Abstract
Inhibition of apoptosis is critical for carcinogenesis. ARC (apoptosis repressor with caspase recruitment domain) is an endogenous inhibitor of apoptosis that antagonizes both intrinsic and extrinsic apoptosis pathways. Although normally expressed in striated myocytes and neurons, ARC is markedly induced in a variety of primary human epithelial cancers and renders cancer cells resistant to killing. The mechanisms that mediate the induction of ARC in cancer are unknown. Herein we demonstrate that increases in ARC abundance are stimulated by Ras through effects on transcription and protein stability. Overexpression of activated N-Ras or H-Ras in normal cells is sufficient to increase ARC mRNA and protein levels. Similarly, transgenic expression of activated H-Ras induces ARC in both the normal mammary epithelium and resulting tumors of intact mice. Conversely, knockdown of endogenous N-Ras in breast and colon cancer cells significantly reduces ARC mRNA and protein levels. The promoter of the Nol3 locus, encoding ARC, is activated by N-Ras and H-Ras in a MEK/ERK-dependent manner. Ras also stabilizes ARC protein by suppressing its polyubiquitination and subsequent proteasomal degradation. In addition to the effects of Ras on ARC abundance, ARC mediates Ras-induced cell survival and cell cycle progression. Thus, Ras induces ARC in epithelial cancers, and ARC plays a role in the oncogenic actions of Ras.
Keywords: Apoptosis, Breast Cancer, Cell Death, Colon Cancer, Ras, ARC (Apoptosis Repressor with CARD), NOL3
Introduction
Apoptosis is a highly regulated cell suicide process that is critical for development, tissue homeostasis and remodeling, and removal of damaged and transformed cells (1). Apoptotic cell death is mediated through two central pathways: the extrinsic pathway involving death receptors and the intrinsic pathway involving the mitochondria/endoplasmic reticulum (2). Several endogenous inhibitory proteins antagonize one or the other of these two pathways. These include FLIP (Fas-associated death domain protein-like interleukin-1β-converting enzyme inhibitory protein), which regulates assembly of the death-inducing signaling complex (DISC)3 (3); B-cell leukemia/lymphoma-2 protein (Bcl-2), which blocks mitochondrial apoptogen release (4); and inhibitor of apoptosis proteins (IAPs), which bind to and inhibit effector caspases by blocking substrate access (5).
In contrast to the above apoptosis inhibitors that act at discrete locations in the extrinsic or intrinsic pathways, ARC (apoptosis repressor with caspase recruitment domain) antagonizes both central apoptosis pathways. ARC suppresses the extrinsic pathway by directly binding to Fas, FADD, and procaspase-8, thereby blocking DISC assembly (6, 7). ARC inhibits the intrinsic pathway through at least two mechanisms. First, ARC interacts directly with Bax, thereby inhibiting Bax conformational activation and translocation to the mitochondria in response to apoptotic stimuli (7, 8). Second, ARC interacts with p53, disrupting p53 tetramerization (9). This disables p53 function as a transcription factor and exposes a nuclear export signal that relocates p53 to the cytoplasm. The ability of ARC to interrupt extrinsic and intrinsic apoptosis pathways through multiple mechanisms makes it a potent cell death inhibitor.
Defects in apoptosis are critical for carcinogenesis in that they allow cancer cells to survive adverse genetic and environmental cues during tumor growth and metastasis (10–13). Moreover, evasion of apoptosis is important for resistance to cancer therapies. One mechanism by which cancer cells escape apoptosis is by overexpressing or activating endogenous inhibitors of apoptosis (14). Whereas ARC is normally present in cardiac and skeletal myocytes and neurons (6, 15), its protein levels are markedly increased in a variety of primary human epithelial cancers including breast, colon, ovary, and cervix compared with corresponding benign tissues (16, 17). Moreover, overexpression of ARC renders breast cancer cells resistant to chemotherapy and γ-radiation (17) and protects melanoma cells from ER stress-induced apoptosis (18).
Despite the biological effects of ARC in cancer cells, the mechanisms responsible for its induction in cancer are unknown. In the heart, the abundance of ARC is regulated both at the level of transcription and protein stability. For example, p53 transcriptionally represses ARC expression in response to hypoxia (19), while oxidative stress triggers ARC degradation via the ubiquitin-proteasome pathway (20). Herein, we investigate the mechanisms responsible for the marked increases in ARC protein in epithelial cancers. Our experiments reveal that Ras is a key inducer of ARC in both breast and colon cancer cell lines and mammary gland tumors in vivo. Ras modulates ARC levels through both transcriptional mechanisms and changes in protein stability. Moreover, ARC is a mediator of certain cellular effects of Ras.
EXPERIMENTAL PROCEDURES
Plasmid and Viral Vectors
ARC-HA and KR3ARC-HA, previously generated in our laboratory (20), were subcloned into pBABE. pBABE H-Ras(V12) was from Dr. Michael Lisanti (Thomas Jefferson University) and pBABE N-Ras(K61) from Addgene. For N-Ras knockdown experiments, shRNAs in the pSHAG-MAGIC 2 backbone were obtained from Open Biosystems: shRNA N-Ras coding region (sh1) (cat. no. RHS1764-9394269), shRNA N-Ras 3′-UTR (sh2) (cat. no. RHS1764–9494042), scrambled shRNA (shScr) (cat. no. RHS1703). Recombinant, replication-deficient retroviruses were generated and used as described (40). Recombinant, replication-deficient adenoviruses expressing shRNA for mouse ARC knockdown were generated using the BLOCK-iT pol II miR RNAi system (Invitrogen, Carlsbad, CA). Multiple shRNA candidates identified in silico were screened for their effectiveness in suppressing expression of a rat ARC cDNA transfected into HEK293A cells or endogenous mouse ARC expression in C2C12 muscle cells. The most effective target sequence was GAACTAGAAGCTGAAGCTACT, corresponding to nucleotides 629–649 in the mouse (NM_030153) and 545–565 in the rat (NM_053516) mRNA sequences. The EmGFP-shARC coding cassette containing this sequence was transferred to pAd/CMV/V5-DEST, transfected into HEK293A cells for adenoviral production, which was purified after a single round of amplification using a commercial kit (Adenopure, Puresyn Inc, Malvern, PA). Human ARC promoter sequences −765 to −1 (with respect to transcription initiation), cloned by Dr. Roger Foo (University of Cambridge), were inserted into the pGL3-basic vector encoding firefly luciferase (Promega). phRL-TK encoding Renilla luciferase was obtained from Promega. Wild-type ERK1, wild-type ERK2, and activated MEK1 (ΔN3+E218+D222 (41)) were from Dr. Chi-Wing Chow (Albert Einstein College of Medicine) and activated Akt1 (D473, D808 (42)) from Dr. Jonathan Backer (Albert Einstein College of Medicine). Human ARC was subcloned into pGEX-6P-2 (GE Healthcare) for production of the GST fusion protein.
Cell Lines
All cell lines were from the American Type Culture Collection. Each line was cultured as specified, except for MCF-10A cells, which were cultured as described (43).
Antibodies and Immunoblotting
Antibodies include rabbit polyclonal antisera against ARC (Cayman), p44/42 MAPK (Cell Signaling), Akt (Cell Signaling), hemagglutinin (Santa Cruz Biotechnology), and mouse monoclonal antibodies against pan-Ras (BD Transduction Laboratory), H-Ras (Santa Cruz Biotechnology), N-Ras (Santa Cruz Biotechnology), K-Ras (Santa Cruz Biotechnology), ubiquitin (Santa Cruz Biotechnology), phospho-p44/42 MAPK (T202, Y204) (Cell Signaling), phospho-Akt (Ser-473) (Cell Signaling), α-tubulin (Sigma), β-actin (Sigma), and GAPDH (Abcam). Whole cell extracts and immunoblotting were performed as described (7). Relative protein levels were quantified by scanning densitometry using Total Lab software.
RNA Isolation, cDNA Synthesis, and Quantitative Real-time RT-PCR
RNA isolation, cDNA synthesis, and assessment of RNA levels were performed as previously described (20). Primers specific for ARC transcripts were: forward 5′-ACTGGCAGCACGTGGGTC-3′ and reverse 5′-TTTAGAGCCCTCAGCTTCCA-3′. Primers specific for N-Ras transcripts were: forward 5′-GAGCTTGAGGTTCTTGC-3′ and reverse 5′-AGTATGTCCAACAAACAGG-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers were: forward 5′-AAATCAAGTGGGGCGATGCTG-3′ and reverse 5′-GCAGAGATGATGACCCTTTTG-3′ primers. Quantitative real-time RT-PCR assays were performed in duplicate, and the number of independent experiments is noted in figure legends.
Luciferase Assay
HEK293T cells were transfected with: 1 μg of firefly luciferase reporter plasmid either lacking a promoter or driven by ARC promoter sequences (−765 to −1); 1 μg of empty pBABE, H-Ras(V12), N-Ras(K61), activated Akt1, or activated MEK1, or a combination of activated MEK1 (0.5 μg) and ERK1 or ERK2 (0.5 μg); and 5 ng of Renilla luciferase reporter plasmid. Cell lysates were harvested 48-h post-transfection and assayed for firefly and Renilla activity using the Dual-Luciferase Reporter Assay System (Promega). In the inhibitor study, cells were treated 24-h post-transfection with either PD98059 (MEK inhibitor) or LY294002 (PI3K inhibitor) (Biomol) at the doses indicated and assayed 24 h later. Luciferase assays were performed in duplicate, and the number of independent experiments noted in figure legends.
Pulse-chase Assay
35S pulse-chase was performed as previously described (20). For each time point, ARC was immunoprecipitated, resolved by SDS-PAGE, autoradiographed, and bands quantified by scanning densitometry using Total Lab software program. ARC protein half-life was determined from four independent pulse-chase experiments for each cell line with R2 values greater than 0.95.
Ubiquitination Assays
To assess ubiquitination in cells, cultures were treated with the proteasome inhibitor MG132 (Calbiochem), 10 μm, for 14 h, following which ARC was immunoprecipitated and immunoblotted as described (20). For the reconstituted cell-free ubiquitination assay, GST-ARC was produced in BL2.1-Star (DE3) (Invitrogen) and purified using glutathione-Sepharose 4B beads (GE Healthcare) as described (7). 5 μg of recombinant ARC was added to the ubiquitination reaction as described (44) with the recombinant E2 as specified. Polyubiquitinated GST-ARC was isolated using GST pull-down as described (7), resolved on 4–20% gradient SDS-PAGE, and immunoblotted for ubiquitin.
Epithelial Cell Isolation and Adenoviral Infection
Epithelial cells were isolated from mouse mammary glands as described (45) except for substitution of collagenase III for collagenase I, and directly lysed for immunoblot analysis. Primary epithelial cell cultures were generated from MMTV-H-Ras transgenic mouse mammary glands in the same manner except that collagenase digestion was carried out overnight without agitation at 37 °C in 5% CO2. Cells were then cultured in MCF-10A growth medium supplemented with 24 mg/ml bovine pituitary extract (Invitrogen). To knockdown murine ARC in primary cultures, three successive rounds of infection with adenoviruses encoding shRNA were carried out at MOI 50 to achieve 80% transduction as described (7).
Immunohistochemistry
Mammary glands were fixed in 10% neutral buffered formalin, paraffinized, sectioned (5 μm), deparaffinized, immunostained as described (46) with α-ARC (Cayman, 1:500) or α-Ras (Cell Signaling, 1:500), and counterstained with hematoxylin. Images were obtained on Nikon Eclipse TE2000-S microscope using Spot R/T CCD camera.
Cell Viability Assay
Cells were plated at a density of 500 cells per cm2 and subjected to increasing concentrations of doxorubicin (Henry Schein) as noted for 20 h. All samples were assayed in quadruplicate. Viability was assessed using CellTiter-Blue (Promega) according to the manufacturer's recommendations.
Cell Cycle Analysis
1 × 106 cells were stained with 25 μg/ml ethidium bromide, and nuclei isolated as described (47). All samples were assayed in triplicate. The fluorescence signal was detected using the Becton Dickinson FACScan flow cytometer, and cell cycle profiles were analyzed with the ModFit LT program (Verity).
Statistical Analyses
Differences between and among groups were compared using Prism 4.0 (GraphPad Software) with Student's paired two-tailed t test or one-way ANOVA as indicated. When significant differences were found by ANOVA, the Tukey or Dunnett's multiple comparison test was used as the post hoc analysis. p < 0.05 was considered significant.
RESULTS
Correlation between Levels of ARC and Ras
ARC protein levels are markedly elevated in primary human epithelial cancers of the breast, colon, cervix, and ovary compared with controls (16, 17). We observed that levels of ARC track with those of N-Ras in a variety of normal and cancer cell lines. This is illustrated by MCF-7 and HCC1419 breast cancer cells that contain high levels of N-Ras and ARC (Fig. 1A). In contrast, MCF-10A and MCF-12A benign breast epithelial cell lines contain low levels of N-Ras and ARC. Even among different breast cancer cell lines that contain varying levels of N-Ras, the correlation of N-Ras and ARC levels is maintained. For example, in contrast to the high levels of N-Ras and ARC in MCF-7 and HCC1419 breast cancer cells, MDA-MB-231 (Fig. 1A) and Hs578T (not shown) breast cancer cell lines contain lower levels of both N-Ras and ARC. A correlation between N-Ras and ARC levels was also observed in normal and colon cancer cell lines (Fig. 1B). HCT 116 colon cancer cells exhibit high levels of N-Ras and ARC, while CCD-112 CoN normal colon cells, as well as HT-29 and Caco-2 colon cancer cells, contain lower levels of N-Ras and ARC. Direct comparison of MCF-10A benign breast epithelial cells with MCF-7 breast cancer cells demonstrates that levels of both N-Ras and ARC proteins are increased in MCF-7 cells (Fig. 1C). Moreover, MCF-7 cells contain 8-fold more N-Ras transcripts (expected from the known N-Ras amplification in these cells (21)) and 3-fold more ARC transcripts (Fig. 1D). These studies demonstrate that ARC levels correlate with those of N-Ras in both normal and cancer cells derived from the breast and colon.
FIGURE 1.
Levels of ARC and Ras in normal and cancer cell lines. A, immunoblot for ARC and N-Ras in benign breast epithelial cell lines MCF-10A and MCF-12A and breast cancer cell lines MDA-MB-231, MCF-7, and HCC1419. ARC antisera against the C terminus were used to avoid detection of other caspase recruitment domain-containing proteins, including Nop30, a putative protein encoded by an alternatively spliced transcript from the Nol3 locus encoding ARC (48). Loading was normalized to α-tubulin. B, immunoblot for ARC and N-Ras in normal colon cell line CCD-112 CoN and colon cancer cell lines HCT116, HT-29, and Caco-2. Loading was normalized to GAPDH. C, direct comparison of ARC protein levels in benign (MCF-10A) and cancer (MCF-7) breast epithelial cells. Immunoblot for ARC and N-Ras (left). Densitometric quantification of ARC protein levels normalized to those of GAPDH (right). D, levels of N-Ras and ARC transcripts normalized to those of GAPDH in the same cells as in C as determined by quantitative real-time RT-PCR using transcript-specific primers. Quantitative data displayed as mean ± S.E. from three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.005 compared with MCF-10A (two-tailed Student's t test).
Ras Induces ARC in a Mouse Model of Mammary Cancer
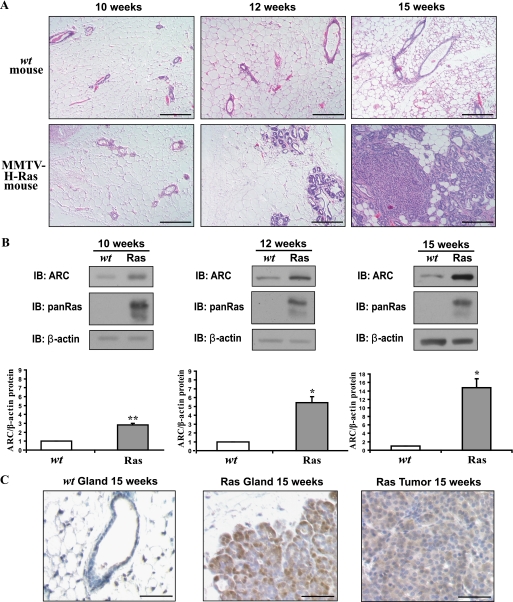
To determine if Ras signaling is sufficient to induce ARC in vivo, we assessed ARC levels in transgenic mice that express activated H-Ras (G12R, A59T) in the breast epithelium (22). We chose to study this mouse model because activated H-Ras expression is known to induce mammary gland carcinogenesis. Hyperplasia is observed at 12 weeks, and overt tumors at 15 weeks (Fig. 2A). Expression of activated H-Ras significantly induces ARC protein in the epithelial cells of the mammary gland (Fig. 2B). ARC protein levels are increased when the gland is histologically normal at 10 weeks and are noted to increase further at 12 and 15 weeks, while H-Ras protein levels remain constant with age (not shown). ARC is expressed in both hyperplastic regions of the mammary epithelium and in overt tumors (Fig. 2C). These results demonstrate that activated H-Ras is sufficient to induce ARC in the normal mammary epithelium in vivo, and the abundance of ARC increases progressively during Ras-driven mammary carcinogenesis.
FIGURE 2.
ARC is induced by Ras in the mammary glands of MMTV-H-Ras (G12R, A59T) transgenic mice in vivo. A, time course of mammary tumorigenesis of nulliparous wild-type and MMTV-H-Ras transgenic mice at the indicated ages. Sections were stained with hematoxylin and eosin. Size bar, 100 μm. B, Ras induces ARC in mammary epithelial cells and tumors in vivo. Immunoblot of ARC and total Ras in the epithelial-enriched fraction of mammary glands from wild-type and transgenic mice of the ages indicated (top). Quantification of ARC levels normalized to those of β-actin (mean ± S.E.) (middle), n = 5 mice of each genotype (littermates) for each age group. *, p < 0.01 and **, p < 0.005 (Ras transgenics compared with wild-type; two-tailed Student's t test). C, ARC immunostaining of mammary glands from 15-week-old wild-type and Ras transgenic mice, and mammary tumors from 15-week-old Ras transgenics (bottom). ARC, brown. hematoxylin counterstain, blue. Size bar, 50 μm.
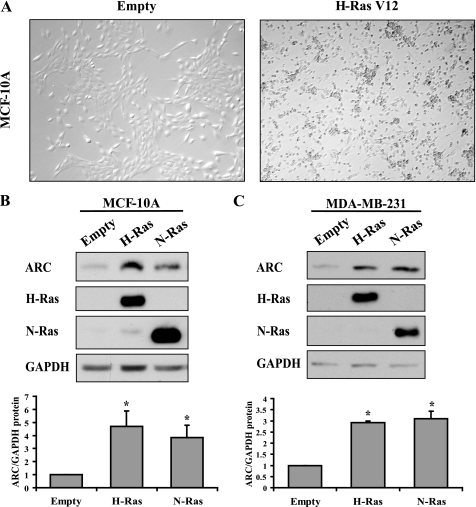
Activated Ras Mutants Are Sufficient to Induce ARC in Normal and Cancer Cells
To determine whether Ras-dependent pathways regulate ARC abundance, we first assessed the sufficiency of Ras to induce ARC in MCF-10A benign breast epithelial cells, which have low levels of N-Ras, H-Ras, and ARC (Fig. 1A and not shown). Expression of either activated N-Ras(K61) or H-Ras(V12) is sufficient to transform MCF-10A cells as evidenced by focus formation (Fig. 3A and not shown). Moreover, both activated N-Ras and H-Ras increase ARC protein levels 3- and 4-fold, respectively, compared with empty vector (Fig. 3B). To assess whether ARC can also be induced in the context of an already transformed cell, we used MDA-MB-231 cells that contain low levels of N-Ras and H-Ras (Fig. 1A and not shown). Expression of either activated N-Ras or H-Ras increases ARC abundance 3-fold (Fig. 3C). These results indicate that activated Ras isoforms are sufficient to induce ARC in both benign and cancer cells.
FIGURE 3.
Induction of ARC by activated Ras. A, phase contrast micrographs demonstrating transformation of benign MCF-10A breast epithelial cells by H-Ras(V12). Cells were stably transduced with retroviruses encoding empty vector (left) and H-Ras(V12) (right). B and C, activated Ras isoforms induce ARC in MCF-10A benign breast epithelial cells (B) and MDA-MB-231 breast cancer epithelial cells (C). Cells were stably transduced with retroviruses encoding empty vector, H-Ras(V12), or N-Ras(K61) (top). Densitometric quantification of ARC protein levels normalized to those of GAPDH (bottom). Quantitative data presented as means + S.E. from three independent experiments. *, p < 0.01 (Ras compared with empty vector; one-way ANOVA followed by Dunnett test).
Knockdown of Endogenous Ras in Cancer Cells Markedly Decreases ARC Levels
To investigate whether the high levels of ARC in cancer cells are dependent on Ras signaling, we knocked down endogenous N-Ras in MCF-7 breast and HCT116 colon cancer cells using two different hairpins corresponding to the coding region (sh1) or the 3′UTR (sh2). In MCF-7 cells, endogenous N-Ras levels were reduced by 50–80% in various clones by each of the N-Ras hairpins compared with scrambled shRNA (shScr) (Fig. 4, A and B). The extent of ARC protein reduction paralleled the magnitude of N-Ras knockdown. Similar results were obtained in HCT116 cells (Fig. 4C). To further test whether endogenous levels of ARC are dependent on Ras, we tested the ability of exogenous Ras to augment ARC levels in the context of the Ras-knockdown. H-Ras, rather than N-Ras, was used for the replacement to evade the N-Ras hairpins. Expression of H-Ras(V12) increased ARC levels 4-fold in the setting of sh1 N-Ras knockdown (Fig. 4D). Similar results were obtained using sh2 N-Ras knockdown cells (not shown). Thus, the reduction of ARC levels that results from N-Ras knockdown is partially reversed when exogenous Ras is expressed. Taken together with the overexpression studies, Ras is necessary and sufficient to induce high levels of ARC in these cell lines.
FIGURE 4.
Knockdown of endogenous N-Ras reduces ARC protein levels. A and B, MCF-7 cells were stably transduced with retroviruses encoding control scrambled hairpin (shScr), N-Ras hairpin (coding region, sh1; A) or N-Ras hairpin (3′-UTR, sh2; B). Individual stable clones are denoted by cl-#. Immunoblots for N-Ras and ARC (top) and quantification of levels of N-Ras and ARC normalized to those of GAPDH (bottom). Data in A are expressed as mean ± S.E. from four independent experiments for each of the two independent clones. *, p < 0.01 and **, p < 0.005 (N-Ras knockdown compared with scrambled hairpin; one-way ANOVA followed by Dunnett test). In B, each of the four clones was assessed in one experiment. C, N-Ras was knocked down in HCT116 colon cancer cells as described for MCF-7 cells in A and B. Lysates of pools of transductants were immunoblotted for N-Ras and ARC (top), and levels of N-Ras and ARC normalized to those of α-tubulin were quantified (bottom). Data expressed as mean ± S.E. from three independent experiments. *, p < 0.01 (N-Ras knockdown compared with scrambled hairpin; one-way ANOVA followed by Dunnett test). D, re-expression of Ras in MCF-7 Ras-knockdown cells increases ARC levels. MCF-7 shScr control cells or N-Ras (sh1) knockdown cells were stably transduced with empty vector (Emp) or H-Ras(V12). Lysates of pools of transductants were immunoblotted for ARC, H-Ras, and N-Ras (top), and ARC levels normalized to those of α-tubulin were quantified (bottom). Data represent mean ± S.E. from two independent experiments. In addition, MCF-7 N-Ras sh2 knockdown cells were studied in two independent analogous experiments with similar results (not shown).
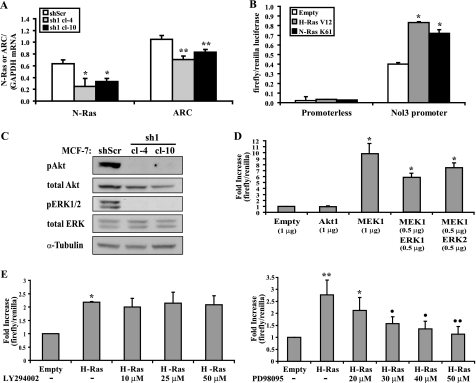
Ras Activates the Promoter of the Nol3 Locus Encoding ARC in a MEK/ERK-dependent Manner
To investigate the mechanisms by which Ras regulates ARC abundance, we first examined the effect of Ras knockdown on ARC mRNA levels assessed by quantitative real-time RT-PCR. Two MCF-7 N-Ras (sh1) stable knockdown clones (cl-4 and cl-10), each with a 5-fold decrease in ARC protein levels (Fig. 4A), demonstrated a 30% reduction in ARC mRNA abundance compared with scrambled shRNA control (p < 0.01, Fig. 5A). Conversely, overexpression of activated N-Ras(K61) or H-Ras(V12) in MCF-10A cells, with a 3- or 4-fold augmentation in ARC protein levels, respectively (Fig. 3B), resulted in a 30% increase in ARC transcript levels (not shown). To assess whether Ras signaling activates the promoter of the Nol3 locus encoding ARC, HEK293T cells, containing low endogenous levels of Ras isoforms and ARC (not shown), were co-transfected with N-Ras(K61) or H-Ras(V12) and a luciferase reporter driven by 765 base pairs of the Nol3 5′ flanking region. Each Ras isoform activated the Nol3 reporter 2-fold (p < 0.01; Fig. 5B), an effect similar in magnitude to their activation of the p21CIP1/WAF1 promoter, a known Ras target (23) (not shown). In contrast, the luciferase activity of a promoterless construct was not activated by either Ras mutant. These studies demonstrate that transcriptional activation is involved in Ras-mediated ARC induction.
FIGURE 5.
Ras activates the promoter of the Nol3 locus encoding ARC. A, Ras knockdown decreases ARC mRNA levels. Quantitative real-time RT-PCR for mRNA levels of N-Ras and ARC (transcript-specific primers) normalized to that of GAPDH in MCF-7 cells stably transduced with retroviruses encoding control scrambled hairpin (shScr) or N-Ras hairpin (sh1, clones 4 and 10). Data expressed as mean ± S.E. from four independent experiments (the same cell preparations used to determine ARC protein levels in Fig. 4A). *, p < 0.05 and **, p < 0.01 (N-Ras knockdown compared with scrambled hairpin; one-way ANOVA followed by Dunnett test). B, activation of the Nol3 promoter by Ras. HEK293T cells were cotransfected with firefly luciferase lacking a promoter or driven by human Nol3 promoter sequences (−765 to −1), H-Ras(V12), N-Ras(K61), or empty vector, and constitutively driven Renilla luciferase for normalization. Promoter activity was assessed by firefly/Renilla luciferase. Data expressed as mean ± S.E. from four independent experiments. *, p < 0.01 (Ras versus empty vector; one-way ANOVA followed by Dunnett test). C, Ras knockdown abrogates ERK and Akt activation. Immunoblots for phosphorylated (Thr-202, Tyr-204) and total ERK1/2 and phosphorylated (Ser-473) and total Akt in lysates of N-Ras (sh1) knockdown MCF-7 cells from A. D, activation of the Nol3 promoter by MEK or MEK/ERK, but not Akt. Phosphomimetic mutants were used for MEK1 and Akt1. Reporter assays as described in B. Data expressed as means + S.E. from four independent experiments. *, p < 0.001 compared with empty vector (one-way ANOVA followed by Tukey test). E, reversal of Ras activation of the Nol3 promoter by inhibition of MEK but not PI3K. Constructs were transfected as described in B. Inhibitors of PI3K (LY294002, left) and MEK (PD98095, right) were added 24 h later, and assayed 24 h after that. Data in each graph expressed as means + S.E. from four independent experiments. *, p < 0.01 and **, p < 0.001 compared with empty vector and •, p < 0.01 and ••, p < 0.001 as compared with H-Ras with no PD inhibitor (one-way ANOVA followed by Tukey test).
Ras signaling is mediated, in part, by PI3K/Akt and MEK/ERK. Consistent with this, knockdown of N-Ras in MCF-7 cells ablated phosphorylation of both Akt and ERK1/2 at critical residues required for their activity (Fig. 5C), correlating with decreases in ARC transcript levels (Fig. 5A). Conversely, overexpression of N-Ras(K61) or H-Ras(V12) in MCF-10A cells enhanced phosphorylation of Akt and ERK1/2, and increased ARC transcript levels (not shown). To investigate if these effectors mediate Ras activation of the Nol3 promoter, we used constitutively active (phosphomimetic) mutants of Akt1 and MEK1, the latter alone or in combination with ERK1 or ERK2, and tested their effects on the Nol3 reporter in HEK293T cells. Whereas activated Akt1 did not induce the Nol3 promoter, MEK1 alone, or in combination with ERK1 or ERK2, resulted in robust activation (Fig. 5D). Moreover, the activation of the Nol3 promoter by H-Ras(V12) was not affected by the PI3K inhibitor LY294002 (Fig. 5E, left), but was substantially reduced in a dose-dependent manner by the MEK inhibitor PD98095 (Fig. 5E, right). Taken together, these results indicate that Ras activates the Nol3 promoter in a MEK/ERK-dependent manner to stimulate production of ARC mRNA.
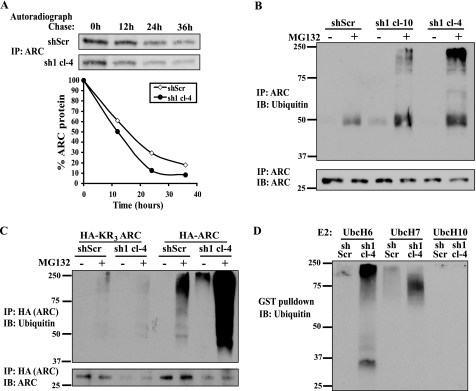
Ras Inhibits ARC Degradation via the Ubiquitin-Proteasome Pathway
The N-Ras knockdown experiments reveal that the magnitude of the reduction in ARC transcript levels does not fully account for the decrease in ARC protein levels (compare Fig. 5A with Fig. 4A). In some contexts, ARC levels are known to be regulated via changes in protein degradation (20). Accordingly, we investigated the effect of Ras knockdown on ARC protein stability using pulse-chase assays. These were performed in MCF-7 cells with stable expression of N-Ras (sh1) or scrambled (shScr) hairpins (Fig. 6A). The half-life of ARC protein in cells expressing scrambled shRNA was ∼16 h, similar to previously published measurements (20). In contrast, Ras knockdown reduced ARC protein half-life to ∼10 h in two independent clones (cl-4, Fig. 6A and cl-10, not shown). Moreover, Ras knockdown stimulated marked ARC polyubiquitination (Fig. 6B), which occurred in a canonical manner as it was ablated by simultaneous mutation of the three lysine ubiquitin acceptor residues in ARC (Fig. 6C). These data suggest that Ras modulates ARC degradation via the ubiquitin-proteasome system. Similar results were obtained with an in vitro reconstituted ubiquitination assay (see Fig. 6 legend). Using this system, lysates from N-Ras knockdown cells, but not control cells, stimulated robust polyubiquitination of ARC (Fig. 6D). Taken together, these results show that Ras suppresses ARC polyubiquitination and degradation.
FIGURE 6.
Ras inhibits ARC degradation via the ubiquitin-proteasome pathway. A, endogenous Ras stabilizes ARC protein. Representative pulse-chase of ARC protein in MCF-7 cells stably transduced with N-Ras hairpins (sh1) or control scrambled hairpin (shScr). Cells were pulsed for 10 h with [35S]cysteine and chased for times indicated, following which ARC immunoprecipitates were resolved by SDS-PAGE. Representative autoradiograph (top) and densitometry (bottom) from four independent experiments using cl-4 and cl-10 (not shown). B, endogenous Ras suppresses ARC ubiquitination. Control and stable N-Ras knockdown (sh1) MCF-7 cells were treated with vehicle or proteasome inhibitor MG132 (10 μm) for 14 h, following which ARC immunoprecipitates were resolved by SDS-PAGE and immunoblotted for ubiquitin (top) or ARC (bottom). C, ARC ubiquitination is primarily canonical. Control and stable N-Ras knockdown (sh1) MCF-7 cells were stably transfected with HA-tagged KR3ARC (in which all 3 ARC lysines are mutated to arginines) or HA-tagged wild-type ARC, and treated with vehicle or MG132 as described in B. HA immunoprecipitates were resolved by SDS-PAGE and blotted for ubiquitin (top) and ARC (bottom). D, in vitro system to assess Ras regulation of ARC polyubiquitination. Bacterially produced and purified GST-ARC, ubiquitin, E1, E2 (UbcH6, UbcH7, or UbcH10 as indicated), and ATP were incubated with lysates from scrambled control or N-Ras knockdown cells (described in panel a) for 2 h. GST-ARC was pulled down with glutathione beads, resolved by SDS-PAGE, and immunoblotted for ubiquitin.
ARC Is a Mediator of Ras-induced Cellular Survival and Cell Cycle Progression
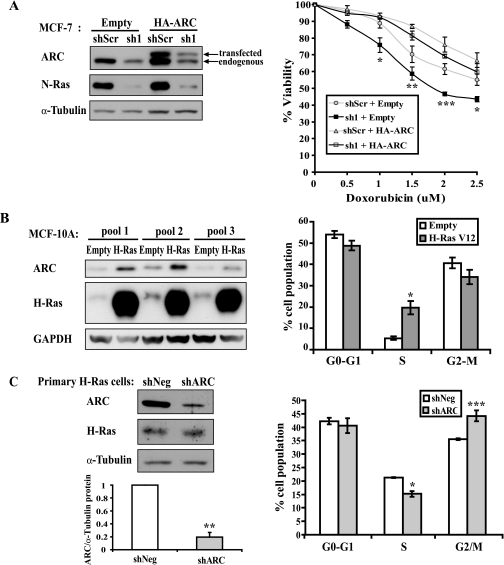
Functionally, Ras regulates an array of cancer-promoting processes including suppression of apoptosis and stimulation of cell cycle progression (24). To determine whether ARC plays a role in mediating Ras-induced cell survival, we restored ARC in MCF-7 cells in which N-Ras had been knocked down. Empty vector or HA-tagged ARC was stably transduced into MCF-7 cells that stably express scrambled (shScr) or N-Ras (sh1) hairpins. These cells were subsequently tested for sensitivity to killing by different doses of doxorubicin. Consistent with the known ability of Ras to suppress cell death, N-Ras knockdown clones were most sensitive to increasing concentrations of doxorubicin (Fig. 7A, right, solid squares). As expected, N-Ras knockdown was accompanied by decreases in endogenous ARC levels (Fig. 7A, left). Replacement of ARC significantly reversed the enhanced sensitivity of N-Ras knockdown cells to doxorubicin (Fig. 7A, right, compare open squares to solid squares). Of note, the level of N-Ras knockdown was unaffected by ARC overexpression (Fig. 7A, left). In addition, doxorubicin treatment did not alter steady state levels of ARC or N-Ras (not shown). These data demonstrate that the resistance to doxorubicin-induced killing conferred by Ras is mediated at least in part through ARC.
FIGURE 7.
ARC is a mediator of Ras-induced survival and cell cycle progression. A, ARC reverses the increased sensitivity to doxorubicin killing caused by Ras knockdown. Immunoblots for N-Ras and ARC (endogenous + transfected) in pools of MCF-7 cells stably transduced with retrovirus encoding control scrambled hairpin (shScr) or N-Ras hairpin (sh1), followed by stable transduction with empty vector or HA-tagged ARC (left). Pools of each of these genetically modified MCF-7 cells were cultured for 24 h in the indicated concentrations of doxorubicin, and cellular viability was assessed using CellTiter-Blue (right). Graph represents mean ± S.E. from five independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (sh1+ARC compared with sh1 + empty; one-way ANOVA followed by Tukey test). B, overexpression of Ras increases S-phase population. Pools of MCF-10A cells stably transduced with retrovirus encoding empty vector or H-Ras(V12) (left) were stained with ethidium bromide, and DNA content analyzed by FACS (right). Data represent mean ± S.E. from three independently generated pools. *, p < 0.05 (Ras compared with empty vector; two-tailed Students t test). C, ARC knockdown decreases S-phase population and increases G2-M population in cultured mammary gland epithelial cells from MMTV-H-Ras transgenic mice. Immunoblots of ARC and H-Ras in primary mammary epithelial cells cultured from 12-week-old MMTV-H-Ras transgenic mice and transduced with adenovirus encoding control hairpin (shNeg) or ARC hairpin (shARC) (left). Cell cycle analysis of these cells (right). Data represent mean ± S.E. from five independent experiments. *, p < 0.01, **, p < 0.005, and ***, p < 0.001 (ARC knockdown compared with scrambled hairpin; two-tailed Student's t test).
Ras is known to stimulate the G1/S transition and G2/M exit (25, 26). In fact, stable expression of H-Ras(V12) in MCF-10A cells significantly increases the proportion of cells in S-phase (Fig. 7B). We wished to knockdown ARC in these cells to determine if it plays a role in Ras-mediated cell cycle progression, but decreased adherence of these cells to tissue culture plates precluded this experiment. Alternatively, ARC knockdown was carried out in primary epithelial cell cultures isolated from mammary glands of five different MMTV-H-Ras transgenics. Adenoviral transduction of an ARC shRNA reduced ARC protein levels by 80% compared with the negative control, while not affecting levels of H-Ras (Fig. 7C, left). Cell cycle analysis of ARC knockdown primary cells showed a decrease in the proportion of cells in the S-phase population and an increase in the proportion of cells in G2-M (p < 0.01 and p < 0.001, respectively, Fig. 7C, right). These data indicate that ARC is involved in cell cycle progression in the mammary epithelium of MMTV-H-Ras mice.
DISCUSSION
Our results show that Ras induces the abundance of ARC through activation of Nol3 transcription and inhibition of ARC protein degradation. These studies employed MCF-7 cells, a commonly utilized model of breast cancer, which contain high levels of N-Ras due to amplification (21) and exhibit enhanced Ras signaling (27). Although Ras mutations are found in only ∼5% of human breast cancers (28), Ras signaling is augmented in 20–50% of cases due to upstream events such as overexpression or activating mutations in the EGF receptor or ErbB2/neu/HER-2 (29–31). Colon cancers, on the other hand, often contain gain of function mutations in K-Ras (32) or in its downstream effector genes such as PIK3CA, which encodes the p110α catalytic subunit of PI3K (33). Thus, regardless of mechanism, enhanced Ras signaling is part of the molecular signature of human breast and colon cancers as well as the various cell culture and mouse models of these malignancies.
We exploited a correlation between cellular levels of Ras and ARC to reveal that Ras is a mediator of ARC abundance. Specifically, overexpression of either activated N-Ras or H-Ras is sufficient to induce ARC in both normal and cancer breast epithelial cells, suggesting the importance of common downstream pathways. Conversely, knockdown studies demonstrate that Ras is necessary for the high levels of ARC in breast and colon cancer cells. These observations extend to intact animals as transgenic expression of activated H-Ras induces ARC in the normal mammary epithelium, and to an even greater extent, in resulting hyperplastic regions and tumors. Taken together, these data indicate the necessity and sufficiency of Ras signaling to induce ARC in cancer cells.
Ras-mediated induction of ARC is due, in part, to activation of Nol3 transcription. Consistent with this transcriptional regulation, knockdown of endogenous N-Ras in MCF-7 cells decreases ARC mRNA. In addition, overexpression of Ras activates the Nol3 promoter, and signaling through MEK/ERK (but not PI3K/Akt) is necessary and sufficient for this effect. While we have not delineated the specific transcription factors involved, ETS family members, AP1, SRF, and RREB1 (34–36) have been implicated in the activation of other genes in response to Ras/MEK/ERK signaling. In fact, the Nol3 promoter sequences we employed contain multiple binding sites for these transcription factors. Of note, multiple ETS proteins contribute to breast carcinogenesis (37).
In addition to regulation at the level of transcription, pulse-chase studies show that Ras increases ARC abundance by stabilizing ARC protein. This results from suppression of ARC polyubiquitination and degradation in the proteasome. Our data suggest two non-mutually exclusive mechanisms by which Ras signaling may regulate ARC stability: (a) Ras-mediated decreases in the abundance and/or activity of E3-ubiquitin ligase(s) that act upon ARC; and (b) Ras-induced post-translational modifications in ARC that decrease its susceptibility to ubiquitination. The RING finger E3-ligase MDM2 (Mouse Double Minute 2) has been reported to be involved in ARC degradation (38). Knockdown of Ras in our studies, however, did not affect protein levels of MDM2 or MDM2 target proteins p21 or p53 (not shown). These results demonstrate that a combination of Ras-mediated stimulation of Nol3 transcription and inhibition of ARC protein degradation contribute to the high steady state levels of ARC protein in breast cancer cells.
The induction of ARC by Ras raises questions as to the functional relationship between these proteins. ARC is best known as an apoptosis inhibitor, and previous studies have demonstrated that it regulates cell death induced by various stressors (17, 18). Ras is also known to confer resistance to apoptotic stimuli (39). Accordingly, we asked whether ARC plays a role in Ras-induced cell survival. To assess this, we tested whether exogenous ARC rescues the increase in doxorubicin-induced death resulting from Ras knockdown. In fact, this increased cytotoxicity is completely reversed by restoration of physiological levels of ARC. Thus, ARC is involved in Ras-induced cell survival. Ras has also been shown to accelerate the cell cycle by promoting the G1/S transition and G2/M exit (25, 26). Knockdown of ARC in primary mammary epithelial cells from MMTV-H-Ras transgenic mice decreases the proportion in S-phase and increases the proportion in G2/M. These data implicate ARC in cell cycle progression in the context of Ras overexpression.
In summary, ARC, an inhibitor of both extrinsic and intrinsic apoptosis pathways that is normally found in terminally differentiated cells, becomes highly induced in a spectrum of primary human epithelial cancers. Our results reveal a new connection between Ras signaling and ARC induction, and demonstrate that Ras regulates ARC levels at both the levels of transcription and protein stability. ARC, in turn, mediates some important oncogenic effects of Ras.
Acknowledgments
We thank Drs. Jonathan Backer, Chi-Wing Chow, and Roger Foo for plasmids. We thank Dr. Cristina Montagna for help with primary mammary epithelial cultures and Min Zheng for assistance with animal maintenance.
This work was supported, in whole or in part, by National Institutes of Health R01 Grants HL60665, HL61550, and HL80607 (to R. N. K.) and institutional National Institutes of Health P30 Cancer Center Grant CA013330. This work was also supported by the Wilf Family.
- DISC
- death-inducing signaling complex
- ARC
- apoptosis repressor with caspase recruitment domain
- UTR
- untranslated region
- GST
- glutathione S-transferase
- ANOVA
- analysis of variance
- ERK
- extracellular signal-regulated kinase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HA
- hemagglutinin
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase.
REFERENCES
- 1.Taylor R. C., Cullen S. P., Martin S. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 231–241 [DOI] [PubMed] [Google Scholar]
- 2.Danial N. N., Korsmeyer S. J. (2004) Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 3.Chang D. W., Xing Z., Pan Y., Algeciras-Schimnich A., Barnhart B. C., Yaish-Ohad S., Peter M. E., Yang X. (2002) EMBO J. 21, 3704–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kluck R. M., Bossy-Wetzel E., Green D. R., Newmeyer D. D. (1997) Science 275, 1132–1136 [DOI] [PubMed] [Google Scholar]
- 5.Salvesen G. S., Duckett C. S. (2002) Nat. Rev. Mol. Cell Biol. 3, 401–410 [DOI] [PubMed] [Google Scholar]
- 6.Koseki T., Inohara N., Chen S., Nuñez G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5156–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nam Y. J., Mani K., Ashton A. W., Peng C. F., Krishnamurthy B., Hayakawa Y., Lee P., Korsmeyer S. J., Kitsis R. N. (2004) Mol. Cell 15, 901–912 [DOI] [PubMed] [Google Scholar]
- 8.Gustafsson A. B., Tsai J. G., Logue S. E., Crow M. T., Gottlieb R. A. (2004) J. Biol. Chem. 279, 21233–21238 [DOI] [PubMed] [Google Scholar]
- 9.Foo R. S., Nam Y. J., Ostreicher M. J., Metzl M. D., Whelan R. S., Peng C. F., Ashton A. W., Fu W., Mani K., Chin S. F., Provenzano E., Ellis I., Figg N., Pinder S., Bennett M. R., Caldas C., Kitsis R. N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20826–20831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frisch S. M., Francis H. (1994) J. Cell Biol. 124, 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrington E. A., Fanidi A., Evan G. I. (1994) Curr. Opin. Genet. Dev. 4, 120–129 [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 13.Mehlen P., Puisieux A. (2006) Nat. Rev. Cancer 6, 449–458 [DOI] [PubMed] [Google Scholar]
- 14.Letai A. G. (2008) Nat. Rev. Cancer 8, 121–132 [DOI] [PubMed] [Google Scholar]
- 15.Geertman R., McMahon A., Sabban E. L. (1996) Biochim. Biophys. Acta 1306, 147–152 [DOI] [PubMed] [Google Scholar]
- 16.Mercier I., Vuolo M., Jasmin J. F., Medina C. M., Williams M., Mariadason J. M., Qian H., Xue X., Pestell R. G., Lisanti M. P., Kitsis R. N. (2008) Cell Cycle 7, 1640–1647 [DOI] [PubMed] [Google Scholar]
- 17.Mercier I., Vuolo M., Madan R., Xue X., Levalley A. J., Ashton A. W., Jasmin J. F., Czaja M. T., Lin E. Y., Armstrong R. C., Pollard J. W., Kitsis R. N. (2005) Cell Death Differ 12, 682–686 [DOI] [PubMed] [Google Scholar]
- 18.Chen L. H., Jiang C. C., Watts R., Thorne R. F., Kiejda K. A., Zhang X. D., Hersey P. (2008) Cancer Res. 68, 834–842 [DOI] [PubMed] [Google Scholar]
- 19.Li Y. Z., Lu D. Y., Tan W. Q., Wang J. X., Li P. F. (2008) Mol. Cell. Biol. 28, 564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nam Y. J., Mani K., Wu L., Peng C. F., Calvert J. W., Foo R. S., Krishnamurthy B., Miao W., Ashton A. W., Lefer D. J., Kitsis R. N. (2007) J. Biol. Chem. 282, 5522–5528 [DOI] [PubMed] [Google Scholar]
- 21.Graham K. A., Richardson C. L., Minden M. D., Trent J. M., Buick R. N. (1985) Cancer Res. 45, 2201–2205 [PubMed] [Google Scholar]
- 22.Sinn E., Muller W., Pattengale P., Tepler I., Wallace R., Leder P. (1987) Cell 49, 465–475 [DOI] [PubMed] [Google Scholar]
- 23.Gartel A. L., Najmabadi F., Goufman E., Tyner A. L. (2000) Oncogene 19, 961–964 [DOI] [PubMed] [Google Scholar]
- 24.McCubrey J. A., Steelman L. S., Chappell W. H., Abrams S. L., Wong E. W., Chang F., Lehmann B., Terrian D. M., Milella M., Tafuri A., Stivala F., Libra M., Basecke J., Evangelisti C., Martelli A. M., Franklin R. A. (2007) Biochim. Biophys. Acta 1773, 1263–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman M. L., Marshall C. J., Olson M. F. (2004) Nat. Rev. Mol. Cell Biol. 5, 355–366 [DOI] [PubMed] [Google Scholar]
- 26.Knauf J. A., Ouyang B., Knudsen E. S., Fukasawa K., Babcock G., Fagin J. A. (2006) J. Biol. Chem. 281, 3800–3809 [DOI] [PubMed] [Google Scholar]
- 27.Eckert L. B., Repasky G. A., Ulkü A. S., McFall A., Zhou H., Sartor C. I., Der C. J. (2004) Cancer Res. 64, 4585–4592 [DOI] [PubMed] [Google Scholar]
- 28.Clark G. J., Der C. J. (1995) Breast Cancer Res. Treat 35, 133–144 [DOI] [PubMed] [Google Scholar]
- 29.Lacroix H., Iglehart J. D., Skinner M. A., Kraus M. H. (1989) Oncogene 4, 145–151 [PubMed] [Google Scholar]
- 30.Iglehart J. D., Kraus M. H., Langton B. C., Huper G., Kerns B. J., Marks J. R. (1990) Cancer Res. 50, 6701–6707 [PubMed] [Google Scholar]
- 31.Moscatello D. K., Holgado-Madruga M., Godwin A. K., Ramirez G., Gunn G., Zoltick P. W., Biegel J. A., Hayes R. L., Wong A. J. (1995) Cancer Res. 55, 5536–5539 [PubMed] [Google Scholar]
- 32.Bos J. L., Fearon E. R., Hamilton S. R., Verlaan-de Vries M., van Boom J. H., van der Eb A. J., Vogelstein B. (1987) Nature 327, 293–297 [DOI] [PubMed] [Google Scholar]
- 33.Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S. M., Riggins G. J., Willson J. K., Markowitz S., Kinzler K. W., Vogelstein B., Velculescu V. E. (2004) Science 304, 554. [DOI] [PubMed] [Google Scholar]
- 34.Oikawa T. (2004) Cancer Sci. 95, 626–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wasylyk B., Hagman J., Gutierrez-Hartmann A. (1998) Trends Biochem. Sci. 23, 213–216 [DOI] [PubMed] [Google Scholar]
- 36.Zhang L., Zhao J., Edenberg H. J. (1999) Nucleic Acids Res. 27, 2947–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner D. P., Findlay V. J., Moussa O., Watson D. K. (2007) J. Cell. Biochem. 102, 549–559 [DOI] [PubMed] [Google Scholar]
- 38.Foo R. S., Chan L. K., Kitsis R. N., Bennett M. R. (2007) J. Biol. Chem. 282, 5529–5535 [DOI] [PubMed] [Google Scholar]
- 39.Cox A. D., Der C. J. (2003) Oncogene 22, 8999–9006 [DOI] [PubMed] [Google Scholar]
- 40.Pear W., Scott M., Nolan G. P. (1997) Methods in Molecular Medicine: Gene Therapy Protocols, Human Press, Totowa, NJ [Google Scholar]
- 41.Mansour S. J., Matten W. T., Hermann A. S., Candia J. M., Rong S., Fukasawa K., Vande Woude G. F., Ahn N. G. (1994) Science 265, 966–970 [DOI] [PubMed] [Google Scholar]
- 42.Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. (1996) EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 43.Debnath J., Muthuswamy S. K., Brugge J. S. (2003) Methods 30, 256–268 [DOI] [PubMed] [Google Scholar]
- 44.Ryo A., Suizu F., Yoshida Y., Perrem K., Liou Y. C., Wulf G., Rottapel R., Yamaoka S., Lu K. P. (2003) Mol. Cell 12, 1413–1426 [DOI] [PubMed] [Google Scholar]
- 45.Sotgia F., Williams T. M., Schubert W., Medina F., Minetti C., Pestell R. G., Lisanti M. P. (2006) Am. J. Pathol. 168, 292–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cosenza M. A., Zhao M. L., Shankar S. L., Shafit-Zagardo B., Lee S. C. (2002) Neuropathol. Appl. Neurobiol. 28, 480–488 [DOI] [PubMed] [Google Scholar]
- 47.Giaretti W., Nüsse M. (1994) Methods Cell Biol. 41, 389–400 [DOI] [PubMed] [Google Scholar]
- 48.Stoss O., Schwaiger F. W., Cooper T. A., Stamm S. (1999) J. Biol. Chem. 274, 10951–10962 [DOI] [PubMed] [Google Scholar]