Abstract
Colorectal cancer is the third most common malignancy in the United States. Modest advances with therapeutic approaches that include oxaliplatin (l-OHP) have brought the median survival rate to 22 months, with drug resistance remaining a significant barrier. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is undergoing clinical evaluation. Although human colon carcinomas express TRAIL receptors, they can also demonstrate TRAIL resistance. Constitutive NF-κB activation has been implicated in resistance to TRAIL and to cytotoxic agents. We have demonstrated constitutive NF-κB activation in five of six human colon carcinoma cell lines; this activation is inhibited by quinacrine. Quinacrine induced apoptosis in colon carcinomas and potentiated the cytotoxic activity of TRAIL in RKO and HT29 cells and that of l-OHP in HT29 cells. Similarly, overexpression of IκBα mutant (IκBαM) or treatment with the IKK inhibitor, BMS-345541, also sensitized these cells to TRAIL and l-OHP. Importantly, 2 h of quinacrine pretreatment resulted in decreased expression of c-FLIP and Mcl-1, which were determined to be transcriptional targets of NF-κB. Extended exposure for 24 h to quinacrine did not further sensitize these cells to TRAIL- or l-OHP-induced cell death; however, exposure caused the down-regulation of additional NF-κB-dependent survival factors. Short hairpin RNA-mediated knockdown of c-FLIP or Mcl-1 significantly sensitized these cells to TRAIL and l-OHP. Taken together, data demonstrate that NF-κB is constitutively active in colon cancer cell lines and NF-κB, and its downstream targets may constitute an important target for the development of therapeutic approaches against this disease.
Keywords: Cell Death, Colon Cancer, NF-κB, Signal Transduction, shRNA, Mcl-1, TRAIL, c-FLIP, Survival Signaling
Introduction
Colorectal cancer continues to be the third most common malignancy in the United States. In 2009 more than 140,000 new cases of colorectal cancer resulting in 50,000 deaths were expected (1, 2). 5-Fluorouracil combined with leucovorin (FUra/LV)2 remains the basis of treatment strategies, targeted to the enzyme thymidylate synthase, thereby inducing DNA damage-dependent cell death in tumor cells.
Topoisomerase I inhibitors such as irinotecan (CPT-11) as well as platinum-based compounds such as oxaliplatin (l-OHP) have more recently been employed in the treatment of colorectal cancer. l-OHP is an analogue of cisplatin that has achieved modest success in the treatment of colorectal cancer. It has been found to have some activity as a single agent, but its anti-tumor effects are further enhanced when it is used in combination with other drugs that include FUra/LV, CPT-11, or taxanes (3–9). In addition to the increased efficacy, these combinations have resulted in decreased gastrointestinal toxicity as well as neurotoxicity, which are usually accompanied with l-OHP treatment (10). Of significant limitation is that these empirically derived current therapeutic approaches result in survival of only 22 months for patients with advanced colorectal cancer, demonstrating the continuing problems with drug resistance, thereby necessitating an urgent need for substantive changes in treatment strategies. New approaches in chemotherapy that combine cytotoxics with anti-tumor agents that modify the function of tumor-dysregulated apoptotic genes are being evaluated using rational research-based design to address these problems in the modification of therapeutic approaches for colon carcinoma.
Our previous studies have demonstrated that the Fas death receptor and its signaling pathway are critical components of thymidylate synthase-dependent DNA damage-induced cell death mediated by FUra/LV (11–16). Fas is frequently down-regulated in human colorectal cancer and is up-regulated after treatment with the cytokine interferon-γ, which is synergistic with Fas-mediated FUra/LV-induced cytotoxicity in cultured human colon carcinoma cell lines and in xenograft models; furthermore, FUra/LV/interferon-γ, in combination, has successfully completed Phase I (17) and Phase II (18) clinical trials. Although stimulation of Fas receptor as well as tumor necrosis factor receptor efficiently induces apoptosis in tumor cells, severe hepatotoxicity of Fas ligand and TNFα has limited their use as anticancer agents (19–22). The discovery of tumor necrosis factor-α-related apoptosis inducing ligand (TRAIL) and its receptors TRAIL-R1 (DR4) and TRAIL-R2 (DR5) has given rise to a new opportunity for therapeutic intervention by specifically targeting death receptors (23, 24). It has been observed that TRAIL selectively induces apoptosis in cancer cells as opposed to normal cells (23–27), resulting in the selective elimination of cancer cells without life threatening toxicity in vivo. The application of TRAIL or TRAIL agonist monoclonal antibodies (targeting DR4 or DR5) are approaches to such therapy, which is currently undergoing intensive clinical evaluation (25–27). Ligation of TRAIL receptors in conjunction with the induction of DNA damage by FUra/LV/interferon-γ results in a highly synergistic combination (FUra/LV/interferon-γ/TRAIL) in xenograft models.3 However, where almost all of the colon carcinoma cell lines express TRAIL receptors DR4 and DR5, they can also demonstrate TRAIL resistance (28).
Current treatment approaches in the clinical management of colorectal cancer is de novo or acquired resistance to the chemotherapeutic agents used in the treatment of this malignancy. It is becoming increasingly evident that several anti-apoptotic pathways including those regulated by nuclear factor κB (NF-κB) are critical in modulating the effects of TRAIL and other chemotherapeutic agents including l-OHP in cancer cells (29–38). NF-κB is a transcription factor that regulates the expression of numerous genes that are critical for survival. It is activated by diverse stimuli (35, 39–42) that include pro-inflammatory cytokines and cellular stress as well as growth factors; its activation is tightly regulated by inhibitor of κB (IκB). Phosphorylation of IκB proteins by the IKK kinase complex leads to proteasomal degradation of the inhibitory protein, thereby allowing active NF-κB (p65-p50 subunits) to translocate into the nucleus where it binds with NF-κB-specific DNA binding sites to transcriptionally activate the expression of several survival genes. In this manner activation of NF-κB leads to increased expression of the inhibitor of apoptosis proteins (IAPs), namely c-IAP1, c-IAP2, XIAP and survivin. Furthermore, NF-κB transcriptionally up-regulates the expression of FLICE-inhibitory protein, c-FLIP, which inhibits caspase-8 activation and the Bcl-2 family of proteins, including Bcl-2, Mcl-1, and Bcl-xL, which counteract the action of pro-apoptotic proteins including Bid, Bad, and Bax (35, 43–47). Constitutive activation of NF-κB has been detected in several cancers including colorectal, prostate, and pancreatic cancers, neuroblastoma, T cell leukemia, multiple myeloma, and hepatocellular carcinoma as well as breast cancer (48–54) and has been implicated in imparting resistance to TRAIL as well as to l-OHP. Furthermore, NF-κB is also known to be activated by chemotherapeutic drugs, including FUra/LV, CPT-11, l-OHP, and carboplatin as well as cisplatin. Therefore, inhibition of NF-κB signaling may serve as a critical target for enhancing the efficacy of these agents. Recently quinacrine, a derivative of 9-aminoacridine and an anti-inflammatory drug used extensively to treat malaria and rheumatoid arthritis, has been identified as a potential anticancer agent that inhibits NF-κB (55). Studies by Gurova et al. (55) have demonstrated that quinacrine up-regulates p53 and down-regulates NF-κB, thereby causing a decrease in the survival of renal cell carcinoma cells. In the current study we have demonstrated high frequency of constitutively active NF-κB expression in human colon carcinoma cell lines and have evaluated the role of quinacrine as an inhibitor of constitutive NF-κB activation as well as for its synergistic interaction with TRAIL and with l-OHP.
Data demonstrate that inhibition of constitutively activated NF-κB by quinacrine is cytotoxic to human colon carcinoma cell lines independent of p53 and that down-regulation of this NF-κB activity by either quinacrine, the IKK inhibitor BMS-345541, or by overexpression of the super-repressor IκBαM sensitizes cells to TRAIL- or to l-OHP-mediated apoptosis, demonstrating the involvement of NF-κB in conferring resistance to these agents in colon carcinoma cells. Inhibition of NF-κB by quinacrine led to a marked decrease in the expression of c-FLIP and Mcl-1 levels within 2 h of exposure with other NF-κB-dependent survival factors, namely, survivin, XIAP, Bcl-2, and Bcl-xL, down-regulated by 24 h. Data demonstrate that the early down-regulation of c-FLIP and Mcl-1 expression alone can account for the quinacrine-induced sensitization to TRAIL and to l-OHP. Thus, constitutively activated NF-κB occurs in high frequency in colon cancers and may constitute an important target for therapeutic intervention in the development of rational research-based targeted approaches in therapy of colorectal cancer.
MATERIALS AND METHODS
Cell Culture and Reagents
HCT8 and HT29 cell lines were obtained from the ATCC; RKO cells were obtained from Dr. Michael Kastan, St. Jude Children's Research Hospital. The GC3/c1 cell line was previously established in our laboratories (14). Cell lines were maintained in the presence of folate-free RPMI 1640 containing 10% dialyzed fetal bovine serum and 80 nm 6-(R,S)-5-methyltetrahydrofolate. Antibodies against p-IKKα/β, IKK, p-IκBα, IκBα, Bcl-xL, c-FLIP, phospho-p65, and p65 were purchased from Cell Signaling Technology (Boston, MA); antibodies against Bcl-2 and Mcl-1 were obtained from BD Biosciences; the survivin antibody was purchased from R&D systems (Minneapolis, MN); the FLIP(γ/δ) antibody was from Calbiochem; the β-actin antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Quinacrine dihydrochloride and BMS-345541 were obtained from Sigma. c-FLIP (CFLAR) and Mcl-1 shRNA were obtained from Origene (Rockville, MD). Recombinant human TRAIL (Apo2L) was produced in our laboratories using cDNA of the extracellular domain of TRAIL corresponding to amino acids 114–281, which was subcloned into the pET17/b bacterial expression vector and expressed in the BL21(DE3)pLysE bacterial host (57). Reagents used for Annexin V/PI assays were purchased from BD Biosciences.
Clonogenic Assay
Cells were plated at a density of 700 (RKO), 1500 (HT29 and HCT8), or 3000 (GC3/c1) cells/well in 6-well plates. After overnight attachment wells were treated in triplicate with quinacrine (0–20 μm) for 24 h followed by replacement with fresh media containing (thymidine 20 μm) for a period of 7 cell doublings (4 days for RKO, 5 days for HCT8, 6 days for HT29, and 7 days for GC3/c1 cells). Cells were washed with 1× Dulbecco's PBS (without calcium or magnesium) and allowed to dry overnight. The following day cells were stained with crystal violet and analyzed using an Alpha Innotech imager and software.
Flow Cytometry for Annexin V/PI (Cell Death)
Cells were treated as described in the figure legends, after which they were collected and incubated with Annexin V/fluorescein isothiocyanate and propidium iodide (PI) for analysis of cell death using a FACSCalibur flow cytometer and CellQuest software (FACS).
Western Blot Analysis
Total cellular lysates were prepared using radioimmune precipitation assay lysis buffer (Cell Signaling Technology); 50 μg of protein were loaded and separated on a 10% or 4–20% gradient Tris-HCl gel. Proteins were transferred to polyvinylidene difluoride membranes through semidry transfer and blotted in blotting buffer for 1 h. Membranes were washed in washing buffer, blotted in primary antibody overnight at 4 °C, and subsequently washed and blotted with the secondary antibody for 1 h. Visualization of the signal was via Super Signal Pico substrate from Pierce.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear lysates were prepared using the nuclear extraction kit from Panomics and were then analyzed (5 μg) by EMSA using the non-radioactive EMSA kit from Panomics as per the manufacturer's protocol.
Reporter Constructs
The NF-κB-dependent reporter plasmid p5XIP10κB (provided by Dr. Bryan Williams, Cleveland Clinic) contains five tandem copies of the NF-κB site from the IP10 gene. HT29 and RKO cells were transfected transiently using Lipofectamine 2000 (Invitrogen) with 4 μg of p5XIP10κB and 0.4 μg of pRLTK (renilla luciferase driven by thymidine kinase promoter). Twenty-four hours post-transfection cells were treated with quinacrine for 2 h or TNFα (10 ng/ml) for 1 h and harvested using the dual luciferase kit (Promega) according to the manufacturer's protocol. Luciferase activity was normalized to Renilla luciferase activity as a control for transfection efficiency.
Transient Transfection with c-FLIP, Mcl-1 shRNA, or IκBαM cDNA
HT29 and RKO cells were plated at 200,000 cells/ml in 6-well plates and allowed to attach overnight. Media were removed, and cells were transfected with shRNA plasmids (c-FLIP or Mcl-1) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were transfected for 72 h with TRAIL or l-OHP added during the last 24 or 48 h of the transfection, respectively. Cell death was subsequently analyzed by Annexin V/PI staining. The conditional expression vector super repressor IκBαM was employed as previously described (42, 58).
Chromatin Immunoprecipitation
HT29 and RKO cells were expanded in 15-cm tissue culture plates to near confluence. Cells were detached by trypsinization and washed ×1 with ice-cold PBS. Cellular proteins were cross-linked to DNA by treating with 1% formaldehyde in PBS for 10 min at 37 °C and terminating the reaction in 125 nm glycine, PBS. Cells were washed in PBS, and nuclear extracts were prepared. Nuclei were sonicated to generate chromatin fragments of ≈500–800 bp. Sonicated chromatin (30 μl) was treated with RNase A and Proteinase K followed by de-cross-linking; the DNA was isolated and quantified. Chromatin was precleared using a mixture of protein A-Sepharose and protein G-Sepharose. The chromatin fragments were immunoprecipitated with 2 μg of rabbit polyclonal antibody (Abcam ab7970) to NF-κB, 2 μg of IgG (negative control), or 2 μg of histone H3 antibody (Abcam, ab1791; positive control). The RNase A and proteinase K treatments and subsequent immunoprecipitation, de-cross-linking, and DNA isolation were performed as described previously (59, 60). The resulting chromatin was resuspended in 200 μl of PCR-grade H2O. Quantitative real-time-PCR was performed by using primers flanking the promoter regions of c-FLIP and Mcl-1 putative NF-κB binding sites using a 96-well plate 7500 real-time PCR system and 1× SYBR Green PCR master mix (Applied Biosystems). The bound DNA was calculated as a fraction of the input.
RESULTS
Human Colorectal Carcinoma Cell Lines Express Constitutively Active NF-κB
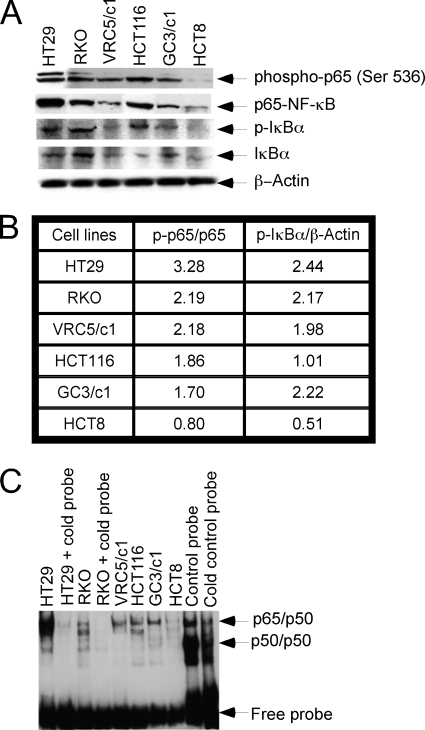
A panel of human colon carcinoma cell lines (HT29, RKO, GC3/c1, VRC5/c1, HCT116, HCT8) was examined for the expression of constitutively active NF-κB by determining the expression of p-p65 and p-IκBα by Western analysis (Fig. 1A), and the ratios of the band intensity were measured by ImageJ software (Fig. 1B). In addition, NF-κB DNA binding in these cell lines was determined by EMSA analysis (Fig. 1B). Constitutive NF-κB activation (p-p65, p-IκBα) was observed in 5 of 6 cell lines (RKO, HT29, VRC5/c1, GC3/c1, HCT116); HT29 exhibited the highest level of constitutive NF-κB expression, whereas negligible expression was determined in HCT8. These results indicate that constitutive NF-κB activation occurs in high frequency in human colon carcinoma cell lines.
FIGURE 1.
Human colorectal carcinoma cell lines express constitutively active NF-κB. A, Western analysis of cell lysates for determination of the expression levels of p-IKKα/β, total IKKα/β, total p65-NF-κB, or activated phospho-p65-NF-κB (S536) as well as p-IκBα and total IκBα is shown. β-Actin was used as the loading control. B, shown is a tabulated form for the ratios of the band intensity for p-p65/p65 and p-IκBα/β-actin from A. C, nuclear lysates of human colon carcinoma cell lines were prepared using a nuclear and cytoplasmic extraction kit from Panomics as described under “Materials and Methods.” Nuclear lysates (5 μg) were labeled with NF-κB oligonucleotides, and DNA-protein binding was analyzed by EMSA (as described under “Materials and Methods”).
Quinacrine Decreases Constitutively Active NF-κB in Human Colon Carcinoma Cell Lines
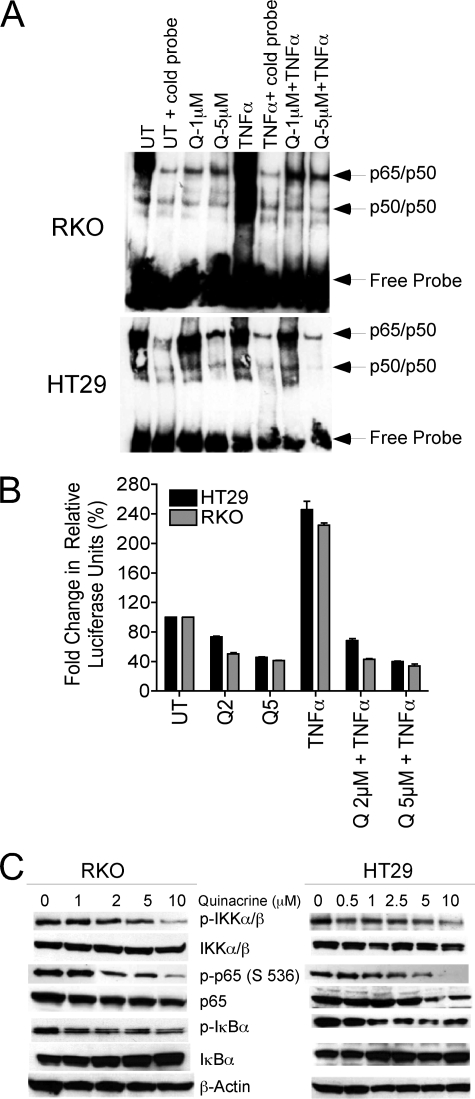
RKO and HT29 cells were treated with their respective IC50 concentrations of quinacrine (5 μm and 1 μm, respectively) for a period of 2 h; TNFα (10 ng/ml) treatment for 1 h was employed as a positive control. Nuclear lysates were prepared and analyzed for NF-κB DNA binding by EMSA (Fig. 2A). In addition, the effects of quinacrine on NF-κB-dependent luciferase reporter activity (Fig. 2B) or on cellular levels of p-IKKα/β, IKK, p-p65 (S536), p65, p-IκBα, and IκBα were also examined. Quinacrine decreased constitutively active as well as TNF-inducible NF-κB DNA binding in both cell lines (Fig. 2A). Corresponding with the down-regulation of DNA binding activity, a significant decrease in constitutive as well as TNF-inducible NF-κB-dependent luciferase reporter activity was also observed (Fig. 2B). Furthermore, cellular levels of p-IKKα/β, p-p65 (Ser 536), and p-IκBα were also significantly decreased after quinacrine treatment (Fig. 2C). Data demonstrate that quinacrine inhibits constitutively expressed (and inducible) NF-κB in human colon carcinoma cell lines.
FIGURE 2.
Quinacrine decreases constitutively active NF-κB in human colon carcinoma cell lines. A, RKO or HT29 cell lines were treated with quinacrine 1 μm (Q1) or 5 μm (Q5), respectively, for 2 h followed by TNFα (10 ng/ml) for 1 h. DNA-protein binding was analyzed by EMSA (as described under “Materials and Methods”). Data are representative of three separate experiments. B, RKO and HT29 were either left untreated (UT) or were transiently transfected with the NF-κB-dependent p5X1P10kb reporter plasmid for 24 h and subsequently treated with quinacrine 2 μm (Q2) or 5 μm (Q5), respectively, for 2 h followed by TNFα (10 ng/ml) for 1 h. Lysates were prepared using a dual luciferase reporter assay kit (Promega), and NF-κB-luciferase activity was measured as per the manufacturer's protocol. Data represent the -fold change in relative luciferase units and are the mean ± S.D. of triplicate determinations. C, RKO and HT29 cells were treated with increasing concentrations of quinacrine (0–10 μm) for 2 h, after which total cellular lysates were prepared and analyzed by Western for the determination of activated NF-κB. β-Actin was used as the loading control. This figure is representative of three separate experiments.
Inhibition of Constitutively Active NF-κB Elicits Cytotoxic Activity in Human Colon Carcinoma Cell Lines
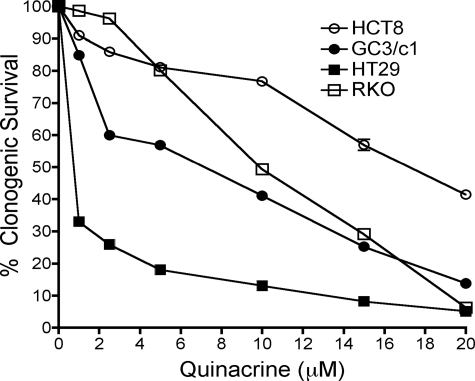
To examine the cytotoxic activity of quinacrine in human colon carcinoma cell lines, HCT8, GC3/c1, HT29, and RKO cells were treated with increasing concentrations of quinacrine (0–20 μm) for a period of 2 h; clonogenic survival was determined as described under “Materials and Methods.” Quinacrine induced a dose-dependent decrease in clonogenic survival (Fig. 3) that correlated with constitutive NF-κB activation. HT29 cells, which strongly express constitutively active NF-κB, exhibited the greatest decrease in clonogenic survival in response to quinacrine, whereas HCT8, which expressed negligible levels of constitutively active NF-κB, demonstrated the smallest decrease in clonogenic survival. Similar results were also observed when the cells were treated with quinacrine for 24 h (data not shown).
FIGURE 3.
Inhibition of constitutively active NF-κB elicits cytotoxic activity in human colon carcinoma cell lines. RKO, HT29, GC3/c1, and HCT8 cell lines were treated with increasing concentrations of quinacrine (0–20 μm) for 2 h. Percent survival was analyzed by clonogenic survival assay (as described under “Materials and Methods”), and the data represent the mean ± S.D. of triplicate determinations.
Quinacrine Sensitizes RKO and HT29 Cells to TRAIL- and l-OHP-induced Cell Death
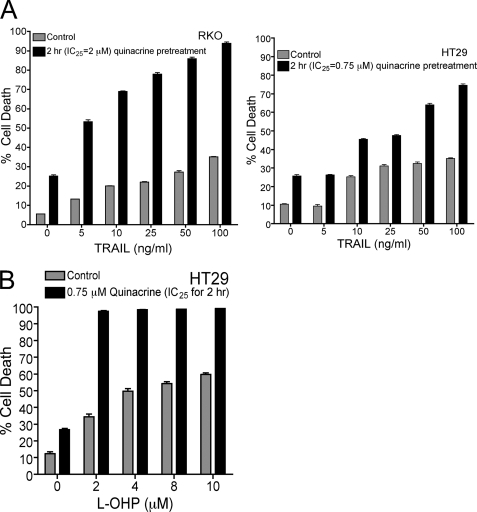
Constitutive NF-κB activation has conferred resistance to TRAIL (61, 62) and to various chemotherapeutic agents including l-OHP in cancer cells (33, 63–65). HT29 and RKO cells, resistant to TRAIL-mediated apoptosis, were selected to elucidate whether inhibition of NF-κB signaling by quinacrine could potentiate TRAIL-induced apoptosis in these cell lines. RKO and HT29 cells were pretreated with IC25 concentrations of quinacrine (2 and 0.75 μm, respectively) for 2 h followed by TRAIL (5–100 ng/ml) for 24 h. Furthermore, HT29 cells, also resistant to l-OHP, were pretreated with IC25 concentrations of quinacrine (0.75 μm) for 2 h followed by l-OHP (0–10 μm) for 72 h. Apoptotic cell death was determined by Annexin V/PI staining as described under “Materials and Methods.” Neither RKO nor HT29 exhibited >30% cell death in response to TRAIL alone (100 ng/ml); however, pretreatment with quinacrine for 2 h followed by TRAIL for 24 h significantly sensitized these cells to TRAIL-induced cell death (∼80–90%; Fig. 4A). HT29 cells, which exhibited ≈45% cell death in response to l-OHP (2 μm), were also significantly sensitized to l-OHP-mediated cell death (∼ 90% cell death; Fig. 4B) after quinacrine pretreatment for 2 h.
FIGURE 4.
Quinacrine sensitizes RKO and HT29 cells to TRAIL- and l-OHP-induced cell death. A, RKO and HT29 cell lines were pretreated with IC25 concentrations of quinacrine (2 or 0.75 μm, respectively) for 2 h followed by treatment with increasing concentrations of TRAIL (0–100 ng/ml) for 24 h, and cell death was analyzed by FACS using Annexin V/PI staining as described under “Materials and Methods.” Data represent the mean ± S.D. of triplicate determinations. B, HT29 cells were pretreated with IC25 concentrations of quinacrine (0.75 μm) for 2 h followed by treatment with increasing concentrations of l-OHP (0.10 μm) for 72 h. Cell death was analyzed by FACS using Annexin V/PI staining as described under “Materials and Methods.” Data represent the mean ± S.D. of triplicate determinations.
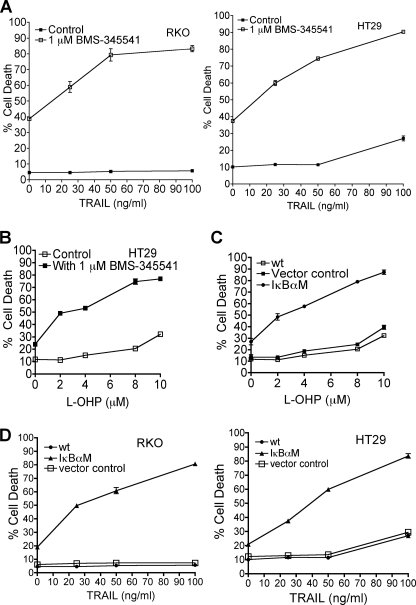
Inhibition of NF-κB by an Inhibitor of IKK or by Overexpression of IκBαM Sensitizes Colon Carcinoma Cell Lines to TRAIL- or l-OHP-induced Cell Death
RKO or HT29 cells were either untreated or treated with the IKK inhibitor BMS-345541 or were transiently transfected with either a control vector or with IκBαM followed by varied concentrations of TRAIL (0–100 ng/ml) or l-OHP (0–10 μm). Similar to the results obtained with quinacrine, pretreatment with the IKK inhibitor BMS-345541 for 1 h sensitized cells to TRAIL-induced apoptosis (Fig. 5A) or to l-OHP-mediated cell death (Fig. 5B). Furthermore, transient overexpression of IκBαM in RKO and HT29 caused a significant increase in cell death in response to TRAIL (Fig. 5C) or to l-OHP (Fig. 5D). These results clearly indicate that inhibition of constitutively expressed NF-κB sensitizes colon carcinoma cells to TRAIL- or to l-OHP- mediated apoptosis, thereby demonstrating that constitutive NF-κB activity confers resistance to TRAIL- or l-OHP-induced apoptosis.
FIGURE 5.
Inhibition of NF-κB by an inhibitor of IKK or by overexpression of IκBαM sensitizes colon carcinoma cell lines to TRAIL- or l-OHP-induced cell death. Inhibition of NF-κB activity was achieved by either using IKK inhibitor, BMS-345541, or by overexpression of IκBαM in RKO and HT29 cell lines as described under “Materials and Methods.” A, RKO and HT29 cells were either left untreated or were treated with 1 μm BMS-345541 for 1 h followed by increasing concentrations of TRAIL (0–100 ng/ml) for 24 h, after which cell death was analyzed by FACS using Annexin V/PI staining as described under “Materials and Methods.” Data represent the mean ± S.D. of triplicate determinations. B, HT29 cells were either left untreated or were treated with 1 μm BMS-345541 for 1 h followed by increasing concentrations of l-OHP (0–10 μm) for 72 h, after which cell death was analyzed by FACS using Annexin V/PI staining as described under “Materials and Methods.” Data represent the mean ± S.D. of triplicate determinations. C, HT29 cells were either left untreated or were transiently transfected with control vector or IκBαM for 72 h with l-OHP added during the last 48 h of the transfection. Apoptotic cell death was analyzed by FACS using Annexin V/PI staining as described under “Materials and Methods.” Data represent the mean ± S.D. of triplicate determinations. D, RKO and HT29 cells were either left untreated or were transiently transfected with control vector or IκBαM for 72 h with TRAIL added during the last 24 h of the transfection. Apoptotic cell death was analyzed by FACS using Annexin V/PI staining as described under “Materials and Methods.” Data represent the mean ± S.D. of triplicate determinations.
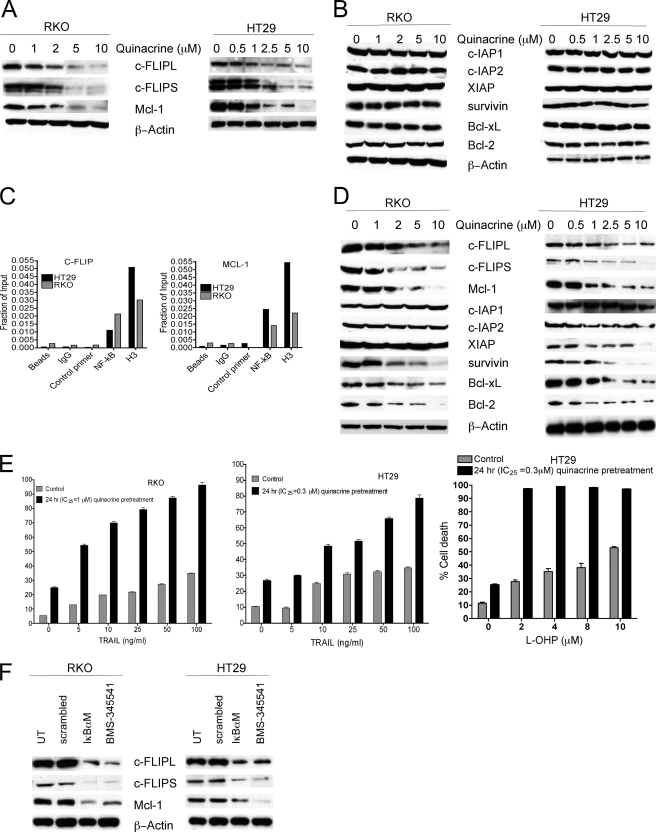
Effect of Inhibition of NF-κB Signaling on Expression of Downstream Survival Genes and TRAIL- and l-OHP-induced Cytotoxicity
NF-κB is a critical transcription factor that regulates the expression of several survival factors, including members of the Bcl-2 family of proteins (Bcl-2, Bcl-xL, Mcl-1) and IAP family (c-IAP1, c-IAP2, XIAP, survivin) as well as the c-FLIP. The effect of inhibition of NF-κB signaling by quinacrine on the expression of these NF-κB-dependent survival genes was subsequently examined further. RKO and HT29 cells were treated with increasing concentrations of quinacrine (0–10 μm) for 2 h after which whole cell lysates were prepared for Western analyses, as described under “Materials and Methods.” Data demonstrate a dose-dependent decrease in expression of c-FLIP (c-FLIPL, c-FLIPS) and Mcl-1 protein (Fig. 6A), whereas no significant changes were observed in the expression of other NF-κB-dependent survival proteins (survivin, Bcl-xL, Bcl-2, XIAP, c-IAP1, c-IAP2; Fig. 6B). Although c-FLIP and Mcl-1 are known to be regulated by NF-κB, direct interaction had not been described before. We identified putative NF-κB-binding sites on both c-FLIP and Mcl-1 promoters using the MatInspector transcription factor binding site analysis tool (Genomatix software). Subsequently, chromatin immunoprecipitation analyses demonstrated the c-FLIP and Mcl-1 are direct NF-κB transcriptional targets and that NF-κB physically interacts with its cognate DNA elements on both c-FLIP and Mcl-1 promoters in colon cancer cells (Fig. 6C). It was also determined that subsequent treatment of cells with quinacrine for 24 h not only down-regulated the expression of c-FLIP and Mcl-1 but also decreased expression of other NF-κB-dependent survival proteins including survivin, Bcl-xL, Bcl-2, and XIAP (Fig. 6D). Although decreased expression of other NF-KB-dependent genes in addition to c-FLIP and Mcl-1 was observed at 24 h after quinacrine exposure, sensitization to TRAIL or to l-OHP after IC25 concentrations of quinacrine pretreatment for 24 h was similar to the sensitization obtained with IC25 concentrations of quinacrine pretreatment for 2 h (Fig. 6E). These results indicate that the early down-regulation of c-FLIP and Mcl-1 by quinacrine plays a primary important role in increasing the efficacy of TRAIL or l-OHP in human colon carcinoma cell lines.
FIGURE 6.
Effect of inhibition of NF-κB signaling on expression of downstream survival genes, TRAIL- and l-OHP-induced cytotoxicity. A and B, RKO and HT29 cells were treated with increasing concentrations (0–10 μm) of quinacrine for 2 h followed by lysis and examined by Western analysis as described under “Materials and Methods.” Data are representative of three separate experiments. C, chromatin immunoprecipitation followed by quantitative real-time-PCR was performed on RKO and HT29 cells as described under “Materials and Methods” to identify putative NF-κB-binding sites on both c-FLIP and Mcl-1 promoters. Data represent the mean ± S.E. of triplicate determinations. D, RKO and HT29 cells were treated with increasing concentrations (0–10 μm) of quinacrine for 24 h followed by lysis and examined by Western analysis as described under “Materials and Methods.” E, RKO and HT29 cell lines were pretreated with IC25 concentrations of quinacrine (1 or 0.3 μm, respectively) for 24 h followed by treatment with increasing concentrations of TRAIL (0–100 ng/ml) for 24 h or l-OHP (0–10 μm) for 72 h. Cell death was analyzed by FACS using Annexin V/PI staining as described under “Materials and Methods.” Data represent the mean ± S.D. of triplicate determinations. F, RKO and HT29 cells were either treated with 1 μm BMS-345541 or were transiently transfected with control vector or IκBαM for 72 h. Total cellular lysates were prepared for the detection of c-FLIP or Mcl-1 protein expression in these cells. β-Actin was used as the loading control. This figure is representative from three separate experiments.
To further confirm the effect of NF-κB inhibition on expression of c-FLIP and Mcl-1, RKO and HT29 cells were treated with the IKK inhibitor BMS-345541 (1 μm) for 2 h, or IκBαM was transiently overexpressed, after which total cellular lysates were examined for the expression of c-FLIP and Mcl-1 by Western analysis (Fig. 6F). Similar to the results obtained with quinacrine-induced inhibition of NF-κB, NF-κB inhibition by either overexpression of IκBαM or by inhibition of IKK by BMS-345541 also demonstrated down-regulated c-FLIP and Mcl-1 expression in these cells.
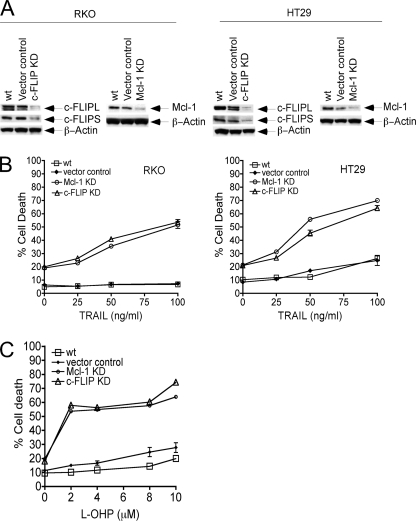
shRNA Knockdown of Endogenous c-FLIP or Mcl-1 with shRNA Significantly Potentiates TRAIL- or l-OHP-induced Apoptosis in Human Colon Carcinoma Cell Lines
To determine the effect of c-FLIP or Mcl-1 down-regulation on TRAIL- or l-OHP-mediated apoptosis in RKO or HT29 cells, transient shRNA knockdown of endogenous c-FLIP or Mcl-1 was performed as described under “Materials and Methods.” RKO or HT29 cells were either untreated or were transiently transfected with the respective shRNA or with a control vector. A significant decrease in the expression of c-FLIP and Mcl-1 was observed in shRNA-transfected RKO or HT29 cells compared with the vector control or wt cells (Fig. 7A). Importantly, this down-regulation of c-FLIP or Mcl-1 significantly enhanced TRAIL-induced cell death (Fig. 7B) as well as l-OHP-mediated cell death (Fig. 7C). Data demonstrate that inhibition of NF-κB and its downstream target genes c-FLIP and Mcl-1 are critical for TRAIL- or l-OHP-induced apoptosis.
FIGURE 7.
shRNA knockdown of endogenous c-FLIP or Mcl-1 facilitates TRAIL-induced apoptosis in RKO and HT29 cells. A, RKO and HT29 cell lines were either untreated or transiently transfected with c-FLIP, Mcl-1, or control vector for 72 h. Total cellular lysates were prepared for the detection of c-FLIP or Mcl-1 protein expression in these cells. β-Actin was used as the loading control. This figure is representative from three separate experiments. KD, knock-down. B, RKO and HT29 cell lines were either untreated or transiently transfected with c-FLIP, Mcl-1 shRNA, or control vector for 72 h followed by treatment with TRAIL (25–100 ng/ml) for 24 h. Cell death was analyzed by FACS using Annexin V/PI staining as described under “Materials and Methods.” Data represent the mean ± S.D. of triplicate determinations. C, HT29 cells were either untreated or transiently transfected with c-FLIP, Mcl-1 shRNA, or control vector for 72 h with l-OHP added during the last 48 h of the transfection. Cell death was analyzed by FACS using Annexin V/PI staining as described under “Materials and Methods.” Data represent the mean ± S.D. of triplicate determinations.
DISCUSSION
Colorectal cancer remains a major contributor to cancer-related deaths in the United States, with currently employed therapeutic regimens yielding an expected median survival rate of 22 months for patients with advanced metastatic colorectal cancer. These regimens have not only been empirically derived, but drug resistance also compounds the problems involved in the discovery and implementation of new and effective therapeutic approaches. Of agents currently under development, TRAIL has gained importance as an anti-tumor agent due to its specificity in the induction of cell death in cancer cells in preference to normal cells, without inducing hepatotoxicity (23–27). Studies conducted in preclinical models using TRAIL have demonstrated potent anti-tumor activity both in vitro and in in vivo cancer models (26, 66, 67). Significant efficacy has also been demonstrated with drug combinations (68–72), and TRAIL is currently being evaluated in clinical trials (73–75). Colon carcinomas express both DR4 and DR5 receptors (76); however, they have demonstrated resistance to TRAIL-induced apoptosis (28, 77). A cytotoxic agent currently employed in standard therapeutic regimens is the platinum-based compound, l-OHP. This is the only platinum analog that is found to demonstrate some activity in colorectal cancer. It is, therefore, of importance to understand the molecular mechanism(s) that imparts resistance to TRAIL and to l-OHP in colon cancers, in particular in light of the findings of expression of constitutively active NF-κB in a wide variety of human cancers (49–52, 54), including colorectal cancer (48, 53).
Several studies have demonstrated aberrant and constitutive activation of NF-κB in hematologic and solid tumors (48–54). Furthermore, chemotherapeutic agents including l-OHP (78, 79) as well as agents that ligate the TRAIL receptors (80–84) can also activate NF-κB in these cells, which may contribute toward imparting resistance against these agents. The current study has investigated the effect of quinacrine on inhibition of constitutively active NF-κB, which we have demonstrated is highly expressed in human colon carcinoma cell lines and which potentiates both TRAIL- and l-OHP-mediated death in these cells. Quinacrine is a derivative of 9-aminoacridine, used extensively for the treatment of malaria, and more recently has been to shown to have activity in cancer cells (55). Thus, it has been shown to activate the tumor suppressor p53 simultaneously with inhibition of NF-κB in renal cell carcinoma cells (55). However, we have demonstrated that HT29 (mp53) and RKO (wtp53) cells, which have very high levels of expression of constitutively active NF-κB, are more sensitive to quinacrine-induced cell death irrespective of the status of the p53 gene, whereas HCT8 cells (wtp53), with significantly lower levels of constitutively active NF-κB, are considerably less sensitive to quinacrine-induced cell death. Thus, the degree of constitutively active NF-κB correlates with quinacrine sensitivity in colon carcinoma cell lines, independent of p53.
NF-κB plays a pivotal role in cell survival and is actively involved in the transcription of >150 genes involved in different cellular functions. NF-κB participates in the regulation of invasion, angiogenesis, and metastasis of tumor cells and is involved in tumor progression through regulation of various chemokines as well as genes that regulate cell proliferation including cyclin D1, c-Myc, TNF, interleukin-1, and MMP-9 (43, 53, 84–86). NF-κB is also a key regulator of apoptosis, where it suppresses apoptotic cell death by inducing the expression of several anti-apoptotic factors that include proteins belonging to the IAP and Bcl-2 families to inhibit caspase-3 activation. NF-κB also transcriptionally regulates the expression of c-FLIP, which inhibits TRAIL- as well as l-OHP-induced apoptosis by inhibiting caspase-8 activation at the receptor complex and can confer resistance to TRAIL or chemotherapeutic agents in human cancers (36, 37). Mcl-1, a survival factor that is also known to be up-regulated by NF-κB (87), may also be a critical component of drug resistance in colon cancer. Mcl-1 inhibits mitochondrial-dependent apoptosis after receptor ligation in death receptor-mediated cell death. These NF-κB target genes are over-expressed in colon carcinoma cells (43, 85, 88) and can confer chemoresistance as well as TRAIL resistance in various tumors (36, 37, 47). Thus, the potential for l-OHP or TRAIL as anti-tumor agents may be significantly enhanced by inhibiting not only NF-κB but also the NF-κB-regulated downstream survival signaling pathways. Numerous reports have shown synergistic anticancer activity when these agents are combined together with each other or with other chemotherapeutic agents (69–71, 89–92), including NF-κB inhibitors (34, 69–71, 93–96). Studies with inhibition of NF-κB by proteasome inhibitors such as NPI-0052 (salinosporamide A) have demonstrated synergistic interaction with various chemotherapeutic drug combinations that include FUra/LV+ CPT-11 (irinotecan) + Avastin (bevacizumab) + oxaliplatin in a colon cancer model (34). Inhibition of NF-κB signaling has also augmented the anti-tumor efficacy of gemcitabine and l-OHP in pancreatic cancer (97). Studies using bortezomib (PS-341), also a proteasomal inhibitor that inhibits NF-κB activation, have demonstrated enhanced sensitization to radiation therapy in the LOVO xenograft model (98), increased chemosensitivity to CPT-11 in colorectal cancer cells (99), and enhancement of the cytotoxic effect of several chemotherapeutic agents including carboplatin and CPT-11 in glioma cells (100). Recently, phase I studies using bortezomib in combination with irinotecan (101) or with gemcitabine (102) or with capecitabine and l-OHP (103) in patients with advanced solid tumors have shown that these drug combinations are safe, with manageable toxicities, and are currently in Phase II trials, indicating that inhibition of NF-κB may play an important role in new developmental therapeutic approaches in cancer. Although very recently Hideshima et al. (104) showed that bortezomib and other proteasome inhibitors can also induce canonical NF-κB signaling in MM cells through phosphorylation of receptor-interacting protein 2 as well as IKKβ (105), which clearly indicates that the action of bortezomib and other proteasome inhibitors is not limited to NF-κB inhibition and that other mechanisms may play a role in its anti-tumor activity.
It has been demonstrated that colorectal cancers have high constitutive NF-κB expression, and the current study has established that the majority of colon carcinoma cell lines also demonstrate constitutive NF-κB expression. Furthermore, data demonstrate that quinacrine is cytotoxic to human colon carcinoma cell lines and that it inhibits this constitutive NF-κB activity independent of p53. Importantly, at IC25 concentrations, quinacrine as well as inhibiting NF-κB activation markedly enhances the anticancer activity of TRAIL and l-OHP in these cell lines. Similarly, a specific IKK inhibitor BMS-345541 or expression of IκBαM, which inhibits constitutively active NF-κB, confers sensitivity to TRAIL- or l-OHP-induced cell death. Several survival factors that are transcriptionally regulated by NF-κB are implicated in conferring resistance to these agents in tumor cells. The current study demonstrates that with a relatively short, 2-h exposure, quinacrine specifically decreases the expression of c-FLIP and Mcl-1 without affecting the expression of other NF-κB-dependent survival proteins (survivin, Bcl-xL, Bcl-2, XIAP, c-IAP1, c-IAP2) in both RKO and HT29 cell lines. However, extended exposure to quinacrine (up to 24 h) resulted in down-regulation of other NF-κB-dependent survival proteins, namely, XIAP, Bcl-2, Bcl-xL, and survivin. Our study also demonstrates for the first time that c-FLIP and Mcl-1 are direct targets of NF-κB. Previous reports have suggested that c-FLIP or Mcl-1 survival factors could be transcriptionally regulated by NF-κB; however, evidence of a direct interaction between the transcription factor and promoters of these putative targets has not been previously described. We have used chromatin immunoprecipitation technology to selectively immunoprecipitate NF-κB from chromatin preparations and determined the associated DNA sequences. Our findings that NF-κB physically binds to its specific DNA elements on the promoters of c-FLIP and Mcl-1 provide strong evidence that NF-κB directly transcriptionally regulates c-FLIP and Mcl-1. Both c-FLIP and Mcl-1 are primarily affected by quinacrine due to their shorter half-lives as compared with the other NF-κB-dependent survival proteins including c-IAP1, c-IAP2, and XIAP as well as Bcl-2 and Bcl-xL (56, 106–108). In addition, shRNA knockdown of c-FLIP or Mcl-1 in HT29 and RKO significantly enhanced TRAIL- and l-OHP-induced cell death. These results, verified by different agents and experimental techniques, clearly demonstrate the importance of constitutively active NF-κB in attenuating apoptosis mediated by these agents and that both c-FLIP and Mcl-1 are the primary contributors of NF-κB-dependent survival factors to the TRAIL and l-OHP resistance phenotype in colon carcinoma cells via an NF-κB-dependent signaling mechanism.
In conclusion, constitutive NF-κB activation contributes significantly to cell survival in human colon carcinoma cells, rendering them resistant to TRAIL as well as to other chemotherapeutic agents including l-OHP and that inhibition of NF-κB signaling by quinacrine is highly synergistic in the potentiation of the cytotoxic effect of TRAIL and l-OHP, supporting the application of NF-κB inhibitors in the development of novel and effective therapeutic approaches for the treatment of colon cancer.
T. S. Jani, J. DeVecchio, T. Mazumdar, A. Agyeman, and J. A. Houghton, unpublished information.
- FUra
- 5-fluorouracil
- LV
- leucovorin
- TNF
- necrosis factor receptor
- TRAIL
- TNF-related apoptosis-inducing ligand
- shRNA
- short hairpin RNA
- l-OHP
- oxaliplatin
- IκB
- inhibitor of κB
- IAP
- inhibitor of apoptosis protein
- IκBαM
- IκBα mutant
- PI
- propidium iodide
- EMSA
- electrophoretic mobility shift assay
- c-FLIP
- FLICE-inhibitory protein
- wt
- wild type
- IKK
- IκB kinase
- PBS
- phosphate-buffered saline
- FACS
- fluorescence-activated cell sorter.
REFERENCES
- 1.American Cancer Society (2008) Colorectal Cancer Facts and Figures 2008–2010, pp. 1–3, American Cancer Society, Atlanta, GA [Google Scholar]
- 2.American Cancer Society (2009) Cancer Facts and Figures 2009, pp. 3–7, American Cancer Society, Atlanta, GA [Google Scholar]
- 3.Hind D., Tappenden P., Tumur I., Eggington S., Sutcliffe P., Ryan A. (2008) Health Technol. Assess. 12, iii–ix, xi–162 [DOI] [PubMed] [Google Scholar]
- 4.Simpson D., Dunn C., Curran M., Goa K. L. (2003) Drugs 63, 2127–2156 [DOI] [PubMed] [Google Scholar]
- 5.Louvet C., de Gramont A. (2003) Curr. Treat. Options Oncol. 4, 405–411 [DOI] [PubMed] [Google Scholar]
- 6.Ragnhammar P., Hafström L., Nygren P., Glimelius B. (2001) Acta Oncol. 40, 282–308 [DOI] [PubMed] [Google Scholar]
- 7.Goldberg R. (2000) Oncology 14, 42–47 [PubMed] [Google Scholar]
- 8.Culy C. R., Clemett D., Wiseman L. R. (2000) Drugs 60, 895–924 [DOI] [PubMed] [Google Scholar]
- 9.Díaz-Rubio E., Sastre J., Zaniboni A., Labianca R., Cortés-Funes H., de Braud F., Boni C., Benavides M., Dallavalle G., Homerin M. (1998) Ann. Oncol. 9, 105–108 [DOI] [PubMed] [Google Scholar]
- 10.Haller D. G. (2000) Oncology 14, 15–20 [PubMed] [Google Scholar]
- 11.Petak I., Tillman D. M., Houghton J. A. (2000) Clin. Cancer Res. 6, 4432–4441 [PubMed] [Google Scholar]
- 12.Petak I., Tillman D. M., Harwood F. G., Mihalik R., Houghton J. A. (2000) Cancer Res. 60, 2643–2650 [PubMed] [Google Scholar]
- 13.Tillman D. M., Petak I., Houghton J. A. (1999) Clin. Cancer Res. 5, 425–430 [PubMed] [Google Scholar]
- 14.Tillman D. M., Harwood F. G., Gibson A. A., Houghton J. A. (1998) Cell Death Differ. 5, 450–457 [DOI] [PubMed] [Google Scholar]
- 15.Houghton J. A., Harwood F. G., Tillman D. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8144–8149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houghton J. A., Harwood F. G., Gibson A. A., Tillman D. M. (1997) Clin. Cancer Res. 3, 2205–2209 [PubMed] [Google Scholar]
- 17.Schwartzberg L. S., Petak I., Stewart C., Turner P. K., Ashley J., Tillman D. M., Douglas L., Tan M., Billups C., Mihalik R., Weir A., Tauer K., Shope S., Houghton J. A. (2002) Clin. Cancer Res. 8, 2488–2498 [PubMed] [Google Scholar]
- 18.Schwartzberg L., Houghton J. A., Phillips D., Stewart C., Onciu M., Geller J., Walker M. (2008) 99th Annual Meeting of the American Association for Cancer Research, San Diego, CA, April 12–16, 2008, Abstract 4456 [Google Scholar]
- 19.Ogasawara J., Watanabe-Fukunaga R., Adachi M., Matsuzawa A., Kasugai T., Kitamura Y., Itoh N., Suda T., Nagata S. (1993) Nature 364, 806–809 [DOI] [PubMed] [Google Scholar]
- 20.Havell E. A., Fiers W., North R. J. (1988) J. Exp. Med. 167, 1067–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.North R. J., Havell E. A. (1988) J. Exp. Med. 167, 1086–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashkenazi A., Herbst R. S. (2008) J. Clin. Invest. 118, 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiley S. R., Schooley K., Smolak P. J., Din W. S., Huang C. P., Nicholl J. K., Sutherland G. R., Smith T. D., Rauch C., Smith C. A. (1995) Immunity 3, 673–682 [DOI] [PubMed] [Google Scholar]
- 24.Pitti R. M., Marsters S. A., Ruppert S., Donahue C. J., Moore A., Ashkenazi A. (1996) J. Biol. Chem. 271, 12687–12690 [DOI] [PubMed] [Google Scholar]
- 25.Rieger J., Naumann U., Glaser T., Ashkenazi A., Weller M. (1998) FEBS Lett. 427, 124–128 [DOI] [PubMed] [Google Scholar]
- 26.Walczak H., Miller R. E., Ariail K., Gliniak B., Griffith T. S., Kubin M., Chin W., Jones J., Woodward A., Le T., Smith C., Smolak P., Goodwin R. G., Rauch C. T., Schuh J. C., Lynch D. H. (1999) Nat. Med. 5, 157–163 [DOI] [PubMed] [Google Scholar]
- 27.Ashkenazi A., Pai R. C., Fong S., Leung S., Lawrence D. A., Marsters S. A., Blackie C., Chang L., McMurtrey A. E., Hebert A., DeForge L., Koumenis I. L., Lewis D., Harris L., Bussiere J., Koeppen H., Shahrokh Z., Schwall R. H. (1999) J. Clin. Invest. 104, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sträter J., Hinz U., Walczak H., Mechtersheimer G., Koretz K., Herfarth C., Möller P., Lehnert T. (2002) Clin. Cancer Res. 8, 3734–3740 [PubMed] [Google Scholar]
- 29.Morales J. C., Ruiz-Magaña M. J., Ruiz-Ruiz C. (2007) Mol. Immunol. 44, 2587–2597 [DOI] [PubMed] [Google Scholar]
- 30.Keane M. M., Rubinstein Y., Cuello M., Ettenberg S. A., Banerjee P., Nau M. M., Lipkowitz S. (2000) Breast Cancer Res. Treat. 64, 211–219 [DOI] [PubMed] [Google Scholar]
- 31.Kim Y. S., Schwabe R. F., Qian T., Lemasters J. J., Brenner D. A. (2002) Hepatology 36, 1498–1508 [DOI] [PubMed] [Google Scholar]
- 32.Rakitina T. V., Vasilevskaya I. A., O'Dwyer P. J. (2003) Cancer Res. 63, 8600–8605 [PubMed] [Google Scholar]
- 33.Wilson C., Purcell C., Seaton A., Oladipo O., Maxwell P. J., O'Sullivan J. M., Wilson R. H., Johnston P. G., Waugh D. J. (2008) J. Pharmacol. Exp. Ther. 327, 746–759 [DOI] [PubMed] [Google Scholar]
- 34.Cusack J. C., Jr., Liu R., Xia L., Chao T. H., Pien C., Niu W., Palombella V. J., Neuteboom S. T., Palladino M. A. (2006) Clin. Cancer Res. 12, 6758–6764 [DOI] [PubMed] [Google Scholar]
- 35.Baeuerle P. A., Baltimore D. (1996) Cell 87, 13–20 [DOI] [PubMed] [Google Scholar]
- 36.Wilson T. R., McEwan M., McLaughlin K., Le Clorennec C., Allen W. L., Fennell D. A., Johnston P. G., Longley D. B. (2009) Oncogene 28, 63–72 [DOI] [PubMed] [Google Scholar]
- 37.Wilson T. R., McLaughlin K. M., McEwan M., Sakai H., Rogers K. M., Redmond K. M., Johnston P. G., Longley D. B. (2007) Cancer Res. 67, 5754–5762 [DOI] [PubMed] [Google Scholar]
- 38.Lagunas V. M., Meléndez-Zajgla J. (2008) Met. Based Drugs 2008, 576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beg A. A., Baltimore D. (1996) Science 274, 782–784 [DOI] [PubMed] [Google Scholar]
- 40.Mayo M. W., Wang C. Y., Cogswell P. C., Rogers-Graham K. S., Lowe S. W., Der C. J., Baldwin A. S., Jr. (1997) Science 278, 1812–1815 [DOI] [PubMed] [Google Scholar]
- 41.Wang C. Y., Mayo M. W., Korneluk R. G., Goeddel D. V., Baldwin A. S., Jr. (1998) Science 281, 1680–1683 [DOI] [PubMed] [Google Scholar]
- 42.Van Antwerp D. J., Martin S. J., Kafri T., Green D. R., Verma I. M. (1996) Science 274, 787–789 [DOI] [PubMed] [Google Scholar]
- 43.Li X., Stark G. R. (2002) Exp. Hematol. 30, 285–296 [DOI] [PubMed] [Google Scholar]
- 44.Stehlik C., de Martin R., Kumabashiri I., Schmid J. A., Binder B. R., Lipp J. (1998) J. Exp. Med. 188, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernandez A., Wang Q. D., Schwartz S. A., Evers B. M. (2001) J. Gastrointest. Surg. 5, 56–65 [DOI] [PubMed] [Google Scholar]
- 46.Kreuz S., Siegmund D., Scheurich P., Wajant H. (2001) Mol. Cell. Biol. 21, 3964–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Micheau O., Lens S., Gaide O., Alevizopoulos K., Tschopp J. (2001) Mol. Cell. Biol. 21, 5299–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lind D. S., Hochwald S. N., Malaty J., Rekkas S., Hebig P., Mishra G., Moldawer L. L., Copeland E. M., 3rd, Mackay S. (2001) Surgery 130, 363–369 [DOI] [PubMed] [Google Scholar]
- 49.Sovak M. A., Bellas R. E., Kim D. W., Zanieski G. J., Rogers A. E., Traish A. M., Sonenshein G. E. (1997) J. Clin. Invest. 100, 2952–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mori N., Fujii M., Ikeda S., Yamada Y., Tomonaga M., Ballard D. W., Yamamoto N. (1999) Blood 93, 2360–2368 [PubMed] [Google Scholar]
- 51.Wang W., Abbruzzese J. L., Evans D. B., Larry L., Cleary K. R., Chiao P. J. (1999) Clin. Cancer Res. 5, 119–127 [PubMed] [Google Scholar]
- 52.Nakshatri H., Bhat-Nakshatri P., Martin D. A., Goulet R. J., Jr., Sledge G. W., Jr. (1997) Mol. Cell. Biol. 17, 3629–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakamoto K., Maeda S., Hikiba Y., Nakagawa H., Hayakawa Y., Shibata W., Yanai A., Ogura K., Omata M. (2009) Clin. Cancer Res. 15, 2248–2258 [DOI] [PubMed] [Google Scholar]
- 54.Tai D. I., Tsai S. L., Chang Y. H., Huang S. N., Chen T. C., Chang K. S., Liaw Y. F. (2000) Cancer 89, 2274–2281 [PubMed] [Google Scholar]
- 55.Gurova K. V., Hill J. E., Guo C., Prokvolit A., Burdelya L. G., Samoylova E., Khodyakova A. V., Ganapathi R., Ganapathi M., Tararova N. D., Bosykh D., Lvovskiy D., Webb T. R., Stark G. R., Gudkov A. V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17448–17453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mufti A. R., Burstein E., Csomos R. A., Graf P. C., Wilkinson J. C., Dick R. D., Challa M., Son J. K., Bratton S. B., Su G. L., Brewer G. J., Jakob U., Duckett C. S. (2006) Mol. Cell 21, 775–785 [DOI] [PubMed] [Google Scholar]
- 57.Petak I., Vernes R., Szucs K. S., Anozie M., Izeradjene K., Douglas L., Tillman D. M., Phillips D. C., Houghton J. A. (2003) Cell Death Differ. 10, 729–739 [DOI] [PubMed] [Google Scholar]
- 58.Harwood F. G., Kasibhatla S., Petak I., Vernes R., Green D. R., Houghton J. A. (2000) J. Biol. Chem. 275, 10023–10029 [DOI] [PubMed] [Google Scholar]
- 59.Weinmann A. S., Farnham P. J. (2002) Methods 26, 37–47 [DOI] [PubMed] [Google Scholar]
- 60.Wells J., Farnham P. J. (2002) Methods 26, 48–56 [DOI] [PubMed] [Google Scholar]
- 61.Sethi G., Sung B., Aggarwal B. B. (2008) Exp. Biol. Med. (Maywood) 233, 21–31 [DOI] [PubMed] [Google Scholar]
- 62.Braeuer S. J., Büneker C., Mohr A., Zwacka R. M. (2006) Mol. Cancer Res. 4, 715–728 [DOI] [PubMed] [Google Scholar]
- 63.Watanabe M., Dewan M. Z., Okamura T., Sasaki M., Itoh K., Higashihara M., Mizoguchi H., Honda M., Sata T., Watanabe T., Yamamoto N., Umezawa K., Horie R. (2005) Int. J. Cancer 114, 32–38 [DOI] [PubMed] [Google Scholar]
- 64.Assef Y., Rubio F., Coló G., del Mónaco S., Costas M. A., Kotsias B. A. (2009) Leuk. Res. 33, 710–716 [DOI] [PubMed] [Google Scholar]
- 65.Dolcet X., Llobet D., Pallares J., Matias-Guiu X. (2005) Virchows Arch. 446, 475–482 [DOI] [PubMed] [Google Scholar]
- 66.Menoret E., Gomez-Bougie P., Geffroy-Luseau A., Daniels S., Moreau P., Le Gouill S., Harousseau J. L., Bataille R., Amiot M., Pellat-Deceunynck C. (2006) Blood 108, 1346–1352 [DOI] [PubMed] [Google Scholar]
- 67.Pukac L., Kanakaraj P., Humphreys R., Alderson R., Bloom M., Sung C., Riccobene T., Johnson R., Fiscella M., Mahoney A., Carrell J., Boyd E., Yao X. T., Zhang L., Zhong L., von Kerczek A., Shepard L., Vaughan T., Edwards B., Dobson C., Salcedo T., Albert V. (2005) Br. J. Cancer 92, 1430–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitsiades N., Mitsiades C. S., Poulaki V., Anderson K. C., Treon S. P. (2001) Expert Opin. Investig. Drugs 10, 1521–1530 [DOI] [PubMed] [Google Scholar]
- 69.Jin H., Yang R., Fong S., Totpal K., Lawrence D., Zheng Z., Ross J., Koeppen H., Schwall R., Ashkenazi A. (2004) Cancer Res. 64, 4900–4905 [DOI] [PubMed] [Google Scholar]
- 70.Zisman A., Ng C. P., Pantuck A. J., Bonavida B., Belldegrun A. S. (2001) J. Immunother. 24, 459–471 [DOI] [PubMed] [Google Scholar]
- 71.Mitsiades C. S., Treon S. P., Mitsiades N., Shima Y., Richardson P., Schlossman R., Hideshima T., Anderson K. C. (2001) Blood 98, 795–804 [DOI] [PubMed] [Google Scholar]
- 72.Gliniak B., Le T. (1999) Cancer Res. 59, 6153–6158 [PubMed] [Google Scholar]
- 73.Wakelee H. A., Patnaik A., Sikic B. I., Mita M., Fox N. L., Miceli R., Ullrich S. J., Fisher G. A., Tolcher A. W. (2010) Ann. Oncol. 21, 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tolcher A. W., Mita M., Meropol N. J., von Mehren M., Patnaik A., Padavic K., Hill M., Mays T., McCoy T., Fox N. L., Halpern W., Corey A., Cohen R. B. (2007) J. Clin. Oncol. 25, 1390–1395 [DOI] [PubMed] [Google Scholar]
- 75.Rowinsky E. K. (2005) J. Clin. Oncol. 23, 9394–9407 [DOI] [PubMed] [Google Scholar]
- 76.Ren Y. G., Wagner K. W., Knee D. A., Aza-Blanc P., Nasoff M., Deveraux Q. L. (2004) Mol. Biol. Cell 15, 5064–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Geelen C. M., de Vries E. G., Le T. K., van Weeghel R. P., de Jong S. (2003) Br. J. Cancer 89, 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu Y., Villalona-Calero M. A. (2002) Ann. Oncol. 13, 1841–1851 [DOI] [PubMed] [Google Scholar]
- 79.Yeh P. Y., Chuang S. E., Yeh K. H., Song Y. C., Ea C. K., Cheng A. L. (2002) Biochem. Pharmacol. 63, 1423–1430 [DOI] [PubMed] [Google Scholar]
- 80.Ammann J. U., Haag C., Kasperczyk H., Debatin K. M., Fulda S. (2009) Int. J. Cancer 124, 1301–1311 [DOI] [PubMed] [Google Scholar]
- 81.Degli-Esposti M. A., Dougall W. C., Smolak P. J., Waugh J. Y., Smith C. A., Goodwin R. G. (1997) Immunity 7, 813–820 [DOI] [PubMed] [Google Scholar]
- 82.Chaudhary P. M., Eby M., Jasmin A., Bookwalter A., Murray J., Hood L. (1997) Immunity 7, 821–830 [DOI] [PubMed] [Google Scholar]
- 83.Schneider P., Thome M., Burns K., Bodmer J. L., Hofmann K., Kataoka T., Holler N., Tschopp J. (1997) Immunity 7, 831–836 [DOI] [PubMed] [Google Scholar]
- 84.Huang S., DeGuzman A., Bucana C. D., Fidler I. J. (2000) Clin. Cancer Res. 6, 2573–2581 [PubMed] [Google Scholar]
- 85.Garg A., Aggarwal B. B. (2002) Leukemia 16, 1053–1068 [DOI] [PubMed] [Google Scholar]
- 86.Lee C. H., Jeon Y. T., Kim S. H., Song Y. S. (2007) Biofactors 29, 19–35 [DOI] [PubMed] [Google Scholar]
- 87.Hall M. A., Cleveland J. L. (2007) Cancer Cell 12, 4–6 [DOI] [PubMed] [Google Scholar]
- 88.Pahl H. L. (1999) Oncogene 18, 6853–6866 [DOI] [PubMed] [Google Scholar]
- 89.Toscano F., Fajoui Z. E., Gay F., Lalaoui N., Parmentier B., Chayvialle J. A., Scoazec J. Y., Micheau O., Abello J., Saurin J. C. (2008) Oncogene 27, 4161–4171 [DOI] [PubMed] [Google Scholar]
- 90.Ballestrero A., Nencioni A., Boy D., Rocco I., Garuti A., Mela G. S., Van Parijs L., Brossart P., Wesselborg S., Patrone F. (2004) Clin. Cancer Res. 10, 1463–1470 [DOI] [PubMed] [Google Scholar]
- 91.Ganten T. M., Koschny R., Sykora J., Schulze-Bergkamen H., Büchler P., Haas T. L., Schader M. B., Untergasser A., Stremmel W., Walczak H. (2006) Clin. Cancer Res. 12, 2640–2646 [DOI] [PubMed] [Google Scholar]
- 92.Xu L., Qu X., Zhang Y., Hu X., Yang X., Hou K., Teng Y., Zhang J., Sada K., Liu Y. (2009) FEBS Lett. 583, 943–948 [DOI] [PubMed] [Google Scholar]
- 93.Koschny R., Holland H., Sykora J., Erdal H., Krupp W., Bauer M., Bockmuehl U., Ahnert P., Meixensberger J., Stremmel W., Walczak H., Ganten T. M. (2010) J. Neurooncol. 97, 171–185 [DOI] [PubMed] [Google Scholar]
- 94.Roué G., Pérez-Galán P., López-Guerra M., Villamor N., Campo E., Colomer D. (2007) J. Immunol. 178, 1923–1930 [DOI] [PubMed] [Google Scholar]
- 95.Nagy K., Székely-Szüts K., Izeradjene K., Douglas L., Tillman M., Barti-Juhász H., Dominici M., Spano C., Luca Cervo G., Conte P., Houghton J. A., Mihalik R., Kopper L., Peták I. (2006) Pathol. Oncol. Res. 12, 133–142 [DOI] [PubMed] [Google Scholar]
- 96.Cusack J. C., Liu R., Baldwin A. S. (1999) Drug Resist. Updat. 2, 271–273 [DOI] [PubMed] [Google Scholar]
- 97.Banerjee S., Kaseb A. O., Wang Z., Kong D., Mohammad M., Padhye S., Sarkar F. H., Mohammad R. M. (2009) Cancer Res. 69, 5575–5583 [DOI] [PubMed] [Google Scholar]
- 98.Russo S. M., Tepper J. E., Baldwin A. S., Jr., Liu R., Adams J., Elliott P., Cusack J. C., Jr. (2001) Int. J. Radiat. Oncol. Biol. Phys. 50, 183–193 [DOI] [PubMed] [Google Scholar]
- 99.Cusack J. C., Jr., Liu R., Houston M., Abendroth K., Elliott P. J., Adams J., Baldwin A. S., Jr. (2001) Cancer Res. 61, 3535–3540 [PubMed] [Google Scholar]
- 100.Weaver K. D., Yeyeodu S., Cusack J. C., Jr., Baldwin A. S., Jr., Ewend M. G. (2003) J. Neurooncol. 61, 187–196 [DOI] [PubMed] [Google Scholar]
- 101.Ryan D. P., O'Neil B. H., Supko J. G., Rocha Lima C. M., Dees E. C., Appleman L. J., Clark J., Fidias P., Orlowski R. Z., Kashala O., Eder J. P., Cusack J. C., Jr. (2006) Cancer 107, 2688–2697 [DOI] [PubMed] [Google Scholar]
- 102.Ryan D. P., Appleman L. J., Lynch T., Supko J. G., Fidias P., Clark J. W., Fishman M., Zhu A. X., Enzinger P. C., Kashala O., Cusack J., Jr., Eder J. P. (2006) Cancer 107, 2482–2489 [DOI] [PubMed] [Google Scholar]
- 103.Cohen S. J., Engstrom P. F., Lewis N. L., Langer C. J., McLaughlin S., Beard M., Weiner L. M., Meropol N. J. (2008) Am. J. Clin. Oncol. 31, 1–5 [DOI] [PubMed] [Google Scholar]
- 104.Hideshima T., Ikeda H., Chauhan D., Okawa Y., Raje N., Podar K., Mitsiades C., Munshi N. C., Richardson P. G., Carrasco R. D., Anderson K. C. (2009) Blood 114, 1046–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McConkey D. J. (2009) Blood 114, 931–932 [DOI] [PubMed] [Google Scholar]
- 106.Poukkula M., Kaunisto A., Hietakangas V., Denessiouk K., Katajamäki T., Johnson M. S., Sistonen L., Eriksson J. E. (2005) J. Biol. Chem. 280, 27345–27355 [DOI] [PubMed] [Google Scholar]
- 107.Adams K. W., Cooper G. M. (2007) J. Biol. Chem. 282, 6192–6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yoshimura F. K., Luo X., Zhao X., Gerard H. C., Hudson A. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5543–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]