Abstract
Multidrug resistance-associated protein 2 (Mrp2, Abcc2) is an ATP-binding cassette transporter localized at the canalicular membrane of hepatocytes that plays an important role in bile formation and detoxification. Prior in vitro studies suggest that Mrp2 can bind to Na+/H+ exchanger regulatory factor 1 (NHERF-1), a PDZ protein that cross-links membrane proteins to actin filaments. However the role of NHERF-1 in the expression and functional regulation of Mrp2 remains largely unknown. Here we examine the interaction of Mrp2 and NHERF-1 and its physiological significance in HEK293 cells and NHERF-1 knock-out mice. Mrp2 co-precipitated with NHERF-1 in co-transfected HEK293 cells, an interaction that required the PDZ-binding motif of Mrp2. In NHERF-1−/− mouse liver, Mrp2 mRNA was unchanged but Mrp2 protein was reduced in whole cell lysates and membrane-enriched fractions to ∼50% (p < 1 × 10−6) and ∼70% (p < 0.05), respectively, compared with wild-type mice, suggesting that the down-regulation of Mrp2 expression was caused by post-transcriptional events. Mrp2 remained localized at the apical/canalicular membrane of NHERF-1−/− mouse hepatocytes, although its immunofluorescent labeling was noticeably weaker. Bile flow in NHERF-1−/− mice was reduced to ∼70% (p < 0.001) in association with a 50% reduction in glutathione excretion (p < 0.05) and a 60% reduction in glutathione-methylfluorescein (GS-MF) excretion in isolated mouse hepatocyte (p < 0.01). Bile acid and bilirubin excretion remained unchanged compared with wild-type mice. These findings strongly suggest that NHERF-1 binds to Mrp2, and plays a critical role in the canalicular expression of Mrp2 and its function as a determinant of glutathione-dependent, bile acid-independent bile flow.
Keywords: ABC Transporter, Liver, Membrane Proteins, Multidrug Transporters, Protein-Protein Interactions, Ezrin-Radixin-Moesin (ERM)-binding Phosphoprotein 50, Multidrug Resistance-associated Protein 2, Na+/H+ Exchanger Regulatory Factor 1, PDZ Proteins
Introduction
The multidrug resistance-associated protein 2 (Mrp2/MRP2,2 Abcc2/ABCC2) is an ATP-binding cassette (ABC) transporter that is predominantly localized at the apical domain of polarized cells such as hepatocytes, renal proximal tubule epithelia, and enterocytes of the small intestine (1–3). In the liver, canalicular Mrp2 plays an important role in bile formation and detoxification by transporting a wide variety of organic anions, including divalent bile salt conjugates, bilirubin-glucuronides, and drug conjugates, into bile (4, 5). Mrp2 is a major determinant of the “bile acid-independent” fraction of bile flow by excreting glutathione and glutathione conjugates (6). In humans, mutations of the MRP2 gene lead to impaired targeting and expression of functional Mrp2 protein at the canalicular membrane, causing Dubin-Johnson Syndrome characterized by conjugated hyperbilirubinemia and defects in the excretion of organic anions (7, 8).
The transcription of MRP2/Mrp2 mRNA is regulated by a network of nuclear receptors that are activated by various ligands, such as bile acids and xenobiotic compounds (4, 9). Mrp2 expression, apical targeting, and localization are also regulated at the post-transcriptional level by a variety of factors (10, 11). In rat hepatocytes, cAMP results in acute enhancement of exocytic insertion of Mrp2 into the apical membrane, accompanied by increased canalicular excretion of Mrp2 substrates (12, 13). In contrast, endocytic internalization of Mrp2 has been observed in animal models of cholestasis induced by bile duct ligation, drugs, endotoxin, and oxidative stress, as well as in human cholestatic liver diseases (14–20).
A growing number of studies suggest that interacting proteins play important roles in this dynamic recycling of Mrp2 between the apical membrane and endosomal sub-apical compartments. Radixin, a member of the ezrin-radixin-moesin (ERM) family of cytoskeletal proteins that functions as a scaffold by cross-linking F-actin cytoskeleton to membrane transporters, has been reported to interact with Mrp2 (21). Deletion of radixin in mice results in selective loss of Mrp2 from the canalicular membrane, leading to hyperbilirubinemia (21). However siRNA-induced suppression of radixin in rat hepatocytes results in dislocation of Mrp2 as well as other apical transporters (22).
The sodium-hydrogen exchanger regulatory factor-1 (NHERF-1), also known as ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 (EBP50) in humans, is a scaffolding protein that contains two tandem PSD-95/Drosophila discs large/ZO-1 (PDZ) protein interaction domains at its N terminus and a C-terminal domain that binds the ERM family of cytoskeletal proteins (23, 24). It is abundantly expressed in the apical membranes of polarized epithelial cells and has been shown to interact with the PDZ-binding motifs in the C terminus of numerous membrane proteins, particularly transporters, ion channels and G-protein-coupled receptors (25–27). Accumulating evidence suggests that NHERF-1 is involved in the membrane retention, endocytic sorting, and regulation of activity of membrane proteins and, therefore, plays important roles in their expression, membrane localization and functional regulation (28, 29). Cross-linking of the cystic fibrosis transmembrane conductance regulator (CFTR) ion channel to the F-actin cytoskeleton via NHERF-1 and ezrin results in a strong reduction in the lateral mobility of CFTR in the membrane (30). Deletion of the PDZ-binding motif at the C terminus of CFTR decreases its endocytic recycling and reduces the half-life of the protein (31). Similarly, the interaction between NHERF-1 and the PDZ-binding motif of the β2-adrenergic receptor is required for the efficient sorting of the internalized receptor protein to recycling endosomes (32). Targeted disruption of the mouse NHERF-1 gene promotes internalization of sodium-phosphate cotransporter type IIa and urate transporter 1 in renal proximal tubule cells, associated with increased urinary excretion of phosphate and uric acid (33, 34). In addition, as a multivalent scaffold, NHERF-1 has been shown to recruit functionally related proteins into macromolecular complexes, facilitating cAMP-mediated PKA phosphorylation of Na+/H+ exchanger 3 (NHE3) and CFTR, providing tight control and efficient inhibition/activation of membrane transport processes (35, 36).
Similar to CFTR, Mrp2 possesses a class I PDZ-binding motif at its C terminus (HTEL in rat/mouse Mrp2, STKF in human MRP2). NHERF-1/EBP50 has been identified on the apical membrane of hepatocytes (37, 38) and has been reported to bind to Mrp2 in vitro (39). However the role of NHERF-1 in the expression, apical localization, and functional regulation of Mrp2 in hepatocytes and other polarized cells remains largely unknown. In the present study, we confirm that NHERF-1 binds to Mrp2 in vitro. Furthermore we demonstrate the importance of this interaction in the apical expression of Mrp2 in hepatocytes and in the function of Mrp2 as a determinant of glutathione-dependent, bile acid-independent bile flow.
EXPERIMENTAL PROCEDURES
Plasmid Constructs and Expression of Recombinant Proteins
Full-length rat NHERF-1/EBP50 cDNA (GenBankTM accession number AF154336) cloned into expression vector p3XFLAG-CMV-7 was described previously (40) and was a gift from Dr. R. Brian Doctor (University of Colorado Health Sciences Center, Denver, CO). Full-length rat Mrp2 (Abcc2) cDNA (GenBankTM accession number NM_012833) cloned into expression vector pcDNA3 was generously provided by Dr. Dietrich Keppler (German Cancer Research Center, Heidelberg, Germany). Full-length bile salt export pump (Bsep) cDNA from rat liver (kindly provided by Dr. Peter Meier, University Hospital, Zurich, Switzerland) was subcloned into a pcDNA3 vector. Rat Mrp2 cDNA with deletion of the sequences coding for the last 3 or 7 amino acids was generated by PCR and cloned into a pcDNA3 vector. cDNA encoding the last 287 amino acids of rat Mrp2 was amplified by PCR and was cloned into a pGEX-3X vector (Amersham Biosciences) for expression of GST-rMrp2 fusion protein in transformed Escherichia coli cells. GST and GST-rMrp2 fusion protein were purified with glutathione-Sepharose 4B beads (GE Healthcare/Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instructions. Purified GST and GST-rMrp2 fusion protein were dialyzed against 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 0.1% 2-mercaptoethanol and were quantified by Bio-Rad protein assay (Bio-Rad).
Cell Culture and Transfection
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum and antibiotics in a 37 °C, 5%CO2, humidified incubator. Cells in 6-well plates (BD Falcon, Franklin Lakes, NJ) were transfected with 4 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen) in Opti-MEM I Reduced Serum Medium (Invitrogen) according to the manufacturer's instructions.
Co-immunoprecipitation (Co-IP) and GST Pull-down Assay
Twenty-four hours following transfection, HEK293 cells were washed with Dulbecco's phosphate-buffered saline (DPBS) and lysed at 4 °C in lysis buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, and Halt Protease Inhibitor Cocktails (Thermo Scientific, Rockford, IL), and the cell lysates were cleared by centrifugation at 10,000 × g for 10 min at 4 °C. For co-immunoprecipitation experiments, 200 μl of the lysates were pre-cleared with normal mouse IgG-agarose (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and then incubated overnight with anti-Flag M2 agarose beads (Sigma-Aldrich) at 4 °C. For GST pull-down assay, 200 μl of cleared cell lysates were incubated overnight with 2.5 μg of purified GST or GST-rMrp2 fusion protein at 4 °C. The samples were then supplemented with glutathione-Sepharose 4B beads and incubated for an additional 2 h at 4 °C. After centrifugation, the beads were washed extensively with lysis buffer and subjected to SDS-PAGE and immunoblotting. For calyculin A or staurosporine treatment, twenty-four hours following transfection, cells were incubated with DMEM in the presence of 100 nm calyculin A (Santa Cruz Biotechnology) for 40 min or 10 μm staurosporine (Sigma-Aldrich) for 1 h and then harvested, washed, and lysed.
Animals
NHERF-1−/− mice bred into a C57BL/6 background for six generations were introduced from the laboratory of Dr. Edward J. Weinman (Departments of Medicine and Physiology, University of Maryland School of Medicine) and were previously described (34). Wild-type control C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Male mice at 18 weeks of age were used for the present studies. Animals were housed in a temperature- and humidity-controlled room under a light cycle with free access to food and water. Upon sacrifice, the liver was flushed with normal saline and quick-frozen in liquid nitrogen. All experimental protocols were approved by the local Animal Care and Use Committee, according to criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences, as published by the National Institutes of Health (NIH Publication 86–23, revised 1985).
Quantitative Real-time Polymerase Chain Reaction
Total RNA was isolated from mouse liver tissue with Trizol reagent (Invitrogen) and was purified by using RNeasy MinElute Cleanup Kit (Qiagen Inc.) according to the manufacturer's instructions. Two micrograms of RNA was used for reverse transcription using the AffinityScriptTM Multiple Temperature cDNA synthesis kit (Stratagene, La Jolla, CA). TaqMan quantitative real-time PCR assay was performed on an ABI 7500 Sequence Detection System (Applied Biosystems, Foster City, CA). The following primers and probes were purchased from Applied Biosystems and were used in the reactions: Gapdh (NM_001001978) forward GCCCAGAACATCATCCCTGC, reverse CCGTTCAGCTCTGGGATGACC, probe TCCACTGGTGCTGCCAAGGCTGTG; Mrp2 (NM_013806) forward CGACCATCCGGAACGAGTT, reverse GCAGCCTGTGTGCGATAGTG, probe. CCCAGTGCACGGTCA. Amplification of cDNA was performed in triplicate in 96-well plates by incubation at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min.
Immunoblotting
Whole cell lysates and membrane-enriched fractions from mouse liver tissue were prepared as previously described (41). Protein samples were quantified by Bio-Rad protein assay and then separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked with 5% nonfat dry milk in TBS-T containing 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 0.1% Tween 20 and then incubated overnight with the appropriate primary antibodies. After extensive washes with TBS-T, the membranes were incubated with goat anti-rabbit or goat anti-mouse IgG conjugated to horseradish peroxidase for 1 h at room temperature. After further washes, immune complexes were detected by Amersham Biosciences ECL Western blotting Detection Reagents and Hyperfilms (GE Healthcare/Amersham Biosciences, Piscataway, NJ). Densitometric scanning of protein bands was performed using TotalLab TL100 software (Nonlinear Dynamics, Newcastle upon Tyne, UK). For membrane stripping, membranes were incubated with Stripping Buffer containing 50 mm Tris-HCl, pH 6.7, 2% SDS and 100 mm 2-mercaptoethanol at 50 °C for 50 min. Membranes were then washed three times with TBS-T and re-blotted.
Antibodies
Polyclonal antibody AB5487 against NHERF-1 was purchased from Millipore (Billerica, MA). Polyclonal antibody EAG15 against Mrp2 was a generous gift from Dr. Dietrich Keppler. Polyclonal antibody K13 against Mrp2 was kindly provided by Dr. Bruno Stieger (University Hospital, Zurich, Switzerland). Monoclonal antibody anti-Flag M2 was purchased from Sigma-Aldrich. Polyclonal antibody anti-SPGP pAb against Bsep was purchased from Kamiya Biomedical Company (Seattle, WA). Polyclonal antibody C-19 against SH-PTP1 was purchased from Santa Cruz Biotechnology. Polyclonal antibody A5060 against actin was purchased from Sigma-Aldrich.
Immunofluorescence
Immunofluorescence was performed on frozen sections of liver and isolated mouse hepatocyte couplets as previously described (41). Briefly, unfixed cryostat sections or isolated mouse hepatocyte couplets were fixed with cold acetone or methanol, respectively, and incubated with primary antibody for 2 h at room temperature, followed by secondary antibodies (Alexa 488/568, Molecular Probes) for 1 h. Images were acquired using a Zeiss LSM 510 confocal microscope (Thornwood, NJ) and processed with Adobe PhotoShop (San Jose, CA).
Bile Analysis
Bile was collected into tared tubes from anesthetized mice by cannulating the gallbladder. Bile samples were collected at 15-min intervals for a total of 60 min. The volume of bile was determined gravimetrically by assuming a density of 1.0 g/ml, and bile flow rate was expressed as microliters per minute per gram of liver. Six percent sulfosalicylic acid was added to minimize oxidation when GSH was determined and samples were protected from light for bilirubin assay. The enzymatic recycling method of Tietze (42) as modified by Shaik and Mehvar (43) was used for the determination of total glutathione. Commercial kits for total bilirubin (Thermo Fisher Scientific Inc., Waltham, MA) and bile acids (Trinity Biotech, St. Louis, MO) were used.
Functional Excretion in Mouse Hepatocytes
Mouse hepatocyte couplets were isolated as previously described (44) and cultured for 4 h on glass coverslips in L15 media with 10% fetal bovine serum. Hepatocytes were incubated with 2.5 μmol/liter 5-chloromethylfluorescein diacetate (CMFDA, Molecular Probes) in HEPES buffer (0.35 g/liter KCl, 0.25 g/liter MgSO4, 0.18 g/liter CaCl2, 0.16 g/liter KH2PO4, 4.8 g/liter HEPES, 7.9 g/liter NaCl, and 0.9 g/liter glucose, pH 7.4) at 37 °C for 15 min. Cells were imaged immediately with a Zeiss LSM 510 confocal microscope (Thornwood, NJ). Cells with visible canaliculi by brightfield optics were then imaged with a 488 nm fluorescent laser at 25% power to reduce bleaching. For each condition, 9 images were taken from each of 4 replicate coverslips. Measurements of total and canalicular fluorescence were acquired using NIH ImageJ, and data are presented as percentage of canalicular excretion as previously described (44).
Statistical Analysis
All data were analyzed using the Student's t test and are expressed as the mean ± S.D. A p value of less than 0.05 was considered significant.
RESULTS
Mrp2 Co-immunoprecipitates with NHERF-1 in Transfected HEK293 Cells
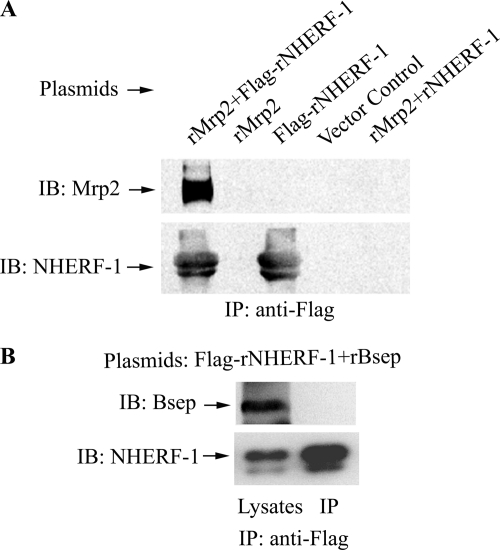
To determine if Mrp2 forms a protein complex with NHERF-1 and other proteins in vitro, we co-transfected HEK293 cells with plasmids expressing rat Mrp2 and Flag-tagged rat NHERF-1. The cell lysates were then incubated with anti-Flag-agarose beads, and the precipitates were analyzed by immunoblotting with anti-Mrp2 antibody. As shown in Fig. 1A, rMrp2 co-precipitated together with Flag-rNHERF-1 from co-transfected HEK293 cells, but was not precipitated from cell lysates transfected with the vector control, the rMrp2 plasmid alone or Flag-NHERF-1 plasmid alone. Neither was Mrp2 detected in the precipitates from cells co-transfected with rMrp2 and a NHERF-1 construct without the Flag tag. Reciprocally, rNHERF-1 co-precipitated with Flag-rMrp2 when cells were co-transfected with Flag-Mrp2 and rNHERF-1 but not from cells transfected with Flag-Mrp2 or rNHERH-1 alone (supplemental Fig. S1). To determine the specificity of the interaction between Mrp2 and NHERF-1, HEK293 cells were co-transfected with Flag-rNHERF-1 and a DNA construct encoding the rat bile salt export pump (rBsep), another liver-specific ABC transporter protein that functions at the apical/canalicular surface of hepatocytes. As demonstrated in Fig. 1B, rBsep did not co-precipitate with Flag-rNHERF-1 from the co-transfected cells. These results demonstrated that NHERF-1 associates with Mrp2 but not Bsep in co-transfected HEK293 cells.
FIGURE 1.
Mrp2 co-immunoprecipitates with NHERF-1 in co-transfected HEK293 cells. A, cell lysates were incubated with anti-Flag M2-agarose beads and the Co-IP complex was immunoblotted with anti-Mrp2 antibody EAG15 and anti-NHERF-1 antibody AB5487. Note that Mrp2 was detected only in the Co-IP complex from cells co-transfected with plasmids expressing both rat Mrp2 and Flag-tagged rat NHERF-1, but was not detected from cells transfected with the vector control, the rMrp2 or Flag-NHERF-1 plasmid alone, or from cells co-transfected with rMrp2 and an NHERF-1 plasmid without the Flag tag. B, HEK293 cells were co-transfected with plasmids expressing Flag-rNHERF-1 and rat Bsep. Cell lysates were incubated with anti-Flag M2-agarose beads, and the Co-IP complex was immunoblotted with anti-Bsep antibody. Note that Bsep was detected in the cell lysates but was absent in the Co-IP complex. IB, immunoblotting; IP, immunoprecipitation.
GST Pull-down Assay Confirms That Both Phosphorylated and Un- or Less Phosphorylated NHERF-1 Interact with Mrp2 in Vitro
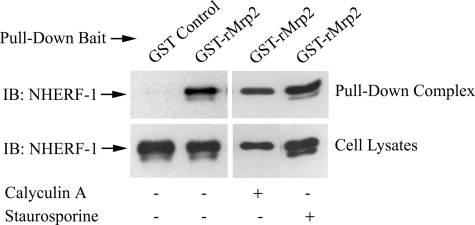
To verify the interaction between Mrp2 and NHERF-1, a GST fusion protein encoding the last 287 amino acids of rMrp2 was used as bait in a GST pull-down assay with lysates of HEK293 cells transfected with Flag-rNHERF-1. Flag-rNHERF-1 was detected in the pull-down complex when GST-rMrp2 was used as bait but was not detected in the pull-down complex by GST alone (Fig. 2), confirming that Mrp2 interacts with NHERF-1 in these in vitro experiments.
FIGURE 2.
GST pull-down assay confirms that both phosphorylated and un- or less phosphorylated NHERF-1 interact with Mrp2 in vitro. Lysates of HEK 293 cells transfected with Flag-rNHERF-1 were incubated with GST control or GST-rMrp2 fusion protein that encodes the last 287 amino acids of rMrp2. The samples were then supplemented with glutathione-Sepharose 4B beads, and the pull-down complex was immunoblotted with anti-Flag M2 antibody (upper panels). The same cell lysates were immunoblotted with anti-NHERF-1 antibody AB5487 to confirm the expression of Flag-rNHERF-1 (lower panels). Twenty-four hours following transfection, cells were incubated with DMEM in the absence (−) or presence (+) of 100 nm calyculin A for 40 min or 10 μm staurosporine for 1 h before harvesting. Note that in both the GST-rMrp2 pull-down complex and the cell lysates, two bands of NHERF-1 protein were detected from untreated cells or cells treated with staurosporine, but only one band of NHERF-1 was detected from cells treated with calyculin A. IB, immunoblotting.
Interestingly, two bands of Flag-rNHERF-1 were detected in the Co-IP complex from HEK293 cells co-transfected with Mrp2 and NHERF-1 constructs (Fig. 1A and supplemental Fig. S1) as well as in the GST-Mrp2 pull-down complex (Fig. 2, upper panels). The upper band, which was detected as the major form of Flag-rNHERF-1 protein in transfected HEK293 cell lysates (Fig. 2, lower panels), represented the phosphorylated NHERF-1; the lower band, which disappeared when the cells were treated with a phosphatase inhibitor calyculin A, and increased in intensity when the cells were treated with a kinase inhibitor staurosporine (Fig. 2), represented unphosphorylated or less phosphorylated NHERF-1 (23, 45, 47, 48). The existence of the two NHERF-1 bands in the Co-IP complex and the GST-Mrp2 pull-down complex demonstrates that both the phosphorylated and the un- or less phosphorylated NHERF-1 interacts with Mrp2 in vitro.
The PDZ-binding Motif in Mrp2 Mediates Its Interaction with NHERF-1
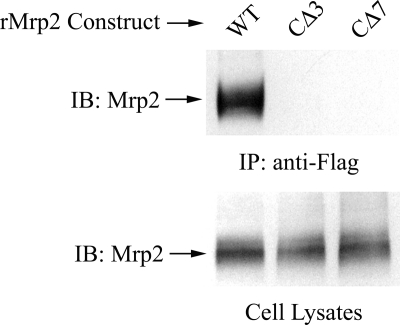
To investigate if the PDZ-binding motif in Mrp2 was required for its interaction with NHERF-1, HEK293 cells were co-transfected with Flag-rNHERF-1 and ΔrMrp2, a truncated rat Mrp2 with deletion of the last 3 or 7 amino acids at its C terminus that constitute the PDZ-binding motif. In contrast to full-length rMrp2, ΔrMrp2 did not co-precipitate with Flag-rNHERF-1 (Fig. 3), indicating that the interaction between rMrp2 and rNHERF-1 was mediated through the C-terminal PDZ binding motif in rMrp2.
FIGURE 3.
The C-terminal PDZ-binding motif in Mrp2 mediates its association with NHERF-1. HEK293 cells were co-transfected with Flag-rNHERF-1 and full-length (WT) or truncated rMrp2 with deletion of the last 3 (CΔ3) or 7 (CΔ7) amino acids that constitute the PDZ binding motif at its C terminus. Cell lysates were incubated with anti-Flag-agarose beads, and the co-precipitation complex was detected with anti-Mrp2 antibody EAG15. The upper panel shows that, in contrast to the result obtained with the full-length control, Mrp2 was not detected in the Co-IP complex when its C-terminal PDZ binding motif was deleted. The lower panel confirms the presence of Mrp2 in the cell lysates used for the Co-IP in the upper panel. IB, immunoblotting; IP, immunoprecipitation.
Mrp2 and NHERF-1 Co-localize at the Apical Membrane of Rat Hepatocytes
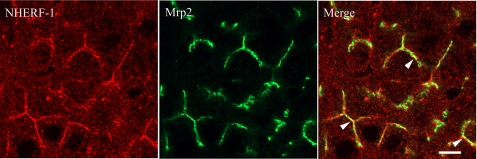
NHERF-1/EBP50 was previously reported to be localized at the apical domain of rat hepatocytes (37, 38). To determine if Mrp2 and NHERF-1 are co-localized in these cells, immunofluorescence microscopy was performed on rat liver sections (Fig. 4). NHERF-1 was highly expressed at the apical/canalicular domain of hepatocytes and in cytoplasm at a much lower level (Fig. 4, left). In some apical/canalicular domains, NHERF-1 overlapped with Mrp2 (Fig. 4, right), suggesting that Mrp2 and NHERF-1 might be associated in a protein complex at the canalicular membrane.
FIGURE 4.
Immunofluorescence microscopy in rat liver demonstrates co-localization of NHERF-1 and Mrp2. NHERF-1 (left) and Mrp2 (middle) were detected at the canalicular membrane of rat liver sections, with some areas of co-localization (right, arrowheads). Scale bar, 5 μm.
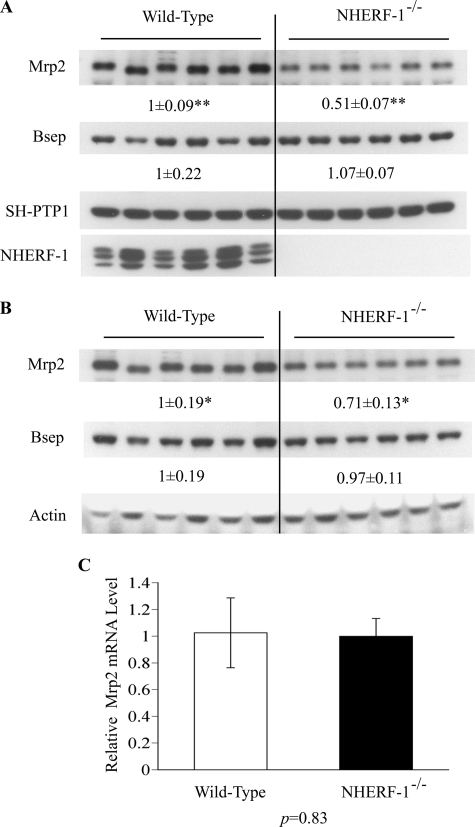
Mrp2 Expression in the Liver of NHERF-1−/− Mice
To determine if NHERF-1 plays a role in the expression of Mrp2, we analyzed liver extracts from NHERF-1−/− mice. Immunoblotting of whole cell lysates with anti-NHERF-1 antibody confirmed that NHERF-1 was not detected in the liver of NHERF-1−/− mice (Fig. 5A), consistent with its absence from kidney extracts reported previously (34). When the same samples were analyzed with anti-Mrp2 antibody, the expression of Mrp2 protein was about 50% of wild-type animals (p < 1 × 10−6; Fig. 5A). The expression of Mrp2 in liver membrane-enriched fractions from the NHERF-1−/− mice also decreased to about 70% of the wild-type controls (p < 0.05; Fig. 5B). In contrast, there was no significant change in the expression of Bsep protein either in liver whole cell lysates or in liver membrane-enriched fractions from NHERF-1−/− mice compared with their wild-type controls (Fig. 5, A and B). This is consistent with the findings that Bsep does not co-precipitate with NHERF-1 in co-transfected HEK293 cells. In contrast to protein expression, no differences were observed in the Mrp2 mRNA levels between NHERF-1−/− mice and wild-type mice (p = 0.83; Fig. 5C), indicating that the decrease of Mrp2 protein expression in the liver of NHERF-1−/− mice was caused by post-transcriptional events.
FIGURE 5.
Expression of Mrp2 protein, but not mRNA, is reduced in liver lysates and membranes from NHERF-1−/− mice. A, representative immunoblots of whole cell lysates from wild-type (n = 6) and NHERF-1−/− (n = 6) mouse liver demonstrate reduction in Mrp2 but not Bsep protein in NHERF-1−/− mice. B, representative immunoblots of membrane-enriched fractions confirm these findings. In both A and B, numbers represent the relative ratio of the respective protein bands in wild-type and NHERF-1−/− mice by densitometry analysis. Data are normalized to SH-PTP1 in whole cell lysates (A) and to actin in membrane-enriched fractions (B). The amount of protein from the wild-type is set as 1. Values represent the means ± S.D. of six individual animals in each group. ** indicates p < 1 × 10−6. * indicates p < 0.05. C, relative levels of Mrp2 mRNA in the liver of wild-type (n = 6) and NHERF-1−/− (n = 6) mice quantified by real-time PCR. Data are normalized to Gapdh, and the amount of Mrp2 mRNA from the wild-type is set as 1. Values represent the means ± S.D. of six individual animals in each group.
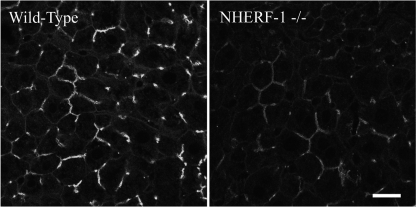
Mrp2 Expression at the Canalicular Membrane Is Reduced in NHERF-1−/− Mouse
The effect of the deficiency of NHERF-1 on the localization of Mrp2 in the liver was assessed by immunofluorescence microscopy in NHERF-1−/− mice. As shown in Fig. 6, Mrp2 was localized at the apical/canalicular membrane of the hepatocytes of NHERF-1−/− mouse, similar to the wild type. However, labeling was noticeably weaker in the NHERF-1−/− mouse than in the wild-type mouse, an observation consistent with the result of the protein expression analysis.
FIGURE 6.
Immunofluorescence microscopy of Mrp2 on mouse liver sections demonstrates reduced expression in NHERF-1−/− mice. Similar to the wild-type (left), Mrp2 remains localized at the canalicular membrane of NHERF-1−/− mouse hepatocytes (right). However, Mrp2 labeling is noticeably weaker in NHERF-1−/− mouse. Images were acquired and processed under identical conditions. Scale bar, 20 μm.
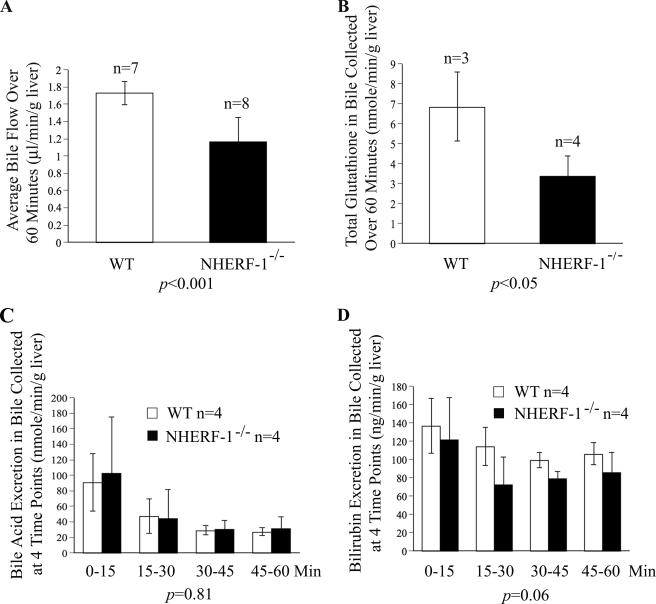
NHERF-1−/− Mice Have Decreased Bile Flow and Glutathione Excretion but Normal Excretion of Bile Acids and Bilirubin
As illustrated in Fig. 7A, bile flow rates in NHERF-1−/− mice measured over 60 min were reduced to ∼70% of wild-type mice (p < 0.001). Similarly, total glutathione (GSH+GSSG) in bile samples collected for 60 min from NHERF-1−/− mice was reduced to about 50% of wild-type mice (p < 0.05; Fig. 7B), indicating reduced Mrp2 function with respect to glutathione excretion. In contrast, bile acid and bilirubin excretion remained unchanged in NHERF-1−/− mice compared with the wild-type mice (Fig. 7, C and D), suggesting that bile acid and bilirubin excretion in the liver was not significantly affected by the deficiency of NHERF-1.
FIGURE 7.
Bile flow and glutathione, but not bile acids and total bilirubin, are reduced in bile samples from bile duct cannulated NHERF-1−/− mice. A, bile flow in NHERF-1−/− mice is reduced to 70% of the wild-type mice. Values represent the means ± S.D. of 7 (WT) or 8 (NHERF-1−/−) independent experiments. B, glutathione excretion in NHERF-1−/− mice is 50% of wild-type mice. Values reported are means ± S.D. of three (WT) or four (NHERF-1−/−) independent experiments. C, bile acid excretion is not significantly different between the two groups. Values are means ± S.D. of four independent experiments. D, bilirubin excretion is also not significantly changed in the NHERF-1−/− mice compared with the wild-type mice. Values are the means ± S.D. of four independent experiments.
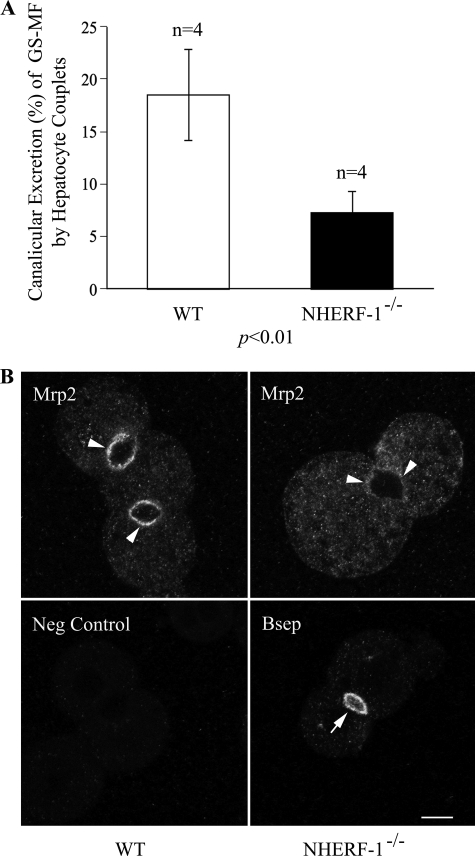
Mrp2 Expression and Function Are Reduced in Isolated Mouse Hepatocyte Couplets from NHERF-1−/− Mice
As shown in Fig. 8A, the canalicular excretion of glutathione-methylfluorescein (GS-MF), the fluorescent product of 5-chloromethylfluorescein diacetate (CMFDA), and a substrate of Mrp2, averaged 18.5% in wild-type hepatocytes, whereas the value was reduced by nearly 60% to 7.3% in NHERF-1−/− hepatocytes (p < 0.01). The expression of Mrp2 was also markedly reduced at the canalicular membrane of the NHERF-1−/− hepatocyte couplets, as demonstrated by confocal microscopy, whereas Bsep labeling remained strong in the NHERF-1−/− mouse hepatocyte (Fig. 8B). Together these findings suggested that the impaired canalicular excretion of GS-MF in NHERF-1-deficient mouse hepatocyte couplets was the result of the reduced canalicular expression of Mrp2 in these cells.
FIGURE 8.
Mrp2 function and expression are reduced in isolated mouse hepatocyte couplets from NHERF-1−/− mice. A, canalicular excretion is expressed as percent of total fluorescence of GS-MF that is excreted into the canalicular lumen. Data were obtained from four independent experiments. B, replicate coverslips were assessed for subcellular localization and expression level of Mrp2 (upper panels) and Bsep (lower right panel) by immunofluorescence microscopy. A negative control (Neg Control, lower left panel) without primary antibody was included to demonstrate the specificity of the labeling. Images were acquired and processed under identical conditions. Note that Mrp2 labeling (arrowheads) is markedly reduced at the canaliculi of NHERF-1−/− mouse hepatocyte couplets compared with the wild type, whereas Bsep labeling (arrow) remains strong in the NHERF-1−/− mouse hepatocyte. Scale bar, 10 μm.
DISCUSSION
The present study provides evidence for a direct interaction of Mrp2 with NHERF-1 via its PDZ-binding motif and establishes a role for this interaction in the expression and functional properties of this ABC transporter at the canalicular membrane of hepatocytes. The interaction of recombinant human MRP2 with PDZ proteins in a GST-EBP50 gel overlay experiment was described a few years ago (39) but the role of NHERF-1/EBP50 in the expression, localization and functional regulation of Mrp2 in hepatocytes and other polarized cells has not been elucidated. Although PDZK1/NHERF-3, another member of the NHERF family of scaffolding proteins, has also been reported to interact with human MRP2 (cMOAT) in a yeast two-hybrid system (49), no change in either the distribution or the expression level of Mrp2 was observed in kidney proximal tubule epithelial cells of PDZK1 knock-out mice, indicating that either the interaction of Mrp2 and PDZK1 is not critical for the localization or expression of Mrp2 in vivo or that functional compensation by other PDZ proteins takes place in PDZK1 knock-out mice (50).
Here we demonstrate that Mrp2 interacts with NHERF-1 via the PDZ-binding motif at its C terminus in transfected HEK293 cells and that NHERF-1 deficiency in mice leads to reduced expression of Mrp2 protein at the canalicular membrane of hepatocytes as well as significant impairment of bile acid-independent bile flow, glutathione secretion, and the canalicular excretion of a Mrp2 substrate.
The role of the C terminus of Mrp2 in its subcellular localization has been controversial. Harris et al. (51) described that deletion of the C-terminal PDZ-binding motif caused MRP2 to be localized predominantly in the basolateral membrane in polarized MDCK cells. However, other studies have shown that the C-terminal sequences are not required for the apical localization of Mrp2 in polarized cells. For example, Nies et al. (52) reported that removal of the C-terminal PDZ-binding motif and stepwise deletion of up to 11 amino acids did not change the predominant apical distribution of MRP2 in polarized liver-derived human HepG2 cells. In another study (15), the exchange of the C-terminal 51 amino acids between Mrp2 and multidrug resistance protein 1 (Mrp1), another member of the ABC transporter superfamily, that in general traffics to basolateral membranes of polarized cells, did not affect the localization of either protein in polarized mammalian LLC-PK1 cells. Recently, by using hybrid proteins in transfected MDCK-1 and LLC-PK1 cells, Bandler et al. (53) reported that all elements necessary for apical targeting of MRP2 reside in an N-terminal MSD0 region and the adjacent cytoplasmic loop (CL) 3, and that this region can target the core of MRP1 to an exclusively apical location. It is noteworthy that all these studies were conducted in model cell lines rather than native animal tissues where Mrp2 expression is regulated by mechanisms of higher complexity.
In the present study, our confocal microscopy data reveal that Mrp2 is localized at the apical/canalicular membrane of hepatocytes in the NHERF-1−/− mouse, similar to the wild-type control. While no mislocalization of Mrp2 was observed, Mrp2 labeling at the apical/canalicular surface was noticeably weaker in the NHERF-1−/− mouse than in the wild type, suggesting that the interaction between the C-terminal PDZ-binding motif of Mrp2 and NHERF-1 is not an absolute requirement for the trafficking of newly synthesized Mrp2 protein from the trans-Golgi network to the apical/canalicular membrane, but that it affects its level of expression. It is also possible that there is functional compensation by other apical PDZ proteins, such as NHERF-2, which is structurally and functionally related to NHERF-1. The C-terminal cytoplasmic domain of human MRP2 has also been reported to interact with radixin in vitro, providing for a direct link between Mrp2 and the actin cytoskeleton (21), which may also contribute to the unaltered apical localization of Mrp2 in NHERF-1−/− mouse hepatocytes. However, knockdown of radixin in rat hepatocytes leads to a more generalized reduction of transporters in the apical canalicular domain (22). Interestingly, the apical localization of NHE3 and CFTR, two other binding targets of NHERF-1, is also unaltered in kidney proximal tubules and jejunal crypts of NHERF-1−/− mice, respectively (34, 45), suggesting common mechanisms involved in the regulation of transporter expression and localization by PDZ proteins in different tissues. In contrast, NHERF-1 has been reported to interact with and regulate the trafficking of multidrug resistance protein 4 (MRP4), another ABC transporter that displays dual membrane localization in polarized cells (54–56). Ectopic expression of NHERF-1 in MDCKI cells redirects the trafficking of endogenous MRP4 from the basolateral to the apical membranes (57). We examined the expression of Mrp4 in the liver of NHERF-1−/− mice by immunoblotting analysis. We found that, unlike Mrp2, the expression level of hepatic Mrp4 protein in NHERF-1−/− mice was not significantly changed compared with wild-type mice (data not shown). This is consistent with studies showing that in hepatocytes Mrp4 is localized at the basolateral membrane (55, 56), whereas NHERF-1 is expressed at the apical membrane (Refs. 37, 38; Fig. 4 of the present study). Future studies on the expression and localization of Mrp4 in other tissues of NHERF-1−/− mice, such as the kidney proximal tubules where both Mrp4 and NHERF-1 are localized at the apical membrane, would be of interest in understanding of the role of NHERF-1 in the trafficking of membrane proteins.
Although mislocalization of Mrp2 in the liver of NHERF-1−/− mice is not detected in the present study, our finding that Mrp2 protein expression is dramatically reduced in both the whole cell lysates and membrane-enriched fractions while the level of liver Mrp2 mRNA remains unchanged in NHERF-1-deficient mice suggests that NHERF-1 plays a critical role in post-transcriptional regulation of Mrp2 expression in mouse liver. Indeed, recent studies suggest that NHERF-1 is required for sorting of internalized membrane proteins between recycling endosomes and degradative lysosomes, thereby regulating the turnover of these membrane proteins (31, 32). In MDCK cells, deletion of the C-terminal PDZ-binding motif of CFTR decreases endocytic recycling of CFTR, leading to a significantly reduced half-life of the protein in its apical membrane (31). Similarly, disrupting the interaction of NHERF-1/EBP50 with either the PDZ domain or the ERM-binding domain causes missorting and enhanced lysosomal degradation of internalized β2-adrenergic receptors in HEK293 cells after agonist stimulation (32). Therefore it seems likely that deficiency of NHERF-1 might lead to decreased endocytic recycling of Mrp2 to the canalicular membrane and result in increased lysosomal degradation. Future studies should examine the half-life and endocytic recycling of Mrp2 in PDZ protein knock-out mice as well as in model cell lines expressing mutant Mrp2 with mutations in the PDZ-binding motif.
Another major finding in the present study is that NHERF-1 is required for the optimal function of Mrp2 in the liver. Our results indicate that there is significant reduction in bile acid- independent bile flow and total glutathione excretion in NHERF-1−/− mice as well as a markedly decreased rate of canalicular excretion of the Mrp2 substrate, GS-MF, in hepatocyte couplets isolated from NHERF-1−/− mice. In contrast, ablation of NHERF-1 apparently has no effect on either the excretion of bile acids or bilirubin. Because bilirubin-glucuronides are also excreted by Mrp2, the lack of effect on bilirubin but not glutathione excretion is likely because there is sufficient Mrp2 expressed at the canalicular domain to excrete the smaller substrate load from bilirubin compared with the much larger flux of glutathione. Whether other apical solute transporter(s) might facilitate the biliary excretion of bilirubin in this study is not known but seems unlikely. Because NHERF-1 does not interact with Bsep and Bsep lacks PDZ binding motifs, Bsep protein expression levels remain unchanged in NHERF-1−/− mouse liver and the excretion of bile acids are unaffected. Taken together, our data strongly supports the view that NHERF-1 deficiency leads to impairment in the function of Mrp2, a major determinant of glutathione-dependent, bile acid-independent bile flow.
Whereas the impairment in Mrp2 function can be explained by its reduced expression in NHERF-1-deficient mice, NHERF-1 may also function as a scaffold to sequester other functionally related signaling proteins into an integrated macromolecule complex and affect the regulation of Mrp2 in this manner. For example, CFTR activity is dynamically regulated by both positive and negative signaling pathways mediated by NHERF-1/EBP50 and its competitor Shank2, another PDZ protein (46). Characterization of the interactions of additional proteins, such as protein kinases and other PDZ proteins in the protein complex associated with Mrp2 should further enhance the understanding of the mechanisms involved in regulation of the apical expression and function of this important ABC transporter.
Supplementary Material
Acknowledgments
We thank Dr. R. Brian Doctor, Dr. Dietrich Keppler, and Dr. Peter Meier for providing the p3XFLAG-CMV-7/NHERF-1 (EBP50) expression vector, the pcDNA3/Mrp2 expression vector, and the Bsep cDNA; Dr. Dietrich Keppler and Dr. Bruno Stieger for the anti-Mrp2 antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grants DK34989 and DK25636 (to J. L. B.) and DK55881 (to E. J. W.). This work was also supported by a grant from the Research Service, Department of Veterans Affairs (to E. J. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- Mrp2
- multidrug resistance-associated protein 2
- NHERF-1
- Na+/H+ exchanger regulatory factor 1
- EBP50
- ezrin-radixin-moesin (ERM)-binding phosphoprotein 50
- ABC
- ATP-binding cassette
- PDZ
- PSD-95/Drosophila discs large/ZO-1
- CFTR
- cystic fibrosis transmembrane conductance regulator
- NHE3
- Na+/H+ exchanger 3
- GST
- glutathione S-transferase
- CMFDA
- 5-chloromethylfluorescein diacetate
- GS-MF
- glutathione-methylfluorescein
- DMEM
- Dulbecco's modified Eagle's medium.
REFERENCES
- 1.Borst P., Zelcer N., van de Wetering K. (2006) Cancer Lett. 234, 51–61 [DOI] [PubMed] [Google Scholar]
- 2.Jedlitschky G., Hoffmann U., Kroemer H. K. (2006) Exp. Opin. Drug Metab. Toxicol. 2, 351–366 [DOI] [PubMed] [Google Scholar]
- 3.Trauner M., Boyer J. L. (2003) Physiol. Rev. 83, 633–671 [DOI] [PubMed] [Google Scholar]
- 4.Geier A., Wagner M., Dietrich C. G., Trauner M. (2007) Biochim. Biophys. Acta 1773, 283–308 [DOI] [PubMed] [Google Scholar]
- 5.Nies A. T., Keppler D. (2007) Pflugers Archiv – Eur. J. Physiol. 453, 643–659 [DOI] [PubMed] [Google Scholar]
- 6.Ballatori N., Truong A. T. (1992) Am. J. Physiol. 263, G617–G624 [DOI] [PubMed] [Google Scholar]
- 7.Kartenbeck J., Leuschner U., Mayer R., Keppler D. (1996) Hepatology 23, 1061–1066 [DOI] [PubMed] [Google Scholar]
- 8.Paulusma C. C., Kool M., Bosma P. J., Scheffer G. L., ter Borg F., Scheper R. J., Tytgat G. N., Borst P., Baas F., Oude Elferink R. P. (1997) Hepatology 25, 1539–1542 [DOI] [PubMed] [Google Scholar]
- 9.Mottino A. D., Catania V. A. (2008) World J. Gastroenterol. 14, 7068–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kipp H., Pichetshote N., Arias I. M. (2001) J. Biol. Chem. 276, 7218–7224 [DOI] [PubMed] [Google Scholar]
- 11.Roma M. G., Crocenzi F. A., Mottino A. D. (2008) World J. Gastroenterol. 14, 6786–6801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roelofsen H., Soroka C. J., Keppler D., Boyer J. L. (1998) J. Cell Sci. 111, 1137–1145 [DOI] [PubMed] [Google Scholar]
- 13.Roma M. G., Milkiewicz P., Elias E., Coleman R. (2000) Hepatology 32, 1342–1356 [DOI] [PubMed] [Google Scholar]
- 14.Kojima H., Sakurai S., Uemura M., Kitamura K., Kanno H., Nakai Y., Fukui H. (2008) J. Gastroenterol. Hepatol. 23, e120–e128 [DOI] [PubMed] [Google Scholar]
- 15.Konno T., Ebihara T., Hisaeda K., Uchiumi T., Nakamura T., Shirakusa T., Kuwano M., Wada M. (2003) J. Biol. Chem. 278, 22908–22917 [DOI] [PubMed] [Google Scholar]
- 16.Kubitz R., Wettstein M., Warskulat U., Häussinger D. (1999) Gastroenterology 116, 401–410 [DOI] [PubMed] [Google Scholar]
- 17.Mottino A. D., Cao J., Veggi L. M., Crocenzi F., Roma M. G., Vore M. (2002) Hepatology 35, 1409–1419 [DOI] [PubMed] [Google Scholar]
- 18.Paulusma C. C., Kothe M. J., Bakker C. T., Bosma P. J., van Bokhoven I., van Marle J., Bolder U., Tytgat G. N., Oude Elferink R. P. (2000) Hepatology 31, 684–693 [DOI] [PubMed] [Google Scholar]
- 19.Sekine S., Ito K., Horie T. (2006) Free Rad. Biol. Med. 40, 2166–2174 [DOI] [PubMed] [Google Scholar]
- 20.Kojima H., Nies A. T., König J., Hagmann W., Spring H., Uemura M., Fukui H., Keppler D. (2003) J. Hepatol. 39, 693–702 [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi S., Hata M., Fukumoto K., Yamane Y., Matsui T., Tamura A., Yonemura S., Yamagishi H., Keppler D., Tsukita S., Tsukita S. (2002) Nat. Genet. 31, 320–325 [DOI] [PubMed] [Google Scholar]
- 22.Wang W., Soroka C. J., Mennone A., Rahner C., Harry K., Pypaert M., Boyer J. L. (2006) Gastroenterology 131, 878–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reczek D., Berryman M., Bretscher A. (1997) J. Cell Biol. 139, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinman E. J., Steplock D., Wang Y., Shenolikar S. (1995) J. Clin. Investig. 95, 2143–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wade J. B., Welling P. A., Donowitz M., Shenolikar S., Weinman E. J. (2001) Am. J. Physiol. - Cell Physiol. 280, C192–C198 [DOI] [PubMed] [Google Scholar]
- 26.Weinman E. J., Cunningham R., Wade J. B., Shenolikar S. (2005) J. Physiol. 567, 27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinman E. J., Hall R. A., Friedman P. A., Liu-Chen L. Y., Shenolikar S. (2006) Annu. Rev. Physiol. 68, 491–505 [DOI] [PubMed] [Google Scholar]
- 28.Brône B., Eggermont J. (2005) Am. J. Physiol. - Cell Physiol. 288, C20–C29 [DOI] [PubMed] [Google Scholar]
- 29.Lamprecht G., Seidler U. (2006) Am. J. Physiol. – Gastrointest. Liver Physiol. 291, G766–G777 [DOI] [PubMed] [Google Scholar]
- 30.Haggie P. M., Kim J. K., Lukacs G. L., Verkman A. S. (2006) Mol. Biol. Cell 17, 4937–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swiatecka-Urban A., Duhaime M., Coutermarsh B., Karlson K. H., Collawn J., Milewski M., Cutting G. R., Guggino W. B., Langford G., Stanton B. A. (2002) J. Biol. Chem. 277, 40099–40105 [DOI] [PubMed] [Google Scholar]
- 32.Cao T. T., Deacon H. W., Reczek D., Bretscher A., von Zastrow M. (1999) Nature 401, 286–290 [DOI] [PubMed] [Google Scholar]
- 33.Cunningham R., Brazie M., Kanumuru S., E X., Biswas R., Wang F., Steplock D., Wade J. B., Anzai N., Endou H., Shenolikar S., Weinman E. J. (2007) J. Am. Soc. Nephrol. 18, 1419–1425 [DOI] [PubMed] [Google Scholar]
- 34.Shenolikar S., Voltz J. W., Minkoff C. M., Wade J. B., Weinman E. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11470–11475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun F., Hug M. J., Bradbury N. A., Frizzell R. A. (2000) J. Biol. Chem. 275, 14360–14366 [DOI] [PubMed] [Google Scholar]
- 36.Weinman E. J., Steplock D., Shenolikar S. (2003) FEBS Letts. 536, 141–144 [DOI] [PubMed] [Google Scholar]
- 37.Fouassier L., Duan C. Y., Feranchak A. P., Yun C. H., Sutherland E., Simon F., Fitz J. G., Doctor R. B. (2001) Hepatology 33, 166–176 [DOI] [PubMed] [Google Scholar]
- 38.Fouassier L., Rosenberg P., Mergey M., Saubaméa B., Clapéron A., Kinnman N., Chignard N., Jacobsson-Ekman G., Strandvik B., Rey C., Barbu V., Hultcrantz R., Housset C. (2009) Am. J. Pathol. 174, 869–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegedus T., Sessler T., Scott R., Thelin W., Bakos E., Varadi A., Szabo K., Homolya L., Milgram S. L., Sarkadi B. (2003) Biochem. Biophys. Res. Commun. 302, 454–461 [DOI] [PubMed] [Google Scholar]
- 40.Fouassier L., Yun C. C., Fitz J. G., Doctor R. B. (2000) J. Biol. Chem. 275, 25039–25045 [DOI] [PubMed] [Google Scholar]
- 41.Ballatori N., Christian W. V., Lee J. Y., Dawson P. A., Soroka C. J., Boyer J. L., Madejczyk M. S., Li N. (2005) Hepatology 42, 1270–1279 [DOI] [PubMed] [Google Scholar]
- 42.Tietze F. (1969) Anal. Biochem. 27, 502–522 [DOI] [PubMed] [Google Scholar]
- 43.Shaik I. H., Mehvar R. (2006) Anal. Bioanal. Chem. 385, 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyer J. L., Soroka C. J. (1995) Gastroenterology 109, 1600–1611 [DOI] [PubMed] [Google Scholar]
- 45.Broere N., Hillesheim J., Tuo B., Jorna H., Houtsmuller A. B., Shenolikar S., Weinman E. J., Donowitz M., Seidler U., de Jonge H. R., Hogema B. M. (2007) J. Biol. Chem. 282, 37575–37584 [DOI] [PubMed] [Google Scholar]
- 46.Lee J. H., Richter W., Namkung W., Kim K. H., Kim E., Conti M., Lee M. G. (2007) J. Biol. Chem. 282, 10414–10422 [DOI] [PubMed] [Google Scholar]
- 47.Voltz J. W., Brush M., Sikes S., Steplock D., Weinman E. J., Shenolikar S. (2007) J. Biol. Chem. 282, 33879–33887 [DOI] [PubMed] [Google Scholar]
- 48.Pietrement C., Da Silva N., Silberstein C., James M., Marsolais M., Van Hoek A., Brown D., Pastor-Soler N., Ameen N., Laprade R., Ramesh V., Breton S. (2008) J. Biol. Chem. 283, 2986–2996 [DOI] [PubMed] [Google Scholar]
- 49.Kocher O., Comella N., Gilchrist A., Pal R., Tognazzi K., Brown L. F., Knoll J. H. (1999) Lab. Investig. 79, 1161–1170 [PubMed] [Google Scholar]
- 50.Kocher O., Pal R., Roberts M., Cirovic C., Gilchrist A. (2003) Mol. Cell. Biol. 23, 1175–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris M. J., Kuwano M., Webb M., Board P. G. (2001) J. Biol. Chem. 276, 20876–20881 [DOI] [PubMed] [Google Scholar]
- 52.Nies A. T., König J., Cui Y., Brom M., Spring H., Keppler D. (2002) Eur. J. Biochem. 269, 1866–1876 [DOI] [PubMed] [Google Scholar]
- 53.Bandler P. E., Westlake C. J., Grant C. E., Cole S. P., Deeley R. G. (2008) Mol. Pharmacol. 74, 9–19 [DOI] [PubMed] [Google Scholar]
- 54.Hoque M. T., Cole S. P. (2008) Cancer Res. 68, 4802–4809 [DOI] [PubMed] [Google Scholar]
- 55.Russel F. G., Koenderink J. B., Masereeuw R. (2008) Trends Pharmacol. Sci. 29, 200–207 [DOI] [PubMed] [Google Scholar]
- 56.Rius M., Nies A. T., Hummel-Eisenbeiss J., Jedlitschky G., Keppler D. (2003) Hepatology 38, 374–384 [DOI] [PubMed] [Google Scholar]
- 57.Hoque M. T., Conseil G., Cole S. P. (2009) Biochem. Biophys. Res. Commun. 379, 60–64 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.