold, Fig. 4A). In contrast, rTau4 and rTau7 showed a significant decrease in MAPK activation compared with EV (Fig. 4B) and PC6-3 (data not shown). In addition, when wild-type human Tau was expressed in rTau4 or rTau7, although the restored level was 43% or 72%, respectively, of the EV level, both showed a significant recovery of reporter activity. These results indicated that, as with AP-1 activation, Tau expression was capable of potentiating MAPK signaling activity (Fig. 4B).
FIGURE 4.
Tau potentiates signaling through the Ras/MAPK pathway. A, PC6-3 and cells stably expressing human Tau (D5) were transfected with MAPK reporter system plasmids. 36 h after transfection, NGF was added for 3 h prior to cell harvest. Fold-reporter increase was calculated relative to minus NGF (naive) control as described under “Experimental Procedures” (*, p < 0.05). B, MAPK activation in response to NGF was assayed in control EV cells, rTau4, and rTau7 as described in A. +hTau indicates the addition of hTau plasmid; other assays received control vector DNA. (**, p < 0.005; ***, p < 0.0001). For panels A and B, data shown are the mean ± S.E. from three independent experiments; for each experiment, transfections were performed in triplicate for each condition. C, cell lysates from PC6-3 and rTau4 were harvested after treatment with NGF (50 ng/ml) for the indicated time points and probed for activated ERK1/2, using anti-phospho-ERK1/2 (pERK1/2), and total ERK1/2. The blot shown is representative of three experiments. 1 μg of lysate was loaded in each lane. D, quantification of immunoblot data shown in C. The ratio of phospho-ERK/total ERK for each condition was calculated and shown relative to the highest value obtained (PC6-3, 5 min), which was assigned 100 (*, indicates p < 0.05 relative to PC6-3). E, control EV and rTau4 cells were transfected with MAPK reporter system plasmids and constitutively active RasV12 (CA-Ras) plasmid. Reporter activity was assayed as described in A 36 h after transfection in the absence of growth factor induction. Fold-increase was calculated relative to control cells that did not receive RasV12.
An analysis of cell lysates harvested after 5, 60, and 180 min of NGF induction showed that the level of active ERK1/2, as detected by anti-phospho-ERK1/2, was consistently reduced in rTau4 compared with PC6-3 (Fig. 4C). Quantification showed significant decreases in ERK activation at all time points tested (average decrease 59.7%), whereas the overall time course of activation was similar in both cell lines (Fig. 4D).
To further probe the interaction of Tau with components of the Ras/MAPK pathway, we expressed a constitutively active Ras protein (G12V mutant) in EV and rTau4 cells in the absence of NGF stimulation, using conditions that would elicit activated MAPK levels in EV similar to those obtained with NGF stimulation. MAPK reporter activity in rTau4 expressing RasV12 was not impaired relative to similarly transfected EV (Fig. 4E), suggesting that the effect of Tau on MAPK activation occurred upstream of Ras activation.
Tau Expression Level Affects MAPK Signaling Induced by EGF
Ras activation results from a cascade of phosphorylation events that can be triggered by a number of growth factors. Because Tau affects MAPK signaling activated by NGF, Tau might also affect signaling induced by other growth factors. To investigate, we treated our cell lines with EGF, which also activates the Ras/MAPK pathway; under our conditions, the magnitude of reporter activity was less than that obtained with NGF (Fig. 5A). Nevertheless, we found that in response to EGF, Tau overexpression caused an increase in MAPK reporter activity, whereas Tau depletion resulted in a significant decrease in reporter activity (Fig. 5A). An analysis of cell lysates collected after EGF induction similarly showed a significant decrease in active ERK1/2 in rTau4 relative to PC6-3 after 5 min (Fig. 5B, with quantification in Fig. 5C). However, the pattern of ERK1/2 activation did not change, showing that the NGF and EGF responses were distinct. Moreover, EGF treatment did not lead to neurite initiation in D5 (data not shown). These findings indicated that whereas Tau is able to enhance the levels of EGF-induced MAPK signaling, it is unable to lengthen the time course of MAPK activation associated with neurite outgrowth. The data suggests that the effects of Tau occurred at a point that is downstream of the receptor and common to both NGF and EGF signaling pathways.
FIGURE 5.
EGF-induced MAPK activity is affected by Tau expression level. A, EV, D5, and rTau4 cells were transfected with MAPK reporter system plasmids. 36 h after transfection, EGF was added 3 h prior to cell harvest. Fold-MAPK reporter increase was calculated as described in the legend to Fig. 4A and the data shown are the mean ± S.E. from three independent experiments; for each experiment, transfections were performed in triplicate for each condition (*, p < 0.05; **, p < 0.005). B, PC6-3 and rTau4 cell lysates were collected after treatment with EGF (25 ng/ml) for the indicated time points and probed for phospho-ERK1/2 and total ERK1/2. The blot shown is representative of three experiments. 1 μg of lysate was loaded in each lane. C, quantification of phospho-ERK1/2 at the 5-min EGF stimulation shown in B. Ratio of phospho-ERK/total ERK was calculated and shown relative to the value obtained for PC6-3, which was assigned 100 (*, p < 0.05 relative to PC6-3).
Effects of Tau Phosphorylation on Its Ability to Potentiate MAPK Activation
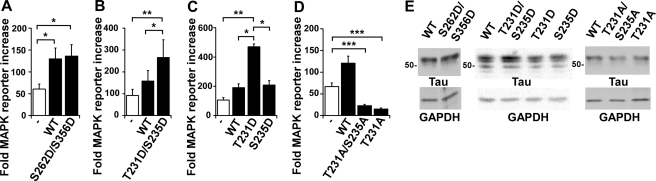
Because the role of Tau in neuronal development is often thought to involve its ability to bind to microtubules, we asked whether this activity would be necessary for Tau to affect MAPK signaling by testing the phosphomimetic Tau mutant, S262D/S356D. We observed that expression of S262D/S356D Tau in rTau4 was able to restore NGF-induced MAPK reporter activity levels to those obtained using wild-type Tau (Fig. 6A), indicating that microtubule binding was not necessary for Tau to affect signaling.
FIGURE 6.
Phosphorylation of Tau at Thr-231 is required for Tau to potentiate MAPK signaling. A, rTau4 was transfected with MAPK reporter system plasmids and either hTau (WT) or S262D/S356D mutant plasmid (rTau4 control received control vector DNA). 36 h after transfection, NGF was added 3 h prior to cell harvest. Fold-MAPK reporter increase was calculated as described in the legend to Fig. 4A (*, p < 0.05 relative to rTau4 control). rTau4 control is indicated by the white bar in A–D. For B–D, rTau4 was transfected with MAPK reporter system plasmids and either hTau (WT) or Tau mutants as indicated (rTau4 control received control vector DNA). NGF treatment and MAPK activation were performed as described in A (*, p < 0.05; **, p < 0.005; ***, p < 0.001). For A–D, data shown are mean ± S.E. from three independent experiments; for each experiment, transfections were performed in triplicate for each condition. E, to assure equal Tau expression in transfections assayed in A–D, lysates from rTau4 cells transfected under identical conditions were probed with Tau13. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels are shown as a loading control. 1 μg of lysate was loaded in each lane.
Phosphorylation of Tau at Thr-231/Ser-235 has also been shown to reduce its affinity for microtubules (37, 38), in addition to reducing its interactions with the SH3 domains of Src family tyrosine kinases (7, 39, 40). Surprisingly, expression of T231D/S235D Tau in rTau4 was not only able to restore MAPK reporter activity but increased the activity level 1.7-fold relative to the level obtained with wild-type Tau (Fig. 6B). This suggested that Tau phosphorylation at Thr-231/Ser-235 might be involved in the interaction of Tau with the MAPK pathway and further confirmed that microtubule interactions were not required. We subsequently tested Tau mutants with single amino acid replacements at either Thr-231 or Ser-235 and found that the T231D mutation was sufficient to reproduce the effects seen with the double mutant T231D/S235D, whereas the S235D mutant resembled wild-type Tau in its ability to restore MAPK reporter activity (Fig. 6C). To corroborate these findings, we tested both T231A/S235A and T231A mutants and found that neither restored NGF-induced MAPK reporter activity (Fig. 6D). In fact, these mutants impaired MAPK signaling relative to rTau4, suggesting that these mutants have a dominant-negative effect. As T231A Tau is able to bind microtubules well (41, 42), this finding also underscored the dissociation between microtubule binding activity and MAPK potentiation activity. Most likely, phosphorylation of Tau at Thr-231 is required for the interaction with the MAPK pathway.
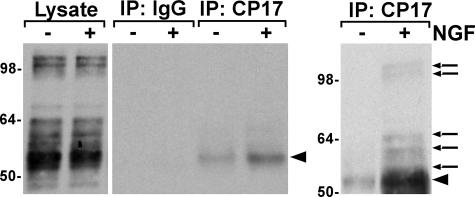
To investigate the phosphorylation of Thr-231 in response to NGF, we carried out immunoprecipitations in D5 cells using CP17, a monoclonal antibody specific for phospho-Thr-231 Tau (30). Cells were treated with or without NGF for 5 min and CP17 immunoprecipitates were probed with the total Tau antibody, Tau5. We detected human Tau at short exposures (Fig. 7, middle panel) as well as endogenous rat Tau, at both high and low molecular weights, after longer exposures (Fig. 7, right panel). The appearance of CP17-positive Tau was consistent with a requirement for phosphorylated Tau in the context of MAPK activation. The CP17-reactive Tau was most likely located in the lipid raft fraction of the cells because solubilization of the cells for the immunoprecipitation required the use of a stringent buffer to recover all of the Tau; solubilization with 1% Triton X-100 resulted in much of the NGF-induced Tau pelleting before the immunoprecipitation (data not shown). Lipid raft-associated Tau has been previously reported (43–46). These data suggested a novel function for Thr-231-phosphorylated Tau in response to NGF.
FIGURE 7.
Tau is phosphorylated at Thr-231 in response to NGF. Serum-starved D5 cells were stimulated with 50 ng/ml of NGF for 5 min. Tau was immunoprecipitated using the anti-phospho-Thr-231 Tau antibody, CP17, and analyzed by probing with horseradish peroxidase-coupled total Tau antibody, Tau5. Input lysate lanes (left panel) were 1% of the total lysate and showed equal loading of protein. The middle and right panels reflect shorter and longer exposures, respectively. The data shown are representative of three independent experiments. Arrows indicate endogenous rat Tau and arrowheads indicate human Tau.
DISCUSSION
Previous investigations of PC12 cells with Tau depletion have reported an effect of Tau on neurite length but effects on neurite initiation have been less clear (18, 19). In the present study, we have used cell lines stably expressing Tau shRNA to show that Tau depletion reduced the number of neurites initiated per cell, suggesting that Tau has a role in the NGF response prior to the stage of neurite elongation. Moreover, our experiments with taxol and the S262D/S356D mutant Tau have revealed that a separate property of Tau, other than microtubule stabilization, was required to aid neurite initiation following NGF stimulation. This finding is consistent with previous studies demonstrating that neurite outgrowth in N2a cells required Tau phosphorylation at Ser-262/Ser-356 (47). In this report, we describe a new function for Tau that is separate from its microtubule binding ability but also affects neurite initiation. It is well established that the AP-1 transcription factors c-Fos and c-Jun are activated during the differentiation response in PC12 cells and are required for neurite outgrowth (48–50). Our results show that during the earliest phase of the NGF response, before Tau levels are up-regulated, Tau served to potentiate NGF-induced AP-1 activity. Tau depletion attenuated AP-1 transcription factor activity, indirectly influencing neurite initiation.
The first hint that Tau expression might be linked to transcription factor activity came from microarray analysis showing that AP-1 transcription factors Fos and Fosb were the two genes most highly up-regulated in mice genetically deficient in Tau (see supplemental data in Ref. 36). However, our results indicated that Tau depletion decreased, rather than increased, the NGF-induced activity of AP-1 transcription factors. Given that the nature of the two experimental systems are vastly different (an undifferentiated neuronal cell line responding to growth factor addition versus adult mouse brain tissue), there could be several explanations for the apparent inconsistency in these results. We speculate that a compensatory up-regulation of Fos and Fosb was required for survival of the mouse during embryonic development, whereas Tau was less critical for the viability of undifferentiated neuronal cells, rendering similar compensatory measures unnecessary. However, in the undifferentiated cells, a critical role for Tau in the induction of AP-1 transcription factor activity was revealed upon growth factor stimulation.
The potentiation of AP-1 activity by Tau raises the issue of nuclear Tau. Nuclear Tau has been reported in a number of cell lines, including PC12 cells (51–53). However, looking at total Tau in our cells, the cells did not display noticeable nuclear Tau nor did the Tau localization significantly change after NGF addition (data not shown). Furthermore, in our previous PC12 work (26), we found transfected Tau mainly in the cytoplasm, rather than in the nucleus. These observations, combined with our present data placing Tau upstream of MAPK activation and possibly even upstream of GTPase activation (Fig. 4), make it less likely that the effect of Tau on AP-1 transcription factors was taking place in the nucleus. Tau phosphorylated at Thr-231 may reside in lipid rafts at the plasma membrane, as has been previously reported for Tau phosphorylated at Ser-396/Ser-404 (43–46). Lipid rafts have established importance in signal transduction (reviewed by Refs. 54–56) and taken together, we speculate that phospho-Tau affects the potentiation of MAPK signaling in lipid rafts.
Our pharmacological tests showed that Tau affected AP-1 transcription factor activity by modulating signaling through the MAPK pathway. In the Tau-depleted rTau4 cells, NGF-induced MAPK reporter activity was reduced 5.5-fold relative to control EV cells, whereas transiently expressing human Tau in rTau4 was able to restore MAPK reporter activity 2.3-fold (Fig. 4B). The inability of the exogenously expressed Tau to totally restore MAPK (or AP-1) reporter activity to control levels resembled the inability of exogenous Tau to completely restore neurite initiation to rTau4 (Fig. 2A). Although it is unclear how many pathways are being affected by Tau depletion, it is also unclear if the same pathways are being affected during both the MAPK and neurite initiation processes. In addition, the difference in activity between EV and human Tau-rescued rTau4 cells suggests that although high levels of exogenous Tau are expressed during the rescue experiment, it may function less efficiently than endogenous Tau. Tau expressed by transfection may differ from endogenous Tau with respect to its subcellular localization (i.e. “mistargeting”). Moreover, Tau expressed by transient transfection in neuronal cells usually migrates faster than the endogenous Tau (data not shown), suggesting that endogenous Tau is more efficiently phosphorylated. Therefore, as phosphorylation at Thr-231 is important for the ability of Tau to enhance MAPK signaling (Fig. 6, B–D), it is unlikely that the exogenously expressed Tau would lead to an increase in MAPK reporter activity proportionate to its level of expression. Exogenous Tau may be a suboptimal substrate and/or binding partner for endogenous proteins. Nonetheless, our data indicate that Tau was able to provide a significant restoration of MAPK signaling to Tau-depleted cell lines, suggesting that Tau has a role in signaling at a point upstream of MAPK activation and that Tau is necessary for normal levels of MAPK to be activated during the NGF response.
The ability of constitutively active-Ras to restore MAPK activity in Tau-depleted rTau4 cells suggests that Tau might impact on signaling upstream of Ras activation. Among the different ways Tau might affect MAPK signaling are interactions with signaling protein complexes or alterations in microtubule-dependent growth factor receptor trafficking to the cell surface. Studies performed in vitro and in cells have suggested a role for Tau in axonal transport and vesicle trafficking where it has been found that the presence of Tau inhibited transport (57–63), whereas another study carried out in mice reported that the Tau level had no effect on transport (64). Based on these findings, one would not predict that Tau depletion would inhibit vesicle transport or lead to decreases in growth factor receptors (e.g. TrkA, EGFR) at the cell surface that would attenuate MAPK signaling. Therefore, because our experiments showed that depleting Tau decreased MAPK signaling and adding Tau increased signaling, the data did not support the involvement of altered receptor trafficking. In addition, our rescue experiments (Fig. 6) suggested that the mechanism did not involve the ability of Tau to associate with microtubules. This also makes it unlikely that a microtubule-dependent mechanism such as vesicle transport is involved.
Our rescue experiments also demonstrated that phosphorylation of Tau at Thr-231 was necessary for its effect on MAPK reporter activity (Fig. 6, B–D). Also, the phosphorylation of Tau at Thr-231 was increased in response to a 5-min NGF treatment (Fig. 7) and both endogenous and exogenously expressed Tau were phosphorylated. Although our study does not identify the kinase responsible for phosphorylation at Thr-231, Thr-231 is a known substrate for several kinases including glycogen synthase kinase-3β (37, 65, 66) and members of the MAPK family such as ERK, c-Jun N-terminal kinase (JNK), and p38 (67–70). Because glycogen synthase kinase-3β is typically inactivated in response to NGF or EGF signaling (71), the phosphorylation of Tau at Thr-231 in the context of growth factor signaling is likely to be mediated by MAPK family members, leading to a positive feedback mechanism that would enhance signaling. In fact, Tau was also found to potentiate MAPK activation in response to EGF (Fig. 5A), suggesting that Tau impacts on MAPK signaling through a mechanism common to both NGF and EGF growth factor signaling pathways. However, the presence of Tau did not change the regulation of MAPK activation that ensued post-NGF or EGF addition. Similarly, the overexpression of Tau in D5 cells did not induce neurite initiation in response to EGF and we anticipate that Tau would not affect the rapid degradation of the EGF receptor that normally follows EGF activation (72, 73). These findings underline our hypothesis, that Tau serves to potentiate the existing levels of NGF- or EGF-activated MAPK, in a phosphorylation dependent manner.
Although we have established a requirement for Thr-231 phosphorylation as Tau participates in MAPK signaling, the mechanism through which this phosphorylation impacts on downstream signaling remains to be elucidated. Tau sequence surrounding Thr-231 contains multiple PXXP motifs capable of binding the SH3 domains of signaling proteins, and phosphorylation at this site may regulate the association between Tau and components of the MAPK pathway (7, 11, 39, 40). Although our results are limited to growth factor-induced MAPK signaling in PC6-3 cell lines, we speculate that these findings may have a functional role in early brain development and might explain the presence of increased levels of Thr-231-phosphorylated Tau in the fetal brain (74). The phosphorylation of fetal Tau at Thr-231 also coincides with an up-regulation of MAPK signaling (reviewed in Ref. 75 and 76), as would be expected if Tau is involved in signaling through this pathway. Further experimentation in primary neurons and neuronal stem cells will provide additional insights.
Phosphorylation of Tau at Thr-231 is one of the earliest phospho-epitopes to appear in Alzheimer disease (77–80), and one of several Tau phosphorylation sites thought to “recapitulate” early brain development during neuropathogenesis (81–83). It will be of interest to determine whether there is a relationship between Tau, Aβ peptide, and the increased levels of AP-1 transcription factors and MAPKs reported in tangle positive neurons (84–86). In addition, in human neuroblastoma cells, the presence of P301L mutant Tau altered the transcription patterns of AP-1-regulated genes (16) and we speculate that this may be due to the effect of Tau on AP-1 activation. Last, a requirement for the amino terminus of Tau in Aβ-mediated signaling has been reported (87), consistent with our finding that the microtubule binding activity of Tau is not required for its role in NGF-activated signal transduction.
In conclusion, we have shown that Tau potentiates signal transduction associated with growth factor stimulation in a manner that does not involve its microtubule binding activity but requires phosphorylation at Thr-231. These findings shed new light on how Tau may be affecting the differentiation and maturation of the developing brain, as well as the pathogenesis of neurodegenerative diseases such as Alzheimer disease.
Supplementary Material
Acknowledgments
We thank Dr. Stefan Strack (University of Iowa) for valuable suggestions and pNTO and FLAG-RasV12 plasmids. We thank Dr. Paul Rothman (University of Iowa) for the AP-1 firefly luciferase reporter plasmid. We thank Dr. Henry Paulson (University of Michigan) and Dr. Victor Miller for the PC6-3 cell line and assistance with dsRNA oligonucleotide screening. We thank Dr. Masaki Kashiwada (University of Iowa) for valuable suggestions and Dr. Bridget Zimmerman (University of Iowa Biostatistics Consulting Center) for assistance with statistical analysis. We also thank Dr. Lester Binder (Northwestern University Medical School) and Dr. Peter Davies (Albert Einstein College of Medicine) for their generous donation of Tau antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grant AG 017753 from the NIA.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- NGF
- nerve growth factor
- MAPK
- mitogen-activated protein kinase
- EGF
- epidermal growth factor
- dsRNA
- double-stranded RNA
- shRNA
- short hairpin RNA
- EV
- empty vector
- ERK
- extracellular signal-regulated kinase
- SH3
- Src homology domain 3
- hTau
- human Tau
- rTau
- rat Tau.
REFERENCES
- 1.Binder L. I., Guillozet-Bongaarts A. L., Garcia-Sierra F., Berry R. W. (2005) Biochim. Biophys. Acta 1739, 216–223 [DOI] [PubMed] [Google Scholar]
- 2.Ballatore C., Lee V. M., Trojanowski J. Q. (2007) Nat. Rev. Neurosci. 8, 663–672 [DOI] [PubMed] [Google Scholar]
- 3.Wolfe M. S. (2009) J. Biol. Chem. 284, 6021–6025 [DOI] [PubMed] [Google Scholar]
- 4.Goedert M., Jakes R. (2005) Biochim. Biophys. Acta 1739, 240–250 [DOI] [PubMed] [Google Scholar]
- 5.Brandt R., Léger J., Lee G. (1995) J. Cell Biol. 131, 1327–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biernat J., Mandelkow E. M. (1999) Mol. Biol. Cell 10, 727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds C. H., Garwood C. J., Wray S., Price C., Kellie S., Perera T., Zvelebil M., Yang A., Sheppard P. W., Varndell I. M., Hanger D. P., Anderton B. H. (2008) J. Biol. Chem. 283, 18177–18186 [DOI] [PubMed] [Google Scholar]
- 8.Yuan Z., Agarwal-Mawal A., Paudel H. K. (2004) J. Biol. Chem. 279, 26105–26114 [DOI] [PubMed] [Google Scholar]
- 9.Jenkins S. M., Johnson G. V. (1998) Neuroreport 9, 67–71 [DOI] [PubMed] [Google Scholar]
- 10.Derkinderen P., Scales T. M., Hanger D. P., Leung K. Y., Byers H. L., Ward M. A., Lenz C., Price C., Bird I. N., Perera T., Kellie S., Williamson R., Noble W., Van Etten R. A., Leroy K., Brion J. P., Reynolds C. H., Anderton B. H. (2005) J. Neurosci. 25, 6584–6593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee G., Newman S. T., Gard D. L., Band H., Panchamoorthy G. (1998) J. Cell Sci. 111, 3167–3177 [DOI] [PubMed] [Google Scholar]
- 12.Souter S., Lee G. (2009) J. Cell. Biochem. 108, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andorfer C., Acker C. M., Kress Y., Hof P. R., Duff K., Davies P. (2005) J. Neurosci. 25, 5446–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schindowski K., Belarbi K., Bretteville A., Ando K., Buée L. (2008) Genes Brain Behav. 7, Suppl. 1, 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delobel P., Lavenir I., Ghetti B., Holzer M., Goedert M. (2006) Am. J. Pathol. 168, 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoerndli F. J., Pelech S., Papassotiropoulos A., Götz J. (2007) Eur. J. Neurosci. 26, 60–72 [DOI] [PubMed] [Google Scholar]
- 17.Drubin D. G., Feinstein S. C., Shooter E. M., Kirschner M. W. (1985) J. Cell Biol. 101, 1799–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanemaaijer R., Ginzburg I. (1991) J. Neurosci. Res. 30, 163–171 [DOI] [PubMed] [Google Scholar]
- 19.Esmaeli-Azad B., McCarty J. H., Feinstein S. C. (1994) J. Cell Sci. 107, 869–879 [DOI] [PubMed] [Google Scholar]
- 20.Pittman R. N., Wang S., DiBenedetto A. J., Mills J. C. (1993) J. Neurosci. 13, 3669–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall G. F., Yao J., Lee G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4733–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.García-Sierra F., Ghoshal N., Quinn B., Berry R. W., Binder L. I. (2003) J. Alzheimers Dis. 5, 65–77 [DOI] [PubMed] [Google Scholar]
- 23.LoPresti P., Szuchet S., Papasozomenos S. C., Zinkowski R. P., Binder L. I. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10369–10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller V. M., Xia H., Marrs G. L., Gouvion C. M., Lee G., Davidson B. L., Paulson H. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7195–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strack S., Cribbs J. T., Gomez L. (2004) J. Biol. Chem. 279, 47732–47739 [DOI] [PubMed] [Google Scholar]
- 26.Léger J. G., Brandt R., Lee G. (1994) J. Cell Sci. 107, 3403–3412 [DOI] [PubMed] [Google Scholar]
- 27.Lee G., Thangavel R., Sharma V. M., Litersky J. M., Bhaskar K., Fang S. M., Do L. H., Andreadis A., Van Hoesen G., Ksiezak-Reding H. (2004) J. Neurosci. 24, 2304–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbacid M. (1987) Annu. Rev. Biochem. 56, 779–827 [DOI] [PubMed] [Google Scholar]
- 29.Sharma V. M., Litersky J. M., Bhaskar K., Lee G. (2007) J. Cell Sci. 120, 748–757 [DOI] [PubMed] [Google Scholar]
- 30.Weaver C. L., Espinoza M., Kress Y., Davies P. (2000) Neurobiol. Aging 21, 719–727 [DOI] [PubMed] [Google Scholar]
- 31.Derry W. B., Wilson L., Jordan M. A. (1995) Biochemistry 34, 2203–2211 [DOI] [PubMed] [Google Scholar]
- 32.Biernat J., Gustke N., Drewes G., Mandelkow E. M., Mandelkow E. (1993) Neuron 11, 153–163 [DOI] [PubMed] [Google Scholar]
- 33.Drewes G., Trinczek B., Illenberger S., Biernat J., Schmitt-Ulms G., Meyer H. E., Mandelkow E. M., Mandelkow E. (1995) J. Biol. Chem. 270, 7679–7688 [DOI] [PubMed] [Google Scholar]
- 34.Haase C., Stieler J. T., Arendt T., Holzer M. (2004) J. Neurochem. 88, 1509–1520 [DOI] [PubMed] [Google Scholar]
- 35.Pellegrino M. J., Stork P. J. (2006) J. Neurochem. 99, 1480–1493 [DOI] [PubMed] [Google Scholar]
- 36.Oyama F., Kotliarova S., Harada A., Ito M., Miyazaki H., Ueyama Y., Hirokawa N., Nukina N., Ihara Y. (2004) J. Biol. Chem. 279, 27272–27277 [DOI] [PubMed] [Google Scholar]
- 37.Cho J. H., Johnson G. V. (2004) J. Neurochem. 88, 349–358 [DOI] [PubMed] [Google Scholar]
- 38.Sengupta A., Kabat J., Novak M., Wu Q., Grundke-Iqbal I., Iqbal K. (1998) Arch. Biochem. Biophys. 357, 299–309 [DOI] [PubMed] [Google Scholar]
- 39.Bhaskar K., Yen S. H., Lee G. (2005) J. Biol. Chem. 280, 35119–35125 [DOI] [PubMed] [Google Scholar]
- 40.Zamora-Leon S. P., Lee G., Davies P., Shafit-Zagardo B. (2001) J. Biol. Chem. 276, 39950–39958 [DOI] [PubMed] [Google Scholar]
- 41.Cho J. H., Johnson G. V. (2003) J. Biol. Chem. 278, 187–193 [DOI] [PubMed] [Google Scholar]
- 42.Lin Y. T., Cheng J. T., Liang L. C., Ko C. Y., Lo Y. K., Lu P. J. (2007) J. Neurochem. 103, 802–813 [DOI] [PubMed] [Google Scholar]
- 43.Hernandez P., Lee G., Sjoberg M., Maccioni R. B. (2009) J. Alzheimers Dis. 16, 149–156 [DOI] [PubMed] [Google Scholar]
- 44.Kawarabayashi T., Shoji M., Younkin L. H., Wen-Lang L., Dickson D. W., Murakami T., Matsubara E., Abe K., Ashe K. H., Younkin S. G. (2004) J. Neurosci. 24, 3801–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein C., Kramer E. M., Cardine A. M., Schraven B., Brandt R., Trotter J. (2002) J. Neurosci. 22, 698–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sui Z., Kovács A. D., Maggirwar S. B. (2006) Biochem. Biophys. Res. Commun. 345, 1643–1648 [DOI] [PubMed] [Google Scholar]
- 47.Biernat J., Wu Y. Z., Timm T., Zheng-Fischhöfer Q., Mandelkow E., Meijer L., Mandelkow E. M. (2002) Mol. Biol. Cell 13, 4013–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leppä S., Eriksson M., Saffrich R., Ansorge W., Bohmann D. (2001) Mol. Cell. Biol. 21, 4369–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenberg M. E., Greene L. A., Ziff E. B. (1985) J. Biol. Chem. 260, 14101–14110 [PubMed] [Google Scholar]
- 50.Eriksson M., Taskinen M., Leppä S. (2007) J. Cell. Physiol. 210, 538–548 [DOI] [PubMed] [Google Scholar]
- 51.Hernández-Hernández O., Bermúdez-de-León M., Gómez P., Velázquez-Bernardino P., García-Sierra F., Cisneros B. (2006) J. Neurosci. Res. 84, 841–851 [DOI] [PubMed] [Google Scholar]
- 52.Loomis P. A., Howard T. H., Castleberry R. P., Binder L. I. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8422–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thurston V. C., Zinkowski R. P., Binder L. I. (1996) Chromosoma 105, 20–30 [DOI] [PubMed] [Google Scholar]
- 54.Allen J. A., Halverson-Tamboli R. A., Rasenick M. M. (2007) Nat. Rev. Neurosci. 8, 128–140 [DOI] [PubMed] [Google Scholar]
- 55.Golub T., Wacha S., Caroni P. (2004) Curr. Opin. Neurobiol. 14, 542–550 [DOI] [PubMed] [Google Scholar]
- 56.Guirland C., Zheng J. Q. (2007) Adv. Exp. Med. Biol. 621, 144–155 [DOI] [PubMed] [Google Scholar]
- 57.Dixit R., Ross J. L., Goldman Y. E., Holzbaur E. L. (2008) Science 319, 1086–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dubey M., Chaudhury P., Kabiru H., Shea T. B. (2008) Cell. Motil. Cytoskeleton 65, 89–99 [DOI] [PubMed] [Google Scholar]
- 59.Seitz A., Kojima H., Oiwa K., Mandelkow E. M., Song Y. H., Mandelkow E. (2002) EMBO J. 21, 4896–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamer K., Vogel R., Thies E., Mandelkow E., Mandelkow E. M. (2002) J. Cell Biol. 156, 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoothoff W., Jones P. B., Spires-Jones T. L., Joyner D., Chhabra E., Bercury K., Fan Z., Xie H., Bacskai B., Edd J., Irimia D., Hyman B. T. (2009) J. Neurochem. 111, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vershinin M., Carter B. C., Razafsky D. S., King S. J., Gross S. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vershinin M., Xu J., Razafsky D. S., King S. J., Gross S. P. (2008) Traffic 9, 882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan A., Kumar A., Peterhoff C., Duff K., Nixon R. A. (2008) J. Neurosci. 28, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishiguro K., Omori A., Takamatsu M., Sato K., Arioka M., Uchida T., Imahori K. (1992) Neurosci. Lett. 148, 202–206 [DOI] [PubMed] [Google Scholar]
- 66.Song J. S., Yang S. D. (1995) J. Protein Chem. 14, 95–105 [DOI] [PubMed] [Google Scholar]
- 67.Reynolds C. H., Nebreda A. R., Gibb G. M., Utton M. A., Anderton B. H. (1997) J. Neurochem. 69, 191–198 [DOI] [PubMed] [Google Scholar]
- 68.Reynolds C. H., Utton M. A., Gibb G. M., Yates A., Anderton B. H. (1997) J. Neurochem. 68, 1736–1744 [DOI] [PubMed] [Google Scholar]
- 69.Roder H. M., Eden P. A., Ingram V. M. (1993) Biochem. Biophys. Res. Commun. 193, 639–647 [DOI] [PubMed] [Google Scholar]
- 70.Blanchard B. J., devi Raghunandan R., Roder H. M., Ingram V. M. (1994) Biochem. Biophys. Res. Commun. 200, 187–194 [DOI] [PubMed] [Google Scholar]
- 71.Kleijn M., Welsh G. I., Scheper G. C., Voorma H. O., Proud C. G., Thomas A. A. (1998) J. Biol. Chem. 273, 5536–5541 [DOI] [PubMed] [Google Scholar]
- 72.Levkowitz G., Waterman H., Ettenberg S. A., Katz M., Tsygankov A. Y., Alroy I., Lavi S., Iwai K., Reiss Y., Ciechanover A., Lipkowitz S., Yarden Y. (1999) Mol. Cell 4, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 73.Waterman H., Levkowitz G., Alroy I., Yarden Y. (1999) J. Biol. Chem. 274, 22151–22154 [DOI] [PubMed] [Google Scholar]
- 74.Watanabe A., Hasegawa M., Suzuki M., Takio K., Morishima-Kawashima M., Titani K., Arai T., Kosik K. S., Ihara Y. (1993) J. Biol. Chem. 268, 25712–25717 [PubMed] [Google Scholar]
- 75.Fukunaga K., Miyamoto E. (1998) Mol. Neurobiol. 16, 79–95 [DOI] [PubMed] [Google Scholar]
- 76.Meloche S., Pouysségur J. (2007) Oncogene 26, 3227–3239 [DOI] [PubMed] [Google Scholar]
- 77.Augustinack J. C., Schneider A., Mandelkow E. M., Hyman B. T. (2002) Acta Neuropathol. 103, 26–35 [DOI] [PubMed] [Google Scholar]
- 78.Hasegawa M., Morishima-Kawashima M., Takio K., Suzuki M., Titani K., Ihara Y. (1992) J. Biol. Chem. 267, 17047–17054 [PubMed] [Google Scholar]
- 79.Luna-Muñoz J., Chávez-Macías L., García-Sierra F., Mena R. (2007) J. Alzheimers Dis. 12, 365–375 [DOI] [PubMed] [Google Scholar]
- 80.Vincent I., Zheng J. H., Dickson D. W., Kress Y., Davies P. (1998) Neurobiol. Aging 19, 287–296 [DOI] [PubMed] [Google Scholar]
- 81.Kanemaru K., Takio K., Miura R., Titani K., Ihara Y. (1992) J. Neurochem. 58, 1667–1675 [DOI] [PubMed] [Google Scholar]
- 82.Jicha G. A., Lane E., Vincent I., Otvos L., Jr., Hoffmann R., Davies P. (1997) J. Neurochem. 69, 2087–2095 [DOI] [PubMed] [Google Scholar]
- 83.Bramblett G. T., Goedert M., Jakes R., Merrick S. E., Trojanowski J. Q., Lee V. M. (1993) Neuron 10, 1089–1099 [DOI] [PubMed] [Google Scholar]
- 84.Anderson A. J., Cummings B. J., Cotman C. W. (1994) Exp. Neurol. 125, 286–295 [DOI] [PubMed] [Google Scholar]
- 85.Pei J. J., Braak H., An W. L., Winblad B., Cowburn R. F., Iqbal K., Grundke-Iqbal I. (2002) Brain Res. Mol. Brain Res. 109, 45–55 [DOI] [PubMed] [Google Scholar]
- 86.Marcus D. L., Strafaci J. A., Miller D. C., Masia S., Thomas C. G., Rosman J., Hussain S., Freedman M. L. (1998) Neurobiol. Aging 19, 393–400 [DOI] [PubMed] [Google Scholar]
- 87.King M. E., Kan H. M., Baas P. W., Erisir A., Glabe C. G., Bloom G. S. (2006) J. Cell Biol. 175, 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.