Abstract
Thrombin elicits functional responses critical to blood homeostasis by interacting with diverse physiological substrates. Ala-scanning mutagenesis of 97 residues covering 53% of the solvent accessible surface area of the enzyme identifies Trp215 as the single most important determinant of thrombin specificity. Saturation mutagenesis of Trp215 produces constructs featuring kcat/Km values for the hydrolysis of fibrinogen, protease-activated receptor PAR1, and protein C that span five orders of magnitude. Importantly, the effect of Trp215 replacement is context dependent. Mutant W215E is 10-fold more specific for protein C than fibrinogen and PAR1, which represents a striking shift in specificity relative to wild-type that is 100-fold more specific for fibrinogen and PAR1 than protein C. However, when the W215E mutation is combined with deletion of nine residues in the autolysis loop, which by itself shifts the specificity of the enzyme from fibrinogen and PAR1 to protein C, the resulting construct features significant activity only toward PAR1. These findings demonstrate that thrombin can be re-engineered for selective specificity toward protein C and PAR1. Mutations of Trp215 provide important reagents for dissecting the multiple functional roles of thrombin in the blood and for clinical applications.
Keywords: Coagulation Factors, Mutagenesis Mechanisms, Mutant, Protease, Thrombin
Introduction
Engineering protease specificity remains an issue of considerable interest and importance (1). Within the same fold, trypsins prefer Arg/Lys side chains at the P1 position (2) of substrate but chymotrypsins prefer Phe/Tyr/Trp residues (3–6). Primary specificity, defined by the nature of the P1 residue, is not the sole determinant of protease function. Many trypsin-like proteases show substrate preference that extends beyond the P1 position of substrate and is dictated by interactions with other structural domains not in contact with the primary specificity pocket. Indeed, inspection of consensus sequences recognized by thrombin for its three primary physiological targets, i.e. the procoagulant substrate fibrinogen, the prothrombotic protease-activated receptor 1 (PAR1)2 and the anticoagulant substrate protein C, reveals an Arg residue at the P1 position in all cases (7). Hence, selection among these targets must depend on interactions beyond the primary specificity pocket of the enzyme. A paradigm widely accepted in the field is that macromolecular specificity in thrombin and related clotting proteases is achieved and controlled by interaction with “exosites,” i.e. domains widely separated from the active site region that kinetically control docking of substrate in ways that restrict choices by the enzyme (8, 9). However, abundant mutagenesis data show that residues within the active site play roles that far exceed in importance those of exosites and, in fact, affect specificity and the choice of which substrate can be cleaved by the enzyme in ways that are unmatched by other domains (10–13). Consistent with these findings, the present study demonstrates that residue 215 within the thrombin active site is the single most important determinant of thrombin specificity.
MATERIALS AND METHODS
Site-directed mutagenesis of human thrombin was performed as described (14–16) using the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA) in a HPC4-modified pNUT expression vector containing the human prethrombin-1 gene. Most of the 97 Ala mutants of thrombin were reported in previous studies (10, 15–23). All mutants of Trp215, except W215M and W215T (see below), were expressed in baby hamster kidney cells and activated overnight at 25 °C with a mixture of a 1:30 dilution of crude venom from Echis carinatus and 1:100 dilution of venom from Agkistrodon acutus (Sigma) in the presence of 10 mm benzamidine and 10 mm CaCl2. All mutants were purified to homogeneity by FPLC using Resource Q and S columns with a linear gradient from 0.04 to 0.5 m choline chloride or NaCl, 5 mm MES, pH 6.0 at room temperature. Active site concentrations were determined by titration with hirudin and found to be >95% for all mutants.
The mutants W215M and W215T failed to express in baby hamster kidney cells and were expressed in Escherichia coli. Wild-type thrombin expressed in E. coli featured functional properties identical to those of thrombin expressed in baby hamster kidney cells and a three-dimensional structure documenting proper folding and activation (data not shown). The cDNA corresponding to the prethrombin-2 sequence of human thrombin was cloned into the pET21a vector (Novagen) using the EcoRI and the XhoI restriction sites. The prethrombin-2 vector was transformed into BL21(DE3) E. coli cells grown overnight in 10 ml of LB medium with 100 μg/ml ampicillin at 37 °C and 200 rpm. The next morning, 3 liters of LB medium with 100 μg/ml of ampicillin was inoculated with 10 ml of overnight culture. Growth was continued at 37 °C and 300 rpm until the cells reached A600 = 0.6. Prethrombin-2 expression was initiated by adding IPTG to a final concentration of 1 mm. E. coli cells were then cultured for an additional 4 h and cultures were spun at 4000 rpm for 15 min at 4 °C. The supernatant was discarded, and the cell paste, from 1 liter of LB medium, was resuspended in 50 ml of a buffer composed of 50 mm Tris, 2 mm EDTA, pH 8.0. The suspension was combined and spun again at 4000 rpm for 15 min at 4 °C. The supernatant was discarded, and the cell paste was stored at −80 °C. The cell paste was thawed at 37 °C and resuspended in 50 ml of 50 mm Tris, pH 8.0 at 25 °C, 20 mm EDTA, 0.5 m NaCl, 0.1% Triton X-100, 20 mm DTE. Cells were further sonicated on ice for 30 s × 5 (∼1 min rest) at constant duty, 5 ½ output. The well-homogenized cells were ultracentrifuged for 1 h at 4 °C, 30,000 rpm, using a 45Ti rotor. The supernatant was discarded, and the pellet was resuspended in 40 ml of 50 mm Tris, pH 8.0 at 25 °C, 20 mm EDTA, 2% Triton X-100, 20 mm DTE using gentle vortexing and a spatula. The homogenate was centrifuged for 30 min at 30,000 rpm at 4 °C. Supernatant was discarded and the pellet was suspended in 40 ml of 50 mm Tris, pH 8.0 at 25 °C, 20 mm EDTA, 1 m NaCl, 20 mm DTE prior to centrifugation for 30 min at 30000 rpm at 4 °C. The supernatant was discarded, and the pellet was resuspended in 25 ml of 50 mm Tris, pH 8.0 at 25 °C, 20 mm EDTA, 6 m GdnHCl, 50 mm DTE overnight. The suspension was spun at 30,000 rpm for 30 min at 4 °C. The supernatant was dialyzed against 20 mm Tris, pH 8.0, 6 m GdnHCl, 5 mm EDTA at 4 °C three times. Postdialysis, the unfolded protein was incubated with 5 mm GSH and 2 mm GSSG for 3 h at room temperature. The supernatant was then dialyzed against two changes of 6 m GdnHCl, 20 mm EDTA, pH 5.0 at 4 °C. Refolding of prethrombin-2 was initiated by adding 2 ml, every 2 h, of the unfolded protein in 500 ml of refolding buffer, 0.6 m l-arginine, 10% glycerol, 25 mm CaCl2, 0.2% Brij58, 50 mm Tris, pH 8.5, at 25 °C. The refolded protein was concentrated from 2 liters to 150 ml using Quickstand, from Amersham Biosciences, with a 10-kDa hollow fiber cartridge. The 150 ml of refolded protein was dialyzed against three changes of 20 mm Tris, pH 7.0 at 25 °C, 0.4 m NaCl. The protein sample was diluted to make the salt concentration less than 75 mm. Precipitate, if any, was removed by centrifugation. Protein solution was loaded onto a 5-ml heparin column, from GE Healthcare, at 2.5 ml/min. The bound protein was extensively washed with 10 mm NaCl, 20 mm Tris, pH 7.0 at 25 °C before elution with a linear gradient of 10 mm to 1.5 m NaCl in 20 mm Tris, pH 7.0 at 25 °C. The elution was monitored by UV spectroscopy and the fractions containing the UV peak were collected and activated. The activation was carried out using 10 μl of ecarin (50 EU/ml) per 1 ml of protein solution. Activation was monitored from the hydrolysis of the chromogenic substrate H-d-Phe-Pro-Arg-p-nitroanilide (FPR). The activated protein was diluted 7-fold and loaded on the heparin column. Bound protein was extensively washed by 10 mm choline chloride, 20 mm Tris, pH 7.0 at 25 °C before elution with a linear gradient of 10 mm to 1.5 m choline chloride in 20 mm Tris, pH 7.0 at 25 °C. Active site concentrations were determined by titration with hirudin and found to be >95% for both the W215M and W215T mutants.
Values of s = kcat/Km for release of fibrinopeptide A (FpA) from fibrinogen, cleavage of the protease-activated receptor PAR1 and activation of protein C in the presence of 50 nm thrombomodulin and 5 mm CaCl2 were obtained as reported elsewhere (14, 17, 24) under experimental conditions of 5 mm Tris, 0.1% PEG8000, 145 mm NaCl, pH 7.4 at 37 °C.
RESULTS
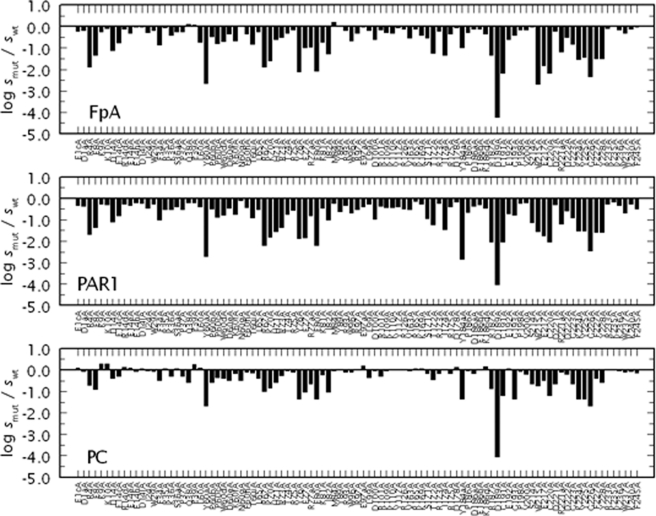
Fig. 1 and Table 1 summarize the results of an Ala scan of 97 residues of thrombin covering >53% of the accessible surface area of the enzyme in terms of the activity toward the procoagulant substrate fibrinogen, the prothrombotic substrate PAR1, and the anticoagulant substrate protein C in the presence of the cofactor thrombomodulin and 5 mm CaCl2. Residues targeted by mutagenesis are highlighted in Fig. 2 and belong to critical regions of the B chain of thrombin defining the active site, the 60-loop above the active site, the autolysis loop below it, exosites I and II, the Na+ site and the A chain. In the case of wild-type, the value of kcat/Km for cleavage of fibrinogen or activation of PAR1 is >100-fold higher than that for activation of protein C in the presence of thrombomodulin (Table 1). A general trend gleaned from the plot is that an Ala mutation of the thrombin scaffold tends to perturb recognition of fibrinogen and PAR1 more than protein C. These findings extend and complement previous mutagenesis studies from our laboratory (10, 15–23) and others (25).
FIGURE 1.
Ala-scanning mutagenesis of thrombin. Effect of Ala substitution of 97 residues of thrombin on the cleavage of fibrinogen (FpA) and activation of PAR1 or PC in the presence of 50 nm thrombomodulin and 5 mm CaCl2. Shown is the change in the specificity constant s = kcat/Km caused by the mutation, expressed as log smut/swt, under experimental conditions of 5 mm Tris, 0.1% PEG, 145 mm NaCl, pH 7.4 at 37 °C. Note the similarity in the profiles for fibrinogen and PAR1 hydrolysis that are distinct from that for activation of protein C. The values of s for wild-type are: 17 ± 1 μm−1 s−1 (FpA), 30 ± 1 μm−1 s−1 (PAR1), and 0.22 ± 0.02 μm−1 s−1 (PC). The values of s for each mutant are listed in Table 1, along with the relative differences for each pair of the three substrates.
TABLE 1.
Properties of the 97 Ala mutants of thrombin toward fibrinogen (FpA), PAR1, and protein C in the presence of Ca2+ and thrombomodulin (PC)
Shown are the values of log kcat/Km in m−1 s−1 and the difference in such values between the three pairs of substrates. Errors are <0.04 log units.
| Mutant | FpA | PAR1 | PC | FpA-PAR1 | PAR1-PC | FpA-PC |
|---|---|---|---|---|---|---|
| WT | 7.23 | 7.48 | 5.34 | −0.25 | 2.14 | 1.89 |
| E1cA | 7.00 | 7.15 | 5.43 | −0.15 | 1.73 | 1.57 |
| D1aA | 7.04 | 7.13 | 5.28 | −0.09 | 1.85 | 1.76 |
| R4A | 5.36 | 5.80 | 4.64 | −0.44 | 1.16 | 0.72 |
| E8A | 5.89 | 6.13 | 4.44 | −0.24 | 1.68 | 1.44 |
| K9A | 6.97 | 7.20 | 5.62 | −0.23 | 1.58 | 1.35 |
| K10A | 7.15 | 7.18 | 5.62 | −0.03 | 1.56 | 1.52 |
| D14A | 6.11 | 6.37 | 4.94 | −0.26 | 1.43 | 1.18 |
| E14cA | 6.46 | 6.68 | 5.04 | −0.22 | 1.64 | 1.42 |
| R14dA | 7.11 | 7.22 | 5.47 | −0.11 | 1.75 | 1.64 |
| E14eA | 6.93 | 7.15 | 5.44 | −0.22 | 1.71 | 1.49 |
| E14hA | 7.20 | 7.29 | 5.28 | −0.08 | 2.01 | 1.93 |
| D14lA | 7.20 | 7.25 | 5.40 | −0.04 | 1.85 | 1.81 |
| I24A | 6.96 | 7.04 | 5.30 | −0.08 | 1.74 | 1.66 |
| W29A | 7.04 | 7.20 | 5.28 | −0.16 | 1.93 | 1.76 |
| F34A | 6.38 | 6.47 | 4.84 | −0.09 | 1.63 | 1.54 |
| R35A | 7.18 | 6.97 | 5.41 | 0.21 | 1.55 | 1.76 |
| K36A | 6.82 | 6.97 | 5.04 | −0.15 | 1.93 | 1.78 |
| S36aA | 6.99 | 7.10 | 5.40 | −0.11 | 1.70 | 1.59 |
| P37A | 6.98 | 6.97 | 5.04 | 0.01 | 1.93 | 1.94 |
| Q38A | 7.34 | 7.27 | 4.78 | 0.08 | 2.49 | 2.57 |
| E39A | 7.30 | 7.29 | 5.59 | 0.02 | 1.70 | 1.71 |
| L60A | 6.49 | 7.10 | 5.43 | −0.61 | 1.67 | 1.06 |
| Y60aA | 4.59 | 4.76 | 3.68 | −0.17 | 1.08 | 0.91 |
| P60bA | 6.75 | 7.00 | 4.78 | −0.26 | 2.23 | 1.97 |
| P60cA | 6.43 | 6.60 | 5.00 | −0.17 | 1.61 | 1.43 |
| W60dA | 6.53 | 6.71 | 4.95 | −0.18 | 1.76 | 1.58 |
| D60eA | 6.88 | 7.04 | 4.84 | −0.16 | 2.19 | 2.04 |
| K60fA | 6.57 | 6.74 | 5.17 | −0.17 | 1.56 | 1.39 |
| N60gA | 7.23 | 7.39 | 4.84 | −0.16 | 2.54 | 2.39 |
| F60hA | 6.89 | 7.07 | 5.25 | −0.18 | 1.82 | 1.63 |
| T60iA | 6.41 | 6.59 | 5.20 | −0.17 | 1.38 | 1.21 |
| L65A | 6.97 | 6.97 | 4.95 | 0.00 | 2.01 | 2.02 |
| R67A | 5.36 | 5.29 | 4.34 | 0.08 | 0.95 | 1.02 |
| K70A | 5.62 | 5.68 | 4.52 | −0.06 | 1.16 | 1.11 |
| H71A | 6.62 | 5.92 | 4.78 | 0.70 | 1.15 | 1.85 |
| R73A | 6.72 | 6.13 | 5.08 | 0.60 | 1.05 | 1.65 |
| T74A | 6.91 | 6.75 | 5.30 | 0.16 | 1.45 | 1.61 |
| R75A | 7.08 | 6.92 | 5.25 | 0.16 | 1.67 | 1.83 |
| Y76A | 5.11 | 5.62 | 4.00 | −0.51 | 1.63 | 1.12 |
| E77A | 6.23 | 5.63 | 4.30 | 0.60 | 1.33 | 1.93 |
| R77aA | 6.28 | 6.68 | 4.70 | −0.40 | 1.98 | 1.58 |
| E80A | 5.15 | 5.29 | 4.00 | −0.14 | 1.29 | 1.15 |
| K81A | 6.50 | 7.04 | 5.11 | −0.53 | 1.93 | 1.39 |
| I82A | 5.96 | 6.48 | 4.30 | −0.52 | 2.18 | 1.66 |
| M84A | 7.43 | 7.25 | 5.32 | 0.18 | 1.93 | 2.11 |
| Y89A | 7.25 | 6.86 | 5.38 | 0.39 | 1.49 | 1.88 |
| R93A | 7.06 | 7.15 | 5.23 | −0.09 | 1.93 | 1.83 |
| W96A | 6.58 | 6.79 | 5.23 | −0.21 | 1.56 | 1.35 |
| R97A | 6.92 | 6.97 | 5.38 | −0.04 | 1.59 | 1.55 |
| E97aA | 7.20 | 7.10 | 5.52 | 0.10 | 1.58 | 1.68 |
| L99A | 7.04 | 7.20 | 5.00 | −0.16 | 2.20 | 2.04 |
| D100A | 6.62 | 6.50 | 5.30 | 0.12 | 1.20 | 1.32 |
| R101A | 7.08 | 7.13 | 5.04 | −0.05 | 2.09 | 2.04 |
| K109A | 6.96 | 7.07 | 5.32 | −0.11 | 1.75 | 1.64 |
| K110A | 6.92 | 7.07 | 5.36 | −0.15 | 1.71 | 1.56 |
| Y117A | 7.18 | 7.10 | 5.36 | 0.08 | 1.74 | 1.82 |
| R126A | 7.11 | 7.00 | 5.36 | 0.11 | 1.64 | 1.75 |
| V163A | 6.69 | 6.97 | 5.30 | −0.28 | 1.67 | 1.39 |
| R165A | 7.12 | 7.34 | 5.40 | −0.22 | 1.94 | 1.72 |
| K169A | 6.81 | 7.22 | 5.41 | −0.42 | 1.81 | 1.40 |
| S171A | 6.71 | 6.53 | 5.17 | 0.18 | 1.35 | 1.53 |
| T172A | 5.98 | 6.25 | 4.90 | −0.27 | 1.35 | 1.07 |
| R173A | 7.01 | 7.25 | 5.17 | −0.23 | 2.07 | 1.84 |
| I174A | 5.88 | 6.04 | 5.34 | −0.16 | 0.70 | 0.54 |
| R175A | 6.89 | 7.10 | 5.17 | −0.21 | 1.93 | 1.72 |
| D178A | 7.19 | 7.30 | 5.47 | −0.11 | 1.83 | 1.72 |
| Y184aA | 6.25 | 4.66 | 4.00 | 1.60 | 0.66 | 2.26 |
| P186A | 6.95 | 6.82 | 5.32 | 0.14 | 1.50 | 1.63 |
| D186aA | 7.11 | 7.13 | 5.17 | −0.01 | 1.95 | 1.94 |
| E186bA | 7.11 | 7.18 | 5.38 | −0.07 | 1.80 | 1.74 |
| K186dA | 6.89 | 7.04 | 5.50 | −0.15 | 1.53 | 1.38 |
| R187A | 5.90 | 5.46 | 4.47 | 0.44 | 0.98 | 1.42 |
| D189A | 3.00 | 3.47 | 1.28 | −0.47 | 2.19 | 1.72 |
| C191A | 5.04 | 5.44 | 4.14 | −0.40 | 1.30 | 0.90 |
| E192A | 6.63 | 6.76 | 5.41 | −0.12 | 1.34 | 1.22 |
| G193A | 6.84 | 6.70 | 4.00 | 0.14 | 2.70 | 2.84 |
| P198A | 7.08 | 7.25 | 5.28 | −0.17 | 1.97 | 1.80 |
| V200A | 7.09 | 7.27 | 5.14 | −0.17 | 2.12 | 1.95 |
| S214A | 7.20 | 6.34 | 4.70 | 0.87 | 1.64 | 2.51 |
| W215A | 4.53 | 5.92 | 4.60 | −1.39 | 1.32 | −0.07 |
| E217A | 5.41 | 5.74 | 4.84 | −0.33 | 0.90 | 0.57 |
| C220A | 5.04 | 5.44 | 4.14 | −0.40 | 1.30 | 0.90 |
| D221A | 7.00 | 7.18 | 4.70 | −0.18 | 2.48 | 2.30 |
| R221aA | 6.04 | 6.29 | 5.23 | −0.24 | 1.06 | 0.81 |
| D222A | 6.74 | 6.91 | 5.14 | −0.17 | 1.77 | 1.60 |
| G223A | 6.40 | 6.57 | 4.70 | −0.17 | 1.87 | 1.70 |
| K224A | 5.69 | 5.97 | 4.00 | −0.28 | 1.97 | 1.69 |
| Y225A | 5.78 | 5.97 | 4.00 | −0.18 | 1.97 | 1.79 |
| G226A | 4.88 | 5.04 | 3.66 | −0.16 | 1.38 | 1.22 |
| F227A | 5.74 | 5.91 | 4.95 | −0.17 | 0.96 | 0.79 |
| Y228A | 5.73 | 5.90 | 4.78 | −0.17 | 1.13 | 0.96 |
| R233A | 7.14 | 7.20 | 5.34 | −0.06 | 1.86 | 1.80 |
| K235A | 7.24 | 7.30 | 5.34 | −0.06 | 1.96 | 1.90 |
| K236A | 7.08 | 7.15 | 5.30 | −0.08 | 1.86 | 1.78 |
| W237A | 6.92 | 6.79 | 5.25 | 0.13 | 1.54 | 1.67 |
| K240A | 7.10 | 7.22 | 5.28 | −0.12 | 1.95 | 1.82 |
| F245A | 7.22 | 7.00 | 5.20 | 0.22 | 1.80 | 2.02 |
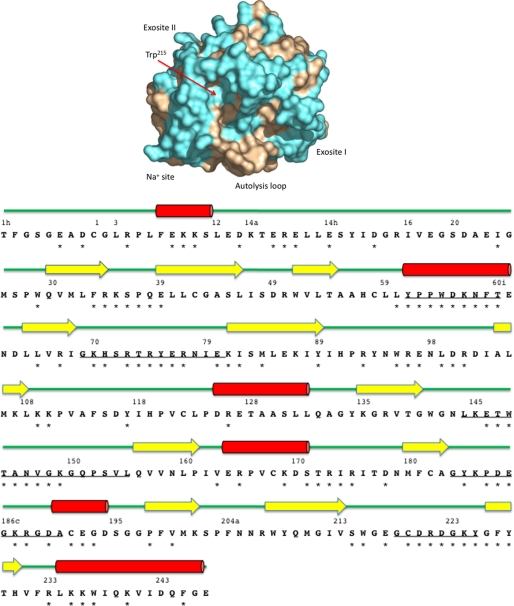
FIGURE 2.
Top, surface representation of thrombin. Shown is the accessible surface area of thrombin in the standard orientation with the active site at the center and other regulatory sites as indicated. The position of Trp215 within the active site is noted by a red arrow. Residues of thrombin subject to Ala-scanning mutagenesis are colored in cyan. These residues cover 53% of the thrombin surface area. Not seen in this orientation are residues of the A chain in the back of the molecule. Bottom, sequence and secondary structure of thrombin. Shown is the sequence of thrombin numbered according to chymotrypsin(ogen) with the position of the 97 Ala-substituted residues (*). Elements of secondary structure are rendered as red cylinders for α-helices, yellow arrows for β-sheets, and green lines for loops. Relevant surface-exposed loops are underlined. The sequence 146–149e in the autolysis loop is also marked by an *.
The double mutant W215A/E217A (WE) was constructed by combining the two single mutations W215A and E217A (11) that stand out for their significant perturbation of fibrinogen and PAR1 cleavage over protein C activation (Fig. 1). WE shows anticoagulant/antithrombotic activity both in vitro and in vivo (10, 11, 26–30). Its antithrombotic effect in non-human primates is more efficacious than the direct administration of activated protein C and safer than the administration of low molecular weight heparins (30). Activated protein C generated in situ with the mutant WE offers cytoprotective advantages over activated protein C administered to the circulation (27). Furthermore, WE acts as a potent and safe antithrombotic by blocking the interaction of von Willebrand factor with the platelet receptor GpIb (26, 30). These intriguing properties of the mutant WE provide proof of principle that a thrombin mutant with exclusive activity toward protein C would be a compelling anticoagulant/antithrombotic agent in vivo. Further mutagenesis of thrombin was therefore undertaken to remove any residual activity toward fibrinogen and PAR1 in preparation for clinical applications.
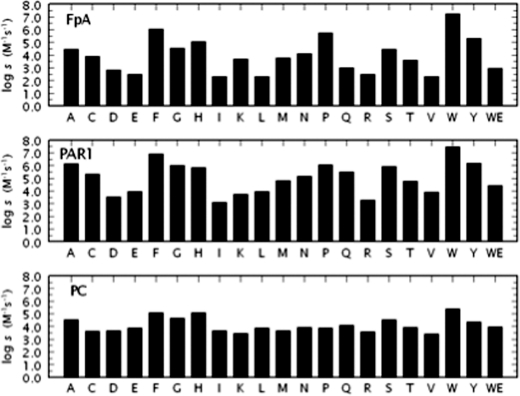
Previous saturation mutagenesis studies of residue 217 yielded an incomplete set of constructs and identified E217K as the most potent anticoagulant mutant (31). Although the properties of E217K provide a slight improvement over those of E217A, they are comparable to those of W215A (10, 11) and make residue 217 of little interest as a target for further mutagenesis. Indeed, the W215A/E217K double mutant features a less pronounced anticoagulant/antithrombotic profile compared with WE (data not shown). Results of saturation mutagenesis of residue 215 are summarized in Fig. 3. All constructs could be expressed to homogeneity and characterized in terms of their interaction with key physiological substrates. Trp215 is highly conserved in trypsin-like proteases (3) and is located within a hydrophobic patch (Fig. 2) essential for recognition of the P4 residue of substrate (15, 32). The role of Trp215 in fibrinogen binding is illustrated directly by the crystal structure (33) and its edge-to-face interaction with residue Phe8 of the Aα chain. Strong hydrophobic interactions involve Trp215 in recognition of the procoagulant substrates factor XIII (34, 35) and PAR4 (17, 36). Trp215 also plays a key role in the conformational transition of thrombin from the inactive E* to the active E form (7, 18) by relocating more than 10 Å within the active site.
FIGURE 3.
Saturation mutagenesis of residue 215. Activity of the 20 possible amino acid substitutions at position 215 of thrombin toward fibrinogen (FpA), PAR1, or PC in the presence of 50 nm thrombomodulin and 5 mm CaCl2. W is the wild-type residue. Shown is the log of the specificity constant s = kcat/Km under experimental conditions of 5 mm Tris, 0.1% PEG, 145 mm NaCl, pH 7.4 at 37 °C. The properties of the anticoagulant mutant WE are also shown for comparison. Average values of log s for the 20 substitutions of residue 215 are 4.0 (FpA), 5.1 (PAR1), and 4.1 (PC), reflecting an average loss in log units relative to wild-type of 3.2 (FpA), 2.4 (PAR1) and 1.2 (PC). A strong correlation (r = 0.91) is observed between the values for fibrinogen and PAR1 cleavage. Correlation between activation of protein C and fibrinogen or PAR1 cleavage is significantly weaker (r = 0.79 and r = 0.84, respectively).
Mutation of Trp215 has a profound effect on thrombin specificity toward macromolecular substrates. Several mutations of Trp215 result in >10,000-fold loss of activity toward fibrinogen and/or PAR1. In some cases (Asp, Glu, Ile, Leu, Arg, Val), the loss of activity toward fibrinogen approaches or exceeds five orders of magnitude and is more pronounced than that observed with the potent anticoagulant mutant WE. For example, the mutant W215I features a kcat/Km value for cleavage of fibrinogen that is >100,000-fold lower than that of wild-type. The kcat/Km value for PAR1 activation is >10,000-fold lower compared with wild-type, but the kcat/Km value for activation of protein C in the presence of thrombomodulin is perturbed <100-fold. The breadth of responses associated with mutations of residue 215 recalls that seen in the saturation mutagenesis of residue 225 (37), but in that case little preference was detected among fibrinogen and protein C and the functional perturbation affected all substrates to similar extent. Perturbation produced by introduction of a charge in the hydrophobic aryl binding site explains the drastic loss in specificity toward fibrinogen and PAR1 for the W215D, W215E, and W215R mutants. However, the effects seen for the W215I, W215L, and W215V mutants are somewhat unexpected and support the need for an aromatic side chain or ring structure at this position. In fact, wild-type and W215F have the highest activity toward fibrinogen and PAR1 (10), with W215Y, W215P, and W215H not far behind. Activity toward PAR1 correlates (r = 0.91) with that of fibrinogen, underscoring the basic similarity of interaction of the two substrates with thrombin (33, 38) and consistent with previous mutagenesis studies (17). Interestingly, activity toward fibrinogen or PAR1 correlates with activity toward the chromogenic substrate FPR (r = 0.88 and r = 0.92, respectively, data not shown) suggesting that perturbation of the active site moiety because of replacement of Trp215 is the dominant factor controlling hydrolysis of the procoagulant and prothrombotic substrates of thrombin. On the other hand, activities toward protein C correlates to lower extent with activity toward FPR (r = 0.77), fibrinogen (r = 0.79), or PAR1 (r = 0.84). The concerted action of thrombomodulin and protein C on thrombin tends to correct the functional deficit associated with a mutation of the enzyme, as demonstrated recently in the case of the double mutant WE (39).
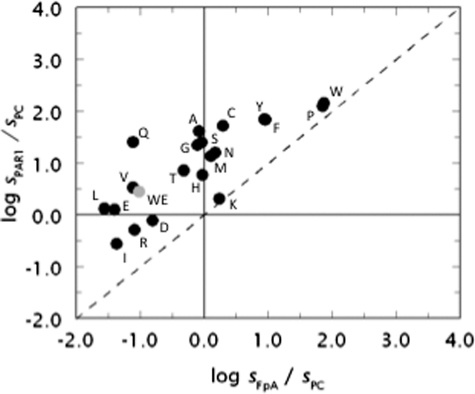
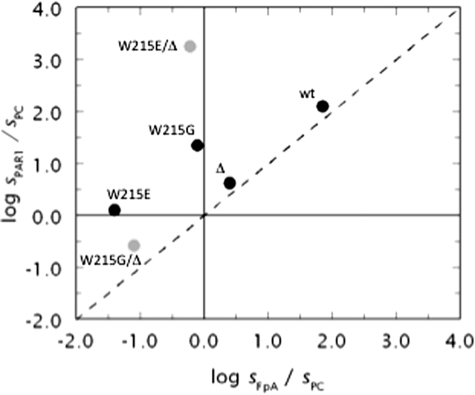
Shift in the specificity of the enzyme is best appreciated in the plot shown in Fig. 4. The plot contains many elements of interest as it incorporates information on three different substrates. The value of sPAR1 for the hydrolysis of PAR1 by thrombin is plotted versus the analogous value for fibrinogen cleavage sFpA and both values are expressed in units of sPC, i.e. the kcat/Km value for the hydrolysis of protein C in the presence of thrombomodulin and Ca2+ (see also Fig. 3). The origin of the axes divides the plot in four regions, with two of them further divided in two regions by the diagonal dotted line reflecting a regime where perturbation of PAR1 cleavage is the same as that of fibrinogen cleavage. The region to the right of the vertical axis denotes activity toward fibrinogen that exceeds that toward protein C, and the reverse is seen to the left of the vertical axis. Likewise, the region above the horizontal axis denotes activity toward PAR1 that exceeds that toward protein C and the reverse is seen below the horizontal axis. Interestingly, all points reflecting the properties of the 20 possible amino acid substitutions of residue 215 map on or above the dotted line, indicating that mutation of Trp215 fails to produce a construct for which the activity toward fibrinogen exceeds that toward PAR1. Distance from the diagonal dotted line quantifies the extent of preferential interaction toward fibrinogen and PAR1. The mutant W215Q emerges as the most specific toward PAR1 relative to fibrinogen, improving on the analogous property seen with the W215A (10) and WE mutants (11). Wild-type maps in the region reflecting procoagulant/prothrombotic propensity almost on the diagonal dotted line, underscoring the similarity of PAR1 and fibrinogen cleavage that both exceed the activity toward protein C by two orders of magnitude. The mutant WE maps in the anticoagulant region of the plot but close to the horizontal line, indicating that although the activity toward protein C exceeds that toward fibrinogen, PAR1 is cleaved with a specificity constant s = kcat/Km higher than that of protein C. This leaves considerable room for optimization by directly interfering with the prothrombotic activity of the mutant. The quadrant in the lower left corner of the plot defines a region where activity toward protein C exceeds that toward fibrinogen and PAR1 and represents a target for protein engineering studies aimed at optimizing the anticoagulant activity of the enzyme at the expense of the procoagulant and prothrombotic activities. A replot of the data shown in Fig. 1 and Table 1 reveals that none of the 97 Ala mutants of thrombin maps into the target region (plot not shown), nor does the mutant E217K. Mutant W215V, bearing a single amino acid substitution, recapitulates the properties of WE in the plot. Five mutants of Trp215 improve on the anticoagulant/antithrombotic profile of WE and map into the target region (Asp, Ile, Arg) or close by (Glu, Leu). These mutations make a compelling case for further mutagenesis.
FIGURE 4.
Multifunctional plot of thrombin activity. Functional properties of the thrombin mutants of residue 215 (W corresponds to wild-type). Shown are the values of s = kcat/Km for the hydrolysis of fibrinogen (sFpA), PAR1 (sPAR1), or protein C (sPC) in the presence of 50 nm thrombomodulin and 5 mm Ca2+ plotted as the logarithm of the ratios sPAR1/sPC versus sFpA/sPC. Experimental conditions are: 5 mm Tris, 0.1% PEG8000, 145 mm NaCl, pH 7.4 at 37 °C. The properties of the anticoagulant mutant WE are also shown for comparison (gray circle). Note how mutagenesis of residue 215 produces constructs (W215V) with properties equivalent to those of WE. In some cases (W215D, W215E, W215I, W215L, W215R) the anticoagulant/antithrombotic profile of WE is significantly improved with a single amino acid substitution.
Having explored the properties of residue 215 by saturation mutagenesis, we turned our attention to the autolysis loop of thrombin that decorates the lower rim of the active site and is strategically positioned between the Na+ site and exosite I (Fig. 2). After being neglected for years because of its intrinsic disorder in crystal structures, the autolysis loop has recently gained attention because deletion of the nine residues 146ETWTANVGK149e produces a potent anticoagulant due to stabilization of the inactive E* form (40, 41). Because the loop is not in contact with the region around residue 215, we surmised that a combined mutation of residue 215 and deletion of 146ETWTANVGK149e in the autolysis loop could afford an additive effect and result in improved anticoagulant/antithrombotic profile relative to the individual mutations. The mutant W215G was initially selected as a basis for the double mutation in view of its properties similar to those of W215A (Figs. 3 and 4). The double mutant W215G/Δ146–149e maps into the target region and features almost complete additivity of the individual W215G and Δ146–149e mutations (Fig. 5). Encouraged by these results, we used W215E as a basis for the combined mutation with Δ146–149e. Our expectation was that the mutant W215E/Δ146–149e would feature no measurable activity toward fibrinogen and PAR1, but would retain appreciable activity toward the anticoagulant protein C. Contrary to this expectation, the mutant W215E/Δ146–149e features a kcat/Km for PAR1 cleavage >1,000-fold higher than that of fibrinogen or protein C and behaves as an “exclusive” PAR1 agonist. An important lesson in protein engineering is learned from these constructs. Although the W215G substitution has similar effects on macromolecular substrate recognition on the wild-type or Δ146–149e scaffolds, the W215E substitution has profoundly different functional consequences on the two scaffolds. On the wild-type scaffold it favors protein C activation over fibrinogen cleavage and equalizes PAR1 and protein C activation. On the Δ146–149e scaffold it produces an almost exclusive agonist of PAR1. Hence, mutation of the same residue in two different thrombin scaffolds returns completely different specificity toward macromolecular substrates and generates a mutant with properties not seen in any of the individual constructs used in the mutagenesis.
FIGURE 5.
Non-additivity of mutational effects in the thrombin scaffold. Functional properties of the double mutants W215G/Δ146–149e (W215G/Δ) and W215E/Δ146–149e (W215E/Δ) (gray circles) obtained by combining the single mutants W215G and W215E (see Figs. 3 and 4) with deletion of residues 146–149e in the autolysis loop (Δ) that produces a thrombin mutant with anticoagulant/antithrombotic profile (40, 41). Note how the double mutant W215G/Δ146–149e has additive properties relative to the single mutations W215G and Δ146–149e, resulting in a construct with preferential activity toward protein C. On the other hand, the double mutant W215E/Δ146–149e has properties that cannot be predicted from additivity of the single mutations W215E and Δ146–149e, resulting in a construct with almost exclusive activity toward PAR1. Experimental conditions are: 5 mm Tris, 0.1% PEG8000, 145 mm NaCl, pH 7.4 at 37 °C. The values of s for the two double mutants are: (W215G/Δ146–149e) 0.43 ± 0.02 mm−1 s−1 (FpA), 1.4 ± 0.1 mm−1 s−1 (PAR1) and 5.3 ± 0.2 mm−1 s−1 (PC); (W215E/Δ146–149e) 0.30 ± 0.01 mm−1 s−1 (FpA), 890 ± 10 mm−1 s−1 (PAR1), and 0.50 ± 0.02 mm−1 s−1 (PC).
DISCUSSION
Although substantial progress has been made in the prevention and treatment of cardiovascular disease and its major risk factors, it has been predicted that thrombotic complications will remain the leading cause of death and disability and will represent a major burden to productivity worldwide well into the year 2020 (42). Indeed, thrombosis is the most prevalent cause of fatal diseases in developed countries. An antithrombotic agent that can be administered to patients with severe acute thrombotic diseases without the risk of causing hemorrhage, as experienced with antithrombotic/thrombolytic therapy in the treatment of acute ischemic stroke (43) or systemic anticoagulants like heparin (44), would likely revolutionize the treatment of cardiovascular and cerebrovascular diseases. Thrombin variants engineered for optimal activity toward protein C and minimal activity toward fibrinogen and PAR1 hold much promise in this connection as shown by their remarkable properties in vitro and in vivo (11, 12, 14, 28–31, 40, 45, 46). The double mutant WE is at the forefront of current engineering efforts and structural biology has elucidated the precise mechanism underscoring its functional properties in ways that are relevant to other thrombin constructs (39, 41, 47). Optimization of the properties of WE to completely abrogate activity toward fibrinogen and PAR1 was deemed necessary to move forward with our engineering efforts.
Mutagenesis studies in proteins are almost always confined to selected substitutions, usually isosteric or Ala. Saturation mutagenesis would not be practical in general cases to establish the role of specific residues. However, once critical residues have been identified from single substitutions, there is value to subject the system to a brute force approach where the entire gamut of naturally occurring amino acids is explored in its functional consequences. The results may be unexpected. In the case of residue 225, the Y225I mutant shows >100,000-fold reduction in the activity toward synthetic substrates and physiological substrates like fibrinogen and protein C (37), although residue 225 makes no contact with substrate based on available crystal structures. Unlike residue 225, Trp215 makes direct interaction with substrate (7, 48) and is absolutely conserved in trypsin-like proteases (3). Hints of the dominant role of residue 215 were gathered from earlier studies on the W215A, W215F, W215Y, and W215P mutants (10, 47), but the saturation mutagenesis presented here demonstrates unequivocally that residue 215 controls thrombin specificity toward macromolecular substrates in ways not seen for any other residue of the enzyme. It should be emphasized that Trp215 is directly involved in the E*-E allosteric equilibrium of thrombin (7, 18, 38, 41, 49, 50) and therefore is expected to play a dominant role in thrombin function. Elucidation of the precise mechanism underlying the functional properties of each mutant of Trp215 will require further investigation. Analysis of Na+ binding to mutants of Trp215 is complicated by the fact that this residue is a major fluorophore reporting the change linked to the E*-E and E-E:Na+ equilibria of the enzyme (18). Structural investigation of the constructs in underway and the mutants W215P, W215S, and W215E show different degrees of collapse of the 215–217 β-strand as documented in the structure of the WE mutant (39) and the inactive E* form of thrombin (38, 41, 49, 50).
Two important results emerge from our study. First, the remarkable properties of WE can be matched and even exceeded by single amino acid substitutions of Trp215, thereby opening new opportunities in the task of completely abrogating activity toward fibrinogen and PAR1 and engineering thrombin variants with exclusive activity toward the anticoagulant protein C for clinical applications. Second, mutations of residue 215 show functional consequences that are context-dependent. In the wild-type scaffold, mutations of Trp215 tend to favor protein C activation over fibrinogen and PAR1 cleavage. Deletion of the entire autolysis loop of thrombin produces anticoagulant propensity but the added mutation of Trp215 in this scaffold produces additive effects for W215G but not W215E. As a result, W215G/Δ146–149e is a construct with dominant specificity toward protein C, but W215E/Δ146–149e is a construct with almost exclusive specificity toward PAR1. It would be of interest to establish if saturation mutagenesis of Trp215 in the Δ146–149e scaffold produces constructs where the three major functions of thrombin, procoagulant, prothrombotic and anticoagulant can be completely dissociated. The constructs already emerged from our mutagenesis studies hold much promise to improve on the anticoagulant/antithrombotic activity of WE in vitro and in vivo (11, 26, 28–30, 39, 47) and to help dissect the role of thrombin and other proteases in the PAR1 signaling pathways (51–57).
The context dependence of mutations of Trp215 in thrombin is of great relevance to protein engineering studies in general. Mutations of residues that participate independently to ligand recognition are expected to produce additive effects (58). The outcome is particularly favorable to protein engineers because it enables facile prediction of the properties of the engineered construct in terms of its building blocks. The additivity observed for the W215G/Δ146–149e mutant may result from combination of two distinct effects, one destabilizing Na+ binding (W215G) and shifting the equilibrium from E:Na+ to E and the other shifting E to the inactive E* form (Δ146–149e). Lack of additivity of mutational effects in thrombin may be caused by the allosteric equilibria involved in the function of the enzyme (18) and the fact that different structural domains are linked to each other through these equilibria. When two mutations interfere with the same mechanism of recognition, say precluding Na+ binding, the double mutant would feature properties no different than those of the individual mutations thereby producing non-additivity of mutational effects. In general, when additivity does not ensue, prediction of the properties of the final construct becomes difficult. Although the lack of additivity of mutational effects limits our ability to rationally design proteins for specific functions, it alerts us about the rich repertoire of functional features often accessible to a protein scaffold. The non-additive effect observed for the W215E/Δ146–149e mutant is caused by the emergence of a new property not observed with either one of the individual mutations, which is the exquisite selectivity toward PAR1 never before encountered in a thrombin construct. In this case, W215E and deletion of the autolysis loop produce a new architecture of the active site that favors interaction with PAR1 over fibrinogen and protein C. Structural investigation of the W215E/Δ146–149e mutant will hopefully reveal the molecular basis of this new property of the thrombin scaffold. The mutant is a striking reminder of the functional plasticity available to trypsin-like proteases and the need to go beyond Ala-scanning mutagenesis in our exploration of the molecular determinants of enzyme specificity, as recently demonstrated by protein engineering studies of the outer membrane protease from E. coli, OmpT (59).
This work was supported, in whole or in part, by National Institutes of Health Research Grants HL49413, HL58141, HL73813, and HL95315 (to E. D. C.).
- PAR
- protease-activated receptor
- PEG
- polyethyleneglycol
- WT
- wild type
- FpA
- fibrinopeptide A
- PC
- protein C
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Di Cera E. (2008) Nat. Chem. Biol. 4, 270–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schechter I., Berger A. (1967) Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 3.Page M. J., Di Cera E. (2008) Cell Mol. Life Sci. 65, 1220–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedstrom L. (2002) Chem. Rev. 102, 4501–4524 [DOI] [PubMed] [Google Scholar]
- 5.Perona J. J., Craik C. S. (1995) Protein Sci. 4, 337–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perona J. J., Craik C. S. (1997) J. Biol. Chem. 272, 29987–29990 [DOI] [PubMed] [Google Scholar]
- 7.Di Cera E. (2008) Mol. Aspects Med. 29, 203–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bock P. E., Panizzi P., Verhamme I. M. (2007) J. Thromb. Haemost. 5, Suppl. 1, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenton J. W., 2nd (1986) Ann. N.Y. Acad. Sci. 485, 5–15 [DOI] [PubMed] [Google Scholar]
- 10.Arosio D., Ayala Y. M., Di Cera E. (2000) Biochemistry 39, 8095–8101 [DOI] [PubMed] [Google Scholar]
- 11.Cantwell A. M., Di Cera E. (2000) J. Biol. Chem. 275, 39827–39830 [DOI] [PubMed] [Google Scholar]
- 12.Gibbs C. S., Coutré S. E., Tsiang M., Li W. X., Jain A. K., Dunn K. E., Law V. S., Mao C. T., Matsumura S. Y., Mejza S. J. (1995) Nature 378, 413–416 [DOI] [PubMed] [Google Scholar]
- 13.Le Bonniec B. F., Esmon C. T. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7371–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang Q. D., Guinto E. R., Di Cera E. (1997) Nat. Biotechnol. 15, 146–149 [DOI] [PubMed] [Google Scholar]
- 15.Pineda A. O., Carrell C. J., Bush L. A., Prasad S., Caccia S., Chen Z. W., Mathews F. S., Di Cera E. (2004) J. Biol. Chem. 279, 31842–31853 [DOI] [PubMed] [Google Scholar]
- 16.Prasad S., Wright K. J., Banerjee, Roy D., Bush L. A., Cantwell A. M., Di Cera E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13785–13790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayala Y. M., Cantwell A. M., Rose T., Bush L. A., Arosio D., Di Cera E. (2001) Proteins 45, 107–116 [DOI] [PubMed] [Google Scholar]
- 18.Bah A., Garvey L. C., Ge J., Di Cera E. (2006) J. Biol. Chem. 281, 40049–40056 [DOI] [PubMed] [Google Scholar]
- 19.Bush-Pelc L. A., Marino F., Chen Z., Pineda A. O., Mathews F. S., Di Cera E. (2007) J. Biol. Chem. 282, 27165–27170 [DOI] [PubMed] [Google Scholar]
- 20.Krem M. M., Prasad S., Di Cera E. (2002) J. Biol. Chem. 277, 40260–40264 [DOI] [PubMed] [Google Scholar]
- 21.Papaconstantinou M. E., Bah A., Di Cera E. (2008) Cell Mol Life Sci 65, 1943–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasad S., Cantwell A. M., Bush L. A., Shih P., Xu H., Di Cera E. (2004) J. Biol. Chem. 279, 10103–10108 [DOI] [PubMed] [Google Scholar]
- 23.Xu H., Bush L. A., Pineda A. O., Caccia S., Di Cera E. (2005) J. Biol. Chem. 280, 7956–7961 [DOI] [PubMed] [Google Scholar]
- 24.Vindigni A., Di Cera E. (1996) Biochemistry 35, 4417–4426 [DOI] [PubMed] [Google Scholar]
- 25.Tsiang M., Jain A. K., Dunn K. E., Rojas M. E., Leung L. L., Gibbs C. S. (1995) J. Biol. Chem. 270, 16854–16863 [DOI] [PubMed] [Google Scholar]
- 26.Berny M. A., White T. C., Tucker E. I., Bush-Pelc L. A., Di Cera E., Gruber A., McCarty O. J. (2008) Arterioscler. Thromb. Vasc. Biol. 18, 329–334 [DOI] [PubMed] [Google Scholar]
- 27.Feistritzer C., Schuepbach R. A., Mosnier L. O., Bush L. A., Di Cera E., Griffin J. H., Riewald M. (2006) J. Biol. Chem. 281, 20077–20084 [DOI] [PubMed] [Google Scholar]
- 28.Gruber A., Cantwell A. M., Di Cera E., Hanson S. R. (2002) J. Biol. Chem. 277, 27581–27584 [DOI] [PubMed] [Google Scholar]
- 29.Gruber A., Fernández J. A., Bush L., Marzec U., Griffin J. H., Hanson S. R., Di Cera E. (2006) J. Thromb. Haemost. 4, 392–397 [DOI] [PubMed] [Google Scholar]
- 30.Gruber A., Marzec U. M., Bush L., Di Cera E., Fernández J. A., Berny M. A., Tucker E. I., McCarty O. J., Griffin J. H., Hanson S. R. (2007) Blood 109, 3733–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsiang M., Paborsky L. R., Li W. X., Jain A. K., Mao C. T., Dunn K. E., Lee D. W., Matsumura S. Y., Matteucci M. D., Coutré S. E., Leung L. L., Gibbs C. S. (1996) Biochemistry 35, 16449–16457 [DOI] [PubMed] [Google Scholar]
- 32.Bode W., Turk D., Karshikov A. (1992) Protein Sci 1, 426–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stubbs M. T., Oschkinat H., Mayr I., Huber R., Angliker H., Stone S. R., Bode W. (1992) Eur J Biochem. 206, 187–195 [DOI] [PubMed] [Google Scholar]
- 34.Isetti G., Maurer M. C. (2007) Biochemistry 46, 2444–2452 [DOI] [PubMed] [Google Scholar]
- 35.Sadasivan C., Yee V. C. (2000) J. Biol. Chem. 275, 36942–36948 [DOI] [PubMed] [Google Scholar]
- 36.Bah A., Chen Z., Bush-Pelc L. A., Mathews F. S., Di Cera E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11603–11608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guinto E. R., Caccia S., Rose T., Fütterer K., Waksman G., Di Cera E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1852–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gandhi P. S., Chen Z., Mathews F. S., Di Cera E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1832–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gandhi P. S., Page M. J., Chen Z., Bush-Pelc L. A., Di Cera E. (2009) J. Biol. Chem. 284, 24098–24105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang Q. D., Sabetta M., Di Cera E. (1997) J. Biol. Chem. 272, 19649–19651 [DOI] [PubMed] [Google Scholar]
- 41.Bah A., Carrell C. J., Chen Z., Gandhi P. S., Di Cera E. (2009) J. Biol. Chem. 284, 20034–20040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerszten R. E., Wang T. J. (2008) Nature 451, 949–952 [DOI] [PubMed] [Google Scholar]
- 43.Padma V., Fisher M., Moonis M. (2005) Exp. Rev. Neurother. 5, 223–233 [DOI] [PubMed] [Google Scholar]
- 44.Busch M., Masuhr F. (2004) Eur. J. Med. Res. 9, 199–206 [PubMed] [Google Scholar]
- 45.Griffin J. H. (1995) Nature 378, 337–338 [DOI] [PubMed] [Google Scholar]
- 46.Grinnell B. W. (1997) Nat. Biotechnol. 15, 124–125 [DOI] [PubMed] [Google Scholar]
- 47.Pineda A. O., Chen Z. W., Caccia S., Cantwell A. M., Savvides S. N., Waksman G., Mathews F. S., Di Cera E. (2004) J. Biol. Chem. 279, 39824–39828 [DOI] [PubMed] [Google Scholar]
- 48.Bode W. (2006) Blood Cells Mol. Dis. 36, 122–130 [DOI] [PubMed] [Google Scholar]
- 49.Niu W., Chen Z., Bush-Pelc L. A., Bah A., Gandhi P. S., Di Cera E. (2009) J. Biol. Chem. 284, 36175–36185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pineda A. O., Chen Z. W., Bah A., Garvey L. C., Mathews F. S., Di Cera E. (2006) J. Biol. Chem. 281, 32922–32928 [DOI] [PubMed] [Google Scholar]
- 51.Brass L. F. (2003) Chest 124, 18S–25S [DOI] [PubMed] [Google Scholar]
- 52.Coughlin S. R. (2000) Nature 407, 258–264 [DOI] [PubMed] [Google Scholar]
- 53.Coughlin S. R. (2005) J. Thromb. Haemost. 3, 1800–1814 [DOI] [PubMed] [Google Scholar]
- 54.Griffin J. H., Zlokovic B., Fernández J. A. (2002) Semin Hematol 39, 197–205 [DOI] [PubMed] [Google Scholar]
- 55.Mosnier L. O., Yang X. V., Griffin J. H. (2007) J. Biol. Chem. 282, 33022–33033 [DOI] [PubMed] [Google Scholar]
- 56.Mosnier L. O., Zampolli A., Kerschen E. J., Schuepbach R. A., Banerjee Y., Fernández J. A., Yang X. V., Riewald M., Weiler H., Ruggeri Z. M., Griffin J. H. (2009) Blood 113, 5970–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riewald M., Petrovan R. J., Donner A., Mueller B. M., Ruf W. (2002) Science 296, 1880–1882 [DOI] [PubMed] [Google Scholar]
- 58.Wells J. A. (1990) Biochemistry 29, 8509–8517 [DOI] [PubMed] [Google Scholar]
- 59.Varadarajan N., Rodriguez S., Hwang B. Y., Georgiu G., Iverson B. L. (2008) Nat. Chem. Biol. 4, 290–294 [DOI] [PMC free article] [PubMed] [Google Scholar]