Abstract
A growing number of studies have investigated the interaction between C1q and PrP, but the oligomeric form of PrP involved in this interaction remains to be determined. Aggregation of recombinant full-length murine PrP in the presence of 100 mm NaCl allowed us to isolate three different types of oligomers by size-exclusion chromatography. In contrast to PrP monomers and fibrils, these oligomers activate the classical complement pathway, the smallest species containing 8–15 PrP protomers being the most efficient. We used Thioflavine T fluorescence to monitor PrP aggregation and showed that, when added to the reaction, C1q has a cooperative effect on PrP aggregation and leads to the formation of C1q-PrP complexes. In these complexes, C1q interacts through its globular domains preferentially with the smallest oligomers, as shown by electron microscopy, and retains the ability to activate the classical complement pathway. Using two cell lines, we also provide evidence that C1q inhibits the cytotoxicity induced by the smallest PrP oligomers. The cooperative interaction between C1q and PrP could represent an early step in the disease, where it prevents elimination of the prion seed, leading to further aggregation.
Keywords: Diseases/Amyloid, Immunology/ Innate Immunity, Prions, Protein/Protein-Protein Interactions, Complement, C1q, Aggregation, Oligomer
Introduction
Prion diseases represent a group of singular transmissible neurodegenerative disorders that affect mammals and occur when the cellular prion protein (PrPc)2 is converted into an abnormal aggregated isoform called PrPSc (1). The host PrPc is an α-helix-rich 30–35-kDa glycoprotein expressed mainly in neuronal tissues in humans and other animals (2). Its expression and membrane attachment through a glycosylphosphatidyl anchor are crucial for transmissible spongiform encephalopathy susceptibility (3). The widely supported protein-only hypothesis stipulates that the infectious agent can replicate by converting the natively folded prion protein (PrPc). During the disease, changes occur in the secondary and tertiary structure of PrPc, resulting in an increased content of the β-sheet, which leads to the formation of aggregates that display a dramatic shift in their physicochemical properties compared with those of the original protein (4). These β-sheet-rich aggregates are resistant to degradation by proteases and tend to form oligomers that can further assemble into amyloid fibrils. Recent investigations support the hypothesis that in protein misfolding and aggregation pathologies like Alzheimer disease, smaller subfibrillar particles may be much more pathological than larger amyloid fibrils or plaques (5). In line with this hypothesis, Silveira and colleagues have reported that the smallest aggregate able to initiate transmissible spongiform encephalopathies pathology is equivalent to ∼14–28 PrP molecules (6). In the past, several in vitro recombinant models have been used to investigate the biochemical and biophysical properties of such oligomeric intermediates (7–10).
C1q is the first component of the classical complement pathway. C1q binds to many non-self and altered-self-materials. These include microorganisms, immune complexes, apoptotic and necrotic cells and their breakdown products, and amyloids. C1q binding to amyloid fibrils found as extracellular deposits in tissues, and subsequent complement activation are involved in the pathology of several amyloid diseases, such as Alzheimer disease (11).
C1q is also expressed in the developing and adult nervous system. The efficient and selective removal of apoptotic cells is an important feature of tissue development, homeostasis, and pathology. In the nervous system, synapses and distal axons are selectively eliminated as part of the remodeling that underpins development and pathology through a process that has some features in common with apoptotic cell removal (12, 13). Recent evidence suggests that the complement components C1q have a role in the selective tagging of supernumerary synapses in the developing visual system and in their efficient removal by as yet unidentified cells (14, 15).
In prion diseases, complement activation is likely to contribute to neuronal damage in the end stages of prion diseases but is also thought to participate in the initial infection, dissemination, and replication stages (16, 17). A recent time course transcriptomic and phenotypic study of mouse prion diseases has shown that the three genes coding for C1q are among the top 10 genes up-regulated in the brain (18).
A growing number of studies are addressing the interaction between PrP and C1q (19–21). In previous works (22, 23), we have characterized the interaction between C1q and several oligomeric PrP species formed in vitro. Our data indicated that this interaction occurs through the globular heads of C1q and only occurs when PrP is converted into β-sheet-rich oligomers. Moreover, this interaction has biological relevance, as it triggers activation of the classical pathway of complement. However, the size of the oligomer, which was the most efficient in complement activation, was unresolved.
In the present work, we identified small sized oligomers as the preferential partners for C1q. These are composed of 8–15 PrP molecules and display the highest complement activation potential. Also, for the first time, we show that C1q can participate to the aggregation of PrP leading to the formation of a complex able to promote complement activation. This outlines a novel role for C1q in prion disease, as a cooperative partner in the PrP aggregation process. We assessed the cytotoxicity potential of PrP oligomers of several sizes using two cell culture models and found that the smaller oligomers trigger cell death. Interestingly, this effect is inhibited through formation of C1q-PrP complexes.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Cloning, expression, and purification of the recombinant full-length (23–230) murine prion protein have been described previously (23). Protein integrity was confirmed by 15% SDS-PAGE, circular dichroism analysis, and aliquots at a concentration of 10 mg/ml were stored at −20 °C until use. Prior to each experiment, protein samples were centrifuged at 13,000 × g for 20 min at 4 °C to eliminate aggregated material. C1q, its globular domain, C1 inhibitor, and the proenzyme form of the C1s-C1r-C1r-C1s tetramer were purified from human serum and quantified as described previously (24, 25). For aggregation kinetics studies, C1q and its globular domain were dialyzed against 20 mm sodium acetate, pH 3.5, prior to use.
Conversion of PrP into β-Oligomers and Aggregation Kinetics
PrP β-oligomers were prepared by incubating freshly purified monomeric recombinant PrP (5 mg/ml in 20 mm sodium acetate, pH 3.5) at 40 °C or 70 °C for 18 h in the presence of 100 mm NaCl. β-Oligomers were separated by size-exclusion chromatography (SEC) using a G4000SW high pressure liquid chromatography column (600 × 7.5 mm, Waters), in 50 mm sodium acetate, 50 mm NaCl, pH 4, at a flow rate of 0.5 ml/min. Elution was monitored at 280 nm, and fractions corresponding to protein species were collected as specified in the text. The weight-average molar mass of oligomers was measured as described in Ref. 10 by static MALLS using a DAWN-EOS detector (Wyatt Technology Corp., Santa Barbara, CA).
The aggregation kinetics was monitored for 18 h by measurement of thioflavin T (ThT) fluorescence, using a fusion alpha FP HT microplate reader (Perkin Elmer, with an excitation filter of 436/20 and emission filter of 520/20). Kinetics experiments were carried out in a 96-well plate format, in a final volume of 75 μl, with 2 drops of mineral oil (Sigma) on top of each well to avoid evaporation during incubation at 40 °C. For all experiments, PrP (5 mg/ml) was incubated in the presence of 10 μm ThT (Sigma) in 20 mm sodium acetate, pH 3.5, in the presence of 100 mm NaCl. When indicated, C1q or its globular domain, after dialysis in 20 mm sodium acetate, pH 3.5, were added to the reaction mixture at concentrations ranging from 0.2 to 0.4 mg/ml. Reaction time point (1, 4, 6, and 18 h) were analyzed by SEC as described above.
Electron Microscopy
Samples of each protein species at a concentration of 0.04 mg/ml were adsorbed onto the clean face of a carbon film, deposited on a mica sheet, and negatively stained with either 2% (w/v) ammonium molybdate (C1q) or 2% (w/v) uranyl acetate (PrP and C1q-PrP). The specimens were examined with a Philips CM12 electron microscope equipped with a LaB6 filament operating at 120 kV. The micrographs were recorded under low dose conditions (<20 electrons/Å2) at a nominal magnification of 45,000. Images were recorded using a Gatan OriusTM CCD camera with a pixel size of 0.2 nm.
Dot Blot Quantification of C1q
One μg of each SEC-purified fraction (calculated from an absorbance at 280 nm using a PrP extinction coefficient of 62,400 m−1 cm−1) was spotted onto a nitrocellulose membrane using a dot blot apparatus (Bio-Rad). The membrane was blocked with 5% nonfat dry milk in phosphate-buffered saline containing 0.1% (w/v) Tween 20 for 1 h at room temperature and then incubated with monoclonal anti-human C1q antibody JL-1 (1:500, Hycult Biotechnology) in blocking buffer. After incubation with anti-mouse horseradish peroxidase antibody (1:5,000, Jackson ImmunoResearch Laboratories) in blocking buffer, membranes were developed by enhanced chemiluminescence (ECL, Amersham Biosciences Pharmacia) and exposed to x-ray film (Hyperfilm ECL, Amersham Biosciences Pharmacia). Films were scanned and treated with the ImageJ software, and the percentage of C1q in each fraction was determined and plotted using GraphPad Prism 4.
C1 Activation Assay
The C1 activation assay was performed as described previously (26). The C1 complex (0.25 μm) was reconstituted from equimolar amounts of C1q and proenzyme C1s-C1r-C1r-C1s, mixed with 1 μm C1 inhibitor, and then incubated in 145 mm NaCl, 1 mm CaCl2, 50 mm triethanolamine-HCl, (pH 7.4) with varying types of PrP oligomers (final concentration, 2.5 μm) for 90 min at 37 °C. The reaction mixtures were submitted to SDS-PAGE analysis under reducing conditions. After electrotransfer to a nitrocellulose membrane (Bio-Rad), C1s was revealed by Western blot analysis using a rabbit polyclonal antibody.
C4 Cleavage Assay
SEC-purified PrP type II oligomers or C1q-PrP complexes were coated on ELISA plates (Maxisorb Nunc) at 1–5 μg/ml in 100 mm Na2C03/NaHC03, pH 9.6, overnight at room temperature. Wells were washed three times in PT (phosphate-buffered saline containing 0.05% (w/v) Tween 20), saturated for 1 h at 37 °C in 1% phosphate-buffered saline (w/v) bovine serum albumin, and washed three times in PT. C1q-depleted serum was diluted to 2% in VB2+ (5 mm Veronal buffer, 145 mm NaCl, 5 mm CaCl2, 1.5 mm MgCl2, pH 7.5), added to the wells, and incubated for 30 min at 37 °C. Plates were washed four times in PT. C1q-depleted serum was obtained by incubating 1.2 ml of normal human serum with 240 μl of ovalbumin anti-ovalbumin Ig complexes (25) for 1 h at 4 °C. The depletion was performed twice. The C1q-deficient serum contained <13 units/liter C1q as measured by nephelometry. However, it comprised all others factors necessary for activation of the complement classical pathway, including C1r and C1s as shown by passive double immunodiffusion assay (supplemental Fig. S1). To detect the C4 cleavage product C4b, a rabbit anti-C4b polyclonal antibody (Siemens, 1:1,000 diluted in PT containing 1% (w/v) bovine serum albumin) was added for 1 h at 37 °C. After washing and addition of peroxidase-conjugated anti-rabbit antibody (diluted 1:20,000 in PT containing 1% (w/v) bovine serum albumin; The Jackson Laboratory) for 1 h at 37 °C, plates were washed and developed with tetramethylbenzidine (Sigma). Reaction was stopped by 1 n H2SO4 and read at 450 nm.
Cell Culture and in Vitro Neurotoxicity Assay
Cell lines were generous gifts from S. Lehmann (CNRS UPR1142, Montpellier, France). N2aD11 cells (PrP+/+) and Npl1 hippocampal cells derived from PrP0/0 mice were maintained in Opti-Mem + Glutamax (Invitrogen) medium supplemented with 10% fetal calf serum and 50 μg/ml gentamycin and grown in 5% CO2 at 37 °C. For toxicity assays, cells were seeded at a density of 10,000 in a 96-well plate. Cells at 90% confluence were incubated with various preparations of oligomeric PrP or C1q-PrP complexes as stated in the text. SEC-purified fractions at pH4 were diluted to the indicated concentrations in 100 μl using fetal calf serum-free and antibiotic-free Opti-MEM medium. After dilution, the final pH of the medium containing oligomers was 7. Each fraction was assayed in three independent experiments. For controls, cells were either untreated or incubated in the presence of an equivalent volume of protein buffer. Twenty-four h after exposure to the proteins, cell viability was measured using the WST-1 assay (Roche), according to the manufacturer's instructions. The assay is based on the reduction of WST-1 by viable cells, which produces a soluble formazan salt. The formazan dye quantified by absorbance correlates directly with cell number. Briefly, 10 μl of the reagent was added to each well and incubated for 5 h. The optical density at 450 nm was measured using an ELISA plate reader. Results were plotted using the GraphPad Prism 4 software, and statistical analysis was performed using Student's t test with Welsh correction.
RESULTS
PrP Oligomers Trigger Differential Complement Activation Depending on Their Size
Direct interaction between C1q and different isoforms (i.e. fibrils or β-oligomers) of PrP has been described (19, 23). However, the precise nature of the PrP isoforms that activate the C1 complex remains unknown. Different PrP species were prepared and purified to answer this question.
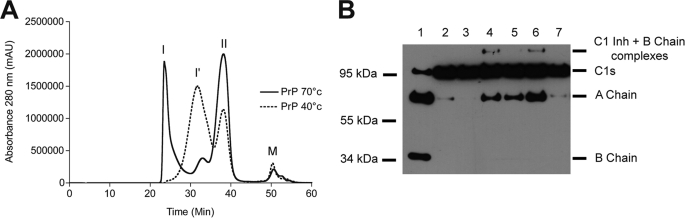
Recombinant murine PrP (23–230) can form soluble oligomers when incubated in 100 mm NaCl at 40 °C or 70 °C (Fig. 1A). Two types of oligomers (I and II) found in different proportions depending on the temperature used were previously identified by SEC (10, 23, 27). In the present study, using a more resolving SEC column, we were able to identify a third oligomeric species (Fig. 1A, I′) of an intermediate size. Whereas type II oligomers are composed of 8–15-mers, I′ oligomers correspond to ∼36-mers (supplemental Fig. S2). Type I, I′, and II oligomers were collected during SEC, and each was tested for its ability to trigger complement activation though the classical pathway.
FIGURE 1.
Small oligomers efficiently trigger C1 activation. A, size-exclusion chromatography elution profile of mouse recombinant PrP after incubation for 18 h at 40 °C (dashed line) or 70 °C (plain line) in the presence of 100 mm NaCl. Three types of oligomers can be purified (type I, I′, and II oligomers). mAu, milliabsorbance units. B, C1 activation assay under different conditions. The C1s-C1r-C1r-C1s tetramer was incubated for 90 min at 37 °C in the presence of C1q alone (lane 1), C1q + C1 inhibitor (lane 2), C1q + C1 inhibitor + PrP monomer (lane 3), C1q + C1 inhibitor + oligomer I (lane 4), C1q + C1 inhibitor + oligomer I′ (lane 5), C1q + C1 inhibitor + oligomer II (lane 6), or C1q + C1 inhibitor + PrP fibrils (lane 7). Each sample was subjected to SDS-PAGE under reducing conditions and C1s was revealed by Western blot. The presence of the C1s A chain indicates complement activation. The gel shown is representative of three independent measurements.
A C1 activation assay (26) was performed using equimolar amounts of C1q and proenzyme C1s-C1r-C1r-C1s, mixed with excess C1 inhibitor to prevent self-activation and incubated in the presence or absence of the oligomeric species at 2.5 μm (Fig. 1B). The generation of the C1s A chain was monitored by Western blot using an anti-C1s antibody. In keeping with our previous findings (23), monomeric PrP (Fig. 1B, lane 3) failed to activate C1. The smaller type II oligomers displayed strong C1 activation (Fig. 1B, lane 6), whereas the larger type I (lane 4) and I′ oligomers (lane 5) were less effective. Aged PrP fibrils (Fig. 1B, lane 7) triggered only very slight C1s cleavage, comparable to that observed for the negative control (lane 2). These fibrils were obtained by leaving monomeric PrP at 4 °C in its storage buffer (20 mm sodium acetate, pH 3.5) for at least 10 weeks, and the presence of fibrils was confirmed by electron microscopy (supplemental Fig. S3). Thus, these experiments provided clear evidence that smaller PrP oligomers are more efficient activators of the classical complement pathway.
C1q Enhances the Formation of PrP Oligomers
Our previous results have indicated that a conformational change in PrP is required for C1q binding (22, 23). This prompted us to assess whether C1q binding can occur during the process of PrP transconformation and aggregation. For this purpose, ThT fluorescence was used to investigate the kinetics of formation of PrP aggregates, in the presence of C1q, or its globular heads (GH) (Fig. 2).
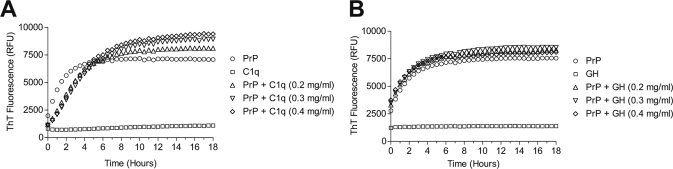
FIGURE 2.
Aggregation kinetics of recombinant mouse PrP monitored by thioflavin T fluorescence. Fluorescence was measured for 18 h as described under “Experimental Procedures.” The PrP concentration was 5 mg/ml in 20 mm sodium acetate, pH 3.5, in the presence of 100 mm NaCl and 10 μm ThT. Aggregation kinetics of PrP alone (empty circles) (A and B), or in the presence of C1q (A), or C1q globular domain (B) at varying concentrations: 0.2 mg/ml (open triangles), 0.3 mg/ml (inverted open triangles) and 0.4 mg/ml (open diamonds). C1q and its globular domain alone do not bind to ThT (open squares) (A and B). All experiments were performed at 40 °C in triplicate using a microplate reader. RFU, relative fluorescence units.
Using our oligomerization conditions (incubation for 18 h at 40 °C in the presence of NaCl), PrP started to aggregate immediately, as no lag phase was observed. The kinetics of β-sheet formation was fast, and fluorescence reached a plateau after ∼6 h of incubation (Fig. 2A). Under the same conditions, incubation of PrP in the presence of 0.2 mg/ml C1q (PrP-C1q molar ratio, 400:1), led to a short lag phase in the aggregation, which reached a plateau at ∼18 h. The fluorescence observed was higher than for PrP alone (Fig. 2A). Increasing C1q concentration to 0.3 and 0.4 mg/ml did not modify the lag phase but led to a dose-dependent increase in ThT fluorescence after 6 h (Fig. 2A). The increased ThT fluorescence at the observed higher plateau levels could be due to the association of C1q with PrP oligomeric structures.
Binding of PrP oligomers to C1q occurs through the globular domain of C1q (23). Therefore, we next investigated whether this domain alone could account for the variations induced by the whole C1q molecule. Using this domain instead of intact C1q only slightly modified the aggregation curve of PrP (Fig. 2B). No lag phase was observed, but a small dose-dependent increase in the ThT fluorescence was seen compared with the curve with PrP only.
In our experimental conditions, no PrP aggregation occurred without the presence of NaCl; optimal oligomerization was achieved at 100 mm NaCl (supplemental Fig. S4). Therefore, we addressed the question whether C1q itself could replace NaCl and thus be sufficient to initiate aggregation. When NaCl concentration was reduced to 5 mm in the reaction mixture, PrP did not aggregate either in the presence or absence of C1q (supplemental Fig. S4). C1q and its globular domain did not bind to thioflavin T, as monitored by kinetic analysis of ThT fluorescence (Fig. 2, A and B). Taken together, these data suggested that C1q can influence the aggregation of PrP in a cooperative manner but only when structurally intact, as a hexameric molecule.
C1q Forms a Complex with PrP Oligomers
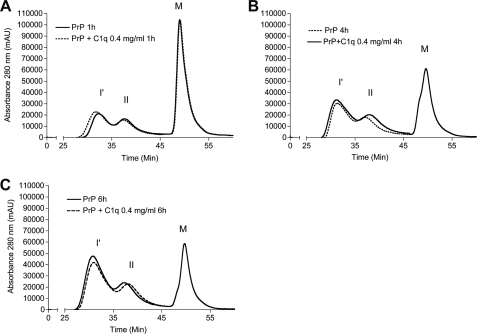
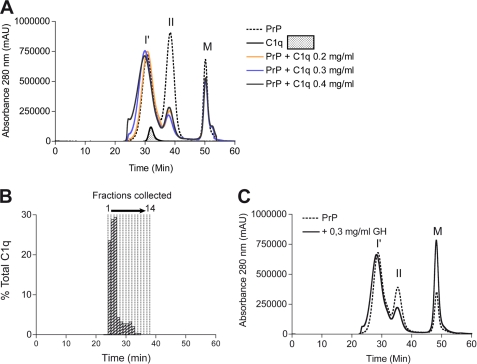
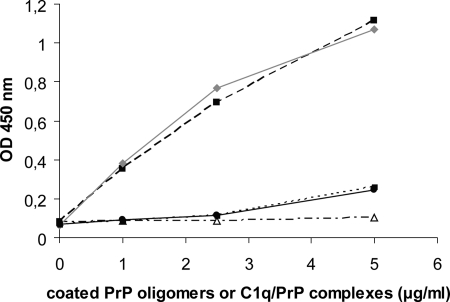
Although C1q seemed enhance formation of β-sheet rich aggregates, the nature of the products formed during aggregation is unknown. Therefore, reaction time points were analyzed by SEC after 1, 4, and 6 h (Fig. 3, A–C) and 18 h (Fig. 4A) of incubation. When PrP was aggregated alone, separation of the reaction products at these time points revealed two major peaks of β-oligomers, I′ and II as described previously (Fig. 1A). The elution profile of the C1q-PrP aggregation mixture showed a similar pattern to that of PrP alone at the early aggregation stages (1, 4, and 6 h). However, specific differences can be seen at 18 h. First, the peak corresponding to type II oligomers was markedly decreased. Second, the peak encompassing I′ oligomers became wider, and a small proportion of larger species was detected in the void volume (elution time of 25 min) (Fig. 4A). The latter observation appears to be C1q dose-dependent. These data suggest that C1q interacts preferentially with type II oligomers. The apparent affinity constant between purified type II oligomers and C1q was high as measured by ELISA (KD = 0.7 10−9 m−1) (supplemental Fig. S5).
FIGURE 3.
Analysis by size-exclusion chromatography of PrP aggregation reaction time points. PrP alone (line) and PrP in presence of 0.4 mg/ml of C1q (dashed line) aggregations are analyzed by SEC over time. A, 1 h after aggregation. B, 4 h after aggregation. C, 6 h after aggregation. I′, oligomer I′; II, oligomer II; M, monomer. Incubation was performed at 40 °C. mAu, milliabsorbance units.
FIGURE 4.
Analysis by size-exclusion chromatography of PrP aggregation endpoint products following incubation at 40 °C as shown in Fig. 2. A, PrP alone (dashed line) aggregated into two types of oligomers, I′ and II. C1q alone eluted as a single peak ∼32 min (line and filled area). PrP aggregation in presence of 0.2, 0.3, and 0.4 mg/ml C1q (orange, blue, and black lines, respectively) led to the formation of two types of oligomers: type II oligomers that were superimposed on their PrP counterpart and a larger heterogeneous peak starting to elute in the void volume of the column. B, 14 500-μl fractions encompassing the C1q-PrP complex peak were collected from 25 to 39 min. Fractions were assayed for C1q by dot blot analysis using an anti-C1q antibody (histogram bars). C, SEC analysis was performed on C1q globular domain/PrP co-aggregation mixtures (line). There are no major differences from the elution profile of PrP alone (dashed line). I′, oligomer I′; II, oligomer II; M, monomer; mAu, milliabsorbance units.
To ascertain the presence of C1q within the elution profiles, the C1q-PrP aggregation mixture was submitted to SEC, and fractions were collected and subjected to dot-blot quantification of C1q using a monoclonal antibody raised against the native collagen moiety (Fig. 4B, elution time of 25–38 min). C1q was found in the first 10 fractions, and fractions 1–4 contained 80% of total C1q. Fractions 5–10 contained only a small amount of unbound C1q, as judged from the retention time of C1q alone (Fig. 4A), whereas fractions 11–15 did not contain any C1q (Fig. 4B). Finally, we analyzed the reaction endpoint of the C1q globular domain/PrP co-aggregation mixture. As seen for the C1q-PrP aggregation products, we observed a decrease in the peak corresponding to type II oligomers (Fig. 4C), suggesting that these oligomers interact with the globular domain of C1q.
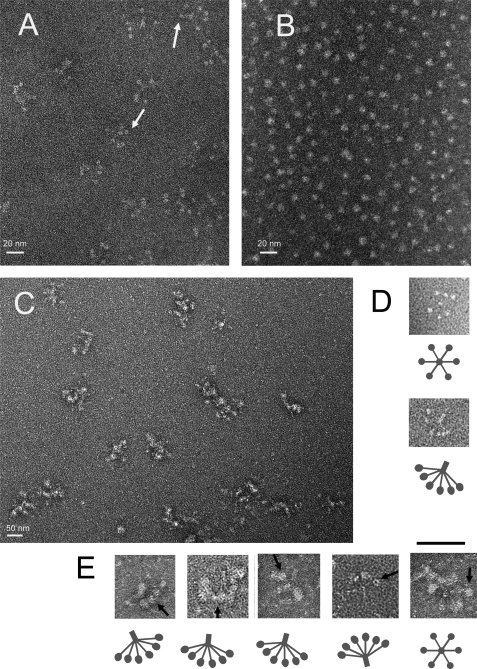
The above observations provided evidence that an oligomeric form of PrP was complexed with C1q, but the nature of this oligomer within the complex remained to be established. To gain insights into the structure of the complexes, C1q, PrP oligomers, and C1q-PrP complexes were analyzed by electron microscopy. PrP type II oligomers (Fig. 5B) appeared as small spherical particles with a diameter of ∼5–8 nm, as described previously (10, 27). The PrP oligomer I′ appeared as annular particles with sizes ranging at ∼15–20 nm (supplemental Fig. S6). The C1q molecule consists of a collagen-like fragment (CLF) and six GHs. The CLF is made up of a stem, which spreads out into six arms, each ending in a GH with a diameter of ∼4 nm (28–30). Images of the C1q molecules (Fig. 5, A and D) appear as clusters of GHs with the CLF visible occasionally in some molecules, as the CLFs are highly sensitive to radiation damage and may also reside out of the plane of the support film where the GHs are attached. In the C1q-PrP fractions collected at the beginning of the elution profile, we found particles strongly reminiscent of the C1q molecules containing bigger globular heads (Fig. 5, C and E). The CLF domain of C1q can be found in some of these particles, ending in a GH with a diameter of ∼8 nm, corresponding to the size of oligomer II. These images suggest that the GHs are bound to some larger particles, likely oligomer II.
FIGURE 5.
Electron microscopy analysis of C1q, PrP type II oligomers, and C1q-PrP complexes. Samples were negatively stained using 2% uranyl acetate or 1% ammonium molybdate. A, overall view for purified C1q. White arrows indicate examples of globular head regions of C1q. B, overall view for SEC-purified PrP type II oligomers (diameter of 5–8 nm). C, overall view of purified C1q-PrP complexes taken from fraction 2 of the SEC shown in Fig. 4B. The proteins are present in clusters. D, detailed top view and side view of C1q. The clear dots represent the GH domains with a diameter of ∼4 nm. E, detailed side views and one top view of purified C1q-PrP complexes. Black arrows indicate examples of PrP bound to C1q GH. The scale bar between D and E represents 50 nm.
Complex-bound C1q Is Functional
As described above, SEC analysis revealed that fractions 5–10 contained a small amount of unbound C1q, most of the C1q molecules being found as larger complexes in fractions 1–4. Although the complexed form of C1q was recognized by a monoclonal antibody to the native protein, its ability to trigger complement activation remained to be established. For this purpose, a C4 cleavage assay was performed using C1q-PrP complexes purified by SEC after co-aggregation or type II oligomers purified after aggregation of PrP alone, as a control. Complement activation was assessed by measuring the extent of C4 cleavage after incubation of the coated proteins with C1q-depleted human serum. No significant activation was observed using type II oligomers (Fig. 6). In contrast, C1q-PrP complexes triggered complement activation in a dose-dependent manner. In a comparative analysis, type II oligomers also triggered a dose-dependent activation when incubated in normal human serum (supplemental Fig. S7). This demonstrated that C1q molecules within the C1q-PrP complexes retained structural and functional integrity after the co-aggregation and purification processes.
FIGURE 6.
PrP-bound C1q retains the ability to trigger C4 cleavage. SEC purified fractions 2 (closed square), 3 (closed diamond), and 4 (closed circle) were subjected to C4 cleavage assay in C1q-depleted serum to assess the structural functionality of C1q in the complexes. As a control, oligomer II alone did not activate the complement in C1q-depleted serum (open triangle).
C1q-PrP Complexes Inhibit PrP Oligomer-induced Cell Toxicity
PrP oligomers recently have been described as neurotoxic (31). To test the cytotoxicity of our protein preparations, we used two neuronal cell lines, N2aD11 (PrP+/+ neuroblastoma cells) and Npl1 (PrP0/0 hippocampal cells). Cells were exposed to different concentrations of purified PrP isoforms, including monomeric PrP, SEC-purified type II and I′ oligomers, aged PrP fibrils, and fraction 3 of purified C1q-PrP complexes (see Fig. 4B). Cell viability was measured 24 h after exposure using the WST-1 assay (Fig. 7).
FIGURE 7.
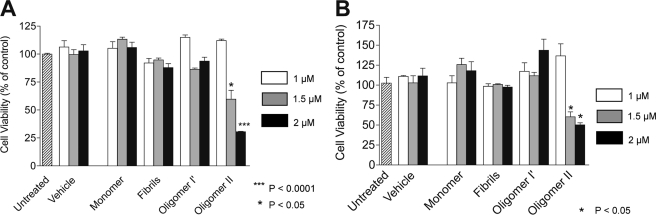
Oligomer II is toxic to cultured cells. Neuronal cells from PrP+/+ or PrP0/0 mice were exposed for 24 h to purified PrP isoforms ranging at 1 μm (white bars), 1.5 μm (gray bars) and 2 μm (black bars). Cell viability was then measured using the WST-1 assay. A, PrP+/+ N2aD11 cells. B, PrP0/0 NplI hippocampal cells. Untreated cells (hatched bars) and cells treated with equivalent volumes of vehicle buffer were used as controls. Statistical analysis was performed using Student's t test with Welsh correction. Error bars represent the means +/− S.E. of three independent experiments.
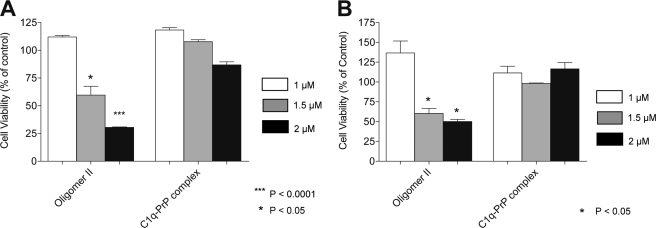
Exposure of N2aD11 cells to 2 μm type II oligomers resulted in a loss of ∼70% of the cells, compared with untreated or to vehicle-treated cells (Fig. 7A). This effect could not be observed at a concentration of 1 μm, but an intermediate effect was seen at 1.5 μm. Monomeric PrP, type I′ oligomers, and aged PrP fibrils had no significant cell toxicity whatever the concentration used. Similar toxic effects were observed using Npl1 hippocampal cells from PrP0/0- defective mice, indicating that the toxicity of type II oligomers was independent of the expression of PrP (Fig. 7B). When C1q-PrP complexes were tested using the same assays, no significant toxicity was observed for these species whatever the concentration used, using either type of cells (Fig. 8, A and B).
FIGURE 8.
C1q-PrP complexes inhibit oligomer II-induced cell toxicity. Cells were treated with SEC-purified C1q-PrP complexes at 1 μm (white bars), 1.5 μm (gray bars), and 2 μm (black bars), and cell viability was measured using the WST-1 assay. Data from cells incubated with PrP type II oligomers are shown for comparison. A, PrP+/+ N2aD11 cells. B, PrP0/0 NplI hippocampal cells.
DISCUSSION
PrP Oligomerization Generates Three Distinct Soluble Oligomers
The ability of monomeric PrP to adopt fibrillar conformations has been studied widely (32, 33). Only recently, efforts have been made toward the identification of other amyloidogenic oligomeric structures (7, 8, 10, 31, 34). PrP oligomers described so far have been generated under different conditions, using various deleted constructions and thus often display distinct biophysical and biochemical characteristics. We previously showed that thermal destabilization of PrP in the presence of NaCl leads to the formation of two discrete oligomeric species named type I oligomers (larger species, heterogeneous in size) and type II oligomers (smaller species, 8–15-mer) (10, 27). In the present study, we were able to separate a third oligomeric species, with an intermediate size. This newly found oligomer was named oligomer I′.
Recently, recombinant ovine PrP has been shown to aggregate and generate three oligomeric forms O1, O2, and O3 (8, 34) which likely correspond to type I, I′, and II oligomers, respectively, obtained with our murine preparations. These authors showed that the best model for ovine PrP oligomerization would be a set of parallel pathways, where O1, O2, and O3 originate from the same partially unfolded monomer but form soluble aggregates independently and are not kinetically related. Interestingly, this work also pointed out that only the O1 oligomer was able to fibrillize. Further biophysical investigations will determine whether our murine oligomers can fit this model.
C1q Binds Preferentially to Small PrP Oligomers
The implication of C1q in prion pathogenesis has been pointed out by showing that the onset of the disease is delayed in C1q- and C3-deficient mice (16, 17). Further studies have described direct interaction between C1q and PrP (19, 20, 22, 23). However, the nature of the PrP isoform (i.e. fibrils or β-oligomers) recognized by C1q remains unclear. In a previous attempt to assess the ability of PrP oligomers to activate the classical complement pathway, we could not use SEC-purified species, because the hemolytic assay used required high protein concentrations (23). In the present work, the use of a specific C1 activation assay requiring lower protein concentrations allowed us to demonstrate that the smaller type II oligomers, comprising 8–15 PrP molecules, are the most efficient activators of complement, compared with larger oligomers (types I and I′).
Triggering of the classical complement pathway results from binding of the C1 complex, via its recognition subunit C1q, to immune and nonimmune activators and leads to activation of its associated proteases C1s-C1r-C1r-C1s. This tetramer is in close contact with all six stems of C1q. Thus binding of each globular head to a target is expected to generate part of the movement of the stems thought to trigger C1r activation, which in turn activates C1s. It is known that in the case of immune complexes, C1q must bind to at least two antibody molecules for effective activation. Larger molecules may induce steric hindrance, thus resulting in a less efficient activation. This could explain why smaller oligomers of PrP are more effective at complement activation than larger ones.
Also, in our study, PrP fibrils did not trigger C1 activation. This finding is in contradiction with previous studies, where fibrillar species from a C-terminal fragment of human PrP were found to activate the classical pathway (19). Because the procedure used for forming PrP fibrils dictates their final structure (35), these discrepancies may arise from the fact that the fibrils used in both studies display different structural features. In addition, the absence of the N-terminal part of PrP in the study by Sjoberg et al. (19) could also account for the observed variations.
C1q in the Aggregation Process of PrP
Investigation of the functional relevance of the C1q-PrP interaction has focused on the ability of PrP-derived ligands to interact with complement factors and trigger complement activation (19, 20, 23). In the present study, we characterized a novel role for C1q, namely its ability to interact with PrP during the process of oligomer formation and to enhance this process in a dose-dependent manner.
ThT is used mostly to monitor fibrillation (36); however, ThT has been shown to interact not only with fibrillar proteins but also with other molecules such as cyclodextrin or acetylcholinesterase (37). Following our experimental conditions, PrP forms only soluble aggregates. Analysis, by SEC and electron microscopy, of aggregation reaction end products confirmed the absence of fibrils.
As no fibrillar structures were observed, this phenomenon cannot account for the variations seen in fluorescence. Because ThT fluorescence is only a global indicator of β-sheet formation, we performed SEC analysis to confirm the relationship between fluorescence and the oligomeric species formed. Although there seems to be a lag phase observed by fluorescence assay, the species formed during early steps (1–6 h) by either PrP alone or PrP in presence of C1q are similar. Late steps of aggregation revealed specific differences in chromatograms as described under “Results.” A plausible hypothesis would be that the hexameric C1q molecule starts interacting with PrP as soon as it reaches a critical oligomeric size. After initial binding, C1q could then bring together PrP molecules more efficiently, thus enhancing oligomer formation (6–18 h). When the C1q globular domain was used instead of the whole C1q molecule, no obvious effect was seen on PrP aggregation. The GH is monovalent, whereas the C1q molecule is a hexavalent structure. Therefore, it is not surprising that GH has no or a little effect on aggregation, whereas multimerization contributes to the functional binding activity of C1q. This reflects the low affinity of purified GH for PrP compared with the whole C1q molecule (16).
It may be concluded from this observation that the cooperative effect of C1q requires the latter to be under a native hexameric form. The effect of C1q on PrP aggregation was only investigated at 40 °C and at C1q concentrations up to 0.4 mg/ml because of the low solubility of C1q at concentrations >0.8 mg/ml and its instability at 70 °C. Also, it would be of interest to test whether C1q can act as a cofactor enhancing PrPSc formation in a protein cyclic misfolding amplification assay (38).
C1q Forms a Complex with PrP Type II Oligomers and Remains Functional
The interaction between PrP and C1q during aggregation led to the formation of a stable complex. Several results indicate that type II oligomers represent the PrP moiety of these complexes. First, the proportion of type II oligomers was strongly decreased in the C1q-PrP aggregation mixture. Secondly, the electron micrographs of the SEC-purified C1q-PrP complexes indicate that most of the oligomers bound to the C1q globular domain resemble type II. When seen from a top view, the complex seems to carry one oligomer II per head; however, it is not as clear when seen from a side view. In this case, it could be argued that one oligomer could span more than one head; however, the small size of a single oligomer (8 versus 4 nm for the GH) may preclude this type of interaction.
We also tried to determine the C1q-PrP complex molar mass by MALLS (supplemental Fig. S2). PrP type II oligomers have a mean molar mass of 2.5 × 105 g/mol, which corresponds to the mass previously calculated for 12-mer PrP, based on a mass of 23 kDa for recombinant PrP (10). Type I′ oligomers are heterogeneous in size, but their mean molar mass could be estimated to ∼8 × 105 g/mol (34-mer). The C1q-PrP molecular mass can be only roughly estimated, as its light scattering is superimposed on that of type I′ oligomers. Nevertheless, taking the apex of the peak as a reference, the mean molar mass could be evaluated to 1.15 × 106 g/mol. C1q is a molecule of ∼460 kDa, which leads to an estimate of 690 kDa for the average mass of the PrP moiety in the complex, corresponding to the size of either one oligomer I′ (800 kDa) or three type II oligomers (780 kDa). This is the minimal estimate for the C1q-PrP complexes because C1q is found mainly in fractions 1 to 3, well before the apex of the peak. In line with electron microscopy data, this strengthens our hypothesis that type II oligomers are recognized by C1q. To be biologically relevant, such complexes must contain functional C1q molecules. Indeed, dot blot analyses indicated that C1q retained its structural integrity, as it was recognized by a monoclonal antibody raised against the native form of the collagen moiety of C1q. In addition, C1q remained functional within the complex and could still bind its partner proteases to activate the classical complement pathway as shown by a C4 cleavage assay. It is interesting to note that C1q ligand binding is efficient at low pH, although most binding experiments described in the literature occur at neutral pH. This result has been confirmed in a control experiment using another known C1q ligand (supplemental Fig. S5).
A Role for C1q-PrP Complexes in the Prevention of Neuronal Cell Death?
Neurotoxicity of prion oligomers and fibrils for both cultured cells and primary neurons has been widely explored (31, 39, 40). Although recent results tend to demonstrate that β-oligomers are cytotoxic to cultured cells, whereas mature fibrils are harmless, the nature of the most neurotoxic species is uncertain. In our hands, only type II oligomers displayed a pronounced dose-dependent cytotoxic effect to cultured cell lines, whereas larger species showed no toxicity. Interestingly, this effect could be observed using both PrP+/+ and PrP0/0 cells. This indicates that expression of endogenous PrP is not required to observe a toxic effect, as previously reported for β-oligomers (31). However, these results must be confirmed using primary neurons from wild-type and PrP0/0 mice. When cells were exposed to C1q-PrP complexes, in contrast, no toxicity was observed, indicating that the toxic effect induced by the type II oligomers present within the complex was inhibited. There are three possible explanations to this phenomenon. (i) PrP oligomers are trapped by the C1q molecules and cannot reach the cell membrane to exert their toxicity. (ii) C1q-PrP complexes bind to C1q receptors at the cell surface, triggering an antiapoptotic signal. This latter scenario is feasible in our cell lines because they express membrane-bound calreticulin, a well known C1q receptor as observed by immunofluorescence using a chicken polyclonal anti-calreticulin antibody (data not shown). (iii) C1q itself has neuroprotective properties, as recently described for β-amyloid and serum amyloid P-induced toxicity to neurons (41), and this seems to be independent of caspase- and calpain-mediated mechanisms. In the brain, C1q is expressed by neurons and thus could protect them against PrP oligomer toxicity.
CONCLUSION
Our findings support the notion that the C1q-PrP interaction could have a broader biological significance in addition to complement activation. Small cytotoxic oligomers such as type II oligomers are transient, thermodynamically unstable species and thus are unable to enter the fibrillization pathway (34). C1q could act as a stabilizer of these species, allowing them to interact with each other to form larger non cytotoxic aggregates. C1q has a broad range of ligands, including non-self-pathogenic motifs and altered self-structures. It is mostly found in serum and is largely expressed by neurons where its role is not fully established. The cooperative interaction between C1q and PrP could represent an early step in the disease where it prevents elimination of the prion seed, leading to further aggregation. It would also be of interest to verify whether our findings on the prion protein could be extended to other oligomers formed by amyloidogenic proteins (42–44).
Supplementary Material
Acknowledgments
We are indebted to Adrienn Biro (Institut de Biologie Structurale, Grenoble, France) for providing us with the C1s-C1r-C1r-C1s tetramer. We thank Marc Jamin (Unit of Virus and Host Cell Interaction, Grenoble, France) for help with MALLS experiments and Camille Rak for technical assistance.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Figs. S1–S8.
- PrPc
- cellular prion protein
- PrPSc
- scrapie prion protein
- CLF
- collagen-like fragment
- SEC
- size-exclusion chromatography
- ThT
- thioflavin T
- GH
- globular heads of C1q
- ELISA
- enzyme-linked immunosorbent assay
- MALLS
- multi-angle laser light scattering.
REFERENCES
- 1.Prusiner S. B. (1982) Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 2.Bailly Y., Haeberlé A. M., Blanquet-Grossard F., Chasserot-Golaz S., Grant N., Schulze T., Bombarde G., Grassi J., Cesbron J. Y., Lemaire-Vieille C. (2004) J. Comp. Neurol. 473, 244–269 [DOI] [PubMed] [Google Scholar]
- 3.Büeler H., Aguzzi A., Sailer A., Greiner R. A., Autenried P., Aguet M., Weissmann C. (1993) Cell 73, 1339–1347 [DOI] [PubMed] [Google Scholar]
- 4.Pan K. M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R. J., Cohen F. E., et al. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10962–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caughey B., Baron G. S., Chesebro B., Jeffrey M. (2009) Annu. Rev. Biochem. 78, 177–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silveira J. R., Raymond G. J., Hughson A. G., Race R. E., Sim V. L., Hayes S. F., Caughey B. (2005) Nature 437, 257–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain S., Udgaonkar J. B. (2008) J. Mol. Biol. 382, 1228–1241 [DOI] [PubMed] [Google Scholar]
- 8.Rezaei H., Eghiaian F., Perez J., Doublet B., Choiset Y., Haertle T., Grosclaude J. (2005) J. Mol. Biol. 347, 665–679 [DOI] [PubMed] [Google Scholar]
- 9.Rezaei H. (2008) Curr. Alzheimer Res. 5, 572–578 [DOI] [PubMed] [Google Scholar]
- 10.Vendrely C., Valadié H., Bednarova L., Cardin L., Pasdeloup M., Cappadoro J., Bednar J., Rinaudo M., Jamin M. (2005) Biochim. Biophys. Acta 1724, 355–366 [DOI] [PubMed] [Google Scholar]
- 11.Velazquez P., Cribbs D. H., Poulos T. L., Tenner A. J. (1997) Nat. Med. 3, 77–79 [DOI] [PubMed] [Google Scholar]
- 12.Boulanger L. M. (2009) Neuron 64, 93–109 [DOI] [PubMed] [Google Scholar]
- 13.Stevens B., Allen N. J., Vazquez L. E., Howell G. R., Christopherson K. S., Nouri N., Micheva K. D., Mehalow A. K., Huberman A. D., Stafford B., Sher A., Litke A. M., Lambris J. D., Smith S. J., John S. W., Barres B. A. (2007) Cell 131, 1164–1178 [DOI] [PubMed] [Google Scholar]
- 14.Fourgeaud L., Boulanger L. M. (2007) Cell 131, 1034–1036 [DOI] [PubMed] [Google Scholar]
- 15.Perry V. H., O'Connor V. (2008) Nat. Rev. Neurosci. 9, 807–811 [DOI] [PubMed] [Google Scholar]
- 16.Klein M. A., Kaeser P. S., Schwarz P., Weyd H., Xenarios I., Zinkernagel R. M., Carroll M. C., Verbeek J. S., Botto M., Walport M. J., Molina H., Kalinke U., Acha-Orbea H., Aguzzi A. (2001) Nat. Med. 7, 488–492 [DOI] [PubMed] [Google Scholar]
- 17.Mabbott N. A., Bruce M. E., Botto M., Walport M. J., Pepys M. B. (2001) Nat. Med. 7, 485–487 [DOI] [PubMed] [Google Scholar]
- 18.Gehlenborg N., Hwang D., Lee I. Y., Yoo H., Baxter D., Petritis B., Pitstick R., Marzolf B., Dearmond S. J., Carlson G. A., Hood L. (August11, 2009) Database 2009, 10.1093/database/bap011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sjöberg A. P., Nyström S., Hammarström P., Blom A. M. (2008) Mol. Immunol. 45, 3213–3221 [DOI] [PubMed] [Google Scholar]
- 20.Sim R. B., Kishore U., Villiers C. L., Marche P. N., Mitchell D. A. (2007) Immunobiology 212, 355–362 [DOI] [PubMed] [Google Scholar]
- 21.Flores-Langarica A., Sebti Y., Mitchell D. A., Sim R. B., MacPherson G. G. (2009) J. Immunol. 182, 1305–1313 [DOI] [PubMed] [Google Scholar]
- 22.Blanquet-Grossard F., Thielens N. M., Vendrely C., Jamin M., Arlaud G. J. (2005) Biochemistry 44, 4349–4356 [DOI] [PubMed] [Google Scholar]
- 23.Dumestre-Pérard C., Osmundson J., Lemaire-Vieille C., Thielens N., Grives A., Favier B., Csopaki F., Jamin M., Gagnon J., Cesbron J. Y. (2007) Cell Microbiol. 9, 2870–2879 [DOI] [PubMed] [Google Scholar]
- 24.Tacnet-Delorme P., Chevallier S., Arlaud G. J. (2001) J. Immunol. 167, 6374–6381 [DOI] [PubMed] [Google Scholar]
- 25.Arlaud G. J., Sim R. B., Duplaa A. M., Colomb M. G. (1979) Mol. Immunol. 16, 445–450 [DOI] [PubMed] [Google Scholar]
- 26.Biró A., Thielens N. M., Cervenák L., Prohászka Z., Füst G., Arlaud G. J. (2007) Mol. Immunol. 44, 1169–1177 [DOI] [PubMed] [Google Scholar]
- 27.Erlich P., Cesbron J. Y., Lemaire-Vieille C., Curt A., Andrieu J. P., Schoehn G., Jamin M., Gagnon J. (2008) Biochem. Biophys. Res. Commun. 365, 478–483 [DOI] [PubMed] [Google Scholar]
- 28.Reid K. B., Porter R. R. (1976) Biochem. J. 155, 19–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaboriaud C., Thielens N. M., Gregory L. A., Rossi V., Fontecilla-Camps J. C., Arlaud G. J. (2004) Trends Immunol. 25, 368–373 [DOI] [PubMed] [Google Scholar]
- 30.Gaboriaud C., Juanhuix J., Gruez A., Lacroix M., Darnault C., Pignol D., Verger D., Fontecilla-Camps J. C., Arlaud G. J. (2003) J. Biol. Chem. 278, 46974–46982 [DOI] [PubMed] [Google Scholar]
- 31.Simoneau S., Rezaei H., Salès N., Kaiser-Schulz G., Lefebvre-Roque M., Vidal C., Fournier J. G., Comte J., Wopfner F., Grosclaude J., Schätzl H., Lasmézas C. I. (2007) PLoS Pathog 3, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baskakov I. V., Legname G., Baldwin M. A., Prusiner S. B., Cohen F. E. (2002) J. Biol. Chem. 277, 21140–21148 [DOI] [PubMed] [Google Scholar]
- 33.Baskakov I. V. (2009) FEBS Lett. 583, 2618–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eghiaian F., Daubenfeld T., Quenet Y., van Audenhaege M., Bouin A. P., van der Rest G., Grosclaude J., Rezaei H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baskakov I. V., Bocharova O. V. (2005) Biochemistry 44, 2339–2348 [DOI] [PubMed] [Google Scholar]
- 36.Graham J. F., Agarwal S., Kurian D., Kirby L., Pinheiro T. J., Gill A. C. (2010) J. Biol. Chem. 285, 9868–9880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groenning M., Olsen L., van de Weert M., Flink J. M., Frokjaer S., Jorgensen F. S. (2007) J. Struct. Biol. 158, 358–369 [DOI] [PubMed] [Google Scholar]
- 38.Saborio G. P., Permanne B., Soto C. (2001) Nature 411, 810–813 [DOI] [PubMed] [Google Scholar]
- 39.Novitskaya V., Bocharova O. V., Bronstein I., Baskakov I. V. (2006) J. Biol. Chem. 281, 13828–13836 [DOI] [PubMed] [Google Scholar]
- 40.Kazlauskaite J., Young A., Gardner C. E., Macpherson J. V., Vénien-Bryan C., Pinheiro T. J. (2005) Biochem. Biophys. Res. Commun. 328, 292–305 [DOI] [PubMed] [Google Scholar]
- 41.Pisalyaput K., Tenner A. J. (2008) J. Neurochem. 104, 696–707 [DOI] [PubMed] [Google Scholar]
- 42.Sörgjerd K., Klingstedt T., Lindgren M., Kågedal K., Hammarström P. (2008) Biochem. Biophys. Res. Commun. 377, 1072–1078 [DOI] [PubMed] [Google Scholar]
- 43.Shin T. M., Isas J. M., Hsieh C. L., Kayed R., Glabe C. G., Langen R., Chen J. (2008) Mol. Neurodegener. 3, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrell R. W., Mushunje A., Zhou A. (2008) FEBS Lett. 582, 2537–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.