Abstract
Desaturase enzymes are composed of two classes, the structurally well characterized soluble class found predominantly in the plastids of higher plants and the more widely distributed but poorly structurally defined integral membrane class. Despite their distinct evolutionary origins, the two classes both require an iron cofactor and molecular oxygen for activity and are inhibited by azide and cyanide, suggesting strong mechanistic similarities. The fact that the soluble desaturase is active as a homodimer prompted us test the hypothesis that an archetypal integral membrane desaturase from Saccharomyces cerevisiae, the Δ9-acyl-Co-A desaturase Ole1p, also exhibits a dimeric organization. Ole1p was chosen because it is one of the best characterized integral membrane desaturase and because it retains activity when fused with epitope tags. FLAG-Ole1p was detected by Western blotting of immunoprecipitates in which anti-Myc antibodies were used for capture from yeast extracts co-expressing Ole1p-Myc and Ole1p-FLAG. Interaction was confirmed by two independent bimolecular complementation assays (i.e. the split ubiquitin system and the split luciferase system). Co-expression of active and inactive Ole1p subunits resulted in an ∼75% suppression of the accumulation of palmitoleic acid, demonstrating that the physiologically active form of Ole1p in vivo is the dimer in which both protomers must be functional.
Keywords: Enzyme Structure, Lipid, Lipid Synthesis, Membrane Enzymes, Membrane Proteins, Yeast Metabolism, Desaturase
Introduction
Unsaturated fatty acids are found in the membranes of prokaryotes and eukaryotes. The balance between saturated and unsaturated fatty acids is a key variable in maintaining membrane fluidity. Postsynthetic desaturation of fatty acids is mediated by desaturase enzymes (1–5). Fatty acid desaturases are derived from two evolutionary lineages, the soluble acyl-ACP desaturases found primarily in the plastids of higher plants (e.g. the castor Δ9-18:0-acyl2 carrier protein (ACP)3 desaturase (6) and the integral membrane desaturases typified by the yeast Ole1 Δ9-desaturase (Ole1p) (7)). The reactions require iron cofactors, molecular oxygen, and reducing equivalents and are both inhibited by azide and cyanide but are insensitive to carbon monoxide. All desaturases investigated to date show remarkable stereo-selectivity in abstracting pro(R) hydrogens from adjacent carbons in the fatty acid substrate (8). As noted by Bloch (1), desaturase selectivity “would seem to approach the limits of the discriminatory power of enzymes” by a mechanism(s) that remain to be determined almost 40 years later.
Ole1p is a bifunctional 53-kDa protein (7), composed of two distinct domains: an N-terminal 42-kDa desaturase domain that has 36% homology to the rat stearoyl-CoA desaturase and a C-terminal 11-kDa domain that has strong homology to cytochromes b5 (9).
Although integral membrane desaturases, like many membrane proteins, have proved difficult overexpress and purify, sequence analysis has provided important information regarding the structure of the enzyme. For example, Martin and co-workers (7) employed hydropathy analysis to predict that the yeast desaturase consists of four transmembrane domains with the N and C termini facing the cytosolic side of the endoplasmic reticulum. Under their model, all three elements of a tripartite motif composed of 8 conserved histidine residues (10) would also be located on the cytoplasmic face. Experimental support for this model came from topology analysis of the closely related mouse stearoyl-CoA desaturase with the use of SCD1 (stearoyl-CoA desaturase 1) fused with epitope tags at various positions along the its sequence, in which the N and C termini, along with the histidine-containing regions, are located on the cytosolic face of the endoplasmic reticulum, and the enzyme is anchored by four transmembrane helices connected by two short hydrophilic loops within the endoplasmic reticulum lumen (11). Similar analysis was performed on the distantly related (11% homology with Ole1) Bacillus subtilis Δ5-desaturase, and although the equivalent structural elements were found on the inner membrane face, six rather than four transmembrane-spanning domains were proposed (12).
Much of what we know about the structure and function of desaturase enzymes comes from studies on the soluble acyl-ACP desaturases, which are readily expressible in Eschericha coli in quantities sufficient for biochemical investigation (13). The crystal structure of the castor and ivy acyl-ACP desaturases confirmed their dimeric structure with deep interdigitation between the monomers adjacent to four helix bundles in which the diiron active site is located (14, 15). A half-of-the-sites mechanism was hypothesized for desaturase function, in which the energy released upon substrate binding is transferred to the second subunit to facilitate product release (16).
To date, no information is available concerning the oligomeric state of the integral membrane class of enzymes. Here we present evidence from co-immunoprecipitation/immunoblot analysis, yeast two-hybrid analysis, and analysis of the co-expression wild type (WT) along with catalytically inactive Ole1p that is consistent with the hypothesis that Ole1p forms dimers in vivo.
EXPERIMENTAL PROCEDURES
Fatty Acid Analysis
Cultures were initiated from single yeast colonies. Yeast cells L8-14C (MATa ole1Δ::LEU2 leu2-3 leu2-112 trp1-1, ura3-52 his4) defective in Δ9-fatty acid desaturase were grown at 30 °C overnight in SD-Leu-Ura (2% glucose, 0.5% Tergitol Nonidet P-40, 0.5 mm oleic acid). Cell pellets were collected by centrifugation and then washed with SD-Leu-Ura lacking dextrose. Cells were induced at an A600 of 1.0 at 30 °C for 16 h in SD-Leu-Ura supplemented with 2% galactose. After centrifugation, the cell pellets were washed with 1% Tergitol Nonidet P-40 and water sequentially and dried under N2. Fatty acid methyl ester was prepared with 1 ml of 1 n HCl-methanol at 80 °C 1 h. After the addition of 1 ml of 0.9% NaCl, fatty acids were extracted with 2 ml of hexane. The resulting fatty acid methyl esters were concentrated by evaporation under a stream of nitrogen. Samples were analyzed on a Hewlett-Packard 6890 gas chromatograph equipped with a 5973 mass-selective detector (gas chromatography/mass spectrometry) and a Supelco 60 m × 250-μm SP-2340 cyanocapillary column. The oven temperature was raised from 100 to 240 °C at a rate of 15 °C/min and held at 240 °C for 5 min. The helium flow rate was 1.1 ml/min.
Yeast Protein Extraction and Co-immunoprecipitation Analysis
Yeast cells were collected by centrifugation, washed once with water, and disintegrated in YEB buffer (50 mm HEPES, pH 7.4, 10 mm EDTA, 0.1% Triton X-100) by vortexing with glass beads. Following 15 min of centrifugation at 12,000 × g, the supernatant was supplemented with 0.5% protease inhibitor (Sigma) and incubated with antibody for 1 h. One-twentieth volume of protein A-Sepharose (Amersham Biosciences) suspension was added, and the mixture was incubated at 4 °C with gentle shaking overnight. Beads were washed twice with 10 volumes of Buffer A (50 mm Tris, pH 8.0, 150 mm NaCl, and 0.1% Triton X-100) and Buffer B (same as buffer A without NaCl and Triton X-100). Bound proteins were eluted with elution buffer (0.1 m glycine, pH 3.0). The eluted proteins were separated by 10% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and analyzed by Western blot. The reactive bands on membranes were visualized after incubation with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate.
Yeast Transformation and Bimolecular Complementation
Yeast transformation was performed with the Yeastmaker Yeast Transformation System 2 (Clontech). Experiments employing the membrane protein split ubiquitin system were performed according to the protocols of the Frommer laboratory (17). The split luciferase system was employed to provide quantitative analysis of the Ole1p-Ole1p interaction in vivo (18). For luminescence detection, yeast cells were grown at 30 °C overnight in SD-Ura supplemented with 2% glucose. Cells were collected by 1 min centrifugation at 16,000 × g, and cell pellets were washed with SD-Ura (lacking dextrose). Cells were induced at an A600 of 0.4) in SD-Ura supplemented with 2% galactose at a temperature of 30 °C for 16 h. Cell cultures (90 μl) were transferred to a 96-well plate, and assays were initiated with the addition of 10 μl of a 60 μm concentration of a coelenterazine derivative (ViviRen live cell substrate, Promega). The assay plate was incubated for 5 min at room temperature in the darkness before luminescence measurement with the use of a TECAN Ultra 384 microplate reader.
RESULTS
C-terminal FLAG- and Myc-tagged Ole1p Retains Activity
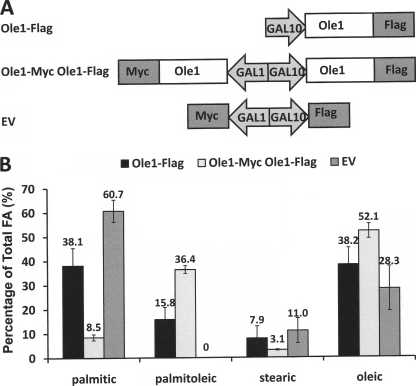
To facilitate the detection and immunocapture of various Ole1 constructs, we explored the consequences of fusing Saccharomyces cerevisiae Ole1p with Myc and FLAG epitope tags (see Fig. 1A). Ole1p was fused with C-terminal tags encoding either FLAG or Myc. Ole1p-FLAG was placed under the control of the GAL10 promoter. A second construct was engineered in which the Ole1p-Myc, under the control of the GAL1 promoter, was added to the plasmid containing the GAL10-Ole1p-FLAG cassette, such that Myc- and FLAG-tagged Ole1p could be co-expressed.
FIGURE 1.
Expression of C-terminally tagged Ole1p in yeast cells. A, schematic representation of the plasmid vector constructs made for expression of Ole1p-Myc under the GAL1 promoter and Ole1p-FLAG under the GAL10 promoter separately. Sequences of oligonucleotide primers used for all constructs are listed in supplemental Table 1. B, fatty acid analysis of transformed L8-14C yeast cells. Cells were supplemented with oleic acid and 2% glucose and then induced with 2% galactose. Lipid metabolites were identified and quantitated by gas chromatography/mass spectrometry.
To evaluate whether C-terminal Myc- or FLAG-tagged Ole1p retains activity, constructs containing either Ole1p-Myc or Ole1p-FLAG were expressed in the yeast ole1 deletion strain L8-14C cultured in medium supplemented with oleic acid (18:1Δ9) as a source of unsaturated fatty acid. Cells were washed to remove exogenous oleic acid, and protein expression was induced for 30 h. Levels of oleate increased from ∼28% in empty vector controls to ∼38% in cells harboring Ole1p-FLAG. Oleate levels increased by a further 14% to ∼52% when Ole1p-Myc was co-expressed along with Ole1p-FLAG, showing that Ole1p-Myc is also active. Because Ole1p is capable of desaturating both 16- and 18-carbon fatty acids, the accumulation of palmitoleic acid (16:1Δ9) is the best indicator of Ole1p function because host cells lack palmitoleic acid, and there is therefore no background to subtract (see Fig. 1B). Ole1p-FLAG results in the accumulation of ∼16% palmitoleic acid, and Ole1p-Myc accounts for ∼20% accumulation of palmitoleic acid. These data confirm that Ole1p is active in the presence of C-terminal FLAG and Myc tags and that both constructs have similar in vivo activities.
Co-immunoprecipitation Provides Immunological Support for Ole1p-Ole1p Association
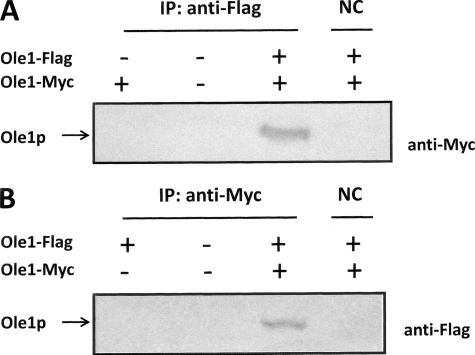
Yeast cells harboring empty vector or Ole1p-Myc alone or co-expressing FLAG- and Myc-tagged Ole1p fusions were subject to immunoprecipitation with anti-FLAG antibodies and protein A capture. The resulting proteins were separated by PAGE and immunoblotted with anti-Myc antibodies after transfer to polyvinylidene difluoride membrane. Fig. 2A shows an immunoreactive signal only upon co-expression both Ole1p-FLAG- and Ole1p-Myc-tagged proteins. Controls show that cells lacking Ole1p-FLAG show negative Western signal, indicating that the capture of anti-Myc-reacting polypeptide was dependent on the co-expression of anti-FLAG-tagged Ole1p. The fact that the signal was lost upon omission of anti-FLAG antibodies for immunoprecipitation further confirms that the anti-Myc-reactive polypeptide was recovered specifically by the anti-FLAG antibody. To validate this result, we performed the same experiment with anti-Myc antibodies for capture and anti-FLAG antibodies for immunodetection (Fig. 2B). Controls confirm that the positive signal is dependent on expression of both tagged forms of Ole1p and the presence of anti-Myc capture antibody. Together, these results provide compelling immunological evidence for Ole1p-FLAG-Ole1p-Myc association.
FIGURE 2.
Ole1p-Myc and Ole1p-FLAG association revealed by co-immunoprecipitation analysis. A, yeast extracts from strain YPH499 expressing combinations of Ole1p-Myc and Ole1p-FLAG, as indicated, were subject to immunoprecipitation (IP) with anti-FLAG antibody or lacking the addition of antibody as negative control (NC). Samples were separated by SDS-PAGE, immunoblotted, and visualized with the use of anti-Myc antibodies. The bands corresponding to Myc- and FLAG-tagged Ole1p are indicated. B, same as A except that immunoprecipitation was performed with anti-Myc antibodies and immunoblotting was performed with anti-FLAG antibodies.
Bimolecular Association Analysis Confirms Ole1p-Ole1p Association in Vivo
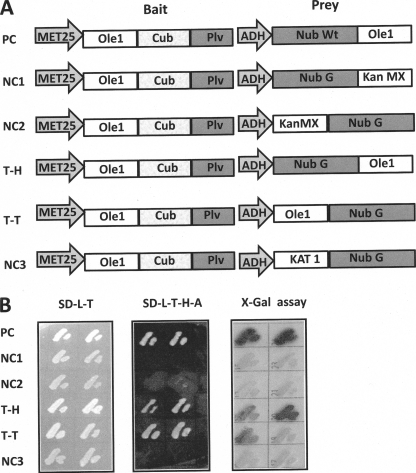
As an independent approach to the immunological experiments described above, we employed split ubiquitin analysis to probe for potential Ole1p-Ole1p interactions in vivo. To achieve this, Ole1p was cloned into a bait vector such that it was expressed with a C-terminal fragment of ubiquitin (Cub) and the artificial transcription factor PLV (protein A-LexA-VP16) at its C terminus. For the split ubiquitin system to function, CubPLV and the Nub peptides must both be located on the cytosolic face of the membrane. In the absence of an empirically tested topology model, two test prey vectors were constructed, a Nub G-Ole1p fusion and an Ole1p-Nub G fusion, the former to test for tail-to-head interaction and the latter to test for tail-to-tail interaction (Fig. 3A) (the designation head or tail is used to refer to the location of the reporter at the N or C terminus, respectively, as fusions to the target protein). Yeasts containing all constructs were able to grow on SD-L-T, but only the positive control, containing a WT Nub in the prey vector and the test prey constructs containing the tail-to-head and tail-to-tail constructs were able to complement growth on SD plates -L-T-H-A by PLV activation of LexA-driven His 3 and Ade 2 in the host genome (Fig. 3B). Growth was not restored in yeast strains containing any of the three negative control prey vectors lacking Ole1p, including the standard membrane protein negative control, the Arabidopsis K+ channel protein, KAT1. As an additional negative control, the plant membrane bound desaturase Fad3 was tested for association with Ole1p, with no interaction being detected (see supplemental Fig. 1). The host strain employed for the experiments shown in Fig. 3 also contains a LexA::lacZ such that the yeast can be plated on X-gal-containing medium and on which β-galactosidase activity can be estimated qualitatively by the degree of blue coloration of the colony. In Fig. 3B (right), dark coloration corresponds to the ability of the colony to grow on -H-A plates (Fig. 3B, middle). The degree of dark coloration is stronger for tail-to-head than for the tail-to-tail orientation. The restoration of His and Ade prototrophy demonstrates in vivo Ole1p-Ole1 association; however, the detection of β-galactosidase activity was performed after SDS lysis of the yeast cells and can thus be considered an in vitro assay. To corroborate the split ubiquitin analysis and to quantitate the observed in vivo interactions, Ole1p was subjected to a second bimolecular fluorescence analysis with the use of the Renilla split luciferase (RLUC) system.
FIGURE 3.
Membrane split ubiquitin yeast two-hybrid analysis shows that Ole1p self-associates in vivo. A, schematic representation of the pairs of bait and prey plasmid constructs. The bait Ole1-Cub-artificial transcription factor PLV (protein-LexA-VP16) construct is driven by the met25 promoter, and prey expression is driven by the ADH promoter. PC, positive control; NC1, NC2, and NC3, negative controls; Kan MX, kanamycin resistance module; KAT 1, Arabidopsis K+ channel protein; T-H, tail to head; T-T, tail to tail. B, challenge of yeast harboring the constructs illustrated in A on nonselective minimal medium, SD-L-T, (left) and selective minimal SD-T-L-H-A medium (middle) and X-gal assay (right) in which LacZ activity with X-gal as substrate is indicated by dark coloration.
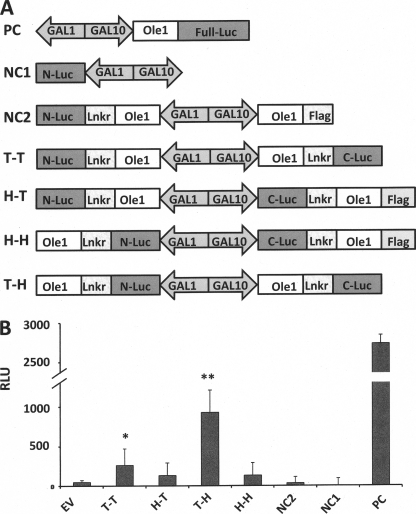
In general, the two complementary fragments of the split luciferase system can be fused to either N or C termini of the target protein. In practice, the functioning of combined luciferase fragments is dependent on their orientation with respect to the protein of interest due to specific steric and/or folding constraints (19). Thus, to optimize the luciferase activity, constructs were engineered to test four relative orientations: tail to tail, head to tail, head to head, and tail to head (Fig. 4A). The tail-head and tail-tail orientations showed a significant increase luminescence (p < 0.001 and p < 0.02 levels, respectively) relative to the negative controls, whereas the head-to-tail and head-to-head orientations showed no significant increase in luminescence (p < 0.2 level). These data reflect the possibility that the two fragments of the RLUC are not sufficiently close to facilitate productive interaction or that their relative orientations are incompatible with productive interaction (19). An alternative interpretation is that the short FLAG tag sequence present on these two constructs results in a reduction of luciferase activity; however, this seems unlikely because the maximally active combination T-H contains a linker plus C-terminal luciferase fragment that represents a much larger C-terminal extension of Ole1p. Similar issues have been reported for homo-FRET (fluorescence resonance energy transfer) interactions (20). The RLUC data are consistent with the qualitative in vitro β-galactosidase activity results presented above.
FIGURE 4.
Quantitation of Ole1p self-association in vivo membrane with the use of the split luciferase yeast two-hybrid system. A, schematic representation of the plasmid constructs made for expression of Ole1 in the RLUC system in which N- and C-terminal luciferase (Luc) fragments are fused to Ole1p via a 10-amino acid linker (-GGGGSGGGGS-). PC, positive control; NC, negative control; T-T, tail to tail; H-T, head to tail; H-H, head to head; T-H, tail to head. B, assay of the activity of reconstituted RLUC by bimolecular complementation. Relative luminescence units (RLU) are plotted for each construct. Stars designate significant differences from the empty vector (EV), negative control value (*, p < 0.05; **, p < 0.01). Error bars, S.D.
Co-expression of Wild Type and Inactive Mutant Desaturase Provides Evidence for Subunit Poisoning of Heterodimers
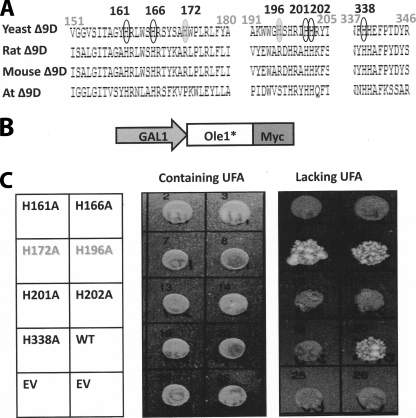
Sequence alignment of integral membrane desaturases identified a series of 8 histidine residues that are strongly conserved between members from evolutionarily distant lineages, such as bacteria, fungi, and humans (10). These conserved histidine residues have been proposed as ligands to a diiron active site cluster identified by Mossbauer spectroscopy in the distantly related alkane ω-hydroxylase enzyme from Pseudomonas oleovorans (21). Functional relevance of the histidine cluster has been determined for rat and cyanobacterial desaturases and also in the alkane ω-hydroxylase (22). However, the relevance of these residues has not been described for Ole1p. We therefore chose several conserved histidines at positions 161, 162, 201, 202, and 338 along with several non-conserved histidines at positions 172 and 196 as targets for alanine-scanning mutagenesis (Fig. 5A). Each of the alanine mutants was introduced behind a GAL1 promoter as a C-terminal Myc-tagged fusion protein (Fig. 5B). The constructs were transformed into the Ole1p mutant unsaturated fatty acid auxotroph yeast strain, which was challenged to grow on medium supplemented with or lacking unsaturated fatty acids (Fig. 5C). The ole1 deletion strain L814C expressing wild type Ole1p or non-conserved substitutions (H172A and H196A) grew on medium lacking unsaturated fatty acid, whereas substitution of conserved histidine residues for alanines resulted in Ole1p variants that were unable to grow in the absence of unsaturated fatty acid-containing medium, demonstrating the essential nature of these five conserved histidine residues for Ole1P function.
FIGURE 5.
Conserved histidine residues are essential for catalytic function of Ole1p. A, comparison of partial sequences of the membrane desaturases from yeast (NP_011460), rat (NP_114029), mouse (NP_033154), and Arabidopsis (AAM63359). Five conserved His residues within Ole1p are indicated in black circles, and two non-conserved His residues are indicated in gray-filled circles; numbers are relative to the Ole1p sequence. B, schematic representation of the plasmid constructs for expression of Ole1p mutant-Myc under the control of the GAL1 promoter. C, growth of yeast Ole1 deletion strain L8-14C lacking or containing either the wild type Ole1 or mutant forms of the gene in which individual His residues were converted to Ala as indicated. Cultures were replica-plated onto media containing unsaturated fatty acids, or media lacking unsaturated fatty acids (UFA), as indicated.
In some enzymes, component protomers act independently, whereas in others, a single inactive subunit can result in an inactive oligomer, a phenomenon that has been referred to as subunit poisoning. The availability of inactive subunits in which conserved histidine residues are replaced by alanines provides a way of assessing whether the desaturase subunits act independently by coexpression of wild type and mutant desaturase subunits and evaluating the resulting fatty acid phenotype. From experiments described above, the tail-to-head orientation results in the highest functionality and is therefore used to test for desaturase subunit poisoning (Fig. 6A). We employed a plasmid pESC-Ura designed for the expression of two proteins simultaneously, one under the control of the GAL1 promoter and the other under the control of the GAL10 promoter. We therefore first evaluated the relative strengths of the GAL1 and GAL10 promoters, comparing the fluorescence generated by expression of an Ole1p-luciferase fusion under each promoter separately (Fig. 6B). Curves of fluorescence versus time showed no significant differences in luminosity between Ole1p-luciferase constructs under the control of the GAL1 and GAL10 promoters, indicating that both promoters are equivalent in strength.
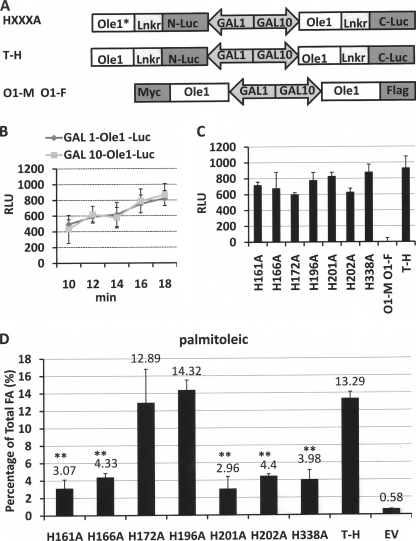
FIGURE 6.
Catalytic activities are reduced when conserved His Ole1p mutants are co-expressed with wild type Ole1p. A, schematic representation of the plasmid constructs made for expression of mutated Ole1 (HXXXA) under the GAL1 promoter along with wild type Ole1 under the GAL10 promoter. The tail to head construct comprising two wild type Ole1-luciferase fragments was used as a positive control construct. An Ole1-Myc, Ole1-FLAG construct (lacking luciferase) was used as negative control. B, time course comparison of the luminosity signal resulting from expression of Ole1-luciferase under the control of the GAL1 and GAL10 promoters in YPH499 yeast cells (in minutes). C, comparison of the luminosity resulting from the YPH499 yeast cells transformed with the indicated individual His-Ala mutations of Ole1 co-expressed and wild type Ole1p in the RLUC bimolecular complementation system; the tail to head construct with two wild type ole1 genes was used as positive control, and the O1-M, O1-F construct was used as negative control (see A). D, palmitoleic acid content of L8-14C yeast cells from the experiment described in C as analyzed by gas chromatography/mass spectrometry. Empty vector (EV) was used as a control. RLU, relative luminescence units.
Each of the mutants was expressed in a tail-to-head orientation along with wild type Ole1p, and the respective strains were subjected to luminescence assay (Fig. 6C). All WT-mutant pairs resulted in a similar magnitude of bimolecular fluorescence complementation, demonstrating that the mutants retain their ability to co-localize and associate with WT Ole1p. Minor differences in fluorescence may reflect minor differences in localization and/or stability of individual mutants.
To evaluate the activity of the co-expressed desaturase constructs, we assayed the accumulation of palmitoleic acid because it is absent from the host strain (Fig. 1B). Co-expression of non-conserved (i.e. active) histidine to alanine mutants (H172A and H196A) with WT Ole1p led to the accumulation of high levels of palmitoleic acid equivalent to those of the cells expressing two WT genes (Fig. 6D). In contrast, co-expression of loss-of-function histidine-to-alanine mutants with WT resulted in a significant (p < 0.001) decrease in accumulation of palmitoleic acid to an average of 24.9% of that of the WT control.
DISCUSSION
In contrast to the soluble desaturases, integral membrane desaturases are notoriously recalcitrant to overexpression in heterologous systems and with several notable exceptions (23, 24) have been very difficult to purify. Several crystal structures of soluble desaturases have been determined, which show that the enzyme exists as a dimer with a deeply interdigitated interface (14, 15, 25). However, the integral membrane desaturases await detailed biophysical characterization, and whether they are active as monomers, dimers, or higher oligomers has yet to be reported. We therefore employed a series of in vitro and in vivo approaches to establish that Ole1p exists as a dimer in vivo.
Several lines of evidence presented here support the hypothesis that Ole1p self-associates, including the following. 1) Co-immunoprecipitation experiments were performed in which a Myc tag was used to precipitate Myc-Ole1p and a FLAG tag was used to visualize a co-expressed FLAG-Ole1p monomer; the experiment yielded equivalent results when either anti-Myc antibodies were used for capture and anti-FLAG was used for visualization or vice versa. 2) Two independent bimolecular complementation approaches, one employing the split ubiquitin and the other employing the split luciferase system, provided evidence for self association. 3) Co-expression of active and inactive Ole1p subunits showed a reduction in activity consistent with a dominant negative effect in which one defective subunit inactivated the oligomer. Equal expression of both subunits gave rise to an ∼75% decrease of the palmitoleic acid accumulation phenotype, consistent with a dimeric organization. The magnitude of the decrease is inconsistent with the formation of higher oligomers because subunit poisoning of the most simple of these (i.e. a tetramer) would be expected to produce an ∼94% decrease in product accumulation (i.e. a level ∼5-fold lower than the experimental observation).
The observed subunit poisoning of Ole1p is consistent with the proposed dominant negative effect as an explanation for the reduction in linoleic acid and accompanying increase in oleic acid upon the expression of a nonfunctional Brassica oleate desaturase (mutant Bnfad2) in the seeds of transgenic cotton (26, 27). The Brassica data also suggest that dimerization of the yeast Ole1p presented herein represents a general structural organization for integral membrane desaturases because the Brassica desaturase lacks the C-terminal cytochrome b5 domain of Ole1p.
It is intriguing that the evolutionarily distinct soluble desaturases and the integral membrane desaturases, typified by the castor Δ9-desaturase and the yeast Ole1p, respectively, have apparently evolved convergently, arriving at similar iron active sites and dimeric organization. This suggests a possible common imperative for this organization with respect to desaturase function. However, a dimeric organization is also common among many enzyme families and may provide several selective advantages (e.g. reducing the genome size for the production of homodimeric proteins and stabilizing proteins because smaller protomers can fold readily prior to assembly into larger oligomeric active structures) (28). In addition to these selective advantages, dimeric organization allows for the biochemical regulation of heterodimers. This can be achieved by the formation of heterodimers from distinct pools of oligomers. It is unknown whether desaturase enzymes form heterodimers in vivo, but it is possible that such a mechanism exists for plants, such as Arabidopsis, in which the soluble desaturase family consists of seven members, each with distinct kinetics and substrate specificities (29), or the ω-3 desaturase family, comprising three enzymes that are evolutionarily related to Ole1. However, such a regulatory role is unlikely for yeast Ole1p because it is encoded by a single gene.
A dimeric organization can also play an important role for homodimers in that the state of component protomers can influence each others' biochemical properties. For instance, the soluble desaturase dimer exhibits high affinity binding for one acyl-ACP substrate, but the affinity for binding a second substrate becomes much reduced, implying that a signal is transmitted from the substrate-bound protomer to the unoccupied protomer that reduces its substrate binding affinity (30). This may have implications for coordinating desaturase-substrate interactions. A half-of-the-sites mechanism was proposed for the soluble desaturase (16) in which the energy of desolvation released upon binding of the hydrophobic substrate to one protomer is transferred to the second protomer, providing a driving force to facilitate the energy-demanding release of the hydrophobic product into the aqueous phase. Binding of substrate to only one of the two protomers would facilitate an unbound substrate that would be available for substrate binding, facilitating the coordination of substrate binding with product release. This is an appealing scenario for the soluble desaturase and acyl-CoA desaturases, such as Ole1p, because the substrates and products exist in the aqueous phase. However, it is not clear if this mechanism would apply to desaturases in which the substrates are lipid-bound acyl chains for which binding of substrate is unlikely to be coordinated with product release.
An alternate imperative for the dimeric organization of desaturases could relate to their requirement for two-electron reduction of the active site iron center for catalysis. In both the soluble and membrane desaturases, electrons are supplied singly by interaction of the desaturase with ferredoxin and cytochrome b5, respectively. In vitro redox titration of the soluble desaturase at ambient temperatures failed to reveal the presence of a meta (Fe2+-Fe3+) state of the iron center that is observed upon reduction at low temperatures (77 K), suggesting that the meta state is unstable at ambient temperatures (31). A dimeric structure of the desaturase would potentially allow for concerted reduction of the iron centers within each protomer to produce two meta state iron centers. These could undergo disproportionation to produce one reduced, Fe2+-Fe2+, and one oxidized, Fe3+-Fe3+, site.
At this time, it is unknown whether the formation of desaturase dimers is related to the general economies associated with the formation of homodimers (described above) or whether it relates to desaturase chemistry-specific imperatives imposed by the energetics of product release or requirement for two-electron reduction. It may be possible to test the desaturase-specific hypotheses empirically. This approach would be analogous to the subunit poisoning experiments presented herein; the activities of heterodimers composed of wild type protomers paired with protomers impaired in either substrate- or redox-partner binding could be evaluated. Inactivation of wild type activity upon pairing of wild type protomers with protomers disrupted in substrate or redox-partner binding would provide empirical support for these hypotheses.
The expression of Ole1p is highly regulated, responding to factors such as carbon source, metal ions, and the availability of oxygen (32). Multiple levels of regulation have been reported, including at the transcriptional, RNA stability, and translational levels. It remains to be determined whether the formation of dimers is a dynamic process that could potentially contribute to this process.
Supplementary Material
Acknowledgment
We thank Dr. C. E. Martin (Rutgers University) for the gift of yeast strain L814C.
Thiswork was supported by the Office of Basic Energy Sciences of the United States Department of Energy and by the Bayer Corp.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
Fatty acid nomenclature is as follows. X:Y indicates that the fatty acid contains X numbers of carbon atoms and Y numbers of double bonds; Δz indicates that a double bond is positioned at the zth carbon atom from the C terminus.
- ACP
- acyl carrier protein
- WT
- wild type
- RLUC
- Renilla split luciferase
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- Cub
- C-terminal ubiquitin fragment
- Nub
- N-terminal ubiquitin fragment.
REFERENCES
- 1.Bloch K. (1969) Acc. Chem. Res. 2, 193–202 [Google Scholar]
- 2.Shanklin J., Cahoon E. B. (1998) Annu. Rev. Plant Physiol. Plant. Mol. Biol. 49, 611–641 [DOI] [PubMed] [Google Scholar]
- 3.Fox B. G., Lyle K. S., Rogge C. E. (2004) Acc. Chem. Res. 37, 421–429 [DOI] [PubMed] [Google Scholar]
- 4.Buist P. H. (2004) Nat. Prod. Rep. 21, 249–262 [DOI] [PubMed] [Google Scholar]
- 5.Shanklin J., Guy J. E., Mishra G., Lindqvist Y. (2009) J. Biol. Chem. 284, 18559–18563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shanklin J., Somerville C. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2510–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stukey J. E., McDonough V. M., Martin C. E. (1990) J. Biol. Chem. 265, 20144–20149 [PubMed] [Google Scholar]
- 8.Behrouzian B., Buist P. H. (2002) Curr. Opin. Chem. Biol. 6, 577–582 [DOI] [PubMed] [Google Scholar]
- 9.Mitchell A. G., Martin C. E. (1995) J. Biol. Chem. 270, 29766–29772 [DOI] [PubMed] [Google Scholar]
- 10.Shanklin J., Whittle E., Fox B. G. (1994) Biochemistry 33, 12787–12794 [DOI] [PubMed] [Google Scholar]
- 11.Man W. C., Miyazaki M., Chu K., Ntambi J. M. (2006) J. Biol. Chem. 281, 1251–1260 [DOI] [PubMed] [Google Scholar]
- 12.Diaz A. R., Mansilla M. C., Vila A. J., de Mendoza D. (2002) J. Biol. Chem. 277, 48099–48106 [DOI] [PubMed] [Google Scholar]
- 13.Lindqvist Y. (2001) Delta Nine Stearoyl-acyl Carrier Protein Desaturase, pp. 725–737, John Wiley & Sons, Chichester, UK [Google Scholar]
- 14.Lindqvist Y., Huang W., Schneider G., Shanklin J. (1996) EMBO J. 15, 4081–4092 [PMC free article] [PubMed] [Google Scholar]
- 15.Guy J. E., Whittle E., Kumaran D., Lindqvist Y., Shanklin J. (2007) J. Biol. Chem. 282, 19863–19871 [DOI] [PubMed] [Google Scholar]
- 16.Broadwater J. A., Ai J., Loehr T. M., Sanders-Loehr J., Fox B. G. (1998) Biochemistry 37, 14664–14671 [DOI] [PubMed] [Google Scholar]
- 17.Obrdlik P., El-Bakkoury M., Hamacher T., Cappellaro C., Vilarino C., Fleischer C., Ellerbrok H., Kamuzinzi R., Ledent V., Blaudez D., Sanders D., Revuelta J. L., Boles E., André B., Frommer W. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujikawa Y., Kato N. (2007) Plant J. 52, 185–195 [DOI] [PubMed] [Google Scholar]
- 19.Massoud T. F., Paulmurugan R., Gambhir S. S. (2004) FASEB J. 18, 1105–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorkin A., McClure M., Huang F., Carter R. (2000) Curr. Biol. 10, 1395–1398 [DOI] [PubMed] [Google Scholar]
- 21.Shanklin J., Achim C., Schmidt H., Fox B. G., Münck E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2981–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avelange-Macherel M. H., Macherel D., Wada H., Murata N. (1995) FEBS Lett. 361, 111–114 [DOI] [PubMed] [Google Scholar]
- 23.Strittmatter P., Spatz L., Corcoran D., Rogers M. J., Setlow B., Redline R. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4565–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt H., Dresselhaus T., Buck F., Heinz E. (1994) Plant Mol. Biol. 26, 631–642 [DOI] [PubMed] [Google Scholar]
- 25.Moche M., Shanklin J., Ghoshal A., Lindqvist Y. (2003) J. Biol. Chem. 278, 25072–25080 [DOI] [PubMed] [Google Scholar]
- 26.Chapman K. D., Austin-Brown S., Sparace S. A., Kinney A. J., Ripp K. G., Pirtle I. L., Rirtle R. M. (2001) J. Am. Oil Chem. Soc. 78, 941–947 [Google Scholar]
- 27.Chapman K. D., Neogi P. B., Hake K. D., Stawska A. A., Speed T. R., Cotter M. Q., Garrett D. C., Kerby T., Richardson C. D., Ayre B. G., Ghosh S., Kinney A. J. (2008) Crop Sci. 48, 1470–1481 [Google Scholar]
- 28.Marianayagam N. J., Sunde M., Matthews J. M. (2004) Trends Biochem. Sci. 29, 618–625 [DOI] [PubMed] [Google Scholar]
- 29.Kachroo A., Shanklin J., Whittle E., Lapchyk L., Hildebrand D., Kachroo P. (2007) Plant Mol. Biol. 63, 257–271 [DOI] [PubMed] [Google Scholar]
- 30.Haas J. A., Fox B. G. (2002) Biochemistry 41, 14472–14481 [DOI] [PubMed] [Google Scholar]
- 31.Davydov R., Behrouzian B., Smoukov S., Stubbe J., Hoffman B. M., Shanklin J. (2005) Biochemistry 44, 1309–1315 [DOI] [PubMed] [Google Scholar]
- 32.Martin C. E., Oh C. S., Jiang Y. (2007) Biochim. Biophys. Acta 1771, 271–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.