Abstract
The Rac1b splice isoform contains a 19-amino acid insertion not found in Rac1; this insertion leads to decreased GTPase activity and reduced affinity for GDP, resulting in the intracellular predominance of GTP-bound Rac1b. Here, using co-precipitation and proteomic methods, we find that Rac1b does not bind to many common regulators of Rho family GTPases but that it does display enhanced binding to SmgGDS, RACK1, and p120 catenin (p120ctn), proteins involved in cell-cell adhesion, motility, and transcriptional regulation. We use molecular modeling and structure analysis approaches to determine that the interaction between Rac1b and p120ctn is dependent upon protein regions that are predicted to be unstructured in the absence of molecular complex formation, suggesting that the interaction between these two proteins involves coupled folding and binding. We also find that directed cell movement initiated by Rac1b is dependent upon p120. These results define a distinct binding functionality of Rac1b and provide insight into how the distinct phenotypic program activated by this protein may be implemented through molecular recognition of effectors distinct from those of Rac1.
Keywords: Cell Migration, Epithelial Cell, Mammary Gland, Protein-Protein Interactions, Signal Transduction
Introduction
The Ras superfamily of small GTPases mediates numerous biological functions through specific binding to a plethora of effector proteins when associated with GTP. The function of the small GTPases in diverse cellular processes, such as signal transduction, cytoskeletal organization, cell migration, transcription, vesicle transport, and cytokinesis, is carefully regulated by regulator proteins: guanine nucleotide exchange factors (GEFs),2 GDP dissociation stimulators (GDSs), and GTPase-activating proteins (GAPs). Rac1b is a splice isoform of Rac1 that results from the inclusion of exon 3b, which contains 57 nucleotides and which leads to a 19-amino acid in-frame insertion (1, 2). Rac1b was first identified in human skin and epithelial tissue of the intestinal tract and has been found to be up-regulated in malignant colorectal (1) and breast cancer (2). When assessed in cultured cells, Rac1b has been found to have many of the properties of GTPase-defective Rac1 mutants. For example, exogenously expressed Rac1b promotes density- and anchorage-independent cell growth of NIH 3T3 cells (3), cell survival and cell cycle progression in mouse fibroblasts (4), and Dishevelled-3-mediated detachment of colorectal cancer cells (5). However, Rac1b also shows distinct activities from Rac1. Unlike Rac1, Rac1b does not promote cadherin-dependent disassembly of adherens junctions in keratinocytes (6), it does not induce cytoskeletal rearrangements in colorectal cancer cells as effectively as GTPase-defective mutants of Rac1 (3), and it is not as effective as Rac1 for activation of RelB (7). Furthermore, Rac1b has distinct positive activities, because expression of Rac1b has been shown to mediate epithelial-mesenchymal transition and genomic instability in mouse mammary epithelial cells exposed to MMP3 (matrix metalloproteinase 3) (8, 9) and to sustain colorectal tumor cell survival (10, 11). These results suggest that Rac1b is capable of acting through pathways quite distinct from those controlled by Rac1.
Structural studies of Rac1b have revealed that the 19-aa insertion confers greatly increased mobility on the switch regions. In crystal structures of Rac1b bound to either GDP or a nonhydrolyzable GTP analog, not only was the insertion not visible in electron density, but the adjacent switch 2 was disordered as well (12). Furthermore, switch 1 residues, although stabilized in an open conformation by crystal packing interactions, exhibited increased B-factors also suggestive of structural destabilization and increased flexibility (12). Biochemical studies have shown that the 19-aa insertion into Rac1b confers accelerated GDP/GTP exchange and impaired GTP hydrolysis, explained by the loss of stabilizing structural elements near the GTP-hydrolyzing active site (12, 13). These impairments in nucleotide binding and hydrolysis result in the predominance of the GTP-bound form in the cellular context, leading to extended signaling activity (12). Because Rac1b is capable of binding with reduced affinity to several Rac1 regulators and effectors (4, 5, 7, 13), previous studies have focused on the physiological consequences mediated by Rac1b through its interaction with a subset of Rac1 binding partners. We hypothesized that the 19-aa insertion and its influence on the conformation and dynamics of the switch regions might confer upon Rac1b the ability to interact with a unique set of binding partners. Here, we report our findings that Rac1b shows markedly different interactions from Rac1 with a variety of regulator and effector proteins and that these interactions are mediated at least in part via interactions with the 19-amino acid insertion. We propose that the 19-amino acid insertion and adjacent switch regions are natively unstructured or sample a multiplicity of conformations in Rac1b but undergo disorder-to-order transition in the molecular recognition of several unique binding partners, distinct from those of Rac1. Such unique interactions may be responsible for the distinct physiological effects of Rac1b, including promotion of malignancy and induction of epithelial-mesenchymal transition.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Murine mammary epithelial (SCp2) cells were cultured as described previously (8). Cells were grown in plastic tissue culture dishes for biochemical analyses and on acid-washed coverslips for fluorescence microscopy. Monoclonal pan-p120ctn and anti-RACK1 antibodies were purchased from Transduction Laboratories (Lexington, KY). Anti-GIT1, -PAK, and -RhoGDI antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-SmgGDS antibodies were a gift from Dr. C. Williams (Department of Pharmacology and Toxicology, Medical College of Wisconsin, Milwaukee, WI). Polyclonal anti-YFP antibodies were from Invitrogen, and horseradish peroxidase-tagged anti-GST antibodies were from Abcam Inc. (Cambridge, MA). Secondary goat anti-rabbit and goat anti-mouse IgG antibodies linked to horseradish peroxidase were from Tago, Inc. (Burlingame, CA). ProLong antifade reagent was from Invitrogen, and glutathione-agarose beads, GDP, and GTPγS were from Sigma. Gene knockdown was performed using Mission shRNA as described previously (14); p120ctn targeting constructs were as follows: MC41, NM_007615.1–1572s1c1; MC42, NM_007615.1–2284s1c1; MC43, NM_007615.1–2545s1c1; MC44, NM_007615.1–3996s1c1; MC45, NM_007615.1–985s1c1. Rac1b quantitation and cell area were calculated as described previously (9).
Expression Constructs
For the formation of GST-tagged Rac1, Rac1V12, Rac1N17, and Rac1b DNA constructs, the corresponding DNAs in YFP expression vectors (8) were cut at BglI and BamHI sites and cloned into the BamHI site of pGEX 4T-1 vector. For the formation of His-tagged Rac1, Rac1V12, Rac1N17, and Rac1b DNA constructs, YFP-tagged Rac1, Rac1V12, Rac1N17, and Rac1b were cut and cloned into pET-28a(+) vector (Novagen, Gibbstown, NJ) using EcoRI and BamHI restriction sites. To generate the 19-amino acid insertion fragment of Rac1b in GST tag and YFP fusion vectors, a DNA sequence coding for the 19-aa insertion was cloned into pGEX 4T-1 vector using the BamHI restriction site and cloned into pYFP-C1 vector (Invitrogen) using EcoRI, BamHI sites. All constructs were confirmed by direct sequencing of the plasmids. p120-GST expression constructs were described previously (15, 16). Transfection of SCp2 cells was performed using Lipofectamine 2000 reagent (Invitrogen). Twenty-four hours after transfection, cells were used for biochemical experiments or fluorescence microscopy.
Mass Spectrometry Analysis
The mass spectrometry analysis was essentially done as previously described (17). Briefly, the protein SDS-polyacrylamide gel was silver-stained using the SilverSNAP stain for mass spectrometry kit (Pierce). Subsequently, the silver-stained gel bands were excised, destained, reduced, and alkylated with dithiothreitol and iodoacetamide. Proteins were digested for 4 h with 0.6 μg of trypsin (Promega) in digestion buffer (20 mm Tris, pH 8.1, 0.0002% Zwittergent 3-16, at 37 °C), followed by peptide extraction with 60 μl of 2% trifluoroacetic acid and then 60 μl of acetonitrile. Pooled extracts were concentrated and then brought up in 0.1% formic acid for protein identification by nano-flow liquid chromatography MS/MS analysis using a ThermoFinnigan LTQ Orbitrap hybrid mass spectrometer (ThermoElectron Bremen) coupled to an Eksigent nano-flow liquid chromatography two-dimensional HPLC system (Eksigent, Dublin, CA). The MS/MS raw data were converted to DTA files using ThermoElectron Bioworks 3.2 and correlated to theoretical fragmentation patterns of tryptic peptide sequences from the Swiss-Prot data base using both SEQUESTTM2 (ThermoElectron, San Jose, CA) and the MascotTM3 (Matrix Sciences, London, UK) search algorithms running on a 10-node cluster (the Swiss-Prot data base was downloaded on May 21, 2006, and 212,425 sequences/77,942,645 residues were actually searched for experiments that identified GIT1, SmgGDS, RhoGDI, and IQGAP1; the Swiss-Prot data base was downloaded on January 9, 2007, and 252,616 sequences/92,372,123 residues were actually searched for experiments that identified p210ctn and RACK1). All searches were conducted with fixed cysteine modifications of +57 for carboxamidomethyl cysteines and variable modifications allowing +16 with methionines for methione sulfoxide and +42 for protein N-terminal acetylation. The search was restricted to trypsin-generated peptides, allowing for two missed cleavages, and was restricted to mice. Peptide mass search tolerances were set to 20 ppm, and fragment mass tolerances were set to ±0.8 Da. Protein identifications were considered when Mascot search results gave at least two unique consensus peptides, each with peptide probability scores at or exceeding the 95% probability cut-off, which was 29 for these searches, and ranking as the number one match for the respective MS/MS spectra. The MS/MS spectra from all of the Mascot-identified peptides were inspected manually for spectrum quality and fragment matching. Matches to poor quality spectra or spectra with numerous unmatched ions were not included. The mass spectrometry analysis was done at the Mayo Proteomics Research center (Mayo Clinic College of Medicine, Rochester, MN). Protein identification data, including accession, number of unique sequences identified, and percentage of sequence coverage are presented in Table 1; a complete list of identified peptides is presented in supplemental Table 1.
TABLE 1.
Protein identification data
Fluorescence Microscopy
Cells were cultured for 24 h, washed in Dulbecco's phosphate-buffered saline, and then fixed with 3% formaldehyde in Dulbecco's phosphate-buffered saline for 10 min. After fixation, the cells were washed three times in Dulbecco's phosphate-buffered saline, permeabilized in 0.1% Triton, and stained with pan-p120 antibody according to immunostaining protocols described previously (18). Fluorescence microscopy was performed using a Zeiss Axiovert 35 microscope with a ×63, 1.4 numerical aperture lens.
Immunoprecipitation Experiments
Immunoprecipitations of YFP-Rac1 fusion proteins were performed using magnetic Dynabeads according to the manufacturer's protocols (Invitrogen). Immunoprecipitations of endogenous proteins was done as previously described (18). Immunoprecipitates were solubilized in an SDS-containing sample buffer and boiled for 5 min. Electrophoresis was performed in 10% acrylamide gels, followed by transfer to polyvinylidene difluoride membranes. SDS-PAGE (19), transfer of proteins to polyvinylidene difluoride membranes (20), and Western blotting (21) were as described. Immunodetection of bound antibodies on nitrocellulose membrane was performed using ECL reagents (Amersham Biosciences). All procedures were carried out according to the manufacturer's instructions. Mass spectrometry analysis of protein bands excised from the silver-stained gels was performed as described previously (17).
GST Pull-down Assay
For GST pull-downs, GST-Rac1, -Rac1V12, -Rac1N17, -19 aa of Rac1b, and -Rac1b or GST-p120ctn in BL21 (DE3) Escherichia coli were plated in 2 ml of 2× YT medium for overnight growth. The next day, the 2-ml cultures were diluted in 500 ml of 2× YT medium and incubated for 3 h at 30 °C. After that, expression of GST fusion proteins was induced by 40 μm isopropyl 1-thio-β-d-galactopyranoside, and bacteria were allowed to grow for another 2 h. Purification of fusion proteins was done using the B-PER GST fusion protein purification kit (Pierce) according to the manufacturer's instructions. Following purification, GST-Rac1, -Rac1V12, -Rac1N17, -Rac1b, and -19 aa of Rac1b fusion proteins were incubated for 1 h with glutathione-agarose beads, thoroughly washed three times in phosphate-buffered saline, and incubated with murine mammary epithelial SCp2 cell lysate for protein binding. For GST-p120ctn pull-downs, GST-p120ctn proteins attached to the glutathione-agarose beads were incubated with cell lysate from SCp2 cells transfected with YFP-Rac1, -Rac1V12, -Rac1N17, -Rac1b, and -19 aa of Rac1b fusion constructs.
In Vitro Direct Binding Assays
Purification of His-tagged fusion proteins from BL21 (DE3) E. coli was performed using the B-PER His6 fusion protein purification kit from Pierce. Direct binding of His-tagged Rac1 and Rac1b to the GST-tagged short isoform of p120ctn-4A was done as recently described (14) with some modifications. Briefly, 80 μl of nickel affinity Ni2+-nitrilotriacetic acid-agarose gel (Qiagen GmbH, Hilden, Germany) was added to purified His-tagged Rac1, Rac1V12, Rac1N17, and Rac1b. After 1 h of incubation at 4 °C, the gel was washed three times with wash buffer (50 mm Tris-HCl, pH 6.8, 300 mm NaCl, 25 mm imidazole, 10% glycerol) and then two times with GDP/GTP binding buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol, 0.1% Triton X-100). After that, the gel was mixed with 500 μm GDP or GTPγS in 1 ml of GDP binding buffer and rotated for 30 min following three washes with GDP/GTP binding buffer. GDP- or GTPγS-bound His-Rac1s were then eluted from the nickel-agarose gel using 100 μl of elution buffer (50 mm Tris-HCl, pH 6.8, 300 mm NaCl, 200 mm imidazole, 10% glycerol), diluted 25 times with protein binding buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 20 mm MgCl2, 1 mm DTT, 1 mm EDTA, 0.1% Triton X-100), and added to GST-p120ctn-agarose beads in protein binding buffer for direct protein binding assay.
Biochemical GDP/GTP Exchange Assay
Biochemical GDP/GTP exchange assay was done using the RhoGEF exchange assay Biochem kit (Cytoskeleton, Inc.) according to the manufacturer's instructions. Assay kinetic reactions were performed in 96-well plates at 22–25 °C on a BioTek fluorometer, and data were analyzed using Gen5 software (BioTek Instruments Inc.).
Disorder Propensity Analyses, Homology Modeling, and Generation of Structural Figures
Disorder propensity sum plots were generated with GlobPlot (version 2.3; Biocomputing Unit, European Molecular Biology Laboratory (available on the World Wide Web)) (22), using Russell/Linding propensities and default parameters, for the amino acid sequences of human p120ctn isoform 1AB (Swiss-Prot entry O60716-2) and mouse Rac1b (NCBI Entrez Protein entry EDL19046). A homology model for mouse p120ctn (Swiss-Prot entry P30999) was generated using Swiss-Model software (Biozentrum Basel/Swiss Institute of Bioinformatics) (23) via the EXPASY Web server (24), using the First Approach mode and default parameters. The template (the ARM (armadillo) domain of plakophilin 1; Protein Data Bank entry 1XM9 chain A) and the modeled residue range (residues 368–830) were selected automatically by the software. The sequence alignment of p120ctn with plakophilin 1 was visually inspected to confirm alignment of conserved residues that define the ARM consensus motif (25), but no manual editing was necessary. Figs. 1 and 8B were generated using PyMOL (version 1.00; DeLano Scientific LLC, Palo Alto, CA). Structure coordinates are from Protein Data Bank entries for Rac1-GDP bound to arfaptin (1I4L), Rac1-GPPNP (1MH1), Rac1b-GDP (1RYF), and Rac1b-GPPNP (1RYH).
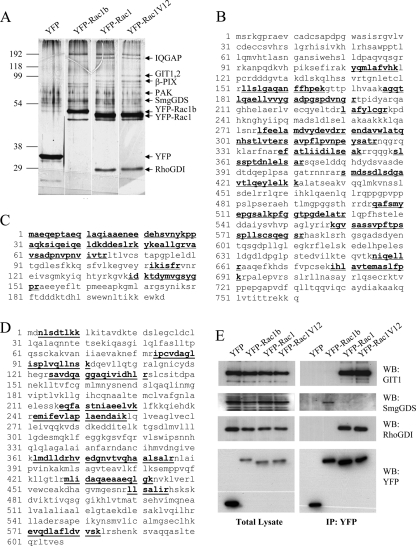
FIGURE 1.
Rac1b has altered binding affinity to common regulators of Rho family GTPases. A, silver-stained protein gel after SDS-PAGE of protein complexes purified from cellular lysates shows proteins that differentially interact with YFP-Rac1b, YFP-Rac1, and YFP-Rac1V12. YFP used as a negative control for protein interactions. B–D, peptide coverage maps of GIT1 (B), RhoGDI (C), and SmgGDS (D). For B–D, the peptides used for identification are underlined and in boldface type. E, Western blot (WB) of total lysates from cells transiently transfected with YFP-Rac1b, YFP-Rac1, and YFP-Rac1V12 shows expression levels of GIT1, RhoGDI, PAK, YFP-Rac1, YFP-Rac1V12, YFP-Rac1b, and YFP as a control. IP, immunoprecipitation.
RESULTS
We previously found that Rac1b could induce breakdown of cell-cell adhesion and increased cell motility in SCp2 mouse mammary epithelial cells (8). In order to compare and identify differences in Rac1 and Rac1b protein complexes, we performed two complementary proteomic screens. In the first, we expressed Rac1b, wild type Rac1, and constitutive active mutant Rac1V12 as YFP fusion proteins so as to produce within the cell context sufficient protein to bind to all available partners. YFP-tagged proteins were immunoprecipitated, and protein complexes were resolved using SDS-PAGE and analyzed by silver stain. We found that Rac1 and Rac1b co-immunoprecipitate with distinct interacting proteins (Fig. 1A). These bands were excised, digested with trypsin, and subjected to mass spectrometry for protein identification. GIT-1, RhoGDI, and IQGAP, were specifically identified in the Rac1 purifications (Fig. 1, B and C; Table 1; supplemental Fig. 1; and supplemental Table 1). None of these proteins were identified in the Rac1b purification, which is also consistent with previous observations demonstrating that PAK and RhoGDI do not bind Rac1b (12, 13). Specific antibodies against GIT1 and RhoGDI were used to validate the mass spectrometry data using Western blot analyses; these experiments confirmed the greater affinity of these effectors for Rac1 and Rac1V12 as compared with Rac1b (Fig. 1E), and similar results were shown for IQGAP and RhoGDI (supplemental Fig. 2). Interestingly, SmgGDS, an atypical GEF with multiple armadillo repeats (26, 27), was identified in this screen as a specific binding partner of Rac1b (Fig. 1D, Table 1, and supplemental Table 1), a finding that was also validated by Western analysis (Fig. 1E).
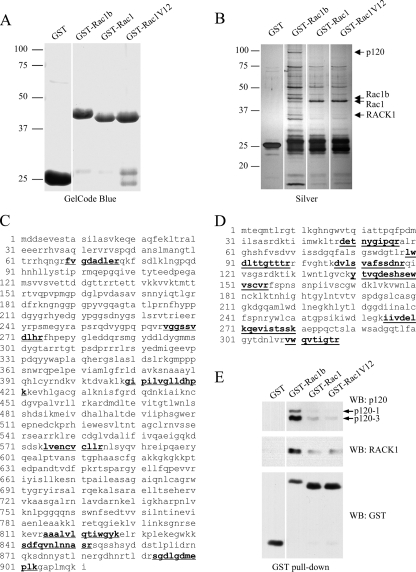
As a complementary screen for Rac1/Rac1b-binding proteins, we generated purified recombinant GST-Rac1, -Rac1V12, and -Rac1b fusion proteins and incubated these with mouse mammary epithelial SCp2 cell lysates, a method that has the benefit of not perturbing the overall protein expression patterns in the cells. Protein complexes were resolved using SDS-PAGE and analyzed using silver staining (Fig. 2A). We found that a number of proteins were enriched in the Rac1b purification compared with the Rac1 purifications (Fig. 2B, Table 1, and supplemental Table 1). Proteomic analysis of two of these bands identified p120ctn, an armadillo domain protein involved in multiple cellular functions (28) (Fig. 2C), and RACK1 (receptor for activated C kinase 1), a scaffolding protein involved in key signaling pathways (29) (Fig. 2D). Specific antibodies against p120ctn and RACK1 were used to validate these data in Western blot analyses (Fig. 2E). These data suggest that the 19-aa insertion found in Rac1b and not Rac1 may act as a novel and specific binding site.
FIGURE 2.
Rac1b preferentially interacts with p120ctn and RACK1. A, protein gel shows the bacterially purified GST fusion proteins used in the GST pull-downs. B, silver-stained gel after SDS-PAGE of proteins precipitated by GST-tagged Rac1b, Rac1, and Rac1V12 from SCp2 mammary epithelial cells demonstrates differential binding of the cellular proteins to GST fusions. GST and beads were used as the negative control. C, peptide coverage map of p120ctn. C, peptide coverage map of RACK1. For C and D, the peptides used for identification are underlined and in boldface type. E, Western blot (WB) of GST pull-downs from mouse SCp2 epithelial cells using anti-RACK1 and pan-p120 antibodies shows that RACK1 and p120ctn preferentially bind to Rac1b.
The identification of p120ctn as a selective binding partner of Rac1b in our pull-down experiment was especially striking, because both Rac1b and p120ctn have been implicated as key players in signaling pathways controlling cell adhesion, migration, and induction of epithelial-mesenchymal transition (3, 8, 9, 12, 15, 28, 30, 31). We wished to determine the structural basis of the interaction between Rac1b and p120ctn and whether it was mediated through direct interaction between the two proteins. At least four isoforms of p120ctn have been described in epithelial cells (p120ctn-1, -2, -3, and -4) (32, 33); all share a central domain with nine tandem ARM repeats (25). Our finding that GST-Rac1b interacts with several isoforms of p120ctn (Fig. 2E) suggests that the central ARM domain contains a Rac1b binding region. Consistent with this expectation, we found that a GST fusion protein of the short isoform p120ctn-4 (GST-p120ctn-4) preferentially binds to Rac1b compared with Rac1 and Rac1-V12 (Fig. 3, A and C).
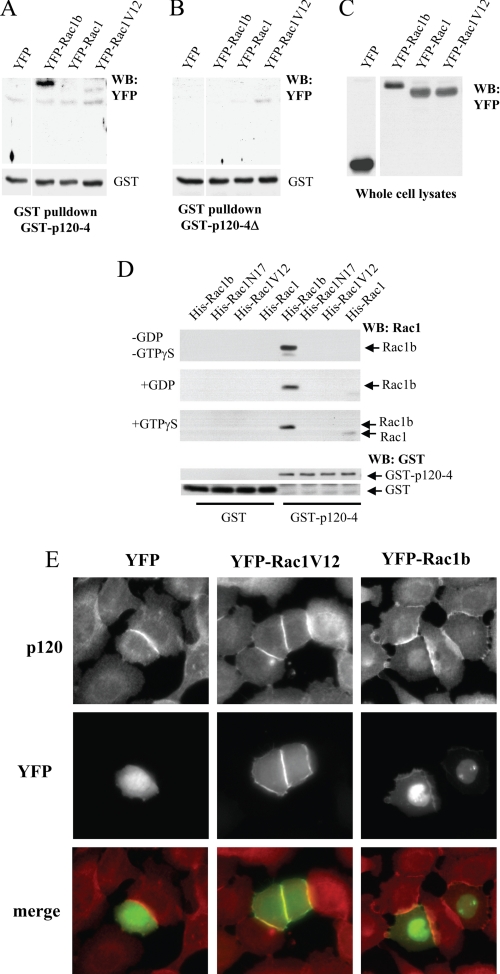
FIGURE 3.
Specificity of Rac1b binding to p120ctn. A–C, SCp2 mammary epithelial cells were transiently transfected with either YFP-Rac1b, YFP-Rac1, YFP-Rac1V12, or YFP as a negative control, and lysates were precipitated with GST-p120ctn-4A (A) or the p120 deletion mutant lacking the polylysine sequence, GST-p120ctn-4A-Δ (B) and Western blotted for YFP. C, Western blot (WB) of total cell lysates from SCp2 cells transiently transfected with YFP fusion proteins. D, GST-p120–4 was used to precipitate His-tagged Rac1, Rac1V12, Rac1N17, and Rac1b proteins that were nucleotide-free (−GDP/−GTPγS) or loaded with GDP or GTPγS, and precipitates were Western blotted and probed with Rac1 antibodies (top three blots) or GST antibodies (bottom two blots). E, immunofluorescence assay of mouse epithelial SCp2 cells transfected with YFP (left column), YFP-Rac1V12 (center column), or YFP-Rac1b (right column) stained with pan-p120 antibody, revealing that endogenous p120ctn co-localizes with Rac1b both at cell-cell junctions and in the nucleus.
Our recent studies have identified a polylysine motif within the p120ctn ARM domain, 622KKGKGKK628, as critical for association with RhoA (15, 34, 35), so we tested whether this domain was also required for association with Rac1b. Using a mutant GST-p120ctn-4 deletion construct lacking this motif (GST-p120–4Δ), we found that deletion of the polylysine motif abolished binding to Rac1b (Fig. 3, B and C). These data suggest that the polylysine motif is involved in the interaction of p120ctn with Rac1b.
To test whether Rac1b and p120ctn bind each other directly or interact via an intermediary, we assessed interaction of purified GST-p120ctn-4 with purified His-Rac1b, His-Rac1, HisRac1V12, and HisRac1N17. We found that GST-p120ctn-4 bound to His-Rac1b in vitro irrespective of the presence of GDP or GTPγS (Fig. 3D). We note that the lack of dependence of this interaction on nucleotide binding is consistent with x-ray structures of Rac1b (12), which showed minimal conformational responsiveness of Rac1b to the nature of the bound nucleotide. We also observed a weak interaction with His-Rac1 that was dependent on the inclusion of GTPγS (Fig. 3D).
Next, we asked whether p120ctn and Rac1b co-localize in vivo. We transiently transfected mouse mammary epithelial cells with expression constructs encoding YFP, YFP-Rac1V12, or YFP-Rac1b and assessed endogenous distribution of p120ctn in these cells. In all cases, p120ctn was localized primarily to cell-cell junctions, which is where the majority of YFP-Rac1V12 and YFP-Rac1b was also found. However, a portion of the localization of YFP-Rac1b was often found in the nucleus, as has been noted previously (5, 36), and in these cases, p120ctn was also found to be colocalized with Rac1b. These results show that the p120ctn and Rac1b interaction exists in cultured cells.
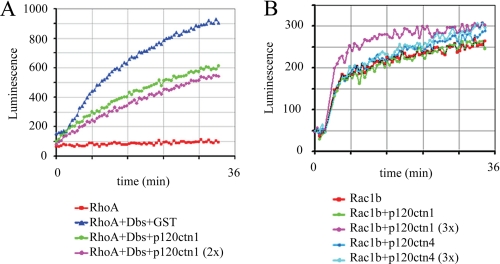
p120ctn acts as an inhibitor of RhoA signaling by inhibiting GDP/GTP exchange (35). Our results show that Rac1b and RhoA both associate with p120ctn through interactions involving a common polylysine motif of p120ctn (Fig. 3, A–C) (15). Previous studies of Rac1b have shown that it can spontaneously exchange GTP for GDP (12). Because a role for p120 as an inhibitor of nucleotide exchange could constitute the basis for a physiological function for the interaction between Rac1b and p120, we tested whether p120ctn association with Rac1b blocks this intrinsic exchange ability, using a fluorescent assay that measures uptake of GTP in GDP-bound proteins. Consistent with previous results in which isoform 1 of p120ctn inhibited GDP release from RhoA (35), we found that RhoA-GDP exhibited no intrinsic GTP uptake and that p120ctn-1 inhibited GTP uptake induced by Dbs, a specific RhoA GEF (Fig. 4A). By contrast, neither isoform 1 nor isoform 4 of p120ctn had any inhibitory effect on the intrinsic GTP uptake by Rac1b (Fig. 4B). These results demonstrate that although p120ctn interacts with RhoA and Rac1b in part using the same binding domain, the consequences of binding are nevertheless quite different.
FIGURE 4.
p120ctn does not inhibit intrinsic GDP/GTP exchange activity of Rac1b. A, p120ctn isoform 1 inhibits Dbs (0.8 μm)-stimulated GDP/GTP exchange of RhoA, tested at 1 and 2 μm (2×). B, biochemical GEF exchange assay for Rac1b shows that neither p120ctn isoform 1 nor p120ctn isoform 4 inhibits Rac1b intrinsic GDP/GTP exchange activity. p120ctn isoform 1 was used at 0.6 and 2 μm (3×); p120ctn isoform 4 was used at 1 and 3 μm (3×).
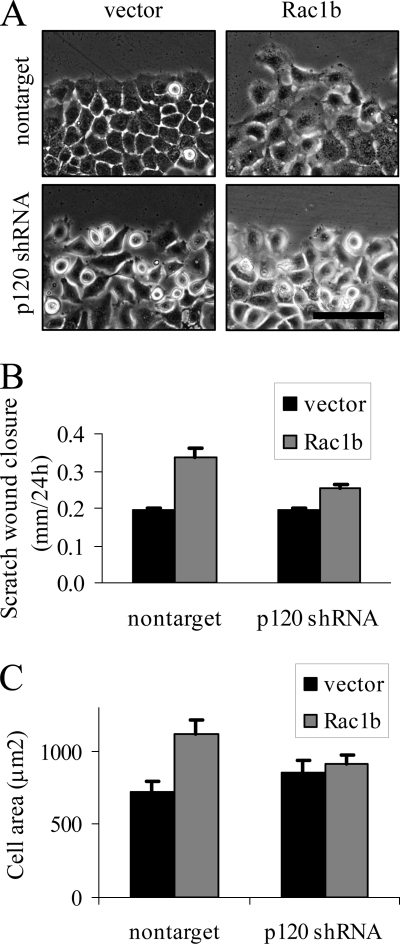
Colocalization of Rac1b and p120ctn at cell-cell junctions is consistent with a function for this interaction in the regulation of cellular processes that are mediated by cytoskeletal structure. Because p120ctn has been implicated in control of lamellipodia formation and cell spreading (28, 37), as well as induction of cell motility (28, 38–41), activities that have been found in mouse mammary epithelial cells expressing Rac1b (8, 9), we evaluated the effect of knockdown of p120 on induction of cell spreading and motility by Rac1b. We assessed the effectiveness of five shRNA constructs for knockdown of p120ctn in SCp2 mouse mammary epithelial cells (supplemental Fig. 3) and used MC41 in these experiments (similar results were seen with other knockdown constructs; data not shown). We infected SCp2 mouse mammary epithelial cells with nontarget control or p120ctn-targeting shRNA and then transfected with YFP (as control) or YFP-Rac1b. The following day, we created 1-mm scratches in the cultures and then assessed motility as rate of scratch wound closure (Fig. 5, A and B) and also quantified cell spreading by measuring the area of cells at the leading edge (Fig. 5C). We found that Rac1b-induced cell motility was inhibited by depletion of p120ctn (Fig. 5B), an effect that could be attributed to altered motility because neither cell proliferation nor viability were affected by knockdown of p120 or overexpression of Rac1b (supplemental Fig. 4). Similarly, although mouse mammary epithelial cells transfected with Rac1b did show increased cell spreading and extensive lamellipodia formation, as noted previously (8, 9), these effects were reduced with knockdown of p120ctn (Fig. 5C). These results are consistent with a role for the p120-Rac1b interaction in control of cell cytoskeletal structure and function.
FIGURE 5.
Rac1b-dependent cell motility and spreading require p120ctn. A, images of SCp2 cells transfected with YFP (vector) or YFP-Rac1b (Rac1b) and infected with nontarget shRNA or shRNA targeting p120ctn. Scale bar, 50 μm. B, scratch wound closure at 24 h, expressed as means ± S.E. (error bars) (p < 0.05) for nontarget/Rac1b versus p120 shRNA/Rac1b. C, area of cells at scratch border at 24 h, expressed as means ± S.E. (p < 0.05) for nontarget/Rac1b versus p120 shRNA/Rac1b.
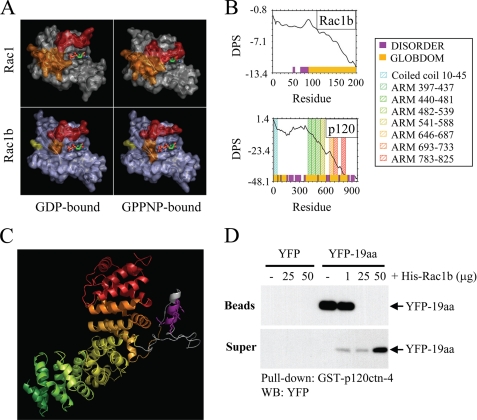
Our results show that Rac1b interacts with p120ctn in part through the 19-aa insertion encoded by exon 3b (Fig. 3, A–C). However, this sequence has been found to be disordered in crystal structures of Rac1b (12). To define how a disordered region of Rac1b may be responsible for a selective interacting with p120ctn, we pursued molecular modeling approaches. Global analysis of structural data from protein complexes has revealed that virtually all of the molecular surfaces of the small GTPases can be used in protein-protein interactions with specific binding partners (including both effectors and regulators); however, the surfaces involved in the greatest number of protein-protein contacts are the so-called “switch” regions (42, 43). Nucleotide binding by the small GTPases is transduced into signaling via these two flexible switches, which border the nucleotide binding site and feature shape-shifting conformations that are sensitive to the nature of the bound nucleotide (Fig. 6A). The residues within the switch regions adapt to interact with multiple partners by demonstrating a substantial amount of structural plasticity (43–45). Using GlobPlot, a Web-based server for computational discovery of unstructured regions within proteins, we find further corroboration of the likelihood that the 19-aa insertion sequence in Rac1b is intrinsically unstructured in solution (Fig. 6B).
FIGURE 6.
p120ctn and Rac1b bind through the interaction of protein regions predicted to be disordered. A, most protein interactions of Rac1 (top panels) involve binding through switch 1 (red) and/or switch 2 (orange); the conformations of these switches are determined by binding to GDP (top left) or GTP (mimicked by uncleavable analog GPPNP; top right). In Rac1b (lower panels), switch 1 (red) fails to adopt a closed conformation upon GPPNP binding. B, disorder propensity sum (DPS) plots predict intrinsically unstructured regions for Rac1b (left) and p120ctn (right). Regions of disorder, characterized by a positive slope on the plot, are represented with magenta blocks along the x axis, whereas predicted globular domains, characterized by negative slope, are represented with gold blocks. Residues 607–644 of p120ctn (containing the polylysine motif) and residues 70–90 of Rac1b (encompassing much of the 19-aa insertion) are predicted to be unstructured. C, homology model of the ARM domain of p120ctn shows that the polylysine motif (magenta) is located in an insertion between ARM repeats 5 and 6. ARM repeats are colored to correspond to the Pfam domains indicated in the plot above. The polylysine motif (magenta) and flanking sequences shown in gray correspond to disordered regions in the crystal structure of plakophilin 1, upon which the homology model is based. D, cells were transfected with YFP or YFP-19aa, and lysates were precipitated with glutathione beads or were preincubated with 1, 25, or 50 μg of His-Rac1b prior to precipitation. Bead precipitates and supernatants were solubilized, Western blotted (WB), and probed with anti-YFP antibodies.
We have shown additionally that the p120ctn-Rac1b interaction involves a polylysine motif 622KKGKGKK628 located within the ARM domain of p120ctn (Fig. 3, A–C). To explore the structural context of this binding motif, we analyzed the p120ctn sequence for predicted globular and unstructured domains using GlobPlot and found that the p120ctn polylysine motif lies within a predicted unstructured region between ARM repeats in the central domain (Fig. 6B). Additional potential disordered regions were located between the N-terminal coiled-coil and ARM domains (Fig. 6B), in a region previously found to modulate binding to RhoA (34).
We also constructed a homology model of the p120ctn ARM domain based on the crystal structure of plakophilin 1, a member of the p120ctn subfamily of ARM domain proteins, which shares 30% sequence identity to p120ctn within this domain. The model features nine trihelical ARM repeats, which form a single superhelical domain (Fig. 6C). The polylysine motif is located in a long insertion loop. Much of the insertion loop is disordered in the corresponding structure of plakophilin A (25), although loop residues that are visible in the plakophilin A structure and make contact with the ARM repeats are conserved in p120ctn. It is probable that the p120ctn insertion loop represents a module for protein-protein recognition that remains unstructured, similar to the 19-aa insertion of Rac1b, in the absence of binding to specific partner proteins. Consistent with this expectation, GlobPlot predicted residues 607–644 of p120ctn (containing the polylysine motif) as unstructured.
Our previous results suggested a highly specific interaction between regions of Rac1b and p120 predicted to be disordered in solution; if the 19-aa region becomes ordered upon interaction with p120, then its association with p120 should show specificity. To test this possibility, we performed a competition experiment, in which we incubated GST-p120ctn-4 immobilized on glutathione beads with cell lysates containing YFP fused to the 19-aa insertion (YFP-19aa), washed thoroughly, and then tested the ability of increasing concentrations of purified His-Rac1b to displace the bound YFP-19aa. We found that YFP-19aa was displaced by high concentrations of His-Rac1b (Fig. 4A), demonstrating a specific interaction between p120ctn and Rac1b that involves the 19-aa insertion sequence.
DISCUSSION
Here we use proteomic methods to show that the 19-aa insertion in Rac1b, a region natively unstructured in the absence of binding (12), confers a markedly altered pattern of protein association. The roles of such intrinsically unstructured protein regions in molecular recognition and signaling represent an emerging area of intense interest (46–48). The ability of such highly flexible regions to adopt multiple conformations may be instrumental in the ability of “hub” proteins in protein-protein interaction networks (46, 48, 49), and specifically the small GTPases (43, 44), to bind multiple, structurally diverse partners. Thus, the alternative splicing of exon 3b in Rac1b, augmenting the already flexible switch 2 region with 19 additional amino acids, may allow presentation of a novel conformational ensemble of binding surfaces for interaction with a distinct set of binding partners. Significantly, regions of intrinsic disorder are found with much greater prevalence in cancer-associated proteins and signaling proteins compared with the set of all eukaryotic proteins in Swiss-Prot (49).
We found that Rac1b associates more effectively than Rac1 with the armadillo repeat-containing proteins SmgGDS and p120ctn. Rac1b has previously been shown to associate with β-catenin, another armadillo-repeat containing protein (5). It may be that the 19-aa insertion associates with multiple ARM-domain proteins using a similar mode of interaction, a possibility that would be consistent with our observation that the 19-aa peptide is able to bind to p120ctn and that this binding is competitive with Rac1b but not Rac1. Structures of β-catenin bound to domains of E-cadherin, Tcf, and ICAT (50–54) and the SmgGDS homolog karyopherin α bound to nuclear localization sequences (55–57) have revealed the binding of ARM domains to intrinsically unstructured protein ligands, which adopt extended conformations within a concave groove of the ARM superhelix. Based on our homology model, it appears possible that an extended portion of the Rac1b insertion could bind within the superhelical groove of p120ctn and also allow contact with the polylysine motif. However, the electrostatic residues of the ARM superhelical grooves responsible for determining ligand specificity differ substantially between the primarily acidic SmgGDS and the primarily basic p120ctn and β-catenin (25, 54). Thus, the structural basis for the intriguing ability of Rac1b to bind such disparate ARM domain proteins awaits future investigation.
Because Rac1b, like other small GTPases, is expected to exert its biological effects through binding to specific effector proteins, the altered binding pattern identified here provides key insight into how Rac1b exerts its unique effects. We found that Rac1 interacts more strongly than Rac1b with GIT1/2, β-PIX, and IQGAP. β-PIX is a specific GEF for Rac1/Cdc42 (58), and GIT1/2 are adenosine ribosylation factor-GAPs that appear to have roles in vesicular trafficking (59). GIT1/2 and β-PIX have been found to associate into signaling complexes with other proteins and to regulate cytoskeletal dynamics by feedback inhibition of Rac1 (reviewed in Refs. 26 and 59). Our observations that GIT and β-PIX do not interact with Rac1b suggest that Rac1b functions most likely cannot be inhibited by this complex.
We found that Rac1b associates with RACK1 more efficiently than does Rac1. RACK1 is a member of the WD40 repeat family involved in many cellular processes, including signal transduction, cell adhesion, and growth (60, 61). Several studies showed that RACK1 functions as an endogenous inhibitor of the Src kinase in suppression of cell proliferation (60, 62) and is involved in cell adhesion by interacting with integrins (63–65). Moreover, RACK1 is an integral core protein of the eukaryotic 40 S ribosomes and regulates protein synthesis in response to cell stimuli (reviewed in Ref. 29). By binding RACK1, Rac1b could potentially sequester RACK1 from Src inhibition, promoting cell cycle progression and cellular growth.
The enhanced association of Rac1b to armadillo repeat-containing proteins, such as SmgGDS and p120ctn, is also likely to have significant functional consequences. We have found that expression of Rac1b in mouse mammary epithelial cells causes breakdown of cell-cell junctions and increased cell motility (8, 9). Many armadillo repeat-containing proteins have been linked with epithelial cell structure and function (28); association with Rac1b may play a key role in the loss of epithelial structure and tumor progression of cancers that express the Rac1b isoform (1, 2). Rac1b-mediated displacement of E-cadherin from cell-cell junctions may also promote nuclear translocation of p120ctn. Activated Rac1 has been demonstrated to provide stimulatory signals for the nuclear accumulation of p120ctn (66), and similarly, Rac1b may regulate p120ctn nuclear translocation. When localized to the nucleus, p120ctn participates in transcriptional regulation by associating with the Kaiso and Glis2 transcription factors. SmgGDS is an atypical GEF with multiple armadillo repeats (26, 27) that interacts with both active and inactive forms of Rac1 (27, 67), although it displays a lower affinity for the GTP-bound form of Rac1 (68). Activated Rac1 has been shown to promote nuclear accumulation of SmgGDS via a polybasic motif present in Rac1 (66); Rac1b also contains this motif and binds even more effectively to SmgGDS and p120ctn to stimulate nuclear accumulation of these proteins. Promotion of nuclear localization of p120ctn by Rac1b could also affect tumor cell growth and metastasis.
In summary, our study has found that Rac1b is more than just an activated mutant of Rac1 but instead shows a markedly different pattern of effector protein association. Rac1b binds less effectively to proteins that act as signal pathway inhibitors (β-PIX and GIT1 and -2) and more effectively to proteins that can promote loss of epithelial cell structure and increased cell proliferation (p120ctn, SmgGDS, and RACK1). These results point to a specific and unexpected role for Rac1b in cell function and in the promotion of tumors that express this splice isoform.
Supplementary Material
Acknowledgment
Proteomic analysis was performed at the Mayo Clinic Proteomic Research Center.
This work was supported, in whole or in part, by National Institutes of Health Grants CA122086 (to D. C. R.), CA128660 (to Celeste M. Nelson and D. C. R.), and CA100467 (to P. Z. A.). This work was also supported by Susan B. Komen Foundation Grant FAS0703855 (to Celeste M. Nelson and D. C. R.) and by the Mayo Clinic Breast Cancer Specialized Program of Research Excellence (SPORE) Grant CA116201 (to James Ingle).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–4.
- GEF
- guanine nucleotide exchange factor
- GDS
- GDP dissociation stimulator
- GAP
- GTPase-activating protein
- aa
- amino acid(s)
- YFP
- yellow fluorescent protein
- YFP-19aa
- YFP fused to the 19-aa insertion
- GST
- glutathione S-transferase
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- shRNA
- short hairpin RNA
- MS/MS
- tandem mass spectrometry.
REFERENCES
- 1.Jordan P., Brazåo R., Boavida M. G., Gespach C., Chastre E. (1999) Oncogene 18, 6835–6839 [DOI] [PubMed] [Google Scholar]
- 2.Schnelzer A., Prechtel D., Knaus U., Dehne K., Gerhard M., Graeff H., Harbeck N., Schmitt M., Lengyel E. (2000) Oncogene 19, 3013–3020 [DOI] [PubMed] [Google Scholar]
- 3.Singh A., Karnoub A. E., Palmby T. R., Lengyel E., Sondek J., Der C. J. (2004) Oncogene 23, 9369–9380 [DOI] [PubMed] [Google Scholar]
- 4.Matos P., Jordan P. (2005) Exp. Cell Res. 305, 292–299 [DOI] [PubMed] [Google Scholar]
- 5.Esufali S., Charames G. S., Pethe V. V., Buongiorno P., Bapat B. (2007) Cancer Res. 67, 2469–2479 [DOI] [PubMed] [Google Scholar]
- 6.Lozano E., Frasa M. A., Smolarczyk K., Knaus U. G., Braga V. M. (2008) J. Cell Sci. 121, 933–938 [DOI] [PubMed] [Google Scholar]
- 7.Matos P., Jordan P. (2006) J. Biol. Chem. 281, 13724–13732 [DOI] [PubMed] [Google Scholar]
- 8.Radisky D. C., Levy D. D., Littlepage L. E., Liu H., Nelson C. M., Fata J. E., Leake D., Godden E. L., Albertson D. G., Nieto M. A., Werb Z., Bissell M. J. (2005) Nature 436, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson C. M., Khauv D., Bissell M. J., Radisky D. C. (2008) J. Cell. Biochem. 105, 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matos P., Jordan P. (2008) Mol. Cancer Res. 6, 1178–1184 [DOI] [PubMed] [Google Scholar]
- 11.Matos P., Oliveira C., Velho S., Goncalves V., Costa L. T., Moyer M. P., Seruca R., Jordan P. (2008) Gastroenterology 135, 899–906 [DOI] [PubMed] [Google Scholar]
- 12.Fiegen D., Haeusler L. C., Blumenstein L., Herbrand U., Dvorsky R., Vetter I. R., Ahmadian M. R. (2004) J. Biol. Chem. 279, 4743–4749 [DOI] [PubMed] [Google Scholar]
- 13.Matos P., Collard J. G., Jordan P. (2003) J. Biol. Chem. 278, 50442–50448 [DOI] [PubMed] [Google Scholar]
- 14.Hirai Y., Bissell M. J., Radisky D. C. (2007) Blood 110, 3082; author reply 3082–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanagisawa M., Huveldt D., Kreinest P., Lohse C. M., Cheville J. C., Parker A. S., Copland J. A., Anastasiadis P. Z. (2008) J. Biol. Chem. 283, 18344–18354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanagisawa M., Kaverina I. N., Wang A., Fujita Y., Reynolds A. B., Anastasiadis P. Z. (2004) J. Biol. Chem. 279, 9512–9521 [DOI] [PubMed] [Google Scholar]
- 17.Dächsel J. C., Taylor J. P., Mok S. S., Ross O. A., Hinkle K. M., Bailey R. M., Hines J. H., Szutu J., Madden B., Petrucelli L., Farrer M. J. (2007) Parkinsonism Relat. Disord. 13, 382–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orlichenko L., Huang B., Krueger E., McNiven M. A. (2006) J. Biol. Chem. 281, 4570–4579 [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 20.Towbin H., Staehelin T., Gordon J. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burnette W. N. (1981) Anal. Biochem. 112, 195–203 [DOI] [PubMed] [Google Scholar]
- 22.Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. (2003) Structure 11, 1453–1459 [DOI] [PubMed] [Google Scholar]
- 23.Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 25.Choi H. J., Weis W. I. (2005) J. Mol. Biol. 346, 367–376 [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto T., Kaibuchi K., Mizuno T., Hiroyoshi M., Shirataki H., Takai Y. (1990) J. Biol. Chem. 265, 16626–16634 [PubMed] [Google Scholar]
- 27.Vikis H. G., Stewart S., Guan K. L. (2002) Oncogene 21, 2425–2432 [DOI] [PubMed] [Google Scholar]
- 28.Anastasiadis P. Z. (2007) Biochim. Biophys. Acta 1773, 34–46 [DOI] [PubMed] [Google Scholar]
- 29.Nilsson J., Sengupta J., Frank J., Nissen P. (2004) EMBO Rep. 5, 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anastasiadis P. Z., Reynolds A. B. (2000) J. Cell Sci. 113, 1319–1334 [DOI] [PubMed] [Google Scholar]
- 31.Stallings-Mann M., Radisky D. (2007) Cells Tissues Organs 185, 104–110 [DOI] [PubMed] [Google Scholar]
- 32.Keirsebilck A., Bonné S., Staes K., van Hengel J., Nollet F., Reynolds A., van Roy F. (1998) Genomics 50, 129–146 [DOI] [PubMed] [Google Scholar]
- 33.Aho S., Rothenberger K., Uitto J. (1999) J. Cell. Biochem. 73, 390–399 [PubMed] [Google Scholar]
- 34.Castaño J., Solanas G., Casagolda D., Raurell I., Villagrasa P., Bustelo X. R., García de Herreros A., Duñach M. (2007) Mol. Cell. Biol. 27, 1745–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anastasiadis P. Z., Moon S. Y., Thoreson M. A., Mariner D. J., Crawford H. C., Zheng Y., Reynolds A. B. (2000) Nat. Cell Biol. 2, 637–644 [DOI] [PubMed] [Google Scholar]
- 36.Esufali S., Charames G. S., Bapat B. (2007) FEBS Lett. 581, 4850–4856 [DOI] [PubMed] [Google Scholar]
- 37.Boguslavsky S., Grosheva I., Landau E., Shtutman M., Cohen M., Arnold K., Feinstein E., Geiger B., Bershadsky A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10882–10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anastasiadis P. Z., Reynolds A. B. (2001) Curr. Opin. Cell Biol. 13, 604–610 [DOI] [PubMed] [Google Scholar]
- 39.Cozzolino M., Stagni V., Spinardi L., Campioni N., Fiorentini C., Salvati E., Alemà S., Salvatore A. M. (2003) Mol. Biol. Cell 14, 1964–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grosheva I., Shtutman M., Elbaum M., Bershadsky A. D. (2001) J. Cell Sci. 114, 695–707 [DOI] [PubMed] [Google Scholar]
- 41.Yanagisawa M., Anastasiadis P. Z. (2006) J. Cell Biol. 174, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corbett K. D., Alber T. (2001) Trends Biochem. Sci. 26, 710–716 [DOI] [PubMed] [Google Scholar]
- 43.Biou V., Cherfils J. (2004) Biochemistry 43, 6833–6840 [DOI] [PubMed] [Google Scholar]
- 44.Sprang S. R. (2000) Sci. STKE 2000, PE1. [DOI] [PubMed] [Google Scholar]
- 45.Vetter I. R., Wittinghofer A. (2001) Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 46.Dunker A. K., Cortese M. S., Romero P., Iakoucheva L. M., Uversky V. N. (2005) FEBS J. 272, 5129–5148 [DOI] [PubMed] [Google Scholar]
- 47.Dyson H. J., Wright P. E. (2005) Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 48.Keskin O., Gursoy A., Ma B., Nussinov R. (2008) Chem. Rev. 108, 1225–1244 [DOI] [PubMed] [Google Scholar]
- 49.Iakoucheva L. M., Brown C. J., Lawson J. D., Obradović Z., Dunker A. K. (2002) J. Mol. Biol. 323, 573–584 [DOI] [PubMed] [Google Scholar]
- 50.Graham T. A., Clements W. K., Kimelman D., Xu W. (2002) Mol. Cell 10, 563–571 [DOI] [PubMed] [Google Scholar]
- 51.Graham T. A., Ferkey D. M., Mao F., Kimelman D., Xu W. (2001) Nat. Struct. Biol. 8, 1048–1052 [DOI] [PubMed] [Google Scholar]
- 52.Graham T. A., Weaver C., Mao F., Kimelman D., Xu W. (2000) Cell 103, 885–896 [DOI] [PubMed] [Google Scholar]
- 53.Huber A. H., Stewart D. B., Laurents D. V., Nelson W. J., Weis W. I. (2001) J. Biol. Chem. 276, 12301–12309 [DOI] [PubMed] [Google Scholar]
- 54.Huber A. H., Weis W. I. (2001) Cell 105, 391–402 [DOI] [PubMed] [Google Scholar]
- 55.Conti E., Kuriyan J. (2000) Structure 8, 329–338 [DOI] [PubMed] [Google Scholar]
- 56.Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. (1998) Cell 94, 193–204 [DOI] [PubMed] [Google Scholar]
- 57.Fontes M. R., Teh T., Kobe B. (2000) J. Mol. Biol. 297, 1183–1194 [DOI] [PubMed] [Google Scholar]
- 58.Rossman K. L., Der C. J., Sondek J. (2005) Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 59.Hoefen R. J., Berk B. C. (2006) J. Cell Sci. 119, 1469–1475 [DOI] [PubMed] [Google Scholar]
- 60.Miller L. D., Lee K. C., Mochly-Rosen D., Cartwright C. A. (2004) Oncogene 23, 5682–5686 [DOI] [PubMed] [Google Scholar]
- 61.McCahill A., Warwicker J., Bolger G. B., Houslay M. D., Yarwood S. J. (2002) Mol. Pharmacol. 62, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 62.Mamidipudi V., Dhillon N. K., Parman T., Miller L. D., Lee K. C., Cartwright C. A. (2007) Oncogene 26, 2914–2924 [DOI] [PubMed] [Google Scholar]
- 63.Liliental J., Chang D. D. (1998) J. Biol. Chem. 273, 2379–2383 [DOI] [PubMed] [Google Scholar]
- 64.Chattopadhyay N., Wang Z., Ashman L. K., Brady-Kalnay S. M., Kreidberg J. A. (2003) J. Cell Biol. 163, 1351–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kiely P. A., O'Gorman D., Luong K., Ron D., O'Connor R. (2006) Mol. Cell. Biol. 26, 4041–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lanning C. C., Ruiz-Velasco R., Williams C. L. (2003) J. Biol. Chem. 278, 12495–12506 [DOI] [PubMed] [Google Scholar]
- 67.Lanning C. C., Daddona J. L., Ruiz-Velasco R., Shafer S. H., Williams C. L. (2004) J. Biol. Chem. 279, 44197–44210 [DOI] [PubMed] [Google Scholar]
- 68.Chuang T. H., Xu X., Quilliam L. A., Bokoch G. M. (1994) Biochem. J. 303, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.