Abstract
Salla disease and infantile sialic acid storage disorder are human diseases caused by loss of function of sialin, a lysosomal transporter that mediates H+-coupled symport of acidic sugars N-acetylneuraminic acid and glucuronic acid out of lysosomes. Along with the closely related vesicular glutamate transporters, sialin belongs to the SLC17 transporter family. Despite their critical role in health and disease, these proteins remain poorly understood both structurally and mechanistically. Here, we use substituted cysteine accessibility screening and radiotracer flux assays to evaluate experimentally a computationally generated three-dimensional structure model of sialin. According to this model, sialin consists of 12 transmembrane helices (TMs) with an overall architecture similar to that of the distantly related glycerol 3-phosphate transporter GlpT. We show that TM4 in sialin lines a large aqueous cavity that forms a part of the substrate permeation pathway and demonstrate substrate-induced alterations in accessibility of substituted cysteine residues in TM4. In addition, we demonstrate that one mutant, F179C, has a dramatically different effect on the apparent affinity and transport rate for N-acetylneuraminic acid and glucuronic acid, suggesting that it may be directly involved in substrate recognition and/or translocation. These findings offer a basis for further defining the transport mechanism of sialin and other SLC17 family members.
Keywords: Carbohydrate Metabolism, Ganglioside, Glycoprotein, Lysosomal Storage Disease, Lysosomes, Cysteine Scanning, Sialic Acid, Sialin, Transporter
Introduction
The lysosomal transporter sialin was first identified through genetic studies of the human free sialic acid storage disorders, Salla disease and infantile sialic acid storage disorder, a group of diseases in which the sialic acid N-acetylneuraminic acid (NANA)2 and glucuronic acid (GlcUA) accumulate in lysosomes (1). These acidic sugars are normally transported out of the lysosome after they are released during degradation of glycolipids and glycoproteins. Loss of sialin function leads to their accumulation in lysosomes and severe neurodevelopmental abnormalities. Studies of the recombinant protein have confirmed that sialin catalyzes the electroneutral proton-coupled transport of NANA and GlcUA (2–4).
Sialin is a member of the SLC17 solute carrier family, a group of structurally related polytopic membrane proteins that is part of the major facilitator superfamily of transporters (5). Also included within the SLC17 family are the vesicular neurotransmitter transporters for glutamate (VGLUT1–3) and purines (VNUT). All of the functionally characterized members of this family mediate organic anion transport, but a common mechanism may not be shared; VGLUT1 transport appears to involve glutamate/Cl− exchange, whereas sialin-mediated transport of NANA and GlcUA is Cl−-independent (6, 7). Further, it has been suggested that members of the SLC17 family have functional properties in addition to organic anion transport, including Na+-dependent phosphate transport and Cl− transport (7–10).
Limited information is available on the structure of sialin or the other SLC17 family members. Genetic studies have identified a number of disease-associated missense mutations in sialin and VGLUT3 (2, 3, 10–13). However, the mutated residues are distributed throughout the protein and thus provide little insight into the structure-function relationship of these proteins. Homology modeling of VGLUT1 and VGLUT2 (10, 14) suggest 12 transmembrane segments (TMs) connected by hydrophilic loops with an overall architecture similar to the distantly related bacterial glycerol 3-phosphate transporter GlpT (15). Site-directed mutagenesis of VGLUT2 driven by this model has been used to demonstrate that a His in TM2 that is specific to the VGLUTs and Arg and Glu residues in TM4 that are conserved in the SLC17 family are required for transport activity (10).
To gain insight into the molecular mechanism of sialin-mediated transport in a structural context, we have applied computational modeling in combination with cysteine scanning mutagenesis and radiotracer flux studies. Our data suggest that sialin has a deep water-filled vestibule that forms part of the substrate permeation pathway. We identified two amino acid positions in the presumed permeation pathway that display substrate-induced alterations in accessibility to thiol-reactive reagents. Further, we found that replacement of Phe-179, which is deep within the aqueous vestibule, with cysteine resulted in a nearly 3-fold increased in VmaxGlcUA along with a KmGlcUA that was increased by more than 1 order of magnitude. In striking contrast, NANA transport remained unchanged. The differential effects on the turnover rate and apparent affinity for GlcUA and NANA transport suggest that Phe-179 may be involved in substrate recognition and/or translocation.
EXPERIMENTAL PROCEDURES
Reagents
Methanethiosulfonate-ethyltrimethylammonium (MTSET) was obtained from Anatrace (Maumee, OH) and N-ethylmaleimide (NEM) from Sigma-Aldrich. 3H-NANA (20 Ci/mmol) and 3H-GlcUA (10 Ci/mmol) were obtained from American Radiolabeled Chemicals (St. Louis, MO). Primers were synthesized by Eurofins MWG Operon (Huntsville, AL). All other reagents were obtained from EMD Chemicals (Gibbstown, NJ).
Bioinformatic Analyses
TMHMM, PredictProtein, and TmPred were used to genereate topology predictions for sialin. Hydrophobicity profiles were generated using the method of Kyte and Doolittle (16) with a sliding window of 19 residues. The three-dimensional model of sialin based on the crystal structure of the glycerol 3-phosphate transporter GlpT was generated using Modeler (17).
Heterologous Expression and Functional Characterization of Rat Sialin Isoforms
Site-directed mutagenesis was carried using the QuikChange kit (Stratagene) according to the manufacturer's directions using oligonucleotide primers listed in supplemental Tables 1 and 2. Heterologous expression of sialin isoforms in HeLa cells, whole cell uptake assays, and immunofluorescence were performed as described previously (2) with minor modifications. Confocal images were collected with a Leica TCS SPE Spectral microscope, and ImageJ-64 software was used to stack five serial images for a final merged image.
Sulfhydryl-reactive Reagent Accessibility
For NEM inhibition assays, cells were washed once with Krebs-Ringer-HEPES (KRH), pH 7.5, and then incubated in the same buffer supplemented with NEM at a concentration of 2 mm for 5 min at 23 °C. Reactions were stopped by adding 4 volumes of KRH, pH 7.5, and the cells were washed once more with the same buffer. Transport was then carried out as indicated above. MTSET inhibition was assayed in an identical manner, but KRM, pH 5.5, was used instead of KRH throughout the protocol. To test the effects of substrates or pH, cells were incubated in KRM, pH 5.5, or KRH, pH 7.0 or pH 8.5, with or without substrates at indicated concentrations for 8 min at 23 °C. Sulfhydryl reagents were added to the final indicated concentration, and incubation was continued for 2 min. The reaction was stopped by dilution, the cells were washed once, and then uptake was measured as indicated above.
Statistics
Statistical analyses were performed using Prism (Graph-Pad Software, San Diego, CA). Reported values are mean ± S.E. from a minimum of four independent experiments. Calculations for p values were done using two-tailed unpaired t test or one-way analysis of variance with Bonferroni correction as appropriate.
RESULTS
Sialin Is Predicted to Have 12 Transmembrane Segments
To develop a structural model for sialin, we analyzed the amino acid sequence of the rat and human isoforms in a multiple sequence file with the web-based topology algorithm PredictProtein. When both human and rat sialin were used together for the analysis, 12 TMs were identified with both the N terminus and the C terminus in the cytoplasm (Fig. 1A). Previous biochemical studies on the closely related VGLUTs have demonstrated that the N- and C-terminal domains are located in the cytoplasm, suggesting an even number of TM segments (18, 19). We further assessed the sialin topology model by determining whether it follows the positive-inside rule. This rule states that positively charged Lys and Arg residues are more commonly clustered in cytoplasmic segments than in extracytoplasmic segments of integral membrane proteins (20). In support of the topology model, we found an enrichment of Lys and Arg in the cytoplasmic segments of the model (18 cytoplasmic and 9 lumenal).
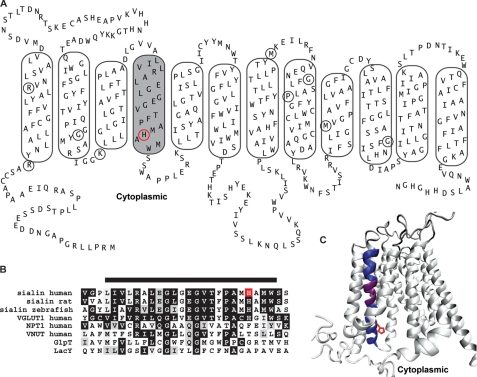
FIGURE 1.
Conserved α-helical structure is predicted to form TM4 in sialin. A, PredictProtein-derived topology model for rat and human sialin sequences with manual adjustments. Putative TMs are outlined with TM4 shaded. Circled residues correspond to those mutated in the lysosomal free sialic acid storage disorders. B, alignment of TM4 and the surrounding residues in sialin of indicated species and human isoforms of VGLUT1, the Na+-dependent phosphate transporter, and VNUT, and the bacterial transporters GlpT and LacY. Bar above the sequence indicates residues predicted to form TM4. C, three-dimensional model of sialin based on crystal structure of GlpT. TM4 is colored dark blue with the conserved GXXXG motif in purple. His-183, a TM4 residue affected by a disease-associated mutation (H183R), is depicted in red in A (circle), B (box), and C (side chain).
To generate a three-dimensional model of sialin, we performed a BLAST search of the Research Collaboratory for Structural Bioinformatics Protein Data Bank, which identified bacterial GlpT as the protein with greatest similarity to sialin for which a crystallographic structure is available. Using GlpT crystal structure as a template for homology modeling, we generated a three-dimensional structure of sialin using Modeller (17). The resulting structure was very similar to the GlpT-based three-dimensional structures of rat VGLUT2 and human VGLUT1 (10, 14). The best structure obtained for sialin (depicted in Fig. 1C) has a z-score of 53 and root mean square deviation of 1.3 Å with a level of identity of 20% based on DaliLite Pairwise Comparison of Protein Structures (21). In this model there is a water-filled cavity opened to the cytoplasmic side with the cavity wall formed by portions of TM1, TM2, TM4, TM5, TM7, TM8, TM10, and TM11. Of these segments, TM4 has the most charged residues (three; Arg-168, Glu-171, and Glu-175) as well as a histidine (His-183). These four residues lie on the same face of TM4. Further, three of the residues (Arg-168, Glu-175, and His-183) are highly conserved in the SLC17 family, suggesting that they may participate directly in substrate binding and/or translocation. In the model TM4 also contains a GXXXG helix packing motif (22) that is conserved in all members of the SLC17 family (Fig. 1B). This motif is also found in TM4 of GlpT and the lactose permease LacY, both of which are members of the major facilitator superfamily family. In the crystal structures of GlpT and LacY (15, 23) and the sialin model, the GXXXG motif in TM4 is located in close proximity to TM2 (Fig. 1C).
Native Cysteines Are Not Required for Sialin Function
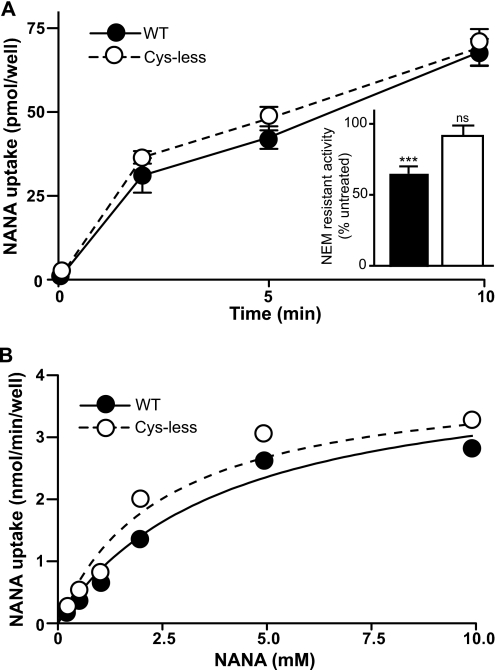
The substituted cysteine accessibility method is a well established approach for obtaining biochemically derived structural information for polytopic membrane proteins and has proved useful in characterizing the role of specific residues in the function of transporters including LacY and the bacterial sugar phosphate transporter UhpT (24–29). A prerequisite is a functional transporter devoid of reactive Cys residues. The rat isoform of sialin has 10 intrinsic Cys residues that we replaced in a conservative manner with Ala, Ser, Phe, or Tyr (supplemental Table 1) in a plasma membrane-targeted isoform of the protein (2). The orientation of the plasma membrane-targeted sialin is such that the lumenal side is facing out so that whole cell uptake corresponds to the physiological direction of substrate transport out of the lysosomes. Cells transfected with the wild-type and Cys-less isoforms transported the sialic acid NANA with similar initial rates and steady-state levels of accumulation (Fig. 2A). After treatment with the small membrane-permeant sulfhydryl-alkylating reagent NEM, we detected ∼35% inhibition of transport by wild-type protein, but no significant inhibition of NANA uptake by Cys-less sialin (Fig. 2A, inset). A detailed kinetic analysis of the initial rates of NANA uptake measured at 2 min, which is within the initial linear portion of transport time course, also revealed a similar KmNANA and VmaxNANA for the two isoforms (Fig. 2B).
FIGURE 2.
Wild-type and Cys-less sialin exhibit similar transport kinetics. A, time course of 3H-NANA uptake (30 nm final concentration) by wild-type (WT) sialin- (filled circles, solid line) and Cys-less sialin- (open circles, dashed line) transfected cells. NEM treatment (2 mm, pH 7.5 for 5 min at 23 °C) inhibits NANA uptake (30 nm final concentration, measured at 5 min) into cells transfected with the wild-type protein, but not Cys-less-transfected cells (inset). B, wild-type- and Cys-less sialin-transfected cells exhibit saturable NANA uptake. Measurement of NANA uptake at 2 min in the presence of increasing substrate concentrations reveals KmNANA values of 4 ± 2 and 5 ± 2 mm for wild-type and Cys-less sialin, respectively, with a VmNANA of ∼5 nmol/min per well for both. ***, p < 0.0001; ns, no significance.
Five Residues in TM4 Do Not Tolerate Cysteine Substitutions
In our three-dimensional model, TM4 is predicted to form an amphiphilic helix with three charged residues (Arg-168, Glu-171, and Glu-175) and a histidine (His-183) facing toward a large aqueous cavity, whereas no other TM segment has more than one charged residue or histidine. Because charged residues and histidines within TM segments can play important roles in substrate and proton translocation (30–33), we suspected that scanning mutagenesis of TM4 would lead to identification of residues with crucial roles in sialin function so we individually mutated each of the residues in TM4 to cysteine. Of the resulting 23 single monocysteine mutants, all but five (R168C, G172C, G176C, P180C, and W186C) mediated measurable transport (Fig. 3A). Immunofluorescence studies indicated that of these only R168C and W186C had patterns of expression similar to the Cys-less protein (supplemental Fig. 1). For the other mutants (G172C, G176C, and P180C) the proteins were expressed at lower levels with much of the protein retained intracellularly. This pattern is similar to that of disease-associated missense mutants G371V and G409E mutants that are believed to be improperly folded or unstable (2, 11) and is consistent with crucial roles for prolines and GXXXG motifs in the structure of TMs and their association with other TMs (22, 34). Monocysteine mutants E171C and E175C exhibited ∼60 and ∼90% of the Cys-less sialin activity, respectively, a surprising observation given their absolute conserved position in sialin across species. To determine whether these residues might be serving redundant functions, we tested the double mutant (E171C/E175C). Again, we found that this mutant catalyzed NANA transport, but at ∼40% of that Cys-less activity (Fig. 3A). We also tested a mutant with the glutamate residues replaced by alanine residues (E171A/E175A) and found that it had a similar level of activity (∼35% of that Cys-less). The uptake mediated by these mutants indicates that the glutamate residues (one or both of which may be negatively charged at pH 5.5) are not essential for NANA transport.
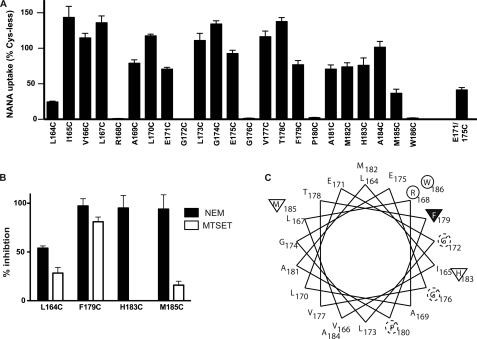
FIGURE 3.
Effects of Cys substitution and subsequent thiol-reactive reagent treatment on residues in TM4. A, NANA transport activity (relative to Cys-less sialin) for each TM4 monocysteine substitution mutant. Transport of 30 nm 3H-NANA was measured after 5 min at pH 5.5. B, inhibition of monocysteine substitution mutants after NEM (filled bars) or MTSET (open bars) pretreatment. Only monocysteine substitution mutants for which NEM inhibited activity by greater than 15% are shown. The effects on the other mutants are presented in supplemental Fig. 2. Cells were pretreated in presence of 2 mm NEM at pH 7.5 or 2 mm MTSET at pH 5.5 for 5 min and transport then measured as in A. C, helical wheel plot of TM4. Circles indicate residues where cysteine substitution was associated with loss of >95% measurable transport. Dashed lines indicate residues where substitution led to markedly decreased expression (see supplemental Fig. 1). Triangles indicate positions where cysteine substitution led to NEM-sensitive transport with the filled triangle at Phe-179 indicating that substitution led to inhibition by MTSET as well as NEM.
TM4 Lines a Deep Outwardly Facing Cavity
To assess our model further, we measured the inhibitory effect of NEM on NANA uptake activity for the 18 functional monocysteine sialin mutants. NEM is small and membrane-permeant, but accessibility to a given cysteine is limited by its proximity to other helices and/or the lipid bilayer (35). To increase the likelihood of identifying positions in TM4 that are readily accessible to NEM, we performed a long incubation (5 min) with a high concentration of NEM (2 mm) in a weakly alkaline buffer (pH 7.5). Transport activities of only four mutants (L164C, F179C, H183C, and M185C) were markedly inhibited by NEM (Fig. 3B), indicating that these residues are not sterically blocked by other parts of the protein or the lipid environment. The activities of the other 14 functional monocysteine mutants were inhibited by less than 15% (supplemental Fig. 2).
To probe further the local environment of the cysteine residues in L164C, F179C, H183C, and M185C, we measured the accessibility of the respective site-directed engineered thiol groups to MTSET. MTSET is larger than NEM, positively charged, and membrane-impermeant (36–38), thus it will only react with thiol groups of cysteines that are accessible from the aqueous phase, i.e. exposed to the outside of the cell in our experiments. Incubating cells expressing the NEM-sensitive monocysteine mutants with a high concentration of MTSET at a low pH (2 mm, pH 5.5, conditions that favor reactivity of MTSET), we found that only L164C and F179C were markedly inhibited (Fig. 3B). Pretreatment of H183C and M185C with MTSET did not reduce the effect of NEM (97.7% and 88.3% inhibition for H183C and M185C, respectively), confirming that these residues are not readily accessible to MTSET. Taken together, the effects of NEM and MTSET on the monocysteine mutants suggest that this region of TM4 lines an outward facing aqueous cavity that reaches at least as deep as Phe-179 (note that Phe-179 is one helical turn above His-183 (side chain in red in Fig. 1C). For the native protein localized to the lysosome, an outwardly open conformation would correspond to the deep cavity of sialin open to the lumen.
Accessibility to F179C Is Altered by pH
Accessibility of thiol-reactive compounds to cysteines can be blocked directly by bound substrate or by substrate-induced conformational changes that occur in close proximity to the reporter position (27, 35, 39). Because F179C and H183C are deep within the outwardly facing vestibule and possibly near the substrate binding site we wondered whether substrates or the co-transported H+ can influence accessibility of these residues to NEM or MTSET.
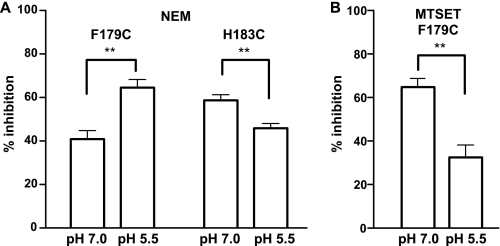
To assess the potential effect of H+ availability, we performed NEM and MTSET pretreatment at pH 5.5, which supports robust transport, and separately at pH 7.0, a H+ concentration at which there is little measurable transport. To increase the likelihood that increases or decreases in accessibility could be detected, we adjusted the concentration of the thiol-reactive reagents and the duration of the incubations for these experiments such that there was ∼40–70% inhibition at pH 7.0. Compared with pH 7.0 at pH 5.5, NEM inhibition was increased for F179C but decreased for H183C (Fig. 4A), whereas MTSET inhibition of F179C was decreased at the lower pH (Fig. 4B). Because NEM is less reactive at lower pH, the effect of pH on NEM reactivity for H183C may not reflect a change in the accessibility of this residue to NEM. The increased NEM inhibition at the lower pH and increased MTSET inhibition at the higher pH for F179C, however, is most consistent with a pH-dependent alteration in the local environment and aqueous accessibility of F179C.
FIGURE 4.
Modulation of thiol-reactive reagent-dependent inhibition by pH. A, effect of NEM incubation (1 mm for 2 min) on activity of F179C and H183C incubated at pH 7.0 and pH 5.5 in absence of substrate. B, effects of MTSET incubation (0.2 mm for 2 min) at pH 7.0 (left) and pH 5.5 (right) in the absence of substrate. For all experiments, pretreatment was for a total of 10 min: the first 8 min at the indicate pH alone and the final 2 min at the same pH with NEM or MTSET. For transport measurements, the concentration of NANA was 30 nm, and the reaction was terminated at 5 min. **, p < 0.01.
NANA and GlcUA Differentially Affect Accessibility of F179C and H183C
Sialin has been shown to mediate both the electroneutral H+ coupled symport of the acidic sugars NANA and GlcUA (2, 3, 6, 40) and the membrane potential-dependent transport of glutamate and aspartate in a reconstituted system (4). Although we detected robust transport of NANA (Fig. 2) and GlcUA (see Fig. 6), we were unable to detect sialin-mediated transport of glutamate or aspartate under several different conditions with the wild-type or Cys-less plasma membrane-targeted proteins, and therefore we limited our studies on accessibility to the effects of NANA and GlcUA.
FIGURE 6.
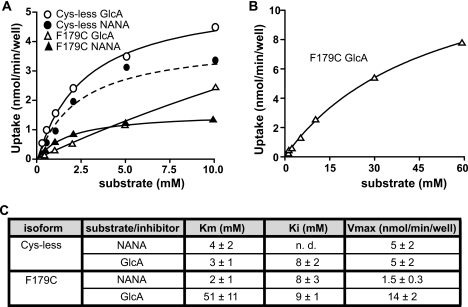
F179C mutation increases the Km but not the Ki value for GlcUA. A, concentration-dependent uptake of NANA (filled symbols) and GlcUA (GlcA; open symbols) by Cys-less (circles) and F179C (triangles) sialin. Values for NANA uptake by Cys-less sialin are also presented in Fig. 2. B, broad range plot of F179C-mediated uptake of GlcUA. C, summary of Km and Ki and Vmax measurements for Cys-less and F179C-mediated transport of NANA and GlcUA. Km and Vmax values were calculated from Michaelis-Menten plots, and Ki values were calculated from IC50 plots using the Cheng-Prusoff equation. For all experiments, uptake was measured for 2 min. Km, Vmax, and Ki values were calculated from four independent experiments and are presented as the mean ± S.E.
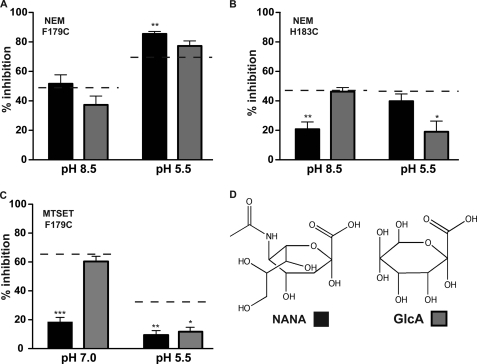
To determine whether substrates influence accessibility in a pH-dependent manner, we tested their effect at pH 5.5, which facilitates robust transport, and a more alkaline pH (pH 8.5 for NEM and pH 7.0 for MTSET because of its short half-life at alkaline pH). Because the pKa values of NANA and GlcUA are 2.6 and 2.9, respectively (41, 42), both substrates are unprotonated throughout the range tested. Again, to increase the likelihood that increases or decreases in accessibility could be detected, we adjusted the concentration of the thiol-reactive reagents and the duration of the incubations for these experiments such that there was ∼40–70% inhibition at specific pH in the absence of substrate. We found that NEM inhibition of F179C (Fig. 5A) was slightly increased at pH 5.5 by the presence of NANA but not GlcUA. Neither substrate had a significant effect on NEM inhibition of F179C at pH 8.5. For NEM inhibition of H183C (Fig. 5B), we found that NANA caused a marked reduction at pH 8.5 with little effect at pH 5.5. In comparison, GlcUA had no measurable effect at pH 8.5, but significantly decreased NEM inhibition at pH 5.5. With MTSET inhibition of F179C (Fig. 5C) when the pretreatment was performed at pH 7.0, NANA reduced the inhibitory effect, but GlcUA failed to protect F179C from MTSET inactivation. At pH 5.5, however, both substrates reduced the extent of MTSET inhibition, indicating that during transport accessibility of MTSET to F179C is reduced.
FIGURE 5.
Modulation of thiol-reactive reagent-dependent inhibition by substrates. A, inhibition of F179C activity by NEM preincubation at pH 8.5 and 5.5 in the presence of NANA (20 mm; black bars) or GlcUA (20 mm; gray bars). Dashed line indicates the level of inhibition in absence of substrate. B, inhibition of H183C activity by NEM as described in A. C, inhibition of F179C by MTEST as described in A. D, structure of NANA and GlcUA (GlcA). Control values were determined for the indicated mutants in the absence of pretreatment. For all experiments, pretreatment was for a total of 10 min: the first 8 min with substrate alone and the final 2 min with substrate and MTSET. For transport measurements the concentration of NANA was 30 nm, and the reaction was terminated at 5 min. *, p < 0.05; **, p < 0.01; ***, p < 0.0001.
Mutation F179C Differentially Affects the Kinetics of GlcUA and NANA Uptake
The differential effect of NANA and GlcUA on accessibility of MTSET to F179C suggested that this residue might be directly involved in substrate recognition. To test this hypothesis we compared the transport of these two substrates by the Cys-less and F179C isoforms (Fig. 6, A and B). Uptake of NANA by the F179C mutant featured Km and Vmax values similar to those determined for the Cys-less isoform. GlcUA transport by the two proteins, however, differed dramatically. The KmGlcUA and VmaxGlcUA were increased ∼15-fold and ∼3-fold, respectively.
NANA and GlcUA have been shown to be competitive substrates for sialin-mediated transport with similar Km and Ki values (6, 40, 43). Given the high KmGlcUA of F179C, we expected that GlcUA would be a low affinity inhibitor of NANA transport for this protein. To test this we first confirmed that GlcUA inhibits NANA transport competitively by measuring the KmNANA in the presence of different concentrations of GlcUA. The observed KmNANA was increased with increasing concentrations of GlcUA whereas VmNANA was unchanged, consistent with competitive inhibition (supplemental Fig. 3). From the IC50 plot and the Cheng-Prusoff equation, we determined the KiGlcUA for NANA transport by F179C to be ∼9 mm, similar to the KiGlcUA for NANA transport by Cys-less sialin and the KmNANA for GlcUA transport by F179C (Fig. 6C). Thus, compared with Cys-less sialin, the F179C mutant exhibited markedly modified kinetics for GlcUA transport, but similar GlcUA-mediated inhibition on NANA transport.
DISCUSSION
Sialin and other members of the SLC17 family of proteins form a group of structurally related transporters with diverse, but crucial physiological functions. Here, using homology modeling and cysteine scanning mutagenesis, we present data that provide new insights into the structure of sialin and mechanisms underlying the transport process it catalyzes.
Our structural model for sialin is derived from the crystal structure of the distantly related GlpT. Sialin and GlpT have limited similarity in their primary sequences, so how valid is this model? A number of our findings suggest that it is a reasonable approximation. The PredictProtein-based secondary structure of sialin (specifically the α-helical TMs) aligns well with the crystallographic structure of GlpT. Within TM4 we also found that the GXXXG helix-packing motif, which is present in all SLC17 family members and GlpT, is required for expression of sialin. This suggests a conserved structure for TM4 that involves interaction with another helix. A number of other GXXXG-like motifs located within TMs (GXXXG/S in TM2, G/AXXXG in TM5, and G/S/AXXXGXXXG/T in TM11) are also conserved in all SLC17 family members and GlpT, indicating conservation of helix-packing structure beyond TM4.
In the GlpT-based model TM4 lines a cavity that likely forms a permeation pathway. Do our data support this? Our findings that Arg-168 and Trp-186 have a limited tolerance for substitution is consistent with TM4 forming a wall of the permeation pathway as are the inhibitory effects of thiol-reactive agents on F179C- and H183C-mediated transport. The modulation of thiol-reactive reagent accessibility to F179C and H183C by the transported substrates also strongly supports a model with these residues in the substrate translocation pathway. Interestingly, all four of these residues are within a narrow stripe along the helical wheel plot of TM4, suggesting that a small but crucial part of helix may line the permeation path.
There is a major discrepancy between the GlpT-based three-dimensional model and our biochemical data. GlpT is closed off to the outside of the cell in the crystal structure (as is our three-dimensional model of sialin; Fig. 1 C), whereas accessibility of F179C sialin to MTSET indicates that at one step in the transport reaction cycle, sialin must have a deep outwardly facing aqueous cavity. This corresponds to a lumenal facing cavity for sialin in its native lysosomal localization, which is not surprising as the physiological role of transporter is to export substrates out of the lysosome. It seems reasonable to assume that sialin undergoes conformational transitions that alternate between inwardly and outwardly open conformations.
Beyond supporting the GlpT-based structural model do our studies also provide insight into the sialin-mediated transport? The intolerance of Arg-168 and Trp-186 to Cys substitution indicates that these residues may be involved in the transport process through substrate binding or facilitating conformational changes. Arg-168 is conserved throughout the entire mammalian SLC17 family and does not even tolerate the conservative substitution to Lys in VGLUT2 (10). Interestingly, Gln is present in the corresponding position of EAT-4, the Caenorhabditis elegans VGLUT ortholog, suggesting that the presence of an Arg at this position is not a universal requirement in the broader family. Trp-186 is also conserved across species in sialin and the VGLUTs, but not in other SLC17 family members. It has been suggested that aromatic residues within sugar transporters such as LacY can be involved in stacking the ring structure of the substrate (44). It is tempting to speculate that this is the case for Trp-186. If so, the corresponding Trp residue in the VGLUTs should not be crucial for glutamate transport.
The limited effect of substituting both Glu-171 and Glu-175 with Cys or Ala was unexpected given the conservation of these residues across species and the prediction that they line the permeation pathway. Although our results indicate that they are not required for H+ coupling, the native Glu residues may underlie, in part, the pH dependence of transport. For example, although not absolutely required, they may facilitate proton translocation in the wild-type protein. Alternatively, they may be involved in substrate binding or influence transporter kinetics. A comprehensive analysis of the pH dependence of the wild-type and mutant proteins may define the role of these glutamate residues and reveal the mechanism of proton translocation.
The analysis of thiol-reactive reagent inhibition as a marker of Cys accessibility provides some further information on H+ and substrate binding. In the absence of substrate, we found an increase in NEM inhibition of F179C when the extracellular pH was dropped and a decrease in MTSET inhibition under similar conditions. This suggests that there is a substrate-independent, H+-mediated conformational change in sialin that occurs in proximity to F179C. We also found that accessibility of F179C to MTSET was markedly reduced by the addition of NANA or GlcUA at the lower pH. The ability of NANA to reduce inhibition was also noted at pH 7.0, suggesting that NANA might bind in the absence of H+ binding (GlcUA may also behave similarly as we did not use saturating concentrations of GlcUA for this mutant). Further supporting a model where substrate binds independently of H+ binding, NANA reduced NEM inhibition of the nearby H183C even when the pH was 8.5, a condition for which there is no measurable transport. These findings together suggest that H+ and sugars likely bind independently of each other and that binding for transport is unordered.
Our finding that the KmGlcUA for F179C was dramatically increased initially suggested to us that this residue is involved in GlcUA binding, but we found little effect of this mutation on the KiGlcUA for NANA transport. How then can the discrepancy between the KmGlcUA and KiGlcUA for NANA transport with F179C be explained? Importantly, transport is a multistep process, and the Km is not equivalent to the Kd (binding dissociation constant) because it also depends upon rate constants of other steps such as translocation and unbinding on the trans side of the membrane. As a result, an increase in the transporter turnover rate can be accompanied by a decrease in the apparent affinity (that is an increase in the Km) with no effect on the binding affinity (or Kd) of the substrate for the transporter. Because the VmaxGluAC for F179C is markedly increased, this could explain the increase in KmGlcUA. Inhibition by competitive substrates reflects competition for binding. The Ki may, therefore, be a better approximation of the actual affinity of the inhibitor to its site. Further supporting this, for F179C the KmNANA and KiNANA for GlcUA transport are reciprocally correlated: the KmNANA is ∼4-fold lower than the KiNANA for GlcUA transport.
A second possibility is that there are two binding sites from which GlcUA can access the permeation pathway from the extracellular space: a high affinity site that is also accessible to NANA and a low affinity site that is accessible only to the smaller GlcUA. This model could explain the different KmGlcUA and KiGlcUA because GlcUA and NANA would only compete at the high affinity site. The low affinity site could be a specific to a transition state, which would also explain the higher turnover rate for GlcUA transport. A two-binding site model with different binding affinities for different substrates is not unprecedented as it has been proposed for the bacterial LeuT transporter (45) and the human dopamine transporter (46). Interestingly, two substrate binding sites have been identified in VGLUT1 through docking studies and molecular dynamic stimulation (14). However, the residue corresponding to Phe-179 is not part of the predicted VGLUT1 binding site. Although this is an intriguing explanation for the discrepancy between the KmGlcUA and KiGlcUA, it would be difficult to prove in the absence of direct structural evidence.
In this structure-function study we have advanced our understanding of the molecular mechanisms underlying substrate recognition and H+-coupled translocation of acidic sugars by sialin. We have demonstrated a crucial role for several residues and have provided evidence for H+- and substrate-dependent conformational changes. We have also identified a single residue, Phe-179, that when mutated to Cys has a substrate-specific influence on transport kinetics. Coupling our findings here and further identification of residues crucial for sialin function with a comparison with the structure of other SLC17 family members may reveal important insights into how these structurally related proteins mediate divergent functions.
Supplementary Material
Acknowledgments
We thank Dr. Merritt Maduke and members of the Reimer laboratory for helpful advice and comments.
This work was supported, in whole or in part, by National Institutes of Health Grants NS050417 and NS045634 (to R. J. R.). This work was also supported by the Fonds de la Recherche en Sante du Quebec and the Stanford University School of Medicine Dean Fellowship Program (to P. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables 1 and 2.
- NANA
- N-acetylneuraminic acid
- VGLUT
- vesicular neurotransmitter transporter for glutamate
- VNUT
- vesicular neurotransmitter for purine
- TM
- transmembrane helix
- MTSET
- methanethiosulfonate-ethyltrimethylammonium
- NEM
- N-ethylmaleimide
- KRH
- Krebs-Ringer-HEPES.
REFERENCES
- 1.Verheijen F. W., Verbeek E., Aula N., Beerens C. E., Havelaar A. C., Joosse M., Peltonen L., Aula P., Galjaard H., van der Spek P. J., Mancini G. M. (1999) Nat. Genet. 23, 462–465 [DOI] [PubMed] [Google Scholar]
- 2.Wreden C. C., Wlizla M., Reimer R. J. (2005) J. Biol. Chem. 280, 1408–1416 [DOI] [PubMed] [Google Scholar]
- 3.Morin P., Sagné C., Gasnier B. (2004) EMBO J. 23, 4560–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyaji T., Echigo N., Hiasa M., Senoh S., Omote H., Moriyama Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11720–11724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reimer R. J., Edwards R. H. (2004) Pflugers Arch. 447, 629–635 [DOI] [PubMed] [Google Scholar]
- 6.Mancini G. M., de Jonge H. R., Galjaard H., Verheijen F. W. (1989) J. Biol. Chem. 264, 15247–15254 [PubMed] [Google Scholar]
- 7.Schenck S., Wojcik S. M., Brose N., Takamori S. (2009) Nat. Neurosci. 12, 156–162 [DOI] [PubMed] [Google Scholar]
- 8.Werner A., Moore M. L., Mantei N., Biber J., Semenza G., Murer H. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9608–9612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni B., Rosteck P. R., Jr., Nadi N. S., Paul S. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5607–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juge N., Yoshida Y., Yatsushiro S., Omote H., Moriyama Y. (2006) J. Biol. Chem. 281, 39499–39506 [DOI] [PubMed] [Google Scholar]
- 11.Myall N. J., Wreden C. C., Wlizla M., Reimer R. J. (2007) Mol. Genet. Metab. 92, 371–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruivo R., Sharifi A., Boubekeur S., Morin P., Anne C., Debacker C., Graziano J. C., Sagné C., Gasnier B. (2008) Biol. Cell 100, 551–559 [DOI] [PubMed] [Google Scholar]
- 13.Ruel J., Emery S., Nouvian R., Bersot T., Amilhon B., Van Rybroek J. M., Rebillard G., Lenoir M., Eybalin M., Delprat B., Sivakumaran T. A., Giros B., El Mestikawy S., Moser T., Smith R. J., Lesperance M. M., Puel J. L. (2008) Am. J. Hum. Genet. 83, 278–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almqvist J., Huang Y., Laaksonen A., Wang D. N., Hovmöller S. (2007) Protein Sci. 16, 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. (2003) Science 301, 616–620 [DOI] [PubMed] [Google Scholar]
- 16.Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 17.Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2006) Curr. Protoc. Bioinformatics, 5.6.1–5.6.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fei H., Karnezis T., Reimer R. J., Krantz D. E. (2007) J. Neurochem. 101, 1662–1671 [DOI] [PubMed] [Google Scholar]
- 19.Jung S. K., Morimoto R., Otsuka M., Omote H. (2006) Biol. Pharm. Bull. 29, 547–549 [DOI] [PubMed] [Google Scholar]
- 20.von Heijne G. (1989) Nature 341, 456–458 [DOI] [PubMed] [Google Scholar]
- 21.Holm L., Kaariainen S., Wilton C., Plewczynski D. (2006) Curr. Protoc. Bioinformatics, 5.5.1–5.5.24 [DOI] [PubMed] [Google Scholar]
- 22.Senes A., Gerstein M., Engelman D. M. (2000) J. Mol. Biol. 296, 921–936 [DOI] [PubMed] [Google Scholar]
- 23.Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. (2003) Science 301, 610–615 [DOI] [PubMed] [Google Scholar]
- 24.Fann M., Davies A. H., Varadhachary A., Kuroda T., Sevier C., Tsuchiya T., Maloney P. C. (1998) J. Membr. Biol. 164, 187–195 [DOI] [PubMed] [Google Scholar]
- 25.Matos M., Fann M. C., Yan R. T., Maloney P. C. (1996) J. Biol. Chem. 271, 18571–18575 [DOI] [PubMed] [Google Scholar]
- 26.Yan R. T., Maloney P. C. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5973–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Javitch J. A. (1998) Methods Enzymol. 296, 331–346 [DOI] [PubMed] [Google Scholar]
- 28.Kaback H. R., Sahin-Tóth M., Weinglass A. B. (2001) Nat. Rev. Mol. Cell Biol. 2, 610–620 [DOI] [PubMed] [Google Scholar]
- 29.Akabas M. H., Stauffer D. A., Xu M., Karlin A. (1992) Science 258, 307–310 [DOI] [PubMed] [Google Scholar]
- 30.Guan L., Nakae T. (2001) J. Bacteriol. 183, 1734–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar R., Bibi E. (1999) EMBO J. 18, 822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahin-Tóth M., Karlin A., Kaback H. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10729–10732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yerushalmi H., Schuldiner S. (2000) J. Biol. Chem. 275, 5264–5269 [DOI] [PubMed] [Google Scholar]
- 34.Brandl C. J., Deber C. M. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 917–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan L., Kaback H. R. (2007) Nat. Protoc. 2, 2012–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu T., Nguyen B., Zhang X., Yang J. (1999) Neuron 22, 571–580 [DOI] [PubMed] [Google Scholar]
- 37.Angelow S., Yu A. S. (2009) J. Biol. Chem. 284, 29205–29217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmgren M., Liu Y., Xu Y., Yellen G. (1996) Neuropharmacology 35, 797–804 [DOI] [PubMed] [Google Scholar]
- 39.Quick M., Yano H., Goldberg N. R., Duan L., Beuming T., Shi L., Weinstein H., Javitch J. A. (2006) J. Biol. Chem. 281, 26444–26454 [DOI] [PubMed] [Google Scholar]
- 40.Havelaar A. C., Mancini G. M., Beerens C. E., Souren R. M., Verheijen F. W. (1998) J. Biol. Chem. 273, 34568–34574 [DOI] [PubMed] [Google Scholar]
- 41.Furuhata K. (2004) Trends Glycosci. Glycotechnol. 16, 143–169 [Google Scholar]
- 42.Wang H. M., Loganathan D., Linhardt R. J. (1991) Biochem. J. 278, 689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mancini G. M., Beerens C. E., Aula P. P., Verheijen F. W. (1991) J. Clin. Invest. 87, 1329–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan L., Hu Y., Kaback H. R. (2003) Biochemistry 42, 1377–1382 [DOI] [PubMed] [Google Scholar]
- 45.Shi L., Quick M., Zhao Y., Weinstein H., Javitch J. A. (2008) Mol. Cell 30, 667–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt K. C., Mamidyala S., Biswas S., Dutta A. K., Reith M. E. (2010) J. Neurochem. 112, 1605–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.