Abstract
Sterol-induced binding to Insigs in the endoplasmic reticulum (ER) allows for ubiquitination of 3-hydroxy-3-methylglutaryl coenzyme A reductase, the rate-limiting enzyme in cholesterol synthesis. This ubiquitination marks reductase for recognition by the ATPase VCP/p97, which mediates extraction and delivery of reductase from ER membranes to cytosolic 26 S proteasomes for degradation. Here, we report that reductase becomes dislocated from ER membranes into the cytosol of sterol-treated cells. This dislocation exhibits an absolute requirement for the actions of Insigs and VCP/p97. Reductase also appears in a buoyant fraction of sterol-treated cells that co-purifies with lipid droplets, cytosolic organelles traditionally regarded as storage depots for neutral lipids such as triglycerides and cholesteryl esters. Genetic, biochemical, and localization studies suggest a model in which reductase is dislodged into the cytosol from an ER subdomain closely associated with lipid droplets.
Keywords: Cholesterol Metabolism, Cholesterol Regulation, Endoplasmic Reticulum (ER), Enzyme Degradation, Proteasome, Ubiquitination
Introduction
The synthesis of cholesterol and nonsterol isoprenoids in mammalian cells is tightly controlled through multiple feedback mechanisms that converge on the rate-limiting enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA)3 reductase, which catalyzes the reduction of HMG-CoA to mevalonate (1). HMG-CoA reductase, a glycoprotein of the endoplasmic reticulum (ER), contains two distinct domains: a hydrophobic N-terminal domain consisting of eight membrane-spanning regions separated by short loops and a hydrophilic C-terminal domain that protrudes into the cytosol where it exerts all catalytic activity (2, 3). Sterol-accelerated ER-associated degradation (ERAD) constitutes one axis of the complex multivalent regulatory system that governs feedback regulation of reductase (4). This degradation requires ubiquitination of reductase, a reaction that results from binding of its membrane domain to one of two ER membrane proteins called Insig-1 and Insig-2 (5, 6). Insigs associate with gp78, a membrane-bound ubiquitin ligase that together with its cognate ubiquitin-conjugating enzyme Ubc7 (or UBE2G2) initiates the ubiquitination of reductase on two cytosolically exposed lysine residues in the membrane domain of the enzyme (7). Once ubiquitinated, reductase is recognized by the gp78-bound VCP/p97, a member of the family of ATPases associated with various cellular activities (AAA-ATPases). VCP/p97 and its associated cofactors Npl4 and Ufd1 have been implicated in the extraction of both soluble and membrane-bound ERAD substrates from ER membranes and their delivery to 26 S proteasomes for degradation (8). Although the precise mechanism for VCP/p97-mediated extraction of ubiquitinated reductase is not completely understood, the reaction appears to be augmented by nonsterol mevalonate-derived end products (9).
ERAD is a highly conserved, multistep process through which misfolded or unassembled proteins, both soluble within the ER lumen and membrane-bound, are selectively degraded by proteasomes in the cytosol (10). Key events in the ERAD pathway involve substrate recognition, polyubiquitination, dislocation from ER membranes into the cytosol, and delivery to proteasomes. It is widely accepted that, after their selection, soluble ERAD substrates are transported across ER membranes into the cytosol through a protein-conducting channel formed by either Sec61, the major component of the translocation channel that imports newly synthesized polypeptides into the ER, or the polytopic Derlin proteins (11–13). Although ER-to-cytosol dislocation of polytopic substrates is an emerging paradigm in the ERAD pathway (14–19), the underlying mechanism for the reaction has not been completely resolved.
In the current study we investigate whether reductase becomes dislocated from ER membranes into the cytosol of sterol-treated Chinese hamster ovary (CHO)-K1 cells. Our results show that reductase becomes dislocated into the cytosol upon sterol treatment; however, this dislocation involves an intermediate step in which reductase is translocated to a buoyant subcellular fraction that co-purifies with lipid droplets. These observations suggest a novel route for delivery of ubiquitinated reductase to cytosolic proteasomes that may be applicable to the ERAD of other polytopic proteins.
EXPERIMENTAL PROCEDURES
Materials
We obtained MG-132 from Boston Biochem (Cambridge, MA) and from Peptide Institute, Inc. (Osaka, Japan); triacsin C was from Sigma; horseradish peroxidase-conjugated donkey anti-mouse, anti-goat, anti-guinea pig, and anti-rabbit IgGs (affinity-purified) were from Jackson ImmunoResearch Laboratories (West Grove, PA); AlexaFluor 568 goat anti-mouse IgG (highly cross-adsorbed) and BODIPY® 493/503 were from Invitrogen; paraformaldehyde 32% solution (EM grade) and Fluoromount G were from Electron Microscopy Sciences (Hatfield, PA); 25-hydroxycholesterol was from Steroloids. Lipoprotein-deficient serum (LPDS, d > 1.215 g/ml) was prepared from newborn calf serum by ultracentrifugation (20), and delipidated fetal calf serum was prepared as described previously (21). All other reagents were obtained from previously described sources (22).
Expression Plasmids
The following plasmids are described in the indicated references: pCMV-Insig-1-Myc, which encodes amino acids 1–277 of human Insig-1 followed by 6 tandem copes of a c-Myc epitope tag under control of the cytomegalovirus promoter (CMV (23)), and wild type and lysine mutant (K89R/K248R) versions of pCMV-HMGCR-(TM1–8)-T7, which encodes amino acids 1–346 of hamster HMG-CoA reductase followed by 3 tandem copies of the T7 epitope tag under control of the CMV promoter (9).
Cell Culture
Stock cultures of CHO-K1 cells were maintained in medium A (1:1 mixture of Ham's F-12 medium and Dulbecco's modified Eagle's medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 5% (v/v) fetal calf serum (FCS). The cells were grown in a monolayer at 37 °C in an 8–9% CO2 incubator.
Transient Transfection
Transfection of CHO-K1 cells in either 60- or 100-mm dishes was carried out using FuGENE 6 (Roche Applied Science) as described previously with minor modifications (5). For transfections in 60-mm dishes, the cells received 1.03 μg of DNA per dish using a ratio of 6 μl of FuGENE 6 to 1.03 μg of DNA in medium A (without antibiotics) in a final volume of 200 μl. For transfections in 100-mm dishes, the cells received 6 μg of DNA per dish using a ratio of 18 μl of FuGENE 6 to 6 μg of DNA in medium A (without antibiotics) in a final volume of 600 μl.
Transfections using the FuGENE HD transfection reagent (Roche Applied Science) were performed in CHO-K1 cells plated in 100-mm dishes. For each transfection, the cells received 6 μg of DNA per dish using a ratio of 18 μl of FuGENE HD to 6 μg of DNA in Opti-MEM I reduced serum medium (Invitrogen) in a final volume of 326 μl. The DNA was diluted in Opti-MEM I before the addition of FuGENE HD. The mixture was incubated for 15 min at room temperature. During this incubation the cells were washed one time with phosphate-buffered saline (PBS) and refed with 5 ml of medium A supplemented with 5% LPDS. The FuGENE HD/DNA mixture (326 μl) was then added to each dish. Conditions of the incubations are described in the legends to Figs. 2 and 6. At the end of the incubations, triplicate dishes of cells were harvested and pooled for analysis.
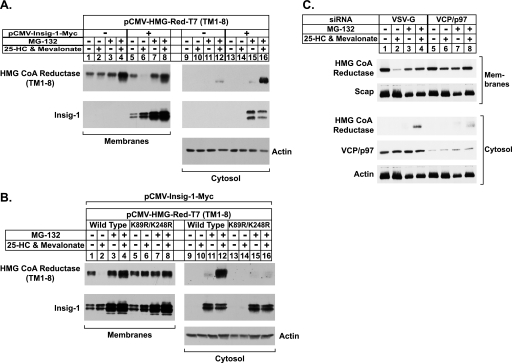
FIGURE 2.
Sterol-mediated dislocation of HMG-CoA reductase into cytosol requires its prior ubiquitination and the actions of Insig-1 and VCP/p97. A and B, CHO-K1 cells were set up for experiments on day 0 at a density of 4 × 105 cells per 60-mm dish in medium A containing 5% FCS. On day 1 cells were transfected in 2 ml of medium A containing 5% LPDS with 1 μg of wild-type or K89R/K248R versions of pCMV-HMG-Red-T7 (TM1–8) in the absence or presence of 30 ng of pCMV-Insig1-Myc as indicated. The total amount of DNA in each dish was adjusted to 1.03 μg per dish by the addition of pcDNA3.1 mock vector. After 3–6 h at 37 °C, the cells received a direct addition of 2 ml of medium A containing 5% LPDS, 20 μm compactin, and 100 μm mevalonate. After incubation for 16 h at 37 °C, cells were switched to medium A containing 5% LPDS and 10 μm compactin in the absence (−) or presence (+) of 1 μg/ml 25-HC, 10 mm mevalonate, and 10 μm MG-132 as indicated. After 5 h at 37 °C, cells were harvested and subjected to cell fractionation as described under “Experimental Procedures.” Aliquots of the membrane (7.5–8 μg of protein/lane) and cytosol (20 μg of protein/lane) fractions were subjected to SDS-PAGE, and immunoblot analysis was carried out with 0.5 μg/ml monoclonal anti-T7 IgG (against reductase), 0.5 μg/ml monoclonal IgG-9E10 (against Insig-1), or 1:2000 dilution of polyclonal anti-actin IgG. C, CHO-K1 cells were set up on day 0 at a density of 2 × 105 cells per 60-mm dish in medium A containing 5% FCS. On day 1 cells were transfected with the indicated siRNA duplexes (100 pmol per dish) as described under “Experimental Procedures.” After transfection, cells were incubated for 16 h at 37 °C in medium A containing 5% LPDS, 10 μm compactin, and 50 μm mevalonate. On day 2 cells were switched to medium A containing 5% LPDS and 10 μm compactin in the absence (−) or presence (+) of 1 μg/ml 25-HC, 10 mm mevalonate, and 10 μm MG-132 as indicated. After 5 h at 37 °C, cells were harvested and subjected to cell fractionation as described under “Experimental Procedures.” Aliquots of the membrane (8 μg protein/lane) and cytosol (30 μg protein/lane) fractions were subjected to SDS-PAGE, and immunoblot analysis was carried out with 5 μg/ml monoclonal IgG-A9 (against reductase), 5 μg/ml polyclonal IgG-R139 (against Scap), 0.625 μg/ml monoclonal IgG-18 (against VCP/p97), and a 1:2000 dilution of polyclonal anti-actin IgG.
FIGURE 6.
Sterol-regulated translocation of HMG-CoA reductase into the lipid droplet fraction precedes its dislocation into the cytosol. A, CHO-K1 cells were set up on day 0 at a density of 5 × 105 cells per 100-mm dish in medium A containing 5% FCS. On day 1 cells were transfected with the indicated duplex of siRNA (120 pmol per dish) as described under “Experimental Procedures.” After transfection, cells were incubated for 16 h at 37 °C in medium A containing 5% LPDS, 10 μm compactin, and 50 μm mevalonate. The cells were then treated in the absence (−) or presence (+) of 10 μm MG-132 in medium A containing 5% LPDS and 10 μm compactin for 1 h at 37 °C followed by treatment in the absence (−) or presence (+) of 1 μg/ml 25-HC, 10 mm mevalonate, and 10 μm MG-132 as indicated. After 5 h at 37 °C, cells were harvested and subjected to cell fractionation as described under “Experimental Procedures.” Aliquots of membrane (5 μg of protein/lane), lipid droplet (5 μg of protein/lane), and cytosol (25 μg of protein/lane) fractions were subjected to SDS-PAGE, and immunoblot analysis was carried out with 4 μg/ml monoclonal IgG-A9 (against reductase) or 0.625 μg/ml monoclonal IgG-18 (against VCP/p97). B and C, CHO-K1 cells were set up for experiments on day 0 at a density of 5 × 105 cells per 100-mm dish in medium A containing 5% FCS. On day 2 cells were transfected in 6 ml of medium A containing 5% LPDS with either 5.82 μg of wild type pCMV-HMG-Red-T7 (TM1–8) and 180 ng of pCMV-Insig1-Myc or 6 μg of pcDNA3.1 mock vector. After 5 h at 37 °C, cells received a direct addition of 2 ml of medium A containing 5% LPDS, 40 μm compactin, and 200 μm mevalonate. After incubation for 16 h at 37 °C, the cells were treated with 10 μm MG-132 for 45 min at 37 °C followed by treatment with 1 μg/ml 25-HC, 10 mm mevalonate, and 10 μm MG-132 for 4 h at 37 °C. Cells were harvested and subjected to subcellular fractionation; the cytosol and lipid droplet fractions were subjected to acetone precipitation, as described under “Experimental Procedures.” In B, aliquots of these fractions (membrane, 0.006% of total; cytosol, 0.06% of total; lipid droplet, 5% of total) were subjected to SDS-PAGE followed by immunoblot analysis with 0.5 μg/ml monoclonal anti-T7 (against reductase). In C, membrane, cytosol, and lipid droplet fractions were dissolved in Buffer B and subjected to incubation in the absence (−) or presence (+) of endoglycosidase Hf (EndoHf; 130 units of enzyme/μg of protein) for 5 h at 37 °C. After incubations, the samples were subjected to SDS-PAGE and immunoblot analysis as in B.
RNA Interference (RNAi)
Duplexes of small interfering RNA (siRNA) were synthesized by Dharmacon/Thermo Fisher Scientific. The siRNA sequence targeting VCP/p97 corresponds to nucleotides 252–270 of mouse VCP/p97 (GenBankTM accession number NM_009503.4) relative to the first nucleotide of its start codon. This sequence is conserved in human, rat, cow, and wolf forms of VCP/p97. The siRNA sequence targeting an irrelevant control gene, vesicular stomatitis virus glycoprotein (VSV-G), was used as a negative control (9).
On day 0, CHO-K1 cells were set up at a density of 2 × 105 or 5 × 105 cells per 60- and 100-mm dish, respectively, in medium A containing 5% FCS. For RNAi experiments conducted in 60-mm dishes, cells were transfected on day 1 with duplexes of siRNA using a ratio of 2 μl of LipofectamineTM RNAiMAX (Invitrogen) to 100 pmol of siRNA duplexes per dish. Cells were washed once with 2 ml of medium A (without antibiotics) and refed 1.6 ml of medium A (without antibiotics) per dish. Duplexes of siRNA were diluted in Opti-MEM I reduced serum medium (Invitrogen) to a final volume of 370 μl per dish. LipofectamineTM RNAiMAX was diluted with Opti-MEM I to a final volume of 30 μl per dish and incubated at room temperature for 5 min. The siRNA and LipofectamineTM RNAiMAX mixtures were combined together and further incubated for 10–20 min at room temperature. Subsequently, the siRNA/LipofectamineTM RNAiMAX mixture (400 μl) was added to each dish. For RNAi experiments in 100-mm dishes, cells were transfected on day 1 with 120 pmol of siRNA duplexes per dish using identical methods, with the exception that all reagent volumes were tripled. Incubation conditions are described in the legends to Figs. 2 and 6. After incubations, triplicate dishes of cells were harvested and pooled for analysis.
Cell Fractionation and Immunoblot Analysis
The pooled cell pellets from triplicate dishes of cells were resuspended in 500–525 μl of buffer A (10 mm HEPES-KOH (pH 7.4), 10 mm KCl, 1.5 mm MgCl2, 5 mm sodium EDTA, 5 mm sodium EGTA, 250 mm sucrose, 5 mm dithiothreitol, and 0.1 mm leupeptin) supplemented with protease inhibitor mixture consisting of 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 0.5 mm Pefabloc, 10 μg/ml leupeptin, 5 μg/ml pepstatin A, 25 μg/ml N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal, and 10 μg/ml aprotinin. The cell suspension was passed through a 22.5-gauge needle 30 times and subjected to centrifugation at 1000 × g for 7 min at 4 °C. The supernatant from this spin (designated post-nuclear supernatant) was then subjected to centrifugation at 100,000 × g (Beckman Coulter TLA-120.1 rotor, 5.5 × 104 rpm) for 45 min at 4 °C. The 100,000 × g supernatant (designated cytosolic fraction) was removed and transferred to a new tube, whereas the 100,000 × g pellet (designated membrane fraction) was resuspended in 100 μl of Buffer B (10 mm Tris-HCl (pH 6.8), 1% (w/v) SDS, 100 mm NaCl, 1 mm EDTA, 1 mm EGTA). The cytosolic fraction was mixed with 5× volume of chilled acetone, incubated at −20 °C overnight, and subsequently subjected to centrifugation at 16,000 × g for 15 min at 4 °C. After removal of the supernatant, the pellet was allowed to air-dry and then resuspended in 40–60 μl of Buffer B.
The buoyant lipid droplet fraction appears as a white opaque material at the meniscus of the supernatant after centrifugation of the post-nuclear supernatant at 100,000 × g; this fraction was isolated as follows. After subjecting the post-nuclear supernatant to centrifugation at 100,000 × g, the lipid droplet fraction was gently mixed back into the supernatant using a pipette tip and transferred together with the supernatant into an Eppendorf tube (400 μl). After centrifugation at 16,000 × g for 8–10 min, the original cytosolic fraction was removed from the bottom of the tube using a gel-loading tip until 40 μl of supernatant enriched for the lipid droplet fraction remained. Both the cytosol and lipid droplet fractions were mixed with a 5× volume of chilled acetone and incubated at −20 °C overnight. Precipitated material was pelleted by centrifugation at 16,000 × g for 15 min at 4 °C. After removal of the supernatant, the pellets were allowed to air-dry, after which they were resuspended Buffer B (25–30 μl for the lipid droplet fraction and 40–60 μl for the cytosolic fraction).
The protein content of the membrane, cytosol, and lipid droplet fractions were determined using the BCA protein assay kit (Thermo Scientific). Aliquots of the fractions were mixed with an equal volume of buffer containing 62.5 mm Tris-HCl (pH 6.8), 15% (w/v) SDS, 8 m urea, 10% (v/v) glycerol, and 100 mm dithiothreitol, after which 4× SDS loading buffer was added to a final concentration of 1×.
All fractions were subjected to SDS-PAGE and immunoblot analysis as described (5). After SDS-PAGE on 8% or 10% gels, proteins were transferred to Hybond C-Extra nitrocellulose filters (Amersham Biosciences) or to Trans-Blot Transfer Medium supported nitrocellulose membranes (Bio-Rad). The filters were incubated with the antibodies described below and in the figure legends. Bound antibodies were visualized with peroxidase-conjugated donkey anti-mouse, anti-rabbit, anti-goat, or anti-guinea pig IgG using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) according to the manufacturer's instructions. Gels were calibrated with prestained molecular mass markers (Bio-Rad). Filters were exposed to x-ray film at room temperature.
Primary antibodies used for immunoblotting were as follows: IgG-A9, a mouse monoclonal antibody against the catalytic domain of hamster reductase (amino acids 450–887) (24); IgG-7D4, a mouse monoclonal antibody against the N-terminal portion of hamster SREBP-2 (25); IgG-9D5, a mouse monoclonal antibody against hamster Scap, and IgG-R139, a rabbit polyclonal antibody against hamster Scap (26); mouse monoclonal anti-Myc (IgG fraction) from the culture medium of hybridoma clone 9E10 (American Type Culture Collection); IgG-740F, a rabbit polyclonal against human gp78 (27); IgG-AP125, a mouse monoclonal against adipose differentiation-related protein (ADRP; Fitzgerald 10R-A117a); a guinea pig polyclonal antibody against ADRP (Fitzgerald 20R-AP002); IgG-18, a mouse monoclonal antibody against VCP/p97 (BD Transduction Laboratories V39620); T7·Tag® mouse monoclonal antibody (IgG2b) against the T7 epitope tag (EMD Biosciences 69522-4); a rabbit polyclonal antibody against actin (Sigma A2066); a rabbit polyclonal antibody against CYP2C9 (Lifespan Biosciences LS-C14768); a rabbit polyclonal antibody against AUP1 (Sigma HPA007674); a goat polyclonal antibody against UBXD8/ETEA (Novus Biologicals, NB100-1296).
RESULTS
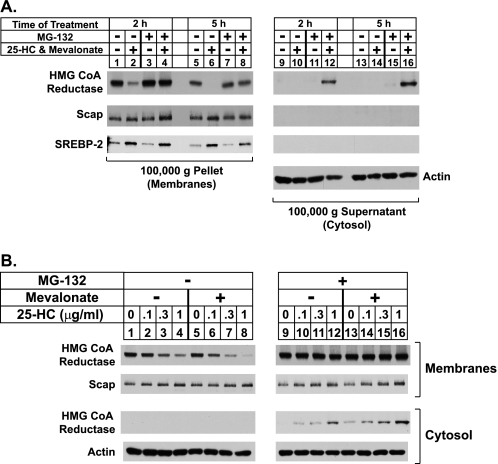
Fig. 1 shows an experiment in which CHO-K1 cells were first depleted of sterols through incubation in medium containing lipoprotein deficient serum, the reductase inhibitor compactin (28), and a low level of mevalonate (50 μm) that allows synthesis of essential nonsterol isoprenoids but not of cholesterol (4). After sterol depletion, the cells were treated with various combinations of the regulatory oxysterol 25-hydroxycholesterol plus a high concentration (10 mm) of mevalonate and the proteasome inhibitor MG-132. The cells were subsequently harvested and lysed in the absence of detergents for the preparation of post-nuclear supernatants, which were subjected to centrifugation at 100,000 × g. The pellet and supernatant fractions of this spin, designated membrane and cytosolic fractions, respectively, were resolved by SDS-PAGE and analyzed by immunoblotting with anti-reductase antibody. Immunoblotting revealed that treatment of the cells with 25-hydroxycholesterol plus mevalonate for 2 and 5 h caused the disappearance of reductase from the pellet fraction (Fig. 1A, top panel, lanes 2 and 6), indicating sterol-accelerated degradation of the protein from ER membranes. Reductase was not recovered in the cytosol regardless of the absence or presence of 25-hydroxycholesterol plus mevalonate (lanes 9, 10, 13, and 14). As expected, MG-132 blocked the sterol-induced disappearance of reductase from membranes (lanes 4 and 8) and caused the appearance of the enzyme in the cytosolic fraction of cells treated with 25-hydroxycholesterol plus mevalonate (lanes 12 and 16). This soluble form of reductase recovered from the supernatant appeared as a sharp band that migrated on SDS-PAGE gels with a molecular weight similar to that of reductase in membranes (data not shown). Control immunoblots revealed that expression of actin in the cytosol was constant under all conditions (lanes 9–16), and ER proteins Scap and sterol regulatory element-binding protein-2 (SREBP-2) were exclusively found in the membrane fraction (lanes 1–16).
FIGURE 1.
Sterol-mediated dislocation of HMG-CoA reductase from ER membranes into the cytosol of CHO-K1 cells. A and B, CHO-K1 cells were set up for experiments on day 0 at a density of 5 × 105 cells per 100-mm dish in medium A containing 5% FCS. On day 2 the cells were washed with PBS and refed medium A supplemented with 5% LPDS, 10 μm sodium compactin, and 50 μm sodium mevalonate. After 16 h at 37 °C, cells were switched to medium A containing 5% LPDS, 10 μm compactin, and the indicated additions. A, cells were treated in the absence (−) or presence (+) of 1 μg/ml 25-HC, 10 mm mevalonate, and 10 μm MG-132 as indicated. After incubation at 37 °C for 2 h (lanes 1–4 and 9–12) or 5 h (lanes 5–8 and 13–16), the cells were harvested and subjected to cell fractionation as described under “Experimental Procedures.” Aliquots of 100,000 × g pellet (7.5 μg protein/lane) and supernatant (20 μg protein/lane) fractions, designated membranes and cytosol, respectively, were subjected to SDS-PAGE and transferred to supported nitrocellulose membranes followed by immunoblot analysis with 5 μg/ml monoclonal IgG-A9 (against reductase), 5 μg/ml polyclonal IgG-R139 (against Scap), 5 μg/ml monoclonal IgG-7D4 (against SREBP-2), or 1:2000 dilution of polyclonal anti-actin IgG. B, cells were incubated with various concentrations of 25-HC in the absence (−) or presence (+) of 10 μm MG-132 and 10 mm mevalonate as indicated. After 5 h at 37 °C, the cells were harvested and subjected to cell fractionation followed by immunoblot analysis as in A.
The experiment shown in Fig. 1B was designed to examine the contribution of 25-hydroxycholesterol and mevalonate-derived nonsterol isoprenoids to the appearance of reductase in the cytosol of cells. In the absence of MG-132, 25-hydroxycholesterol caused a modest decrease in the amount of reductase recovered from the membrane fraction (Fig. 1B, lanes 1–4); this decrease was significantly enhanced by the addition of 10 mm mevalonate (lanes 5–8). Reductase was not recovered from the supernatant fraction in the absence of MG-132 (lanes 1–8). Treatment with MG-132 prevented the sterol-dependent disappearance of reductase from membranes, which was accompanied by the appearance of the enzyme in the cytosol of cells treated with the highest level (1 μg/ml) of 25-hydroxycholesterol (lane 12). Similar to the results for degradation, mevalonate enhanced the 25-hydroxycholestrol-induced appearance of soluble reductase in the cytosolic fraction (compare lanes 9–12 with 13–16). Thus, nonsterol mevalonate-derived products enhance not only sterol-induced degradation of reductase from membranes but also the sterol-induced appearance of the enzyme in the cytosol of cells.
To examine the role of Insig in dislocation of reductase from ER membranes into the cytosol, cells were transfected with the expression plasmid pCMV-HMG-Red-T7 (TM1–8) encoding the entire membrane domain of hamster reductase followed by three tandem copies of the T7 epitope tag. This protein resisted sterol-induced ERAD when transfected alone (Fig. 2A, top panel, compare lanes 1 and 2); the addition of MG-132 had no effect on this (lanes 3 and 4). Co-expression of pCMV-Insig-1-Myc encoding amino acids 1–277 of human Insig-1 followed by 6 tandem copies of the c-Myc epitope tag conferred sterol-accelerated ERAD upon the membrane domain of reductase (lanes 5 and 6). This degradation was blocked when the cells were also treated with MG-132 (lanes 7 and 8). A faint band corresponding to the membrane domain of reductase was detected in the cytosol of cells that were treated with 25-hydroxycholesterol/mevalonate and MG-132 (lane 12). The appearance of this band was markedly enhanced when Insig-1 was co-expressed (lane 16).
Insig-1 is known to be subject to proteasome-mediated ERAD from membranes (29). When overexpressed to high levels in cells, this degradation is constitutive and blocked by MG-132. Consistent with this, Insig-1 became stabilized by MG-132 (Fig. 2A, compare lanes 5 and 6 with lanes 7 and 8) and was dislocated into the cytosol of cells (lanes 15 and 16).
Substitutions of arginine for lysines 89 and 248 in the membrane domain prevent sterol-mediated ubiquitination of reductase and block its degradation, implicating these residues as sites for sterol-regulated modification (9). To examine a role for ubiquitination in the sterol-regulated appearance of reductase in the cytosol, cells were transfected with wild type or K89R/K248R versions of pCMV-HMG-Red-T7 (TM1–8) together with pCMV-Insig-1-Myc (Fig. 2B). As expected, the membrane domain of reductase disappeared from the membrane fraction when the cells were treated with the combination of 25-hydroxycholesterol plus mevalonate (Fig. 2B, lanes 1 and 2). This disappearance was blocked by MG-132 (lanes 3 and 4), which also caused the appearance of the membrane domain of reductase in the cytosolic fraction when cells were also treated with 25-hydroxycholesterol and mevalonate (lane 12). In contrast, the K89R/K248R mutant of the membrane domain of reductase resisted sterol-induced degradation (lane 6), and the protein was not found in the supernatant (lane 16). Note that Insig-1 was present in the cytosol upon MG-132 treatment regardless of whether the wild type or mutant version of the membrane domain of reductase was co-expressed (lanes 11, 12, 15, and 16).
We next conducted an RNAi experiment to demonstrate a role for VCP/p97 in the cytosolic dislocation of reductase. In cells transfected with control siRNAs targeting VSV-G, a control gene that is not expressed in the cells, 25-hydroxycholesterol plus mevalonate caused endogenous reductase to become degraded from membranes (Fig. 2C, lanes 1 and 2). This degradation was blocked by MG-132 (lanes 3 and 4), which caused the appearance of reductase in the cytosol of 25-hydroxycholesterol plus mevalonate-treated cells (lane 4). RNAi-mediated knockdown of VCP/p97 blunted both the sterol-accelerated ERAD of reductase from membranes (lane 6) and cytosolic dislocation of the enzyme into the cytosol (lane 8).
Having established that sterols trigger cytosolic dislocation of reductase, we contemplated potential mechanisms for the reaction. Several lines of evidence suggest that cytosolic organelles called lipid droplets, which consist of neutral lipids such as triglycerides and cholesteryl esters enclosed by a phospholipid monolayer, may play a role in ERAD. For example, apolipoprotein B-100, the primary protein of very low density lipoproteins, is subjected to ERAD without appropriate lipidation (30). When proteasome activity is blocked, degradation of apolipoprotein B-100 is blunted, and the protein accumulates on lipid droplets (31). Proteomic studies have identified several chaperones as well as VCP/p97 and its membrane receptor UBXD8 as lipid droplet-associated proteins (32, 33). Finally, a recent genome-wide RNAi screen revealed a potential role for the ubiquitin-proteasome pathway in the formation of lipid droplets in insect cells (34).
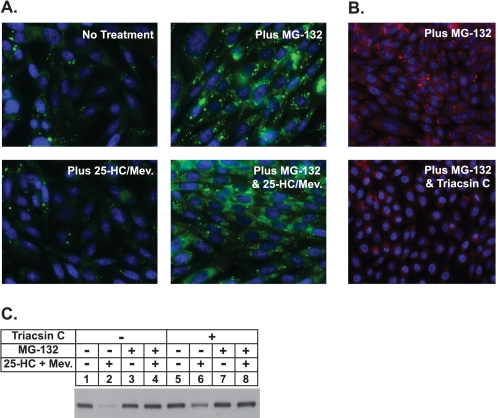
In Fig. 3A, cells treated with various combinations of 25-HC plus mevalonate and MG-132 were fixed, permeabilized, and subjected to staining with Hoechst reagent to visualize nuclei and with the neutral lipid-specific dye BODIPY to visualize lipid droplets. The results show very few lipid droplets scattered throughout the cytosol of untreated cells. MG-132 treatment caused a significant intracellular accumulation of lipid droplets that was inhibited by triacsin C (Fig. 3B), an acyl-CoA synthetase inhibitor that prevents incorporation of free fatty acids into triglycerides (35). Significantly, triacsin C also blunted sterol-accelerated degradation of reductase (Fig. 3C, lanes 4 and 6), indicating that the presence of lipid droplets is necessary for the reaction.
FIGURE 3.
Proteasome inhibition stimulates formation of lipid droplets in CHO-K1 cells. A, on day 0 CHO-K1 cells were set up on glass coverslips at a density of 2 × 105 cells per well of 6-well plates in medium A containing 5% FCS. On day 1 cells were refed medium A supplemented with 5% LPDS, 10 μm compactin, and 50 μm mevalonate (Mev.). After 16 h at 37 °C, cells were switched to medium A containing 5% LPDS and 10 μm compactin in the absence or presence of 10 μm MG-132 and 1 μg/ml 25-hydroxycholesterol plus 10 mm mevalonate as indicated. After incubation for 5 h at 37 °C, cells were washed twice with PBS, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and stained with 10 μg/ml BODIPY 493/503 (green) to visualize lipid droplets or Hoechst reagent (blue) to visualize nuclei using standard procedures. Images were acquired with Zeiss AxioPlan 2E (Thornwood, NY) and a Hamamatsu monochrome camera (63× objective, Bridgewater, NJ). B and C, on day 0 CHO-K1 cells were plated at 2 × 105 cells per well of 6-well plates with glass coverslips or at 4 × 105 cells per 100 mm dish in medium A containing 5% FCS. On day 2 cells were refed medium A supplemented with 5% delipidated fetal calf serum, 10 μm compactin, and 50 μm mevalonate. After 16 h at 37 °C, cells were switched to medium A containing 5% LPDS and 10 μm compactin without (−) or with (+) 9.6 μm triacsin C. After 3.25 h at 37 °C, cells were treated in the absence (−) or presence (+) of 9.6 μm triacsin C, 10 μm MG-132, and 1 μg/ml 25-HC plus 10 mm mevalonate as indicated. B, after 4 h at 37 °C cells on the coverslips were stained with Oil Red O and Hoechst by standard methods. C, cells on the 100-mm dishes were harvested after 4 h at 37 °C, and membrane fractions were obtained as described under “Experimental Procedures.” Aliquots of the membrane fractions (10 μg protein/lane) were subjected to SDS-PAGE and transferred to supported nitrocellulose membranes followed by immunoblot analysis with 5 μg/ml monoclonal IgG-A9 (against reductase).
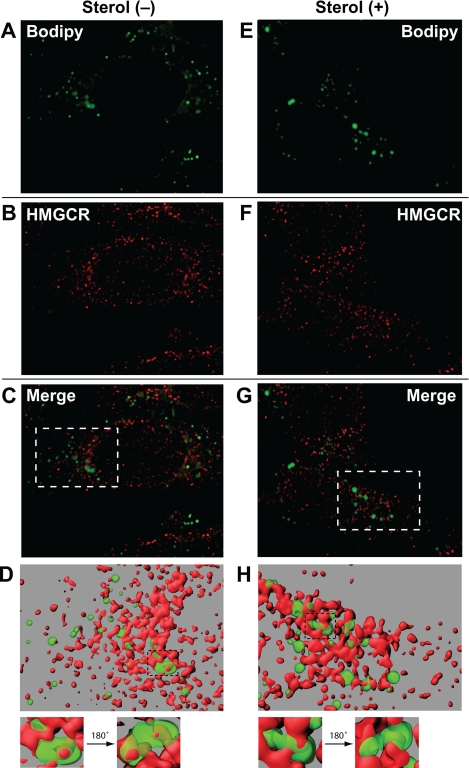
The results of Fig. 3 prompted us to next determine whether physical association exists between reductase and MG-132-induced lipid droplets. Immunofluorescence microscopy revealed that almost all lipid droplets in MG-132-treated cells were in close apposition with reductase-positive ER membrane structures (Fig. 4). In fact, several reductase-positive membrane structures protruding into the interior of lipid droplets were observed along with a smaller number of instances in which membrane structures passed completely through lipid droplets (panels D and H, magnified areas, supplemental movies). However, treatment with 25-hydroxycholesterol plus mevalonate did not lead to detectable changes in the association between reductase and lipid droplets.
FIGURE 4.
Association of MG-132-induced lipid droplets with ER membranes containing HMG-CoA reductase. On day 0 CHO-K1 cells were plated at 5 × 104 cells per well of 12-well plates with glass coverslips. On day 1 cells were refed medium A supplemented with 5% LPDS, 10 μm compactin, and 50 μm mevalonate. After 16 h at 37 °C, cells were switched to medium A containing 5% LPDS, 10 μm compactin, and 10 μm MG-132 in the presence or absence of 1 μg/ml 25-hydroxycholesterol plus 10 mm mevalonate as indicated. After incubation for 5 h at 37 °C, cells were washed twice with PBS, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and stained with 10 μg/ml BODIPY 493/503 to visualize lipid droplets (green) and with 50 μg/ml IgG-A9 with 5 μg/ml AlexaFluor 568 goat anti-mouse IgG to visualize reductase (HMGCR; red) using standard procedures. Images were taken using a Deltavision pDV deconvolution microscope (Bitplan, Inc., St. Paul, MN) with an Olympus UPlanApo 100× oil immersion objective. Z slices were taken at 0.2-μm intervals through the thickness of the cell (∼25 slices) and were deconvolved using SoftWoRx software using the default settings. The middle five slices of the deconvolved Z stack were used to generate surface volume renderings using Imaris software (Bitplane, Inc.). Segmentation was carried out interactively to obtain surfaces that most closely matched the localization of lipid droplets and reductase staining. The green surface was rendered at 45% transparency to reveal red structures contained within green structures. The maximum intensity projection of the Z slices are shown in A–C and E–G. The surface volume renderings are shown in D and H.
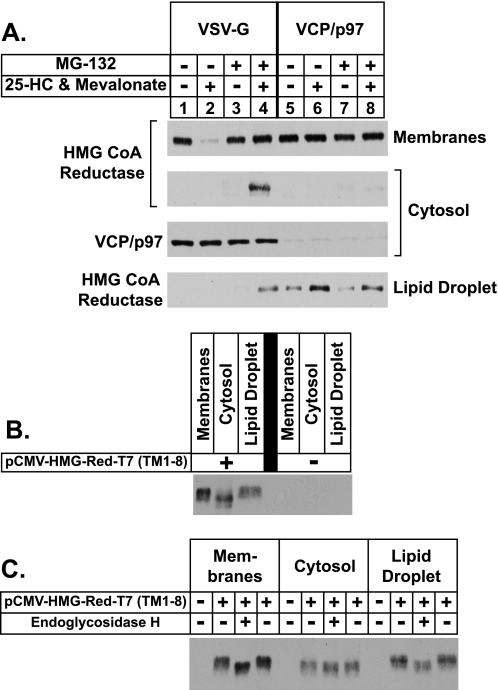
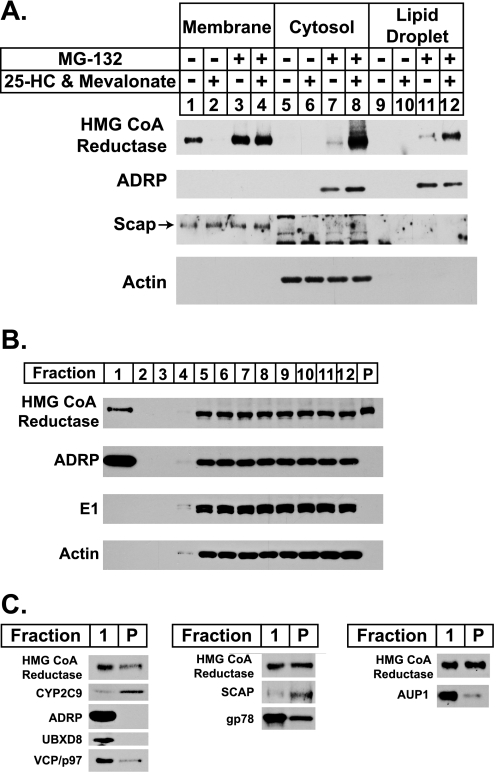
We next investigated whether any sterol-mediated changes in the association between reductase and lipid droplets could be detected through biochemical approaches. Lipid droplets contain a relatively low protein content compared with their high content of neutral lipids (36). This property renders lipid droplets more buoyant than other organelles such as the ER, Golgi, and plasma membrane, thereby allowing their isolation by high speed centrifugation. In the experiment of Fig. 5A, cells were treated with various combinations of 25-hydroxycholesterol plus mevalonate and MG-132, after which they were harvested and lysed, and post-nuclear supernatants were subjected to 100,000 × g centrifugation. Aliquots of the resulting membrane, cytosol, and lipid droplet fractions were subjected to SDS-PAGE and analyzed by immunoblotting with anti-reductase antibody. As expected, 25-hydroxycholesterol plus mevalonate stimulated the degradation of reductase (Fig. 5A, lanes 1 and 2); this degradation was inhibited by the inclusion of MG-132 in the treatment medium (lanes 3 and 4). In the presence of the proteasome inhibitor, reductase was detected not only in the cytosol of sterol-treated cells but also in the lipid droplet fraction (lanes 8 and 12). The lipid droplet marker, ADRP, was distributed in the cytosol and lipid droplet fractions but only when cells were treated with MG-132 (lanes 7, 8, 11, and 12). This result is consistent with previous observations that ADRP is subject to proteasome-mediated degradation (37, 38). The lack of detectable ADRP in the absence of MG-132 correlates with the low levels of lipid droplets observed in the cells by microscopy under equivalent conditions (see Fig. 3A). The control proteins Scap and actin were exclusively localized to the membrane and cytosolic fractions as expected (lanes 1–4 and 5–8, respectively).
FIGURE 5.
Sterol-regulated localization of HMG-CoA reductase to the buoyant, ADRP-enriched lipid droplet fraction. A and B, CHO-K1 cells were set up for experiments on day 0 at a density of 5 × 105 cells per 100-mm dish in medium A containing 5% FCS. On day 2 cells were washed with phosphate-buffered saline and refed medium A with 5% LPDS, 10 μm compactin, and 50 μm mevalonate. A, after incubation for 16 h at 37 °C, cells were switched to medium A containing 5% LPDS and 10 μm compactin in the absence (−) or presence (+) of 1 μg/ml 25-HC, 10 mm mevalonate, and 10 μm MG-132 as indicated. After 5 h at 37 °C, cells were washed with PBS, harvested, and lysed by Dounce homogenization (40 strokes). The resulting lysates were subjected to centrifugation at 1000 × g for 10 min at 4 °C to obtain a post-nuclear supernatant, which was transferred to a new tube and subjected to an additional centrifugation step at 250,000 × g for 30 min. The top 100 μl of the resulting supernatant was designated the lipid droplet fraction, 700 μl of the underlying supernatant was designated the cytosol, and the pellet (P) was designated the membrane fraction. Aliquots of membrane (0.5 μg of protein/lane), cytosol (20 μg of protein/lane), and lipid droplet (4 μg of protein/lane) fractions were subjected to SDS-PAGE and analyzed by immunoblot analysis with 5 μg/ml monoclonal IgG-A9 (against reductase), 1:500 dilution of monoclonal IgG-AP125 (against ADRP), 5 μg/ml polyclonal IgG-R139 (against Scap), and 1:2000 dilution of polyclonal anti-actin IgG. B, 60 plates of CHO-K1 cells were treated with 10 μm MG-132 for 45 min at 37 °C followed by treatment with 1 μg/ml 25-HC, 10 mm mevalonate, and 10 μm MG-132 for an additional 4 h at 37 °C. Cells were then harvested, and lysates were prepared in 10 ml of Buffer A as described under “Experimental Procedures.” The resulting lysates were centrifuged at 1000 × g for 7 min to obtain a post-nuclear supernatant, which was transferred to an ultracentrifuge tube, overlaid with Buffer A containing a reduced concentration of sucrose, and subjected to centrifugation at 150,000 × g for 1 h. The top 2 ml of the supernatant (fraction 1) containing of the lipid droplet fraction was isolated using a tube slicer. The remainder of the supernatant was fractionated by removing 1-ml aliquots from the top of the tube (fractions 2–12). The lipid droplet fraction was concentrated through centrifugation at 16,000 × g for 8 min and removal of the translucent infranatant. All supernatant fractions were precipitated with acetone and resuspended in Buffer B. The membrane pellet fraction (P) was resuspended in 2 ml of Buffer B. Aliquots of each fraction (fraction 1: 4.4% of total; fraction 2–12: 1.58% of total; fraction P: 0.0075% of total) were subjected to SDS-PAGE, and immunoblot analysis was carried out with 4 μg/ml monoclonal IgG-A9 (against reductase), 1:2000 dilution of guinea pig polyclonal anti-ADRP antibody, 1.67 μg/ml rabbit polyclonal anti-E1 IgG, and 1:2000 dilution of rabbit polyclonal anti-actin IgG (Sigma A2066) as indicated. C, the lipid droplet fraction (Fraction 1) and the membrane pellet fraction (P) from B were loaded for SDS-PAGE such that the two would yield equivalent signal intensity when transferred to nitrocellulose filters and immunoblotted with 4 μg/ml of monoclonal IgG-A9 (against reductase). The nitrocellulose filters were also immunoblotted with 1:500 dilution of rabbit polyclonal anti-CYP2C9 antibody, 1:2000 dilution of guinea pig polyclonal anti-ADRP antibody, 0.5 μg/ml goat polyclonal anti-UBXD8/ETEA IgG, 0.625 μg/ml monoclonal IgG-18 (against VCP/p97), 20 μg/ml monoclonal IgG-9D5 (against Scap), 2 μg/ml rabbit polyclonal IgG-740F (against gp78), and 0.05 μg/ml rabbit polyclonal anti-AUP1 antibody.
The subcellular localization of reductase in cells treated with 25-hydroxycholesterol, mevalonate, and MG-132 was next examined using a single-step, discontinuous sucrose gradient (36). Immunoblotting revealed the presence of reductase in the top, lipid droplet-enriched fraction of the gradient (Fig. 5B, lane 1) as well as the cytosol (fractions 5–12) and membrane pellet (P) fractions. Reductase was absent in the fractions composed of the buffer overlay (fractions 2–4). The ubiquitin-activating enzyme (E1) and actin were found in the cytosol fraction and excluded from the lipid droplet, membrane pellet, and buffer overlay fractions. ADRP exhibited a similar distribution to that of reductase except that it was not found in the pellet fraction. Relative to reductase, the buoyant lipid droplet fraction was enriched for UBXD8, VCP/p97, AUP1, and gp78 (Fig. 5C), which are known to play key roles in ERAD (39). Scap and the P450 enzyme CYP2C9 (40) were found in the membrane pellet and were largely excluded from the lipid droplet fraction.
The role of VCP/p97 in the sterol-regulated appearance of reductase in the lipid droplet fraction was addressed in the RNAi experiment shown in Fig. 6A. In control-transfected cells, 25-hydroxycholesterol plus mevalonate stimulated reductase degradation from membranes (Fig. 6A, lane 2). This degradation was blocked by MG-132 treatment, which also led to the appearance of reductase in the cytosol and lipid droplet fractions (lane 4). Knockdown of VCP/p97 blocked reductase degradation (lane 6) and prevented dislocation of the protein into the cytosol (lane 8). However, reductase was present in the lipid droplet fraction in a sterol-regulated manner even in the absence of MG-132 (lane 5–8). This finding indicates that reductase is translocated to a lipid droplet-associated region of the ER before its VCP/p97-mediated extraction into the cytosol for degradation.
Throughout our studies we noticed that cytosolic reductase migrated at a slightly lower apparent molecular weight on SDS-PAGE gels compared with reductase in the membrane and lipid droplet fractions. Fig. 6B shows that, indeed, the membrane domain of reductase in the membrane and lipid droplet fractions has a decreased mobility compared with that of the protein found in the cytosol. Like many secretory and membrane proteins, reductase in N-glycosylated, a single high mannose N-glycan, localizes to the lumenal loop between transmembrane helices 7 and 8 of the enzyme (41). Removal of N-glycans by a cytosolic N-glycanase is a hallmark of ERAD substrates that have become dislocated into the cytosol (42). To determine whether cytosolic reductase is deglycosylated, we subjected subcellular fractions of cells transfected with the membrane domain of reductase and treated with 25-hydroxycholesterol, mevalonate, and MG-132 to incubation with endoglycosidase H, an enzyme that removes high mannose N-glycans from proteins (43). The results show that reductase was sensitive to endoglycosidase H digestion when localized to the membrane and lipid droplet fractions but not when the protein was present in the cytosol (Fig. 6C).
DISCUSSION
Dislocation of ERAD substrates from ER membranes into the cytosol is a well established phenomenon that occurs not only for soluble substrates contained entirely within the ER lumen but also for substrates with single or multiple membrane-spanning segments (10, 11). This dislocation event is thought to allow full access of the ubiquitinated ERAD substrate to proteasomes for proteolytic degradation. The current study examines the polytopic HMG-CoA reductase, which is subjected to accelerated ERAD when intracellular levels of certain sterols rise (5). In our studies sterol treatment caused the appearance of full-length reductase in the 100,000 × g supernatant of cells incubated with the proteasome inhibitor MG-132 (Fig. 1A). Biochemical analysis revealed this subcellular fraction to exhibit characteristics of the cytosol, namely, the absence of ER proteins Scap and the uncleaved precursor of SREBP-2 and the presence of known cytosolic proteins actin and the E1 ubiquitin-activating enzyme (Figs. 1 and 5B). Importantly, sterol-induced dislocation of reductase into the cytosol was found to be augmented by nonsterol isoprenoids derived from mevalonate (Fig. 1B), and the reaction exhibited an absolute requirement for ubiquitination of reductase and the actions of Insig-1 and VCP/p97 (Fig. 2). Taken together, these observations strongly argue that sterol-accelerated ERAD and cytosolic dislocation of reductase are causally linked events.
The data summarized above confirm previous observations in the yeast Saccharomyces cerevisiae and in mammalian cells that reductase becomes dislocated from ER membranes into the cytosol (18, 19). However, these data extend previous observations by providing biochemical evidence that lipid droplets play a role in sterol-accelerated ERAD of reductase. Based on our studies, we envision a model in which sterols trigger translocation of reductase to a buoyant, lipid droplet-associated subdomain of the ER from which the protein becomes extracted into the cytosol through the action of VCP/p97. Once in the cytosol, reductase becomes deglycosylated and subsequently degraded by proteasomes. The current study provides three lines of evidence that support this model. First, inhibition of proteasome function with MG-132 caused accumulation of lipid droplets (Fig. 3A). These lipid droplets were composed of triglycerides and cholesteryl esters (supplemental Fig. 1 and Table 1); their formation was blocked by triacsin C (Fig. 3B). Importantly, triacsin C also blocked sterol-accelerated ERAD of reductase (Fig. 3C), indicating that the presence of lipid droplets is required for the reaction. Second, although knockdown of VCP/p97 prevented sterol-induced degradation and cytosolic dislocation of reductase (Fig. 2C), the treatment did not prevent its sterol-regulated translocation to the lipid droplet fraction (Fig. 6). This translocation occurred even in the absence of proteasome inhibition, indicating that lipid droplet-associated reductase is not accessible to proteasomes in the absence of VCP/p97 and that the lipid droplet fraction may be the true locus of VCP/p97 activity. This notion is supported by the enrichment of VCP/p97 and its membrane recruitment factor UBXD8 in the lipid droplet fraction (Fig. 5C). Third, reductase localized to the membrane, and lipid droplet fractions retain its N-linked glycan, whereas the protein in the cytosol does not (Fig. 6C). Considering that deglycosylation of proteins by the cytosolic N-glycanase is an irreversible reaction, this observation provides strong evidence that translocation of reductase to a lipid droplet-associated subdomain of the ER precedes dislocation of the enzyme into the cytosol.
The precise subcellular structure the MG-132-induced lipid droplet fraction represents remains to be fully elucidated. Nearly all lipid droplets in the MG-132-treated cells were in close association with ER membranes that contain reductase (Fig. 4). This finding is consistent with numerous studies that report close association between lipid droplets and ER membranes across a variety of mammalian cell lines (for review, see Refs. 45 and 46). A variety of distinct models regarding the association between ER membranes and lipid droplets are suggested in these studies. Some simply report the spatial association of lipid droplets with ER membranes, implicitly assuming them to be discrete entities (47, 48). Other studies provide evidence for continuity between the surface phospholipid monolayer of the lipid droplet and the cytosolic leaflet of the ER membrane (49, 50), suggesting that lipid droplets can be viewed as specialized subdomains of the ER (45). Protrusions of membrane structures into the outer edges of lipid droplets have been reported (51), and remarkably, ribosomes as well as extensive ER-like membranes have been detected within the core of lipid droplets (52). Our imaging studies reveal that the ER membranes are not simply in close apposition to lipid droplets but, rather, protrude into them. Portions of the ER membranes in close proximity to lipid droplets are quite often partially embedded within them in a fist-in-palm-like conformation (Fig. 4, D and H). In some cases the ER even passes completely through the interior of a lipid droplet (Fig. 4, D and H, magnified areas, supplemental movies). Fluorescence imaging of biochemically isolated lipid droplets may aid in further characterization of the nature of association between ER membranes and lipid droplets. Further investigation will also reveal whether this type of association between ER membranes and lipid droplets, made particularly visible in our study through the generation of three-dimensional models from optical slices, is a common theme in the subcellular localization of lipid droplets or whether it is a peculiarity of lipid droplets in MG-132-treated CHO-K1 cells.
The results of imaging studies summarized above coupled with the presence of reductase in the lipid droplet fraction of cells supports a model in which sterol repletion induces the partitioning of reductase to lipid droplet-associated subdomains of the ER. Presumably, these structures are revealed in our fluorescence imaging data (see Fig. 4), and they appear to be enriched for components of the ERAD pathway (see Fig. 5C). Imaging techniques that afford higher resolution, such as immunogold electron microscopy, will be utilized in future studies to fully elucidate the nature of association between reductase and lipid droplets.
In a previous study MG-132-mediated inhibition of proteasomes stabilized ADRP and led to a significant increase of intracellular triglycerides, key components of lipid droplets (38). This finding may help to explain the unexpected observation in this study that proteasome inhibition leads to the accumulation of lipid droplets in cells (see Fig. 3). Whether this accumulation is reflective of increased synthesis or suppressed catabolism of triglycerides and/or other lipid droplet components (e.g. phospholipids, cholesteryl esters, etc.) remains to be determined. An intriguing alternative possibility is that MG-132-induced accumulation of lipid droplets results from the disruption of an intermediate step in the ERAD pathway. This would be consistent with the finding that gp78, VCP/p97, UBXD8, and AUP1 are all enriched in the lipid droplet fraction. It should be noted that the enrichment of gp78 in lipid droplets indicates that this subcellular compartment may be the site for ubiquitination of reductase and other substrates. Future studies will focus on determining whether translocation to the lipid droplet fraction as an intermediate step in proteasomal delivery is a peculiarity of reductase or whether it represents a general phenomenon in the degradation of ERAD substrates as has been proposed by Ploegh (44).
Supplementary Material
Acknowledgments
We thank Drs. Michael S. Brown and Joseph L. Goldstein for continued encouragement and insightful advice. We also thank Tammy Dinh, Moriah Scarbrough, and Kristi Garland for excellent technical assistance, Dr. Lina Abi-Mosleh for assistance with neutral lipid analysis of lipid droplets, and Shomanike Head for help with tissue culture.
This work was supported, in whole or in part, by National Institutes of Health Grant HL20948. This work was also supported by the Perot Family Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental two movies, Fig. 1, and Table 1.
- HMG-CoA
- 3-hydroxy-3-methylglutaryl coenzyme A
- CHO
- Chinese hamster ovary
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- SREBP
- sterol regulatory element-binding protein
- 25-HC
- 25-hydroxycholesterol
- LPDS
- lipoprotein-deficient serum
- FCS
- fetal calf serum
- VSV-G
- vesicular stomatitis virus glycoprotein
- ADRP
- adipose differentiation-related protein
- CMV
- cytomegalovirus
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Brown M. S., Goldstein J. L. (1980) J. Lipid Res 21, 505–517 [PubMed] [Google Scholar]
- 2.Roitelman J., Olender E. H., Bar-Nun S., Dunn W. A., Jr., Simoni R. D. (1992) J. Cell Biol. 117, 959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liscum L., Finer-Moore J., Stroud R. M., Luskey K. L., Brown M. S., Goldstein J. L. (1985) J. Biol. Chem. 260, 522–530 [PubMed] [Google Scholar]
- 4.Nakanishi M., Goldstein J. L., Brown M. S. (1988) J. Biol. Chem. 263, 8929–8937 [PubMed] [Google Scholar]
- 5.Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. (2003) Mol. Cell 11, 25–33 [DOI] [PubMed] [Google Scholar]
- 6.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 7.Song B. L., Sever N., DeBose-Boyd R. A. (2005) Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 8.Vij N. (2008) J. Cell. Mol. Med. 12, 2511–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., DeBose-Boyd R. A. (2003) J. Biol. Chem. 278, 52479–52490 [DOI] [PubMed] [Google Scholar]
- 10.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 12.Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 13.Lilley B. N., Ploegh H. L. (2004) Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 14.Huppa J. B., Ploegh H. L. (1997) Immunity 7, 113–122 [DOI] [PubMed] [Google Scholar]
- 15.Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L. (1996) Cell 84, 769–779 [DOI] [PubMed] [Google Scholar]
- 16.Wiertz E. J., Tortorella D., Bogyo M., Yu J., Mothes W., Jones T. R., Rapoport T. A., Ploegh H. L. (1996) Nature 384, 432–438 [DOI] [PubMed] [Google Scholar]
- 17.VanSlyke J. K., Musil L. S. (2002) J. Cell Biol. 157, 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leichner G. S., Avner R., Harats D., Roitelman J. (2009) Mol. Biol. Cell 20, 3330–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garza R. M., Sato B. K., Hampton R. Y. (2009) J. Biol. Chem. 284, 14710–14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein J. L., Basu S. K., Brown M. S. (1983) Methods Enzymol. 98, 241–260 [DOI] [PubMed] [Google Scholar]
- 21.Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S. (2001) J. Biol. Chem. 276, 4365–4372 [DOI] [PubMed] [Google Scholar]
- 22.DeBose-Boyd R. A., Brown M. S., Li W. P., Nohturfft A., Goldstein J. L., Espenshade P. J. (1999) Cell 99, 703–712 [DOI] [PubMed] [Google Scholar]
- 23.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. (2002) Cell 110, 489–500 [DOI] [PubMed] [Google Scholar]
- 24.Liscum L., Luskey K. L., Chin D. J., Ho Y. K., Goldstein J. L., Brown M. S. (1983) J. Biol. Chem. 258, 8450–8455 [PubMed] [Google Scholar]
- 25.Yang J., Brown M. S., Ho Y. K., Goldstein J. L. (1995) J. Biol. Chem. 270, 12152–12161 [DOI] [PubMed] [Google Scholar]
- 26.Sakai J., Nohturfft A., Cheng D., Ho Y. K., Brown M. S., Goldstein J. L. (1997) J. Biol. Chem. 272, 20213–20221 [DOI] [PubMed] [Google Scholar]
- 27.Lee J. N., Song B., DeBose-Boyd R. A., Ye J. (2006) J. Biol. Chem. 281, 39308–39315 [DOI] [PubMed] [Google Scholar]
- 28.Brown M. S., Faust J. R., Goldstein J. L., Kaneko I., Endo A. (1978) J. Biol. Chem. 253, 1121–1128 [PubMed] [Google Scholar]
- 29.Lee J. N., Gong Y., Zhang X., Ye J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4958–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher E. A., Ginsberg H. N. (2002) J. Biol. Chem. 277, 17377–17380 [DOI] [PubMed] [Google Scholar]
- 31.Ohsaki Y., Cheng J., Fujita A., Tokumoto T., Fujimoto T. (2006) Mol. Biol. Cell 17, 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartz R., Zehmer J. K., Zhu M., Chen Y., Serrero G., Zhao Y., Liu P. (2007) J Proteome Res. 6, 3256–3265 [DOI] [PubMed] [Google Scholar]
- 33.Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., Anderson R. G. (2004) J. Biol. Chem. 279, 3787–3792 [DOI] [PubMed] [Google Scholar]
- 34.Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J. S., Vale R. D., Walter P., Farese R. V. (2008) Nature 453, 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omura S., Tomoda H., Xu Q. M., Takahashi Y., Iwai Y. (1986) J. Antibiot. 39, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 36.Brasaemle D. L., Wolins N. E. (2006) Current Protocols in Cell Biology, Chapter 3, pp. 3.15.1–3.15.12, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 37.Masuda Y., Itabe H., Odaki M., Hama K., Fujimoto Y., Mori M., Sasabe N., Aoki J., Arai H., Takano T. (2006) J. Lipid Res 47, 87–98 [DOI] [PubMed] [Google Scholar]
- 38.Xu G., Sztalryd C., Lu X., Tansey J. T., Gan J., Dorward H., Kimmel A. R., Londos C. (2005) J. Biol. Chem. 280, 42841–42847 [DOI] [PubMed] [Google Scholar]
- 39.Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zehmer J. K., Bartz R., Liu P., Anderson R. G. (2008) J. Cell Sci. 121, 1852–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liscum L., Cummings R. D., Anderson R. G., DeMartino G. N., Goldstein J. L., Brown M. S. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 7165–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirsch C., Blom D., Ploegh H. L. (2003) EMBO J. 22, 1036–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robbins P. W., Trimble R. B., Wirth D. F., Hering C., Maley F., Maley G. F., Das R., Gibson B. W., Royal N., Biemann K. (1984) J. Biol. Chem. 259, 7577–7583 [PubMed] [Google Scholar]
- 44.Ploegh H. L. (2007) Nature 448, 435–438 [DOI] [PubMed] [Google Scholar]
- 45.Goodman J. M. (2008) J. Biol. Chem. 283, 28005–28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujimoto T., Ohsaki Y., Cheng J., Suzuki M., Shinohara Y. (2008) Histochem. Cell Biol. 130, 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novikoff A. B., Novikoff P. M., Rosen O. M., Rubin C. S. (1980) J. Cell Biol. 87, 180–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Targett-Adams P., Chambers D., Gledhill S., Hope R. G., Coy J. F., Girod A., McLauchlan J. (2003) J. Biol. Chem. 278, 15998–16007 [DOI] [PubMed] [Google Scholar]
- 49.Blanchette-Mackie E. J., Dwyer N. K., Barber T., Coxey R. A., Takeda T., Rondinone C. M., Theodorakis J. L., Greenberg A. S., Londos C. (1995) J. Lipid Res. 36, 1211–1226 [PubMed] [Google Scholar]
- 50.Ohsaki Y., Cheng J., Suzuki M., Fujita A., Fujimoto T. (2008) J. Cell Sci. 121, 2415–2422 [DOI] [PubMed] [Google Scholar]
- 51.McGookey D. J., Anderson R. G. (1983) J. Cell Biol. 97, 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan H. C., Melo R. C., Jin Z., Dvorak A. M., Weller P. F. (2007) FASEB J. 21, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.