Abstract
Bem1 is a scaffold protein essential for the establishment of cell polarity in Saccharomyces cerevisiae. This work reports the solution structure of a Cdc42 binding module of Bem1 comprising the second SH3 domain (SH3b) and its C-terminal flanking region termed Cdc42 interacting (CI). First, the structure of Bem1 SH3b-CI was determined by NMR spectroscopy, which shows that Bem1 SH3b-CI is a structurally and functionally related domain that binds Cdc42. Next, the solution structure of Bem1 SH3b-CI in complex with the proline-rich region of p21-activated kinase Ste20 (Ste20 PRR) was determined. Finally, the interaction surface of Bem1 SH3b-CI with Cdc42 was identified based on chemical shift perturbation studies which reveals that Bem1 SH3b-CI interacts simultaneously with both Ste20 PRR and Cdc42 using the opposite surfaces. Thus, Bem1 can tether Cdc42 and Ste20 in close proximity so that Cdc42 can efficiently interact with Ste20 Cdc42 and Rac interactive binding (CRIB). Based on the present results together with the previous biochemical studies (Lamson, R. E., Winters, M. J., and Pryciak, P. M. (2002) Mol. Cell. Biol. 22, 2939–2951 and Winters, M. J., and Pryciak, P. M. (2005) Mol. Cell. Biol. 25, 2177–2190), a model was suggested that the autoinhibition of Ste20 kinase activity by CRIB is released through the Cdc42-CRIB interaction, which is mediated by Bem1, and Ste20 is subsequently activated, an initial step for the establishment of the cell polarity.
Keywords: Cdc42, NMR, Protein Domains, Protein Kinases, Protein Structure, Protein-Protein Interactions, SH3 Domains, Bem1, Ste20, Scaffold
Introduction
Cell polarity is essential for the physiological functions of eukaryotic cells. Yeast cells polarize during budding in vegetative growth and mating (1), and it is now recognized that a variety of proteins are recruited in this process. Especially, the Rho-type small GTPase, Cdc42, well conserved from yeasts to humans, plays a crucial role for the cell polarity establishment (2). In Saccharomyces cerevisiae, Cdc24, a guanine nucleotide exchange factor of Cdc42, activates Cdc42 (GTP-bound Cdc42) (3). The GTP-bound Cdc42 can activate p21-activated kinase (PAK) Ste20 through interaction with the Cdc42- and Rac-interactive binding (CRIB)2 domain of Ste20 in mating (4). In budding, PAK Cla4 is similarly activated by Cdc42 (5).
It is well established that Bem1 interacts with Cdc24, Ste20, Cla4, and Cdc42, which facilitates efficient cell signaling (6–8) and is therefore considered to work as a scaffold protein in Saccharomyces cerevisiae. The Bem1 PB1 domain binds to the Cdc24 PB1 domain through electrostatic interaction (9–11). As a disruption of this PB1-PB1 interaction leads to defects in budding and mating, the Bem1 PB1 domain is considered to be crucial for the cell polarity establishment (6). The second SH3 domain (SH3b) of Bem1 interacts with the Ste20 proline-rich region (Ste20 PRR) and this interaction contributes to the localization of both Ste20 and Cdc42 in mating (7). The Cla4 proline-rich region (Cla4 PRR) was also reported to bind to SH3b in budding (8).
The GTP-bound form of Cdc42 was reported to bind Bem1 in vivo and in vitro (8, 12). Recently, the Cdc42 binding region of Bem1 has been identified, that interacts directly with GTP-bound Cdc42 (7, 13, 20). This region is comprised of the second SH3 domain (SH3b), and its flanking C-terminal sequence, named CI (Cdc42 interacting) (13). Because SH3b alone was insoluble and CI alone did not interact with Cdc42, both regions appear to be required to form a stable structural and functional domain. Therefore, this Cdc42 binding region was designated as SH3b-CI (13). Considering that the sequence of the CI region in Bem1 homologues is well conserved but exhibits no similarity to CRIB, SH3b-CI appears to be a novel Cdc42 binding module.
Mutational studies using in vitro binding assays have verified that the N253D mutation in the CI moiety blocks Cdc42 binding, but it does not block the Ste20 PRR binding. Whereas the W192K mutation in the SH3b moiety inhibits the Ste20 PRR binding, it does not inhibit the Cdc42 binding, indicating that the binding surfaces of Bem1for Ste20 PRR and Cdc42 are distinct (13).
The biological role of the Bem1-Cdc42 interaction has been studied in vivo using Bem1 mutants; although Bem1 N253D exhibits slight mating defects around 50%, the W192K mutant is highly defective in mating (less than 5%), whereas the Bem1 W192K/N253D double mutant completely loses mating activity. These results indicate that to conduct efficient mating signaling, SH3b anchors Ste20 and Cdc42 in collaboration with CI (13).
Here, we determined the three-dimensional structures of Bem1 SH3b-CI and its complex with Ste20 PRR using NMR. The Bem1 SH3b-CI has a SH3 domain that has a canonical SH3 structure (22) and the CI region comprising of two α-helices and a flexible tail. Because a number of hydrophobic interactions between SH3b and CI are observed, it is possible to conclude that Bem1 SH3b and CI are structurally and functionally related to each other. In addition, the structure of the Bem1 SH3b-CI/Ste20 PRR complex and the chemical shift perturbation study of Bem1 SH3b-CI with Cdc42 showed that Bem1 SH3b-CI can bind both Ste20 PRR and Cdc42 simultaneously. These results provide insights into the signal transduction mechanism mediated by Bem1.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification for NMR Samples
The Bem1 SH3b-CI-(156–260) was cloned into the pGEX-6P-1 vector (GE Healthcare), expressed in Escherichia coli strain BL21 (DE3) as a glutathione S-transferase (GST) fusion protein and purified using glutathione-Sepharose 4B (GE Healthcare). The GST tag was removed with PreScission protease (GE Healthcare). Finally, Bem1 SH3b-CI was purified by gel filtration on a HiLoad 26/60 Superdex 75 prep grade column (GE Healthcare). The Ste20 PRR (463–486) was cloned into the pGEX-6P-1 vector and expressed in E. coli strain BL21 (DE3) as a glutathione S-transferase (GST) fusion protein. The Ste20 PRR was first purified using glutathione-Sepharose 4B and the GST tag was removed with PreScission protease. Finally, Ste20 PRR was purified by reverse phase chromatography on a Resource RPC column (GE Healthcare).
NMR Experiments
For NMR samples, the purified 13C/15N-labeled Bem1 SH3b-CI was concentrated to 0.3 mm in 20 mm MOPS buffer in 90% H2O/10% D2O with 150 mm NaCl at pH 6.5. In addition, the purified 13C/15N-labeled Bem1 SH3b-CI complexed with unlabeled Ste20 PRR and 13C/15N-labeled Ste20 PRR complexed with unlabeled Bem1 SH3b-CI were prepared, and separately concentrated to 0.3 mm in 20 mm MOPS buffer in 90% H2O/10% D2O, with 150 mm NaCl at pH 6.5. All NMR experiments were carried out at 25 °C on Unity Inova 800 and 600 spectrometers (Varian) each equipped with three radiofrequency channels and a triple resonance pulsed filed gradient probe. The 15N-labeled Bem1 SH3b-CI was used for the steady state heteronuclear 1H-15N NOE experiments. The sequential assignments of 1H, 13C, and 15N were carried out using the following spectra: HNCO, HN(CO)CA, HNCA, CBCA(CO)NH, HNCACB, HBHA(CO)NH, CCH-TOCSY, HC(C)H-TOCSY, HBCBCGCDHD, and HBCBCGCDCEHE (14). Interproton distance information for structure calculations was obtained from 15N-edited and 13C-edited NOESY spectra recorded with a 100 ms mixing time (14). These spectra were processed using the NMRPipe program (15), and the data analysis was made using the Sparky program (16).
Structure Calculations
The NOE peak assignments and the structure calculations were made using the CYANA 2.1 program (17). The dihedral angle restraints were estimated by the TALOS program (18). A total of 100 structures were calculated by simulated annealing and the mean structure was obtained by averaging the coordinates of the 20 lowest CYANA energy structures. The overlay of the 20 lowest energy structures were generated using the MOLMOL program (19), and a ribbon diagram of the mean structure was generated using the PyMOL program.
Chemical Shift Perturbation Experiments
The Bem1 SH3b-CI and Cdc42 (1–179, Q61L) were cloned into the pGEX-6P-1 vector and expressed in E. coli strain BL21 (DE3) as GST fusion proteins. The 13C/15N-labeled Bem1 SH3b-CI and unlabeled Cdc42 were first purified using glutathione-Sepharose 4B. After these GST-fusion proteins were mixed and the GST tag was removed with PreScission protease, Bem1 SH3b-CI complexed with Cdc42 was purified by anion exchange chromatography on a Resource Q column (GE Healthcare), and then Cdc42 was activated by loading GTPγS. Finally, the complex was purified by gel filtration on a HiLoad 26/60 Superdex 75 prep grade column (GE Healthcare). The purified 13C/15N-labeled Bem1 SH3b-CI complexed with unlabeled Cdc42 was concentrated to 0.3 mm in 20 mm MOPS buffer in 90% H2O/10% D2O, with 150 mm NaCl at pH 6.5. The sequential assignments of 1H, 13C, and 15N of Bem1 SH3b-CI were carried out as was described in NMR experiments. Chemical shift perturbations in Bem1 SH3b-CI upon complex formation with Cdc42 were monitored in the HSQC spectra. Normalized chemical shift changes were calculated using the equation, Δδ = [(ΔδHN)2 + (ΔδN/5)2]1/2.
In Vitro Pull-down Binding Assays
The Bem1 SH3b-CI, Ste20 PRR, and Ste20 CRIB domain (328–375) were cloned into the pGEX-6P-1 vector and expressed in E. coli strain BL21 (DE3) as GST fusion proteins. The Cdc42 (1–179, Q61L) were cloned into a modified pET-28a (Novagen) where the thrombin protease cleavage site was changed to the PreScission protease cleavage site and were expressed in E. coli strain BL21 (DE3) as a His-tagged protein. The GST fusion proteins were purified using glutathione-Sepharose 4B and the His-tagged protein was purified using Ni-NTA Superflow resin (Qiagen). For in vitro pull down binding assays, the GST fusion and His-tagged proteins were mixed in phosphate-buffered saline (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, and 1.5 mm KH2PO4, pH 7.4) or TBS containing 5 mm MgCl2, 0.5 mg/ml bovine serum albumin, and 0.1% Triton X-100 and incubated at 4 °C for 30 min. After the glutathione-Sepharose 4B resin was washed with phosphate-buffered saline containing 0.1% Triton X-100 or TBS containing 5 mm MgCl2, 0.5 mg/ml bovine serum albumin, and 0.1% Triton X-100, bound proteins were eluted with 10 mm glutathione in 20 mm Tris-HCl, pH 8.0. The eluates were subjected to 15% SDS-PAGE and stained by Coomassie Brilliant Blue or immunoblotting using anti-GST and anti-His antibodies.
RESULTS AND DISCUSSION
Identification of the SH3b-CI Structural Region
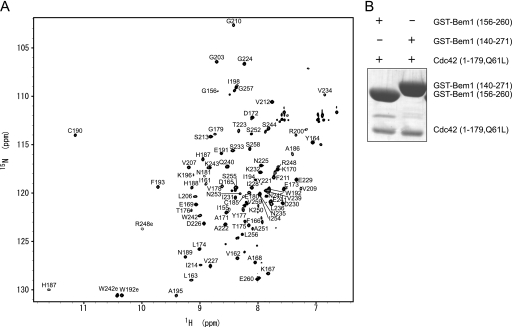
For the structure determination, we first expressed a construct of Bem1-(140–271) in E. coli strain BL21 (DE3) cells as a GST fusion protein, which was originally reported as a Cdc42-binding region and has been named SH3b-CI (13). However, an HSQC spectrum showed that this construct was not suitable for the structure determination because of the presence of a highly disordered region (data not shown). Therefore, we prepared several constructs based on the sequence alignment of other SH3 domains and the secondary structure prediction. Here, Tyr-159 was found to be the N terminus of SH3b (data not shown) and Ala-251 was estimated to be the C terminus of CI from the secondary structure prediction using the PSIPRED program (23). Consequently, we expressed a construct of Bem1-(156–260) in E. coli strain BL21 (DE3) cells as a GST fusion protein. The HSQC spectrum of Bem1-(156–260) was highly dispersed, showing that this construct corresponds to a structural domain (Fig. 1A). Further, Bem1-(156–260) could interact with Cdc42 to an extent similar to Bem1-(140–271) in in vitro pull-down binding assays (Fig. 1B). We concluded that Bem1-(156–260) is a structurally and functionally related domain, and it is designated as SH3b-CI in the following.
FIGURE 1.
Identification of the SH3b-CI structural region. A, 1H-15N HSQC spectrum of Bem1-(156–260). The cross peaks are labeled with one-letter codes of the amino acids and residue numbers. B, in vitro pull-down binding assay. GST-Bem1-(156–260) and GST-Bem1-(140–271) were incubated with Cdc42-(1–179, Q61L). The proteins pulled down with glutathione-Sepharose 4B were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue.
Structure of Bem1 SH3b-CI
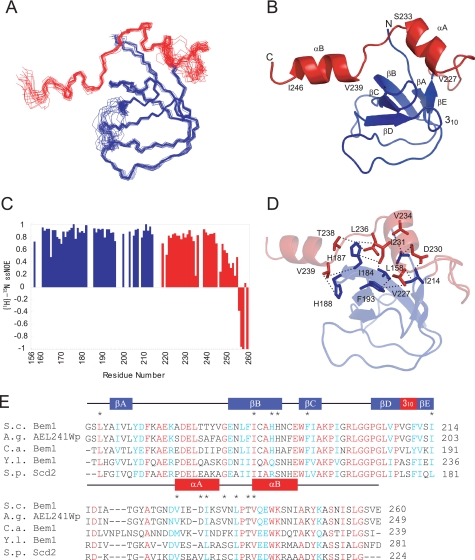
The assignments of 1H, 15N, and 13C nuclei in Bem1 SH3b-CI were made using a suite of NMR spectra as described under “Experimental Procedures.” The solution structure of SH3b-CI was determined on the basis of 1553 distance and 95 dihedral angle restraints. A total of 100 structures were calculated, and the 20 lowest CYANA energy structures were selected (Table 1). The residues 158–196, 204–214, and 227–246, are well defined with the root mean square deviation (r.m.s.d.) from the mean structure of 0.40 Å for the backbone atoms as illustrated in Fig. 2A, while residues 156–157, 197–203, 215–226, and 247–260 in SH3b-CI were not well defined due to a lack of long range NOEs.
TABLE 1.
Structural statistics of Bem1 SH3b-CI
| NOE distance constraints | 1553 | |
| Short range (intra-residue and sequential) | 823 | |
| Medium range (2 ≤ i-j ≤ 4) | 249 | |
| Long range (i-j > 4) | 481 | |
| Structural coordinates rmsd (158–196, 204–214, 227–246) | ||
| Backbone atoms | 0.40 Å | |
| All heavy atoms | 0.91 Å | |
| Ramachandran plot | ||
| Most favored regions | 75.1% | |
| Additionally allowed regions | 24.3% | |
| Generously allowed regions | 0.7% | |
| Disallowed regions | 0.0% | |
FIGURE 2.
Solution structure of Bem1 SH3b-CI. A, overlay of the backbone atoms (N, Cα, and C′) of the 20 lowest energy structures of Bem1 SH3b-CI. The SH3b and CI moieties are shown in blue and red, respectively. B, ribbon diagram of Bem1 SH3b-CI. The C-terminal flexible regions (248–260) are omitted. C, main chain dynamics of Bem1 SH3b-CI. The {1H}-15N heteronuclear NOE intensities are plotted against the residue number of Bem1 SH3b-CI. D, intramolecular interactions in Bem1 SH3b-CI. The residues involved in the hydrophobic interactions between SH3b and CI are shown in stick models. Bem1 SH3b and CI are shown in blue and red, respectively. E, sequence alignment among Bem1 SH3b-CI homologs. The secondary structural elements are indicated above the alignment. The conserved residues are marked in red, whereas type-conserved residues are marked in blue. The residues involved in the interactions between Bem1 SH3b and CI are marked with asterisks above the alignment. The atomic coordinates of SH3b-CI were deposited in the Protein Data Bank (PDB code: 2RQV).
The solution structure of Bem1 SH3b-CI is comprised of two α-helices (αA:227–234, αB:239–246), one 310helix (209–211), and five β-strands (βA:160–162, βB:180–188, βC:192–195, βD:205–208, and βE:212–214) (Fig. 2B). The structure of the SH3b moiety is similar to those of the canonical SH3 domains (22), composed of five antiparallel β-strands and a single 310 helix. The CI moiety contains two α-helices, αA and αB, but has a flexible C-terminal tail (247–260) as can be deduced from the small or negative {1H}-15N heteronuclear steady-state NOEs (Fig. 2C), including Asn-253, an important residue for Cdc42 binding (13). The flexible C-terminal tail of the CI moiety appears to form an ordered structure only when Cdc42 binds Bem1. A number of interactions between SH3b and CI were observed, suggesting that SH3b-CI is a structural unit, and it is a novel Cdc42 binding domain.
The interface between SH3b and CI, has an extensive hydrophobic core which is formed by Leu-158, Ile-184, His-187, His-188, Phe-193, and Ile-214 of SH3b together with Val-227, Asp-230, Ile-231, Val-234, Leu-236, Thr-238, and Val-239 of CI (Fig. 2D). These residues are conserved or type-conserved among Bem1 homologs implying that CI in Bem1 homologs also interacts with SH3b in a manner similar to Saccharomyces cerevisiae Bem1 (Fig. 2E). This is consistent with the previous reports (13, 20, 21) that the SH3b domain alone in the Bem1 homologs was insoluble (data not shown), and Bem1 CI-(214–260) failed to interact with Cdc42, indicating that SH3b and CI are structurally and functionally related and synergistically bind Cdc42. In contrast, the linker between SH3 and CI were not assigned because of broadening, and were not defined in the structure.
Structure of Bem1 SH3b-CI and Ste20 PRR Complex
The assignment of 1H, 15N, and 13C nuclei in Bem1 SH3b-CI complexed with Ste20 PRR-(463–486) was made using a suite of NMR spectra as described under “Experimental Procedures.” The solution structure of the complex was determined on the basis of 1558 distance and 90 dihedral angle restraints. A total of 100 structures were calculated, and the 20 lowest CYANA energy structures were selected (Table 2). The residues 158–196, 204–214, and 227–246 had a r.m.s.d. from the mean structure of 0.48 Å for the backbone atoms, indicating that the backbone Bem1 SH3b-CI complexed with Ste20 PRR is well defined, while residues 156–157, 197–203, 215–226, and 247–260 in Bem1 are not well defined due to a lack of long range NOEs as illustrated in Fig. 3, A and B. There are no appreciable backbone structure differences in Bem1 SH3b-CI between the free and complexed forms.
TABLE 2.
Structural statistics of Bem1 SH3b-CI and Ste20 PRR complex
| NOE distance constraints | 1509 | |
| Short range (intraresidue and sequential) | 821 | |
| Medium range (2 ≤ i-j ≤ 4) | 214 | |
| Long range (i-j > 4) 474 | ||
| (Intermolecular NOEs) | (104) | |
| Structural coordinates rmsd (158–196, 204–214, 227–246) | ||
| Backbone atoms | 0.46 Å | |
| All heavy atoms | 0.89 Å | |
| Ramachandran plot | ||
| Most favored regions | 66.7% | |
| Additionally allowed regions | 32.0% | |
| Generously allowed regions | 1.4% | |
| Disallowed regions | 0.0% | |
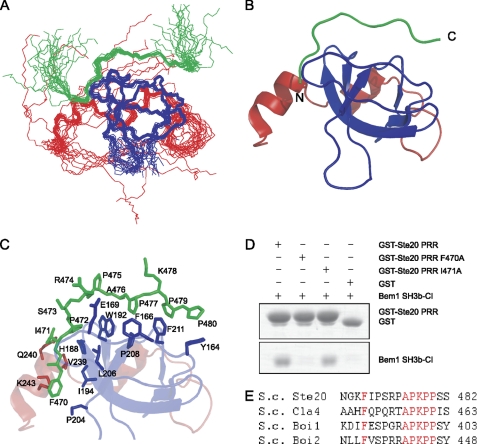
FIGURE 3.
Solution structure of Bem1 SH3b-CI and Ste20 PRR complex. A, overlay of the backbone atoms (N, Cα, and C′) of the 20 lowest energy structures of Bem1 SH3b-CI in complex with Ste20 PRR. Bem1 SH3b, Bem1 CI, and Ste20 PRR are shown in blue, red, and green, respectively. B, ribbon diagram of Bem1 SH3b-CI complexed with Ste20 PRR. Flexible regions in the N and C termini are omitted. C, interface between Bem1 SH3b-CI and Ste20 PRR. Bem1 SH3b, Bem1 CI, and Ste20 PRR are shown in blue, red, and green, respectively. The residues involved in interactions between Bem1 SH3b-CI and Ste20 PRR are shown in stick models. D, mutational analysis of Ste20 PRR. The GST-Ste20 PRR, GST-Ste20 PRR F470A, GST-Ste20 PRR I471A, or GST was incubated with Bem1 SH3b-CI. The proteins pulled down with glutathione-Sepharose 4B were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue. E, sequence alignment of Bem1 SH3b-CI-binding proteins. The conserved residues are marked in red. The atomic coordinates of SH3b-CI complexed with Ste20 PRR were deposited in the Protein Data Bank (PDB code: 2RQW).
Interaction Mode between Bem1 SH3b-CI and Ste20 PRR
The detailed molecular interactions observed between Bem1 SH3b-CI and Ste20 PRR-(463–486) are shown in Fig. 3C. The 470–480 residues in Ste20 PRR were well defined, although the flanking residues 463–469 and 481–486 were disordered. In a manner similar to other SH3 domains, Arg-474 interacts with an acidic residue in the RT loop, and this interaction determines the ligand orientation as Class I (22). The Ala-476—Pro-480 interact with the hydrophobic pockets formed by the conserved residues in Bem1 SH3b. The Pro-475–Pro-480 forms a PPII helix: the side-chains of Pro-475 and Lys-478 are exposed, whereas Ala-476, Pro-477, Pro-479, and Pro-480 fit into hydrophobic pockets of Bem1SH3b-CI. The Pro-479 and Pro-480 in PPII are surrounded by Tyr-164 and Phe-211, while Ala-476 and Pro-477 fit into the pocket lined with Phe-166, Trp-192, Pro-208, and Phe-211. These interactions between Bem1 and Ste20 PRR are very similar to those observed in the canonical SH3-PRR complex (22).
Although Ser-473 has no intermolecular NOEs with Bem1 SH3b-CI, Phe-470, Ile-471, and Pro-472 form a hydrophobic cluster together with His-188, Trp-192, Ile-194, Pro-204, and Leu-206 in the SH3b moiety and Val-239, Gln-240, and Lys-243 in the CI moiety. These results are consistent with the previous mutation study using the yeast two-hybrid assay (7), showing that the mutations of F470G, I471G, and P472A in Ste20 inhibit the binding between Bem1 SH3b-CI and Ste20 PRR, whereas the S473A mutation retains its binding activity.
To identify the residues critical to the interaction between Bem1 SH3b-CI and Ste20 PRR in vitro, we performed pull-down binding assays (Fig. 3D). The I471A mutant was bound to Bem1 SH3b-CI but the F470A mutant completely lost binding affinity to Bem1 SH3b-CI, although the core PRR sequence of Ste20 remained intact. These results together with the conserved phenylalanine at this position among Bem1 SH3b-CI binding proteins (Ste20, Cla4, Boi1, and Boi2) indicate that Phe-470 is critical for the interaction between Bem1 SH3b-CI and Ste20 PRR (Fig. 3, D and E). Thus, Phe-470 located at the N-terminally flanking region of Ste20 PRR is essential for the binding to Bem1 SH3b-CI.
Identification of the Binding Region of Bem1 SH3b-CI for Cdc42
Because of the high affinity binding of Cdc42 to Bem1 SH3b-CI, Cdc42 was mixed with excess amounts of 13C/15N-labeled SH3b-CI and the complex was purified by size-exclusion chromatography as described under “Experimental Procedures.” The backbone signals of 13C/15N labeled SH3b-CI complexed with unlabeled Cdc42 were assigned and the chemical shift difference between Cdc42 bound and unbound forms of Bem1 SH3b-CI was obtained as is plotted in Fig. 4A.
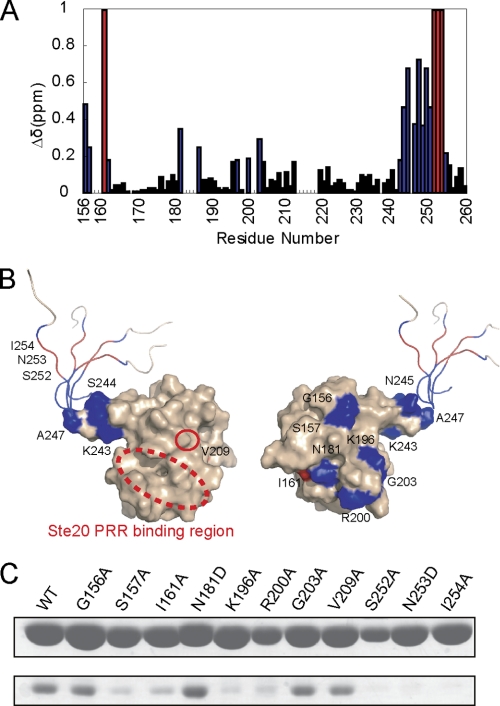
FIGURE 4.
Identification of the Cdc42 binding surface of SH3b-CI. A, backbone chemical shift changes of SH3b-CI complexed with Cdc42. The chemical shift changes (Δδ ≥ 0.20) upon Cdc42 binding are plotted in blue. The residues disappearing upon Cdc42 binding are plotted in red. B, surface and ribbon diagram of SH3b-CI. Residues are colored as noted in A. C, effect of amino acid substitutions of SH3b-CI on the binding to Cdc42 by in vitro pull-down binding assays. GST-Bem1-(156–260) mutants were incubated with Cdc42 (1–179, Q61L). The proteins pulled down with glutathione-Sepharose 4B were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue.
Chemical shift changes of more than 0.20 ppm upon Cdc42 binding were observed for seventeen residues (Gly-156, Ser-157, Val-162, Asn-181, Ala-186, Lys-196, Arg-200, Gly-203, Lys-243, Ser-244, Asn-245, Ala-247, Arg-248, Tyr-249, Lys-250, Ala-251, and Ser-255). In addition, four residues (Ile-161, Ser-252, Asn-253, and Ile-254) disappeared upon complex formation. The residues with disappearing peaks or appreciable chemical shift changes were mapped on the surface of SH3b-CI (Fig. 4B). It is to be noted that the residues on the C-terminal flexible region of CI were highly affected, suggesting that this region is critical for the Cdc42 binding. This is consistent with the result that the N253D mutation in CI inhibited the Cdc42 binding in vivo and in vitro (13). Although the C-terminal region is disordered in the free state, this region may form an ordered structure upon complex formation with Cdc42. To confirm that the C-terminal region of CI is critical for Cdc42 binding, mutation studies using an in vitro pull-down binding assay was performed using S252A, N253D, and I254A mutants. Compared with wild-type Bem1 SH3b-CI, these mutants completely lost binding affinity with Cdc42 (Fig. 4C). Thus, the C-terminal region of CI plays a critical role for complex formation with Cdc42.
Next, the mutation analyses using in vitro pull-down binding assay were performed to elucidate whether the SH3b moiety contributes to the binding to Cdc42. The residues that showed considerable chemical shift changes upon complex formation with Cdc42 were mutated to Ala or Asp and these mutants were applied for the pull-down binding assay with Cdc42. The G156A, N181D, and G203A mutants showed binding affinity similar to that of the wild type, supporting that these residues were not involved in the interaction with Cdc42, but the binding affinity to Cdc42 decreased for the Ser-157, Ile-161, Lys-196, and Arg-200 mutants. Interestingly, these residues are located on the particular surface (Fig. 4B, right), but the opposite surface (Fig. 4B, left) has no residues involved in the interaction. Therefore, Bem1 SH3b-CI uses both this particular SH3b surface and the C-terminal region of CI for the interaction with Cdc42. It should be noted that the Ste20 PRR binding surface (dotted circle in Fig. 4B, left) is opposite to this expected Cdc42 binding surface, facilitating the possible formation of the ternary complex, Bem1 SH3b-CI, Cdc42, and Ste20 PRR.
A Model for Ste20 Activation Mediated by Bem1
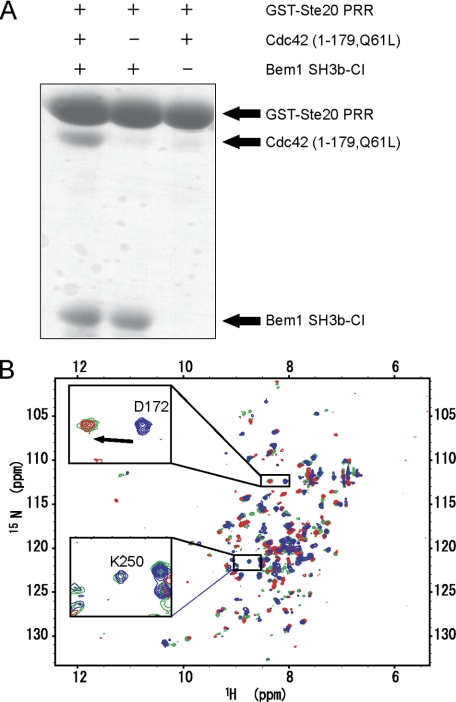
To investigate the possibility of a ternary complex formation among Bem1 SH3b-CI, Cdc42, and Ste20 PRR, we performed the in vitro pull-down binding assay, using these proteins (Fig. 5A). The GST-Ste20 PRR was bound to Bem1 SH3b-CI (Fig. 5A, mid) but not to Cdc42 (Fig. 5A, right). However, Cdc42 was pulled-down with GST-Ste20 PRR only in the presence of Bem1 SH3b-CI (Fig. 5A, left), indicating that the ternary complex formation is mediated by Bem1 SH3b-CI. Thus, Bem1 SH3b-CI appears to serve as a scaffold tethering both Ste20 and Cdc42 simultaneously. This is consistent with the NMR observations (Fig. 5B). Fig. 5B shows the overlay of the 1H-15N HSQC spectra of 15N-labeled Bem1 SH3b-CI in the presence of Cdc42 (blue), in the presence of Ste20 PRR (red), and in the presence of both Cdc42 and Ste20 PRR (green). In the presence of the three proteins, Asp-172 located on the Ste20 PRR binding surface of Bem1 SH3b-CI showed the chemical shift characteristic to a complex with Ste20 PRR, while, Lys-250 located on the C-terminal region of the CI moiety exhibited a chemical shift similar to that of the complex with Cdc42. The in vitro pull-down binding assay and the NMR study taken together support that Bem1 SH3b-CI, Cdc42, and Ste20 PRR are able to form a ternary complex.
FIGURE 5.
Ternary complex formation among Bem1 SH3b-CI, Cdc42, and Ste20 PRR. A, GST-Ste20 PRR was incubated either with Bem1 SH3b-CI or Cdc42 (1–179, Q61L), or both. The proteins pulled down with glutathione-Sepharose 4B were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue. B, 1H-15N HSQC spectra of Bem1 SH3b-CI. 15N-Bem1 SH3b-CI complexed with unlabeled Cdc42 (blue), complexed with unlabeled Ste20 PRR (red), and complexed with both unlabeled Cdc42 and Ste20 PRR (green) are overlaid. The chemical shift changes of Asp-172 (located on the PRR binding surface of Bem1 SH3b-CI) and Lys-250 (located on the Cdc42 binding surface of Bem1 SH3b-CI) are shown in the insets.
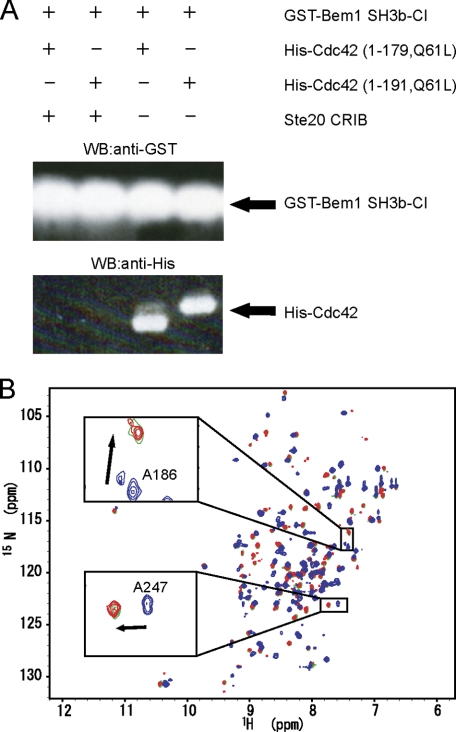
Next, we performed an in vitro pull-down binding assay using GST-Bem1 SH3b-CI, His-Cdc42, and Ste20 CRIB (Fig. 6A). It is to be noted that Bem1 SH3b-CI binds Cdc42 (Fig. 6A, the third and fourth lanes) but cannot bind Cdc42 in the presence of Ste20 CRIB (Fig. 6A, the first and second lanes). An NMR titration experiment with Ste20 CRIB showed that cross peaks in the HSQC spectrum of 15N-labeled Bem1 SH3b-CI complexed with Cdc42 (blue) shifted to those of free Bem1 SH3b-CI (red) upon addition of 1.0 eq. molar ratio of Ste20 CRIB to Bem1 SH3b-CI (green) (Fig. 6B). These results suggest that Ste20 CRIB deprives Bem1 SH3b-CI of Cdc42 because Cdc42 has a much higher binding affinity to Ste20 CRIB than to Bem1 SH3b-CI.
FIGURE 6.
The interactions among Bem1 SH3b-CI, Cdc42, and Ste20 CRIB. A, His-Cdc42 (1–179, Q61L) or His-Cdc42 (1–191,Q61L) was incubated with GST-Bem1 SH3b-CI in the presence or absence of Ste20 CRIB. The proteins pulled down with glutathione-Sepharose 4B were subjected to SDS-PAGE and stained by immunoblotting using appropriate antibodies. B, overlay of 1H-15N HSQC spectra of 15N-Bem1 SH3b-CI (red), 15N-Bem1 SH3b-CI in the presence of unlabeled Cdc42 (blue), and 15N-Bem1 SH3b-CI in the presence of both unlabeled Cdc42 and Ste20 CRIB (green) are shown. Chemical shift changes of Ala-186 and Ala-247 are shown in the insets.
Based on the present experimental results, we propose the following model for the Ste20 activation mechanism mediated by Bem1 and Cdc42. First, GTP-bound Cdc42 binds Bem1 through SH3b-CI. Next, Ste20 binds to the Bem1-Cdc42 complex through interaction between the Ste20 PRR moiety and Bem1 SH3b-CI. Now, Ste20 CRIB is located in close proximity to Cdc42 because of the ternary complex formation and thus Ste20 CRIB can interact with Cdc42 and deprive Bem1 SH3B-CI of Cdc42. Because Ste20 CRIB interacts with the Ste20 kinase domain to repress its enzymatic activity (4), the interaction between Cdc42 and Ste20 CRIB can release this repression and enhance the Ste20 kinase activity, essential for the cell polarity establishment. In order to verify the last step in the model for the Ste20 kinase activation in vitro, we expressed intact Ste20, constructs including CRIB and the kinase domain and only the kinase domain, respectively, using wheat germ cell-free system. Although we confirmed the expression of these constructs, we could not detect kinase activity even for the kinase only construct using myelin basic protein as a substrate. We could not verify the last step for the Ste20 activation in the present study. However, in in vivo assay, the relief of auto-inhibition of Ste20 kinase activity has been already demonstrated that is mediated through interaction of Cdc42 with Ste20 CRIB (4, 7). Thus, the present NMR evidence together with the previous in vivo experiments indicated that Bem1 is a scaffold protein which binds both Cdc42 and Ste20 to facilitate efficient binding of Cdc42 with Ste20 CRIB, leading to Ste20 kinase activation.
CONCLUSION
We determined the solution structure of a novel Cdc42 binding domain, Bem1 SH3b-CI. The study shows that Bem1 SH3b and CI are structurally and functionally related. Further, we determined the solution structure of Bem1 SH3b-CI complexed with Ste20 PRR. In addition to the core region of Ste20 PRR, its N-terminally flanking region participates in the interaction with Bem1. This enhances the binding affinity between Bem1 SH3b-CI and Ste20 PRR. The chemical shift perturbation study determined the Cdc42 binding surface of Bem1 SH3b-CI. In vitro pull-down binding assay together with the NMR titration study shows that Bem1, SH3b-CI, Ste20 PRR, and Cdc42 forms a ternary complex but Ste20 CRIB disrupts the ternary complex by removing Cdc42 from Bem1 SH3b-CI. This suggests the activation mechanism of Ste20 mediated by Bem1, which facilitates an efficient interaction of Ste20 CRIB and Cdc42 to release auto inhibition of Ste20 kinase activity.
This work was supported by a grant-in-aid for Scientific Research (Kiban S) and the National Projects on Targeted Proteins Research Program (TPRP) from Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
The atomic coordinates and structure factors (codes 2RQV and 2RQW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CRIB
- Cdc42- and Rac-interactive binding
- Bem
- bud emergence
- SH
- Src homology
- Cdc
- cell cycle division
- CI
- Cdc42 interacting
- PRR
- proline-rich region
- Ste
- sterile
- PAK
- p21-activated kinase
- GST
- glutathione S-transferase
- TOCSY
- total correlation spectroscopy
- NOE
- nuclear Overhauser enhancement effect
- NOESY
- NOE correlation spectroscopy
- MOPS
- 4-morpholinepropanesulfonic acid
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1.Chant J. (1999) Annu. Rev. Cell Dev. Biol. 15, 365–391 [DOI] [PubMed] [Google Scholar]
- 2.Etienne-Manneville S. (2004) J. Cell Sci. 117, 1291–1300 [DOI] [PubMed] [Google Scholar]
- 3.Zheng Y., Cerione R., Bender A. (1994) J. Biol. Chem. 269, 2369–2372 [PubMed] [Google Scholar]
- 4.Lamson R. E., Winters M. J., Pryciak P. M. (2002) Mol. Cell. Biol. 22, 2939–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benton B. K., Tinkelenberg A., Gonzalez I., Cross F. R. (1997) Mol. Cell. Biol. 17, 5067–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito T., Matsui Y., Ago T., Ota K., Sumimoto H. (2001) EMBO J. 20, 3938–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winters M. J., Pryciak P. M. (2005) Mol. Cell. Biol. 25, 2177–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bose I., Irazoqui J. E., Moskow J. J., Bardes E. S., Zyla T. R., Lew D. J. (2001) J. Biol. Chem. 276, 7176–7186 [DOI] [PubMed] [Google Scholar]
- 9.Terasawa H., Noda Y., Ito T., Hatanaka H., Ichikawa S., Ogura K., Sumimoto H., Inagaki F. (2001) EMBO J. 20, 3947–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshinaga S., Kohjima M., Ogura K., Yokochi M., Takeya R., Ito T., Sumimoto H., Inagaki F. (2003) EMBO J. 22, 4888–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogura K., Tandai T., Yoshinaga S., Kobashigawa Y., Kumeta H., Ito T., Sumimoto H., Inagaki F. (2009) J. Biochem. 146, 317–325 [DOI] [PubMed] [Google Scholar]
- 12.Butty A. C., Perrinjaquet N., Petit A., Jaquenoud M., Segall J. E., Hofmann K., Zwahlen C., Peter M. (2002) EMBO J. 21, 1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi Y., Ota K., Ito T. (2007) J. Biol. Chem. 282, 29–38 [DOI] [PubMed] [Google Scholar]
- 14.Cavanagh J., Fairbrother W. J., Palmer A. G., III, Skelton N. J. (1996) Protein NMR Spectroscopy, Academic Press, San Diego, CA [Google Scholar]
- 15.Delaglio F., Grzesiek S., Vuister G. W., Zhu W., Pfeifer J., Bax A. (1995) J. Biomol. NMR. 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 16.Kneller D. G., Goddard T. D. (1997) SPARKY 3.105 Ed., University of California, San Francisco, CA [Google Scholar]
- 17.Güntert P., Mumenthaler C., Wüthrich K. (1997) J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 18.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR. 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 19.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 20.Endo M., Shirouzu M., Yokoyama S. (2003) J. Biol. Chem. 278, 843–852 [DOI] [PubMed] [Google Scholar]
- 21.Wheatley E., Rittinger K. (2005) Biochem. J. 388, 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer B. J. (2001) J. Cell Sci. 114, 1253–1263 [DOI] [PubMed] [Google Scholar]
- 23.Jones D. T. (1999) J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]