Abstract
Small ubiquitin-like modifier (SUMO) is conjugated to its substrates via an enzymatic cascade consisting of three enzymes, E1, E2, and E3. The active site of the E2 enzyme, Ubc9, recognizes the substrate through binding to a consensus tetrapeptide ΨKXE. However, recent proteomics studies suggested that a considerable part of sumoylation occurs on non-consensus sites. Current unbiased sumoylation site identification techniques typically require high stoichiometry in vitro sumoylation, mass spectrometry, and complex data analysis. To facilitate in vivo analysis, we have designed a mass spectrometric method based on an engineered human SUMO-1 construct that creates a signature tag on SUMO substrates. This construct enables affinity purification by covalent binding to cysteine residues in LysC/trypsin-cleaved peptides and site identification by diglycyl lysine tagging of sumoylation sites. As a proof of concept, site-specific and substrate-unbiased in vivo sumoylation analysis of HeLa cells was performed. We identified 14 sumoylation sites, including well known sites, such as Lys524 of RanGAP1, and novel non-consensus sites. Only 3 of the 14 sites matched consensus sites, supporting the emerging view that non-consensus sumoylation is a common event in live cells. Six of the non-consensus sites had a nearby SUMO interaction motif (SIM), which emphasizes the role of SIM in non-consensus sumoylation. Nevertheless, the lack of nearby SIM residues among the remaining non-consensus sites indicates that there are also other specificity determinants of non-consensus sumoylation. The method we have developed proved to be a useful tool for sumoylation studies and will facilitate identification of novel SUMO substrates containing both consensus and non-consensus sites.
Keywords: Mass Spectrometry (MS), Post-translational Modification, Proteomics, Sumoylation, Ubiquitin-conjugating Enzyme (Ubc)
Introduction
Sumoylation is a post-translational modification that consists of covalent conjugation of the small ubiquitin-like modifier (SUMO)5 to substrate proteins and results in altered activity of the substrate. Sumoylation influences a plethora of cellular processes, including transcriptional regulation of gene expression and genome integrity (1). SUMO conjugation involves an enzymatic cascade employing three enzymes: activating E1, conjugating E2, and ligating E3 (2). Because the active site of the single E2, Ubc9, recognizes the substrate through binding to a consensus tetrapeptide ΨKXE (Ψ, a hydrophobic amino acid; K, the target lysine, X, any amino acid, and E, glutamic acid), SUMO acceptor sites are to date predominantly identified through mutagenesis of target lysine residues on consensus tetrapeptides (3). Recent proteomics studies by us and others have shown that a considerable proportion of sumoylated proteins do not contain the consensus sites (4–6) and are unreachable by the conventional mutagenesis approach. Furthermore, a model for non-consensus SUMO targeting has been proposed, where SUMO-Ubc9 thioester is recruited by a SUMO interaction motif (SIM) located on the substrate (7). However, the mechanisms for targeting non-consensus substrates remain largely unknown. Thus, novel tools that allow unbiased identification of sumoylation sites are urgently needed.
A proteomics approach, such as tryptic digestion of proteins followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and database search, has often been used for identification of post-translational modification sites in an unbiased manner (8–10). In the case of sumoylation, the site identification has relied upon the high stoichiometry obtained by in vitro sumoylation and target-specific data inspection (11–13). Tryptic digestion of mammalian sumoylated proteins results in peptides with a long side chain on the target lysine residues (see Fig. 1A). The commonly used proteomics techniques are not readily applicable for identification of these branched peptides, which show highly complicated fragmentation patterns in MS/MS analyses when compared with linear peptides. One way to circumvent this problem is a threonine-to-arginine substitution (T95R) on the C terminus of human SUMO-1 (14). The arginine residue leads to a tryptic cleavage of the long side chain, whereas two glycine residues remain on the lysine residue as a signature tag (+114.0429 Da), which represents the sumoylation site. This approach is still challenging due to poor detection sensitivity and lack of specific purification techniques for the peptides containing the sumoylation sites (sumoylated peptides) in complex tryptic digests. To facilitate the in vivo identification of sumoylation sites, we have developed a novel combined strategy using a further engineered human SUMO-1 construct. In addition to the easily detectable signature tag, this strategy also provides enrichment capability of sumoylated peptides. Application of our substrate-unbiased approach on HeLa cells resulted in identification of both consensus and non-consensus sumoylation sites, providing novel insights into SUMO target recognition.
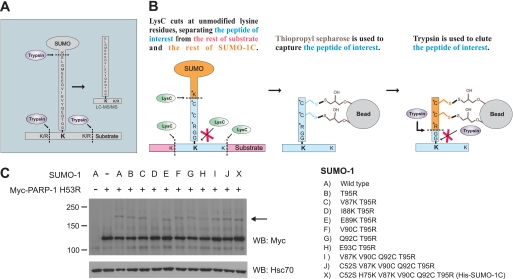
FIGURE 1.
Cysteine-tagged human SUMO-1 for enrichment of sumoylated peptides. A, tryptic digestion of mammalian sumoylated peptides results in peptides with a long side chain on the target lysine residues. B, identification of sumoylation sites using SUMO-1C. Altered residues are marked with asterisks, and the SUMO target lysine is shown in bold. The peptide of interest is different between subsequent purification steps and is indicated with blue color. Left panel, LysC cleavage results in a release of the cysteine tag from the rest of SUMO-1C. The substrate is also digested, but the sumoylated lysine is not cleaved. Middle panel, using Thiopropyl-Sepharose, the cysteine-containing peptides are covalently retained. Right panel, target peptides are eluted with trypsin, and the diglycine (GG)-modified internal lysine is identified as a sumoylation site by LC-MS/MS. C, construction of SUMO-1C. PARP-1 H53R was expressed together with WT SUMO-1 or the indicated mature forms of SUMO-1 mutants in HeLa cells and blotted against the Myc tag to detect sumoylation. SUMO-1 I88K,T95R has lost its ability to conjugate to PARP-1, whereas other mutants, which had no effect on sumoylation, were combined to create His-SUMO-1C, marked with an X. The arrow indicates the sumoylated form of PARP-1. Molecular masses in kDa are indicated on the left side of the blot. Hsc70 was used as a loading control. WB, Western blot.
EXPERIMENTAL PROCEDURES
Plasmid Constructs, Cell Culture and Transfection, Western Blotting, His Purification of Sumoylated Proteins, and Verification of Novel Sumoylation Site
The experimental procedures are described in the supplemental Experimental Procedures.
LysC Digestion
The purified sumoylated proteins (92 μg in 470 μl) were reduced for 1 h at 37 °C by the addition of 20 mm dithiothreitol (DTT; one sample volume), basified by the addition of 200 mm Tris base (one sample volume), and then digested overnight (∼20 h) at 37 °C by the addition of LysC (4.6 μg; Wako) in H2O (one sample volume). The resulting solution consisted of 50 mm Tris, 25% phosphate-buffered saline, 2 m urea, 5 mm DTT (pH ∼9). To quench the digestion, the solution was acidified with 10% trifluoroacetic acid to a final concentration of 1%. The resulting LysC digest was desalted with three pieces of OMIX pipette tips C18 (Varian) according to the instructions of the manufacturer. Briefly, an aliquot of the digest was loaded onto the OMIX tip that was pretreated with acetonitrile (ACN; 2 × 100 μl) and 1% trifluoroacetic acid (2 × 100 μl). After washing the OMIX tip with 1% trifluoroacetic acid (2 × 100 μl), the LysC digest was eluted with 0.1% trifluoroacetic acid, 80% ACN (100 μl). Three eluates were combined in a 1.5-ml tube and then evaporated.
Enrichment of Cysteine-tagged Sumoylated Peptides
The enrichment method using Thiopropyl-Sepharose 6B beads (GE Healthcare) was developed based on previous reports by Liu et al. (15, 16). The desalted LysC digest was reduced for 1 h at 37 °C by the addition of 5 mm DTT in a Tris-HCl/EDTA buffer (50 mm Tris-HCl, 1 mm EDTA, pH 7.4; 10 μl). To capture cysteine-containing peptides, the reduced digest was shaken for 1 h with the Thiopropyl-Sepharose 6B beads that were pre-equilibrated and suspended in the Tris-HCl/EDTA buffer (∼50 μl of beads in 100 μl). After spinning the tube, unbound peptides were removed as a supernatant. The beads were washed by rotation for 10 min with the Tris-HCl/EDTA buffer (500 μl), 2 m NaCl (500 μl), 0.1% trifluoroacetic acid, 80% ACN (500 μl), and then the Tris-HCl/EDTA buffer (500 μl). After removal of a supernatant, the beads were shaken for 1 h at 37 °C with modified trypsin (920 ng; Promega) in the Tris-HCl/EDTA buffer (50 μl) and then incubated at 37 °C overnight (∼20 h) to perform on-bead digestion. The resulting tryptic digest, which was expected to contain sumoylated peptides, was collected as the supernatant, further eluted by shaking the beads for 10 min with the Tris-HCl/EDTA buffer (2 × 50 μl), and then combined in a tube. Although the tryptic digest was not expected to include cysteine-containing peptides, this digest was reduced for 1 h at 37 °C by the addition of 100 mm DTT in 50 mm Tris-HCl buffer (pH ∼9; 15 μl) and then alkylated for 30 min at room temperature in the dark by the addition of 500 mm iodoacetamide in the Tris-HCl buffer (15 μl). The alkylation was quenched by the addition of 500 mm DTT in the Tris-HCl buffer (15 μl) to prevent side reaction from excess iodoacetamide (17). After acidification with 10% trifluoroacetic acid (15 μl), the treated tryptic digest was desalted according to Rappsilber et al. (18) with slight modification. Briefly, a C18 microcolumn was made with three pieces of Empore C18 disk (3M) packed into a 200-μl pipette tip. The tryptic digest was loaded onto the C18 microcolumn that was pretreated with ACN (50 μl) and 0.1% formic acid (50 μl). After washing the column with 0.1% formic acid (5 × 50 μl), the tryptic digest was eluted with 0.1% formic acid, 80% ACN (50 μl) and then evaporated. After reconstitution with 0.1% formic acid, an aliquot (8.5% v/v) and another aliquot (85% v/v) of the desalted tryptic digest were evaporated for LC-MS/MS analysis and fractionation, respectively.
Strong Cation Exchange Chromatography
The aliquot (85% v/v) of the desalted tryptic digest was fractionated by strong cation exchange (SCX) chromatography according to Ishihama et al. (19) with modification. Briefly, an SCX microcolumn was made with a piece of Empore styrenedivinylbenzene-reverse phase sulfonate (SDB-RPS) disk (3M) packed into a 200-μl pipette tip. The SCX microcolumn was pretreated with ACN (20 μl), 0.1% trifluoroacetic acid, 5% ACN (20 μl), 500 mm AcONH4, 15% ACN (20 μl), and then 0.1% trifluoroacetic acid, 5% ACN (3 × 100 μl). The desalted tryptic digest was reconstituted in 0.1% trifluoroacetic acid, 5% ACN (50 μl) and loaded onto the SCX microcolumn. The pass-through and the following washing with 0.1% trifluoroacetic acid, 5% ACN (50 μl) were collected and combined as a pass-through fraction. Elution was performed stepwise with 2, 4, 20, 100, and then 500 mm AcONH4 in 15% ACN (50 μl), and the eluates were collected separately. To ensure the elution, the SCX microcolumn was treated with 0.1% trifluoroacetic acid, 80% ACN (10 μl), and the eluate was collected as an ACN fraction. All the seven fractions were diluted by the addition of 1% formic acid to a final volume of 200 μl and then desalted with the Empore C18 microcolumn as described above.
LC-MS/MS Analysis
Each half of the samples was analyzed by LC-MS/MS using either a QSTAR Pulsar hybrid quadrupole-time-of-flight tandem mass spectrometer (Applied Biosystems) or an LTQ Orbitrap XL hybrid linear ion trap-orbitrap mass spectrometer (Thermo Fisher Scientific). The detailed procedures are described in the supplemental Experimental Procedures.
Mascot Database Search and Manual Data Inspection
The peak lists were subjected to database searches using an in-house Mascot server (version 2.2; Matrix Science) against the Swiss-Prot database (version 57.3, Homo sapiens) supplemented with His-SUMO-1C, LysC (API_ACHLY), trypsin (TRYP_PIG), and bovine serum albumin (ALBU_BOVIN) (total 20,336 sequences). The search criteria allowed for two missed tryptic cleavage sites and specified protein N-terminal acetylation (+42.010559 Da), cysteine carbamidomethylation (+57.021469 Da), cysteine N-ethylmaleimide modification (+125.047684 Da), methionine oxidation (+15.994919 Da), phosphorylation of serine/threonine/tyrosine (+79.966324 Da), and diglycine modification of lysine (+114.042923 Da) as variable modifications. Mass tolerances in MS and MS/MS modes were 0.05 and 0.15 Da for the QSTAR data, respectively, whereas they were 5 ppm and 0.8 Da for the LTQ Orbitrap data, respectively.
MS/MS spectra of sumoylated peptides suggested with a Mascot expectation value less than 0.05 were inspected manually for identification. The QSTAR MS/MS spectra were centroided before the manual inspection. In case sumoylated peptides were identified with only one of the mass spectrometers, the identical peptides suggested with the other mass spectrometer were inspected manually although the expectation value was not less than 0.05. A sumoylated peptide containing a known sumoylation site was manually inspected although the expectation value was high. Sumoylated peptides, which showed slightly high expectation values (i.e. a Mascot score >15), were considered as candidates and inspected manually. One of the candidate peptides, which showed well annotatable fragment ions and reasonable fragmentation pattern, was subjected to biological validation.
RESULTS AND DISCUSSION
Enrichment Method for Sumoylated Peptides
To facilitate in vivo identification of sumoylation sites of human proteins, we have developed an enrichment method based on a modified, cysteine-tagged form of SUMO-1 (Fig. 1B). Two proteases are used. LysC digestion provides a cysteine-containing branched peptide that can be specifically retained by covalent binding to a thiol-specific resin (Thiopropyl-Sepharose 6B, GE Healthcare), and trypsin releases the target peptide from the immobilized cysteine tag. As a result, peptides containing an extra mass of two glycine residues on a lysine residue are obtained, allowing identification of sumoylation sites by mass spectrometry. To create cysteine-tagged SUMO-1 (SUMO-1C), we mutated several residues of His-SUMO-1 and tested these mutants using human PARP-1 as a model substrate (4, 20, 21). PARP-1 is a stress-inducible SUMO substrate, and for practical reasons, we used a DNA-binding mutant PARP-1 H53R that is moderately sumoylated also under non-stress conditions.6 Mutation of some residues, i.e. Ile88 and Glu93, in SUMO-1 resulted in a loss or severely compromised PARP-1 sumoylation, whereas others left its sumoylation intact. These mutations were combined to create His-SUMO-1C containing C52S, H75K, V87K, V90C, Q92C, and T95R, which still retained the sumoylation capability and is marked with X in Fig. 1C.
Identification of Human Sumoylation Sites in Vivo
The applicability of our enrichment method was tested without substrate overexpression using the following experimental setup. The His-SUMO-1C construct was expressed in HeLa cells followed by metal affinity purification under denaturating conditions and removal of unconjugated His-SUMO-1C by a 30-kDa regenerated cellulose cut-off membrane (supplemental Fig. 2). Purified sumoylated proteins were subjected to the cysteine tag peptide enrichment as described above. The resulting samples were fractionated with an SCX microcolumn and analyzed by LC-MS/MS with QSTAR Pulsar and LTQ Orbitrap XL mass spectrometers. The obtained MS/MS data were searched using Mascot against the Swiss-Prot human database, with variable modifications including the diglycyl lysine residue as a sumoylation site.
We found 14 sumoylated peptides, which were derived from 12 substrate proteins (Table 1, supplemental Tables 1 and 2). According to the Swiss-Prot database, 7 of the 12 substrate proteins were assigned to the nuclear or nuclear membrane compartment, 2 were unassigned, and 3 were assigned to reside outside the nucleus, in membranes, or even cellular projections. Among the sumoylated peptides, 3 and 11 were sumoylated on consensus and non-consensus sites, respectively. This result supports our earlier presented hypothesis that SUMO substrate recognition mechanisms would be more versatile than earlier anticipated and that numerous SUMO substrates would be targeted by yet unknown mechanisms, independent of the consensus tetrapeptide (4). The identified sites included two well known consensus sites, Lys524 of Ran GTPase-activating protein 1 (RanGAP1) and Lys11 of SUMO-2 (22, 23), and two non-consensus sites, Lys7 of SUMO-1 and Lys779 of transcription intermediary factor-1β (TIF-1β) (13, 24) (Table 1). SIM sequences were found in close proximity in 6 of the 11 non-consensus sites (data not shown). Our results are consistent with the model of the SIM-dependent non-consensus targeting (7), and we propose that individual non-consensus sumoylation sites, at least on small proteins, could be searched for by mutating SIM-like residues. The lack of SIM residues nearby the remaining identified non-consensus sites indicates that there are also other, yet undefined, specificity determinants that govern the targeting of non-consensus sumoylation. Hundreds of SUMO substrates have been reported in recent proteomics studies (4, 5). However, without a verified sumoylation site assignment, the insights of non-consensus sumoylation sites mentioned here are clearly beyond reach.
TABLE 1.
Sumoylated peptides observed in vivo by cysteine tag peptide enrichment and LC-MS/MS
| Swiss-Prot accession number | Entry name | Protein name | SUMO site | Peptide sequencea | QSTARb |
LTQ Orbitrapc |
||
|---|---|---|---|---|---|---|---|---|
| Mascot score | Expectation value | Mascot score | Expectation value | |||||
| P46060 | RAGP1_HUMAN | Ran GTPase-activating protein 1 | Lys524 | 516LLVHMGLL(GG-)KSEDK528 | 16.1 | 1.5 | 29.5 | 0.011 |
| Lys8 | Ac-2ASEDIA(GG-)KLAETLAK15 | 48.5 | 0.00025 | |||||
| Q13263 | TIF1B_HUMAN | Transcription intermediary factor 1-β | Lys779 | 775LTED(GG-)KADVQSIIGLQR790 | 28.2 | 0.21 | 41.6 | 0.0012 |
| P63165 | SUMO1_HUMAN | Small ubiquitin-related modifier 1 | Lys7 | Ac-2SDQEA(GG-)KPSTEDLGDKK17 | 43.0 | 0.012 | 32.5 | 0.026 |
| Lys25 | 24L(GG-)KVIGQDSSEIHFK37 | 30.2 | 0.018 | |||||
| P61956 | SUMO2_HUMAN | Small ubiquitin-related modifier 2 | Lys11 | 8EGV(GG-)KTENNDHINLK21 | 14.3 | 6.6 | 16.8 | 0.72 |
| Q16666 | IF16_HUMAN | γ-Interferon-inducible protein Ifi-16 | Lys561 | 560L(GG-)KTEPEEVSIEDSAQSDLK578 | 58.9 | 0.00006 | ||
| Q9UPN6 | RBM16_HUMAN | Putative RNA-binding protein 16 | Lys18 | 6pTFNSELYSLNDY(GG-)KPPISKAK25 | 37.0 | 0.012 | ||
| Q9UG01 | IF172_HUMAN | Intraflagellar transport protein 172 homolog | Lys4 | Ac-1MHL(GG-)KHLR7 | 30.0 | 0.014 | ||
| Q99518 | FMO2_HUMAN | Dimethylaniline monooxygenase (N-oxide-forming) 2 | Lys492 | 486NAIFTQ(GG-)KQRILKPLK500 | 16.3 | 0.023 | ||
| Q96QD9 | FYTD1_HUMAN | Forty-two-three domain-containing protein 1 | Lys140 | 139R(GG-)KANLLR145 | 22.8 | 0.028 | ||
| Q9UBD0 | HSFX1_HUMAN | Heat shock transcription factor, X-linked | Lys215 | 211RVGV(GG-)KSAPR219 | 21.1 | 0.036 | ||
| Q9UBG0 | MRC2_HUMAN | C-type mannose receptor 2 | Lys1142 | 1139LLQ(GG-)KPLR1145 | 21.9 | 0.043 | ||
| Q8IYA6 | CKP2L_HUMAN | Cytoskeleton-associated protein 2-like | Lys198 | 198(GG-)KPDPKLpYTR206 | 26.8 | 0.33 | 17.9 | 0.36 |
a (GG-)K, diglysyl Lys (sumoylation site); Ac-, acetylated protein N terminus; pT/pY, phosphorylated Thr/Tyr.
b Mass tolerances for precursor and fragment ions were 0.05 and 0.15 Da, respectively.
c Mass tolerances for precursor and fragment ions were 5 ppm (Orbitrap) and 0.8 Da (LTQ), respectively.
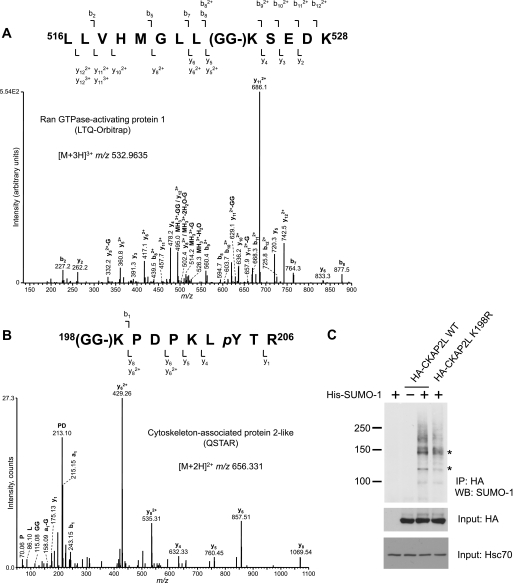
As examples of MS/MS spectra inspected manually, we present Lys524 of RanGAP1 obtained with LTQ Orbitrap XL and Lys198 of cytoskeleton-associated protein 2-like (CKAP2L) obtained with QSTAR Pulsar (Fig. 2, A and B). The remaining MS/MS spectra are found in supplemental Fig. 3. Low energy collision-induced dissociation of LTQ and QSTAR provided losses of glycine residue(s) from the sumoylation sites (Fig. 2, supplemental Fig. 3). The losses accompanied dominant fragment ions and especially precursor-related ions. When compared with the QSTAR Pulsar, the modern LTQ Orbitrap XL provided a larger number of sumoylated peptides due to the superior sensitivity and precursor mass accuracy (Table 1). LTQ MS/MS used in this study provided many fragment ions, which should result in the good Mascot scores for identification. However, due to the relatively low mass accuracy and resolution provided by LTQ MS/MS, annotation of fragment ions with charge states was ambiguous in some cases (e.g. an ion at m/z 502.4 in Fig. 2A), and the MS/MS spectra of sumoylated peptides were hard to analyze manually.
FIGURE 2.
In vivo identification of sumoylation sites by cysteine tag peptide enrichment and LC-MS/MS. His-SUMO-1C was expressed in HeLa cells, and the resulting sumoylation sites were identified by His tag protein purification, cysteine tag peptide enrichment, and LC-MS/MS analysis. A, MS/MS analysis of RanGAP1 sumoylated peptide 516LLVHMGLL(GG-)KSEDK528 (LTQ Orbitrap XL). This peptide shows a known consensus sumoylation site, Lys524. (GG-)K, diglycyl lysine (sumoylation site). B, MS/MS analysis of CKAP2L sumoylated peptide 198(GG-)KPDPKLpYTR206 (QSTAR Pulsar). This peptide shows a non-consensus sumoylation site, Lys198. pY, phosphotyrosine. C, biological validation of the CKAP2L sumoylation site. HA-CKAP2L WT and HA-CKAP2L K198R were expressed together with WT His-SUMO-1 in HeLa cells. Samples were immunoprecipitated (IP) with anti-HA antibody, and sumoylated forms were detected by blotting with SUMO-1 antibody. Asterisks indicate sumoylated bands that are lost upon mutation of Lys198. The full-length HA blot is presented in supplemental Fig. 1. WB, Western blot.
Confirmation of CKAP2L Novel Sumoylation Site
To our knowledge, sumoylation of CKAP2L has not been reported previously. By manual inspection of the MS/MS spectrum obtained with QSTAR (Fig. 2B), most of the predominant ion peaks were annotated as peptide fragment ions. Because QSTAR MS/MS provides isotope-resolved ion peaks, charge states of the fragment ions could be verified. Intensive fragment ions, y6, y8, and their doubly charged forms corresponded to the proline-induced fragment ions. Diglycine modification of the ϵ-amino group of the peptide N-terminal lysine residue apparently induced b1 and a1 ions instead of the b2 and a2 ions that are frequently observed by QSTAR MS/MS. An ion observed at m/z 115.08 corresponds to fragment GG (theoretical m/z 115.05). Because none of the ion peaks matched to the threonine-containing fragment ions with neutral loss of H3PO4, this peptide should be phosphorylated at the tyrosine residue (25).
The novel site of CKAP2L was confirmed by expressing HA-CKAP2L WT and HA-CKAP2L K198R together with WT SUMO-1 in HeLa cells. After HA immunoprecipitation and Western blotting against SUMO-1, a complex pattern of several sumoylated bands corresponding to several sumoylation sites of HA-CKAP2L WT was detected (Fig. 2C, supplemental Fig. 1). The finding that two sumoylated bands marked with asterisks were lost in the HA-CKAP2L K198R mutant, i.e. the upper band on top of a smear and the lower band on top of another band (Fig. 2C), demonstrates that CKAP2L is sumoylated on Lys198 and that our method can be used to identify novel non-consensus sites of sumoylation.
Recently, Hsiao et al. (6) reported an informatics tool to facilitate identification of endogenous sumoylated peptides, which are branched with the long side chains, i.e. C termini of SUMOs (Fig. 1A). This tool linearizes expected sumoylated peptides in silico to simplify the MS/MS database search. However, the peptide identification was achieved mainly with detection of predominant fragment ions originating from the C termini of SUMOs, and sufficient numbers of the fragment ions from the substrate peptides, required for confident identification of the sumoylation sites, were not observed (6). Our enrichment strategy using the removable cysteine tag facilitates considerably the in vivo identification of sumoylation sites. Similarly to the recent advances in phosphopeptide enrichment (25–28) that have significantly promoted phosphoproteomics studies, the concept of sumoylated peptide enrichment is likely to expand the field of the site-specific SUMO proteomics.
Taken together, the results of our study validate a useful tool for sumoylation studies that already in this study gave novel insight into non-consensus sumoylation motifs. As the method is readily available for a broad spectrum of research environments, it will certainly be helpful in unraveling SUMO-targeting mechanisms and facilitate identification of novel SUMO consensus and non-consensus sites.
Supplementary Material
Acknowledgments
We thank Robert Gourlay and Helena Saarento for excellent technical assistance and the Eriksson and Sistonen laboratories for valuable comments. We are grateful to the Proteomics Core Facility at the Turku Centre for Biotechnology for instrument support.
This work was supported by grants from the Academy of Finland (to J. E. E., L. S., and S. Y. I.), the Sigrid Jusélius Foundation, the Finnish Cancer Organizations and the Åbo Akademi University (to J. E. E. and L. S.), the Turku Graduate School of Biomedical Sciences, the Magnus Ehrnrooth Foundation, and the Stiftelsens för Åbo Akademi forskningsinstitut (to H. A. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. 1–3, and Tables 1 and 2.
H. A. Blomster, unpublished observations.
- SUMO
- small ubiquitin-like modifier
- SUMO-1C
- cysteine-tagged SUMO-1
- SIM
- SUMO interaction motif
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- DTT
- dithiothreitol
- ACN
- acetonitrile
- SCX
- strong cation exchange
- RanGAP1
- Ran GTPase-activating protein 1
- CKAP2L
- cytoskeleton-associated protein 2-like
- WT
- wild type
- HA
- hemagglutinin.
REFERENCES
- 1.Makhnevych T., Sydorskyy Y., Xin X., Srikumar T., Vizeacoumar F. J., Jeram S. M., Li Z., Bahr S., Andrews B. J., Boone C., Raught B. (2009) Mol. Cell 33, 124–135 [DOI] [PubMed] [Google Scholar]
- 2.Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 3.Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. (2002) Cell 108, 345–356 [DOI] [PubMed] [Google Scholar]
- 4.Blomster H. A., Hietakangas V., Wu J., Kouvonen P., Hautaniemi S., Sistonen L. (2009) Mol. Cell. Proteomics 8, 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golebiowski F., Matic I., Tatham M. H., Cole C., Yin Y., Nakamura A., Cox J., Barton G. J., Mann M., Hay R. T. (2009) Sci. Signal. 2, ra24. [DOI] [PubMed] [Google Scholar]
- 6.Hsiao H. H., Meulmeester E., Frank B. T., Melchior F., Urlaub H. (2009) Mol. Cell. Proteomics 8, 2664–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J., Zhu S., Guzzo C. M., Ellis N. A., Sung K. S., Choi C. Y., Matunis M. J. (2008) J. Biol. Chem. 283, 29405–29415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen O. N. (2006) Nat. Rev. Mol. Cell Biol. 7, 391–403 [DOI] [PubMed] [Google Scholar]
- 9.Jeram S. M., Srikumar T., Pedrioli P. G., Raught B. (2009) Proteomics 9, 922–934 [DOI] [PubMed] [Google Scholar]
- 10.Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 11.Cooper H. J., Tatham M. H., Jaffray E., Heath J. K., Lam T. T., Marshall A. G., Hay R. T. (2005) Anal. Chem. 77, 6310–6319 [DOI] [PubMed] [Google Scholar]
- 12.Matic I., van Hagen M., Schimmel J., Macek B., Ogg S. C., Tatham M. H., Hay R. T., Lamond A. I., Mann M., Vertegaal A. C. (2008) Mol. Cell. Proteomics 7, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedrioli P. G., Raught B., Zhang X. D., Rogers R., Aitchison J., Matunis M., Aebersold R. (2006) Nat. Methods 3, 533–539 [DOI] [PubMed] [Google Scholar]
- 14.Knuesel M., Cheung H. T., Hamady M., Barthel K. K., Liu X. (2005) Mol. Cell. Proteomics 4, 1626–1636 [DOI] [PubMed] [Google Scholar]
- 15.Liu T., Qian W. J., Strittmatter E. F., Camp D. G., 2nd, Anderson G. A., Thrall B. D., Smith R. D. (2004) Anal. Chem. 76, 5345–5353 [DOI] [PubMed] [Google Scholar]
- 16.Liu T., Qian W. J., Gritsenko M. A., Xiao W., Moldawer L. L., Kaushal A., Monroe M. E., Varnum S. M., Moore R. J., Purvine S. O., Maier R. V., Davis R. W., Tompkins R. G., Camp D. G., 2nd, Smith R. D.the Inflammation and the Host Response to Injury Large Scale Collaborative Research Program (2006) Mol. Cell. Proteomics 5, 1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boja E. S., Fales H. M. (2001) Anal. Chem. 73, 3576–3582 [DOI] [PubMed] [Google Scholar]
- 18.Rappsilber J., Ishihama Y., Mann M. (2003) Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 19.Ishihama Y., Rappsilber J., Mann M. (2006) J. Proteome Res. 5, 988–994 [DOI] [PubMed] [Google Scholar]
- 20.Martin N., Schwamborn K., Schreiber V., Werner A., Guillier C., Zhang X. D., Bischof O., Seeler J. S., Dejean A. (2009) EMBO J. 28, 3534–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messner S., Schuermann D., Altmeyer M., Kassner I., Schmidt D., Schär P., Müller S., Hottiger M. O. (2009) FASEB J. 23, 3978–3989 [DOI] [PubMed] [Google Scholar]
- 22.Mahajan R., Gerace L., Melchior F. (1998) J. Cell Biol. 140, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., Hay R. T. (2001) J. Biol. Chem. 276, 35368–35374 [DOI] [PubMed] [Google Scholar]
- 24.Mascle X. H., Germain-Desprez D., Huynh P., Estephan P., Aubry M. (2007) J. Biol. Chem. 282, 10190–10202 [DOI] [PubMed] [Google Scholar]
- 25.Imanishi S. Y., Kochin V., Ferraris S. E., de Thonel A., Pallari H. M., Corthals G. L., Eriksson J. E. (2007) Mol. Cell. Proteomics 6, 1380–1391 [DOI] [PubMed] [Google Scholar]
- 26.Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jørgensen T. J. (2005) Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 27.Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y. (2007) Mol. Cell. Proteomics 6, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 28.Thingholm T. E., Jensen O. N., Robinson P. J., Larsen M. R. (2008) Mol. Cell. Proteomics 7, 661–671 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.