Abstract
The PQBP1 (polyglutamine tract-binding protein 1) gene encodes a nuclear protein that regulates pre-mRNA splicing and transcription. Mutations in the PQBP1 gene were reported in several X chromosome-linked mental retardation disorders including Golabi-Ito-Hall syndrome. The missense mutation that causes this syndrome is unique among other PQBP1 mutations reported to date because it maps within a functional domain of PQBP1, known as the WW domain. The mutation substitutes tyrosine 65 with cysteine and is located within the conserved core of aromatic amino acids of the domain. We show here that the binding property of the Y65C-mutated WW domain and the full-length mutant protein toward its cognate proline-rich ligands was diminished. Furthermore, in Golabi-Ito-Hall-derived lymphoblasts we showed that the complex between PQBP1-Y65C and WBP11 (WW domain-binding protein 11) splicing factor was compromised. In these cells a substantial decrease in pre-mRNA splicing efficiency was detected. Our study points to the critical role of the WW domain in the function of the PQBP1 protein and provides an insight into the molecular mechanism that underlies the X chromosome-linked mental retardation entities classified globally as Renpenning syndrome.
Keywords: Brain, Peptide Arrays, Protein Domains, Protein-Protein Interactions, RNA Splicing, RNA Processing, WW Domain, Mental Retardation, Modular Protein Domain, Point Mutation
Introduction
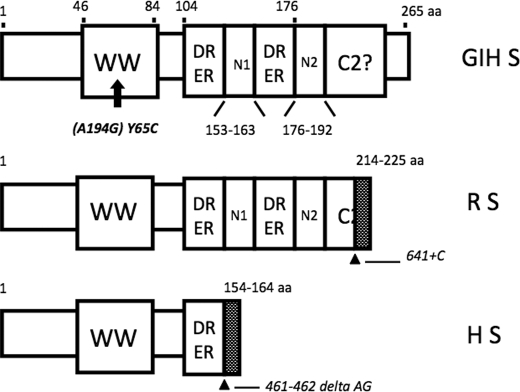
The PQBP1 (polyglutamine tract-binding protein 1)2 gene encodes a nuclear protein of 38 kDa that is abundantly expressed in the central nervous system (1–5). Several studies have provided evidence for a role of the PQBP1 protein in the pathogenesis of polyglutamine expansion diseases, including spinocerebellar ataxia type 1 (6, 7). More direct evidence for the contribution of PQBP1 to neurological disorders has come from clinical genetics. Mutations in the PQBP1 gene were reported in several X chromosome-linked mental retardation (XLMR) disorders, such as Renpenning, Sutherland-Haan, Hamel, Porteous, and Golabi-Ito-Hall syndromes (8–14). Interestingly, although caused by different mutations within the PQBP1 gene (e.g. frame shifts that result in truncated protein products, missense point mutation; see Fig. 1 for three examples), these syndromes share similar clinical features. In addition to severe mental retardation, the patients also have a short stature, lean body, small head, and are frequently diagnosed with cardiac abnormalities (atrial septal defects) (11).
FIGURE 1.
Schematic representation of predicted protein products encoded by the mutated PQBP1 gene in the Golabi-Ito-Hall syndrome (GIH S), the original family of Renpenning syndrome (R S), and the Hamel cerebropalatocardiac (H S) syndrome. The presence of the WW domain in PQBP1 was reconfirmed between amino acids 46 and 84. However, the C2 domain, another module that was proposed at the C-terminal region of PQBP1, could not be confirmed. Neither SMART nor PROSITE domain resource could detect the C2 domain in PQBP1. Therefore, this domain is considered as questionable (C2?). The region of PQBP1 that contains multiple, di-amino acid repeats, DR and ER, was demarcated between amino acids 104 and 176. A potential nuclear localization signal (N1), flanked by amino acids at positions 53–163, was predicted by the PredictNLS program (Columbia University, New York). The second potential nuclear localization signal (N2) was characterized previously and maps to amino acids 176–192 (3). In GIH syndrome, the transition mutation at the nucleotide 194 of PQBP1 (NCBI accession number NM_005710) causes missense substitution of Tyr to Cys at the amino acid position 65 in the conserved aromatic core of the WW domain. In Renpenning syndrome, the insertion of cytidine at the nucleotide position 641 caused a frameshift mutation that terminated the WT protein at amino acid position 213 and added new amino acids 214–225. In Hamel cerebropalatocardiac syndrome, a deletion of AG dinucleotide at position 461–462 caused a frameshift that resulted in the deletion of the putative nuclear localization signal (N1). Mutations are shown in italics (modified from Refs. 11 and 12).
The missense mutation in the GIH syndrome is unique among the PQBP1 mutations reported so far because it maps within a functional region of the protein known as the WW domain and because the lesion does not affect the size of the mutated protein (12). The WW domain is a well characterized protein module that mediates specific protein-protein interactions with ligands that contain short proline-rich motifs (15–17). The domain is composed of 38 amino acids, and it is one of the smallest among modular protein domains. The structure is a three β-strand meander that forms a shallow binding pocket for proline-rich ligands (for review, see Ref. 18). In general, the folding of WW domains does not require cognate ligands or co-factors.
The WW domain of PQBP1 was shown to interact with the pre-mRNA splicing factor WBP11, also known as SIPP1, and with RNA polymerase II (2, 7). Consequently, the PQBP1 protein was suggested to regulate pre-mRNA splicing and transcription. The WW domain mutation implicated in GIH syndrome results in a chemically significant substitution by changing the conserved tyrosine at position 65 within the aromatic core of the domain to cysteine. Mutations that locate within modular domains or in their cognate ligands have been shown to affect the complexes and result in diseases (for review, see Refs. 18 and 19). Therefore, we hypothesized that the Y65C mutation of the PQBP1-WW domain would lead to a loss or gain of function by affecting the binding of the domain, and ultimately of the protein, to cognate ligands. We also considered that the mutation could affect the folding and stability of the protein. We tested our hypotheses by a number of complementary biochemical, biophysical, and cell biology techniques. Our data show that the binding function of the mutated domain is affected, as assayed by in vitro screens of peptide repertoires that represent the complete proteomic complement of PPXY motif-containing human proteins. Selected proline-rich peptides derived from known protein partners of PQBP1 WW domain, including that of WBP11, were used in binding assays and showed that the Y65C mutation resulted in decreased binding of the domain to these peptides. Moreover, we documented that the compromised complex between PQBP1 and WBP11 resulted in the pre-mRNA splicing defect in GIH-derived lymphoblasts but not in the control cells. We also found suggestive evidence that the Y65C mutation affected folding of the mutated protein, but apparently it did not change the rate of protein degradation. Our report sheds light on the molecular consequence of the missense mutation that causes GIH syndrome and provides guides for further molecular analysis of PQBP1 function.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant WT and Y65C mutant WW domains span residues 36–94 of PQBP1 from Homo sapiens (7). They were expressed in Escherichia coli BL21 [DE3] pLysS cells transformed with a pGEX-2TK vector to produce a glutathione S-transferase (GST) fusion protein. The protein was purified using a glutathione-Sepharose affinity matrix (Amersham Biosciences). The protein purity was evaluated by SDS-PAGE and mass spectrometry. Protein concentrations were determined using UV absorption, with calculated extinction coefficients at 280 nm of 22,460 and 20,970 m−1 cm−1 for WT and Y65C mutant WW domains, respectively. Two synthetic polypeptides of WT and Y65C mutant WW domains were synthesized spanning residues 47–79 and 46–82 of human PQBP1. Peptide concentrations were determined by UV absorption. The molar absorption value for the WT 47–79 and 46–82 was taken as 20,970 m−1 cm−1 at 280 nm. The molar absorption value of the (Y65C) mutants 47–79 and 46–82 was taken as 19,480 m−1 cm−1 at 280 nm. The untagged recombinant WW domain was isolated by gel filtration chromatography on a (1.6 × 60 cm) HR-Superdex-75 from GE Healthcare, eluted at a flow rate of 0.5 ml/min. The effluent was monitored at 280 nm. To prevent cysteine oxidation, protein purification conditions and buffers in all experiments contained 2–5 mm β-mercaptoethanol or 2–5 mm Tris(2-carboxyethyl)phosphine hydrochloride reducing agents. The buffer used was phosphate-buffered saline (20 mm phosphate, pH 7.4, 5 mm KCl, and 100 mm NaCl). EGFP-WBP11 was obtained as described (20). In parallel, we obtained WBP11 cDNA from Open Biosystems. PCR was conducted by using the Open Biosystems plasmid as template. The PCR product was inserted into p2×FLAG-CMV2 vector using KpnI and XbaI sites. The sequences of primers are 5′- agaagaggtacccatgggacggagatctacatc-3′ and 5′-agaagatctagactacagtagcccttccatctc-3′. HA- PQBP1 was a kind gift of Dr. E. Golemis (21). PCR site-directed mutagenesis was used to create the HA-PQBP1-Y65C mutant. The source of rabbit polyclonal antibodies against WBP11 and PQBP1 was previously described (7, 22). Anti-hemagglutinin antibodies were purchased from Santa Cruz Biotechnology. FLAG Atrophin 1 cDNA construct was obtained from Dr. S. Tsuji (23).
Immunoprecipitations of protein complexes were conducted as described before (24). Briefly, human embryo kidney 293 (HEK293) cells were transfected with expression vectors that encode the protein of interest using Lipofectamine (Invitrogen). 24 h later cells were lysed with modified immune precipitation assay buffer (50 mm Tris-HCl, pH 7.45, 5 mm EDTA, 300 mm NaCl, 1% glycerol, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) and immunoprecipitated using anti-FLAG M2 affinity gel (Sigma). The immunoprecipitates were washed with the modified immune precipitation assay buffer, and bound proteins were separated by SDS-PAGE followed by immunoblotting.
Evaluating the Stability of PQBP1 Proteins
HEK293 cells were seeded in 24-well plates in Dulbecco's modified Eagle's medium containing 10% serum for 12 h before transfection. 1 μg of pcDNA-HIS-MAX-A-PQBP1 was transfected with 1 μl of Lipofectamine 2000. An empty vector was used as control. Medium was changed every 24 h after transfection (i.e. 24, 48, 72, and 96 h). Transfected cells were lysed with immune precipitation assay buffer without SDS (10 mm Tris-HCl, pH 7.4, 5 mm EDTA, 300 mm NaCl, 10% glycerol, 1% Triton X-100, 1% sodium deoxycholate). Immune precipitation assay buffer was added directly to the 24-well plates after washing with phosphate-buffered saline, and the cells were incubated on ice for ∼10 min. The protein concentration of total lysate was measured by Bio-Rad protein assay, and equal amounts were loaded into each lane of the SDS-PAGE gel. Human full-length PQBP1 cDNA tagged with His at the C-terminal end was detected at the protein level with anti-His monoclonal antibody from Clontech. The dilution of the primary antibody was 1:5000. The secondary antibody, anti-mouse IgG conjugated to horseradish peroxidase, was also used at a 1:5000 dilution. The membranes were probed with anti-His antibody, exposed, and subsequently stripped with the stripping buffer (62.5 mm Tris-HCl at pH 6.7, 2% SDS, 100 mm β-mercaptoethanol) and then reprobed with anti-glyceraldehyde-3-phosphate dehydrogenase monoclonal antibody. The ratio of the antibody was 1:10,000. The relative intensity of the protein bands was detected by ECL and autoradiography.
Optical Spectroscopy
Circular dichroism (CD) measurements were performed on a Jasco J-715 spectropolarimeter equipped with a PTC-348 Peltier system for temperature control. The instrument was calibrated with d-(+)10-camphorsulphonic acid. Protein concentrations of 60 and 150 μm and quartz cuvettes with path lengths of 1 and 2 mm were used for recording far- and near-UV spectra, respectively. Thermal unfolding experiments were performed by recording the dichroic signal at 230 nm in the temperature range of 5–95 °C. The samples were heated at a rate of 1 °C/min and successively cooled down to 10 °C to determine reversibility. Steady-state fluorescence measurements were performed on a SPEX Fluoromax spectrometer by exciting protein samples at 295 nm (slit width, 0.4 nm) and recording the emission intensity from 300 to 450 nm (slit width, 1.5 nm). All data were evaluated using the ORIGIN program package (Micro-Cal Software).
NMR Measurements
NMR spectra were recorded on the non-labeled recombinant proteins at temperatures of 25 °C on a 600-MHz proton frequency Varian spectrometer. Experiments acquired included the one-dimensional spectrum and two-dimensional 1H-1H two-dimensional NOESY (nuclear Overhauser effect spectroscopy), TOCSY (two-dimensional total correlation spectroscopy) spectra in H2O and in D2O.
Isothermal Titration Calorimetry (ITC) Measurements
ITC experiments were performed on a Microcal VP-ITC instrument, and data were acquired and processed using fully automized features in Microcal ORIGIN software (25). Twelve-mer peptides corresponding to WBP11 (sequence PPGPPPRGPPPR) and to Atrophin 1 (SPOT-1221) (sequence SPPGPPPYGKRA) were synthesized by Biosynthesis, Inc., (Lewisville, TX). Purified GST-PQBP1 WW domain (WT and Y65C mutant) fusion proteins were used.
Surface Plasmon Resonance (SPR) Analysis
SPR analysis was conducted on a BiacoreX instrument. A 38-mer peptide from human WBP11 (residues 429–466) containing the binding site for the WW domain of human PQBP1 was chemically synthesized with an N-terminal 3×Lys tag preceded by a β-alanine spacer so as to promote its immobilization onto a carboxyl-modified dextran-coated sensor chip (BIAcore CM5 sensor chip). Immobilization of the WBP11 peptide domain on the sensor chip was achieved using the standard amine-coupling strategy. The WT and mutant (Y65C) WW domains of PQBP1, corresponding to residues 47–79, were also chemically synthesized and used as analytes.
Synthesis and Purification of Soluble WW Domain Variants
Solid-phase peptide synthesis was performed according to the Fmoc strategy on the TentaGel S RAM resin (0.25 mmol/g; Rapp Polymere, Tübingen, Germany) using the multiple peptide synthesizer (SYRO II, MultiSynTech GmbH, Witten, Germany). The coupling reagents (benzotriazol-1-yloxy)tripyrrolidino-phosphonium hexafluorophosphate (PyBOP) and N-methylmorpholine were used for activation and for Fmoc deprotection, and a solution of 20% piperidine in N,N-dimethylformamide was applied. The requested peptide dye-labeling was achieved by coupling 5- (and 6-)carboxytetramethylrhodamine (TAMRA) at the N terminus via PyBOPB/N-methylmorpholine activation. Upon completion of the automated synthesis, the resin was washed 5 times each with 5 ml of dichloromethane and the TAMRA-labeled, and unlabeled peptides were cleaved from the resin and deprotected with 3 ml of 90% trifluoroacetic acid, 4% methylphenylsulfide, 4% water, 2% 1,2-ethanedithiol, and 7.5% w/v phenol. Peptide precipitation was carried out by the addition of 30 ml of tert-butyl methyl ether (cold −20 °C) followed by centrifugation. The precipitated solid was resuspended in 10 ml of diethyl ether and centrifuged again, repeating the cycle 3 times to remove all cleavage and deprotection reagents from the precipitate. The white solid was dissolved in 10% acetic acid and purified via high performance liquid chromatography (HPLC).
Preparative HPLC was carried out on a 2700 Sample Manager (Waters GmbH, Eschborn, Germany) using a 10–15-μm C18 polymeric reversed-phase column (Vydac 218TP152022, 300 Å pore size) and a linear gradient (5–90% B) over the course of 25 min at a flow rate of 12 ml/min. Solvents consisted of 0.05% trifluoroacetic acid in water (solvent A) or acetonitrile (solvent B). Detection was accomplished by absorbance at 214 nm or, additionally, in the case of the TAMRA-labeled peptides, at 540 nm. Fractions containing pure peptide were pooled and lyophilized to yield a white powder, which was analyzed by analytical HPLC and electrospray mass spectrometry. Purity was >95%, and identity did not vary greater than 1 Da from the expected values (monoisomeres).
Synthesis and Incubation of Peptide Arrays
Specific peptides were synthesized in situ on different array positions on a cellulose membrane according to the standard SPOT synthesis protocol as described (26, 27). The deprotection step of the standard protocol was slightly modified. The N-terminal free sequences were deprotected with 50% trifluoroacetic acid in dichloromethane with 3% triisobutylsilane and 2% water and 1% w/v phenol as scavengers for 1.5 h after 30 min of shock treatment with an equivalent 90% trifluoroacetic acid solution. Cellulose-supported peptides were washed three times with dichloromethane, three times with N,N-dimethylformamide, three times with ethanol, two times with ether, and dried under normal conditions.
In arrays in Fig. 2, SPOT membranes were incubated for 24 h in 10 mm Tris-HCl at pH 7.5, 150 mm NaCl, 0.1% Triton X-100, 0.1 mm reduced glutathione, 1× Sigma blocking buffer with of 32P-labeled GST-WW domain of PQBP1 (WT or Y65C mutant). For labeling of the probes, we used 10 μg each of the fusion proteins following the previously published protocol without modifications (28, 29). Three 20-min-long washes with phosphate-buffered saline containing 1% Triton X-100 were used. Dried membranes were exposed to Kodak x-ray films without using screens.
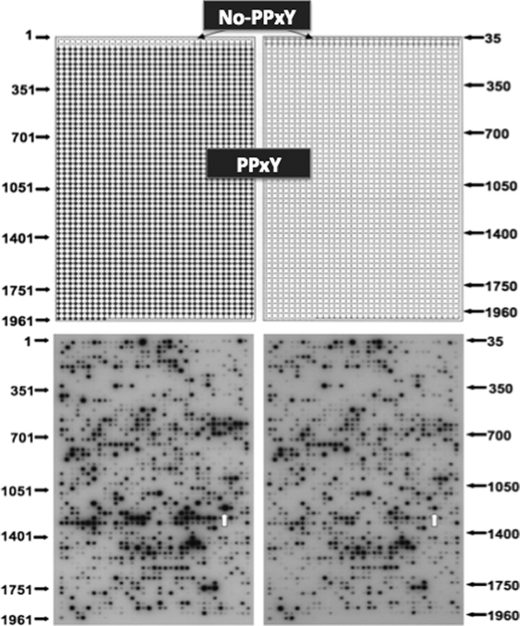
FIGURE 2.
SPOT peptide array probed with WT and Y65C PQBP1-WW domain. Upper panel, a scheme of grid used to construct the arrays is shown. Selected 59 peptides on the top of the array are 12-mer peptides without the PPXY motif. Peptides 1, 7, 13, 19, 25, 31, 37, 43, 48, and 54 were previously shown as in vitro ligands of the human PQBP1 WW domain. The remaining peptides in that set were selected as closely related and were derived from known human proteins. Positions 60–70 did not contain any peptide. The complete set of PPXY-containing 12-mer peptides, which represents all known coding sequences of the human proteome, is on the remaining part of the array from position 71 to 1965. Supplemental Figs. 1–3 provide the list of peptide sequences and names of the host proteins and gene accession numbers. Lower panel, shown is results of probing the repertoire of proline-rich peptides with the radioactive (32P-labeled) WT WW domain of PQBP1 (left panel) or with the radioactive Y65C mutant of the PQBP1 WW domain (right panel) expressed as a GST protein fusion. The arrays were incubated overnight in a standard SPOT membrane buffer that contained 0.1 mm reduced glutathione. Signals from the GST protein alone were negligible. Background binding was barely detectable and found on only a few spots (data not shown). Red arrows point to SPOT 1221, which contains Atrophin 1-derived peptide.
In arrays in supplemental Figs. 2 and 3 before incubation with dye-labeled synthetic or recombinant WW domains, dried cellulose-immobilized peptides were swelled shortly in ethanol and then washed three times with Tris-buffered saline. Background reactive groups were blocked for 3 h with 1× blocking buffer (Sigma) in Tris-buffered saline (TBS) containing 0.5% (w/v) sucrose. Cellulose-immobilized peptides were incubated for 2 h with 50 μm WW domain (WT or the Y65C mutant) solutions in 1× blocking buffer in TBS containing 0.5% (w/v) sucrose. Before detection of binding, the arrays were washed three times with TBS. Binding of WW domains to specific peptides was recorded by fluorescent detection of the TAMRA dye on a Lumi-Imager (Roche Applied Science). Signal intensity was quantified with the software Genespotter (Microdiscovery, Berlin, Germany).
Molecular Modeling
Three-dimensional structures of WT and Y65C mutant WW domains of PQBP1 were modeled using the MODELLER software based on homology modeling (30). In each case the NMR structure of Pin1 WW domain (with a PDB code of 1I6C) was used as a template. Briefly, MODELLER employs molecular dynamics and simulated annealing protocols to optimize the modeled structure through satisfaction of spatial restraints derived from amino acid sequence alignment with a corresponding template in Cartesian space. For amino acid sequence identity between 30–40% between the template and target, MODELLER can generate three-dimensional structures with accuracy comparable with NMR and x-ray structures for small proteins such as WW domains of around 30–40 amino acids. It is of worthy note that the WW domain of PQBP1 shares greater than a 40% sequence identity with the WW domain of Pin1. Thus, the modeled structures of the WT and mutant WW domains of PQBP1 should be expected to bear high accuracy. In each case a total of 100 structural models were calculated, and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for further energy minimization in MODELLER before analysis. The modeled structures were rendered using RIBBONS (31). All calculations were performed on the lowest energy structural model.
Cell Culture and Immunoprecipitations
Human lymphoblast cell lines were obtained from a patient suffering from the Golabi-Ito-Hall syndrome and a healthy control subject matched for age, gender, and race (12). The cells were grown in RPMI 1640 medium with 15% fetal calf serum and 1% penicillin-streptomycin. HEK293T cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum and 1% penicillin-streptomycin. Transfections of HEK293T cells were carried out with polyethyleneimine obtained from Polyplus-TransfectionTM, whereas lymphoblast cells were transfected using Lipofectamine LTX (Invitrogen). For immunoprecipitation experiments, exponentially growing lymphoblast cells and transfected HEK293T cells were washed twice with phosphate-buffered saline and harvested in a lysis buffer containing 50 mm Tris-HCl at pH 7.5, 0.3 m NaCl, 0.5% (v/v) Triton X-100, 0.5 mm phenylmethylsulfonyl fluoride, 0.5 mm benzamidine, and 5 μm leupeptin. The homogenates were centrifuged for 10 min at 10,000 × g, and the supernatants (cell lysates) were used for immunoprecipitation. Anti-WBP11 or anti-HA antibodies were added for 1 h at 10 °C followed by incubation with Protein-A-TSK-SepharoseTM for another hour. Before immunoblotting, the precipitates were washed once with TBS (20 mm Tris-HCl at pH 7.4 plus 150 mm NaCl) containing 0.1 m LiCl, twice with TBS supplemented with 0.1% Nonidet P-40, and once with 20 mm Tris-HCl at pH 7.4.
Splicing Assays
The double-reporter assays were performed as described by Nasim et al. (45). Lymphoblast cells were transfected with the pTN24 reporter plasmid. After 48 h the cells were harvested in passive lysis buffer (Promega), and the lysates were used for the assay of luciferase and β-galactosidase activities. The luciferase activities were measured with a Luminoskan Ascent luminometer (Labsystem) using the Promega Luciferase kit. The β-galactosidase activities were measured using ortho-nitrophenyl-β-galactosidase as substrate. The splicing efficiency was also determined by RNA analysis.
RESULTS
Binding Profiles of the Mutated and WT WW Domains on Peptide Arrays
To assess the effect of the Y65C mutation within the WW domain of PQBP1 on the binding function of the domain, we screened the repertoire of PPXY motif containing 12-mer peptides that represent the entire human proteome. In addition, we included 59 proline-rich peptides that did not contain the PPXY motif but scored positively in the proteomic map of the WW domain completed by the AxCell Co. (32). The total of 1958 peptides was synthesized on cellulose membranes using the SPOT technique (33) and probed with radioactively labeled GST-PQBP1-WW-WT or GST-PQBP1-WW-Y65C mutant (the list of the arrayed peptides is in supplemental Fig. 1 and Tables 1 and 2). As seen in Fig. 2, no dramatic differences in the intensity of spots detected by binding of the WT versus the Y65C WW domain were detected. We did see that the majority of peptides bound more strongly to the WT domain than to the mutant WW domain. Using densitometric scanning, we measured the relative intensity of the signal for each peptide spot and categorized all spots into three categories; those whose ratio of signal between WT and mutant were below 1, those that were equal (or very close) to 1, and those with the WT/mutant ratio bigger than 1 (supplemental Table 3). Of 1958 peptides assayed with WT and Y65C mutant WW domain probes labeled to the same specific activity, 1116 displayed relatively lower binding to the Y65C mutant than to the WT WW domain.
To make sure that the observed differences were not due to experimental variations (e.g. the efficiency of peptide synthesis, peptide coupling to the membrane) or due to errors intrinsic in the binding assay, we arbitrarily selected 46 different peptides, re-synthesized them on SPOT membranes, and probed them with GST-free synthetic WW domain (WT or mutant) labeled with the fluorescent dye TAMRA. As seen in supplemental Fig. 2, the pattern of binding to this array was qualitatively similar between the WT and the mutant domains. Although as in the initial array, we reproducibly saw a slightly less intense signal of binding by the mutant domain, when compared with that for the WT domain, the overall variations observed between the first and the second array made it difficult for us to reach firm conclusions.
Because WW domains were shown to interact with the C-terminal domain (CTD) of RNA polymerase II (17, 34, 35) and PQBP1 was suggested to interact functionally with the RNA polymerase II protein (7), we synthesized a SPOT array of 100 peptides, each representing two consecutive hexamer repeats from the CTD end of RNA polymerase II and probed the arrays with the WT and mutant PQBP1 WW domains. We used synthetic phosphoserine, phosphothreonine, and phosphotyrosine to generate a repertoire of putative combinations of modified repeats. As seen in supplemental Fig. 3, no major differences between the WT and the mutant WW domains could be observed in the binding patterns to the CTD RNA polymerase II peptide arrays. Although we could not exclude the existence of cognate, proline-rich ligands or specifically modified CTD repeats, which we did not include in our screens and which could represent still unknown physiological targets of PQBP1 WW domain, we tentatively concluded that the Y65C mutation of the WW domain may have an effect on binding to CTD repeat peptides, but the differences observed for Y65C mutant in screens of PPXY containing peptides were more significant.
As an interesting corollary of our screens, we found that the WW domain of PQBP1 recognizes both the PPXY motif-containing peptides and proline-rich peptides without aromatic residues. Therefore, the WW domain of PQBP1 is versatile and belongs to those domains that show ligand predilections of Class I, II, and III WW domains (17, 36, 37). From 1895 PPXY peptides probed with the WT WW domain of PQBP1 (Fig. 2) we selected 20 that bound most strongly to the domain. From 59 proline-rich peptides that did not contain the PPXY consensus, we selected 5 strong binders (supplemental Fig. 1, bold sequences). The sequence logos (38, 39) of PQBP1 WW domain are shown in Fig. 3. The 25 strong binders of the PQBP1 WW domain represent gene products of diverse ontology. This repertoire of gene products represents potential physiological targets of the PQBP1 protein and could be further narrowed for genes that are expressed in the brain and whose products are localized in the nucleus.
FIGURE 3.
“Logo” representation of consensus sequences of peptides that bind strongly to the WW domain of PQBP1. Twenty “super binders” from the PPXY motif-containing peptides were selected and submitted to the WebLogo generator of consensus (upper panel). When an additional five “super binders” were added from the non-PPXY motif-containing set, the logo consensus was slightly changed (lower panel). The x axis shows the position of amino acids from the N- to C-terminal end. The y axis represents “bites” of information as specified by the Logo algorithm (26).
Among the further-narrowed set of PQBP1 WW strongly interacting peptides (Fig. 2), which act in neural tissues and localize in the nucleus, we selected one that represents Atrophin 1 (SPOT 1221 peptide = SPPGPPPYGKRA (GenBankTM accession number AAH51795)). The binding to this peptide was clearly diminished for the mutant PQBP1 WW domain (white arrows, Fig. 2). In addition, a recent report showed that Atrophin 1 mutant mice were growth-retarded (40). We also chose a peptide from the WBP11 protein (WBP11-PR peptide = PPGPPPRGPPPR (NCBI accession number NM_016312)), which was shown as a cognate partner of PQBP1 in several unbiased protein interaction and functional assays (2, 37, 41, 42). Functionally, WBP11 acts as a nucleocytoplasmic shuttle that regulates pre-mRNA splicing (20). These peptides were synthesized and further analyzed for their binding potential to the WT and Y65C mutant WW domains of PQBP1 using ITC.
However, even though we employed saturating conditions for both the WW domain and the peptides in ITC measurements, the WW constructs, particularly the Y65C mutant construct, were unstable and aggregated at concentrations greater than 100 μm. In addition, the SPOT-1221 and WBP11-PR peptides showed limited solubility as well. Although the ITC data provided suggestive evidence that the Y65C mutation resulted in decreased binding to cognate peptides, when compared with the wild type domain (data not shown), we decided to use an alternative approach of SPR analysis.
It should be noted that the rather low affinity of the WW domain of PQBP1 for its cognate ligands observed in our assays is the hallmark of many WW domains. This is because the reversible nature of cellular signaling cascades favors weak interactions between the various underlying components (43, 44).
SPR Analysis Suggests a Loss of Function for the Y65C Mutant of PQBP1 WW Domain
Because, in contrast to Atrophin 1, WBP11 protein was implicated in the function of PQBP1 (42) and the proline-rich peptide used in the ITC study was arbitrarily selected from a long proline-rich region of WBP11 with many potential, overlapping WW domain binding motifs, we decided to use a longer proline-rich polypeptide and perform a SPR binding assay with WW domains. The 38-amino acid long human WBP11 peptide (429GLPPGPPPGAPPFLRPPGMPGLRGPLPRLLPPGPPPGR466) was modified at the N-terminal region with three lysine and one β-alanine residues for oriented binding. Synthetic WT or Y65C WW domains (34 residues long) was used for binding studies employing a standard SPR assay. The SPR results provided suggestive evidence on the weaker binding of the Y65C mutant WW domain than the WT WW domain to WBP11 polypeptide (data not shown). As was the case with the ITC assay, we could not determine absolute Kd values for the complexes in the SPR assay.
PQBP1-Y65C Has a Deficient WW Domain
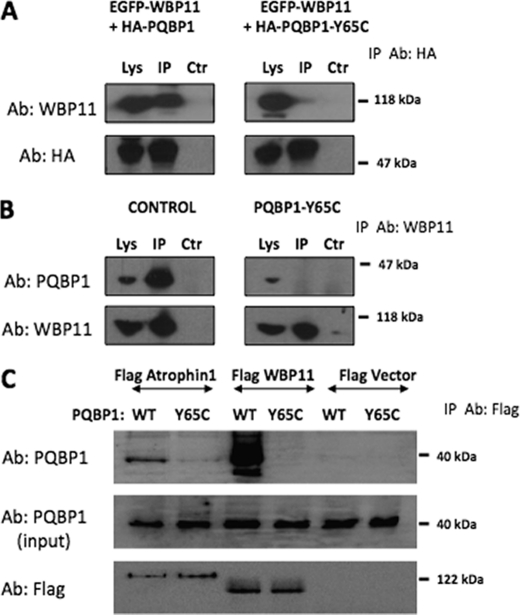
Because the missense mutation (Y65C) in the WW domain of PQBP1 resulted in compromised binding to cognate peptides, we decided to test if this holds true for the full-length proteins expressed in cells. We examined whether the PQBP1-Y65C mutant would be affected in its ability to interact with WBP11 and Atrophin 1. After the transient co-expression of EGFP-WBP11 and HA-PQBP1 in HEK293T cells, a clear co-immunoprecipitation between both proteins was detected (Fig. 4A). However, no such interaction was seen when the HA-PQBP1-Y65C mutant was co-expressed with EGFP-WBP11. Likewise, an interaction between endogenous PQBP1 and WBP11 could not be detected by co-immunoprecipitation analysis in a lymphoblast cell line from a patient with GIH but could be readily visualized in lymphoblasts from a healthy person matched for age, gender, and race (Fig. 4B). We also detected diminished interaction between PQBP1 Y65C mutant and Atrophin 1 compared with controls (Fig. 4C).
FIGURE 4.
PQBP1-Y65C shows a deficient interaction with WBP11. A, EGFP-WBP11 was transiently expressed in HEK293T cells with either HA-PQBP1 or HA-PQBP1-Y65C. Subsequently, the Lys, anti-HA immunoprecipitates (IP) and control immunoprecipitates (Ctr) were used for immunoblotting with anti-WBP11 antibodies (Ab). B, immunoblots show that PQBP1 but not PQBP1-Y65C co-immunoprecipitates with WBP11 from control- and GIH patient-derived lymphoblast cell lysates, respectively. C, PQBP1 forms a complex with Atrophin1 and WBP11. HEK293 cells were transiently co-transfected with PQBP1 and the indicated FLAG-tag plasmids. Cell lysates were immunoprecipitated with FLAG antibodies, resolved on SDS-PAGE, and immunoblotted with PQBP1 antibody (upper panel). Because WBP11 interacted strongly with PQBP1, only 10% of the total immunoprecipitates were applied on the gel for WBP11 samples (middle and lower panels show the expression of transfected proteins).
Deregulation of Pre-mRNA Splicing in GIH Lymphoblasts
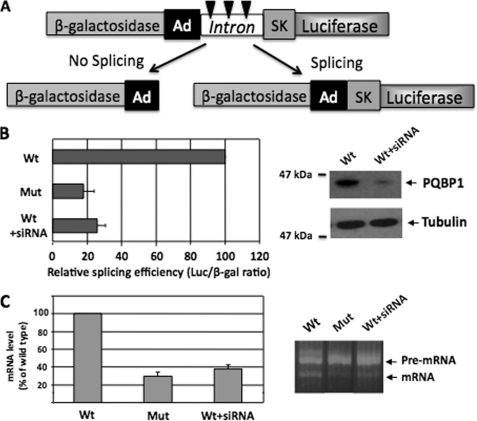
To provide a functional read-out of the compromised complex between PQBP1-Y65C mutant and WBP11 ligand, we decided to investigate differences in pre-mRNA splicing in GIH cells. We used the dual reporter assay developed by Nasim et al. (45). The reporter construct contains sequences encoding β-galactosidase as well as luciferase that are separated by an intronic sequence with multiple stop codons (Fig. 5A). When the primary transcript is not spliced, it generates β-galactosidase, whereas a fusion of β-galactosidase and luciferase is generated only after splicing. Thus, the luciferase/β-galactosidase ratio reflects the splicing efficiency. Lymphoblasts from a GIH patient showed a more than 80% lower splicing efficiency, as compared with the control cells (Fig. 5B). A small interfering RNA-mediated knock-down of PQBP1 in the control lymphoblasts, as verified by immunoblotting, had a similar inhibitory effect on the splicing efficiency. These splicing differences were also independently confirmed by quantitative real-time-PCR (Fig. 5C), ruling out effects on the translation or stability of the reported proteins.
FIGURE 5.
Impaired splicing of a reporter construct in GIH cells. A, shown is the structure of the TN24 reporter construct. The β-galactosidase and luciferase reporter genes are fused in-frame but separated by an intronic sequence derived from the adenovirus (Ad) and the skeletal muscle isoform of human tropomyosin (SK). The intron contains three in-frame translation stop signals (indicated by three arrowheads). In the absence of splicing, β-galactosidase is generated. Splicing generates an active fusion of β-galactosidase and luciferase. B, lymphoblasts from a healthy person before (Wt) or after the small interfering RNA-mediated knockdown of PQBP1 (Wt + siRNA) or from a GIH patient (Mut) were transiently transfected with the reporter plasmid TN24. The splicing efficiency of this reporter was assessed using the luciferase/β-galactosidase ratio. The data represent the means ± S.E. (n = 3) and are expressed as a percentage of the values obtained in WT cells. The right panel shows the level of PQBP1 in the cell lysates, as verified by immunoblotting. Tubulin was used as a loading control. C, relative transcript levels of the TN24 reporter plasmid, quantified by quantitative real-time-PCR (left panel). ACTIN was used as a normalization control for quantitative real-time-PCR. The data represent the means ± S.E. (n = 3). The right panel shows the pre-mRNA and mRNA levels of the TN24 reporter plasmid in a representative experiment, as visualized after RT-PCR and electrophoresis on 2.5% agarose gels.
Y65C Mutation of the WW Domain Destabilizes the Folding of the Protein
Four conserved hydrophobic amino acids within the WW domain were shown to form a canonical hydrophobic core that is critical for the structural stability of the three β-strand meander of the domain (46–48). In the human PQBP1 WW domain, the equivalent positions of the four conserved amino acids are Trp-52, Tyr-65, Asn-67, and Pro-78 (numbering is as used in Fig. 1). Individually, Ala mutations at these specific positions in several WW domains, including YAP1, PIN1, and TCERG1, yielded unfolded proteins (48). Because the Y65C mutation in the PQBP1 WW domain affects one of the four critical positions, the very Tyr-65 residue of the hydrophobic core, we expected a compromised folding of the mutant.
Spectroscopic Analyses Suggest Changes between the WT and Y65C WW Domain Fold
To evaluate the effect of the Y65C mutation on protein folding we used recombinant and synthetic WW domains and their mutants and subjected them to comparative analyses by fluorescence, CD and one- and two-dimensional NMR. Three different constructs of the WW domain of PQBP1 were used (see “Experimental Procedures”). Two synthetic WW polypeptides were 34 or 38 amino acids long. A short version of the WW domain was 34 amino acids long, whereas the longer, 38-amino acid-containing version had the originally defined length of the domain (15). We also used a 74-amino acid recombinant polypeptide that was expressed in bacteria as GST fusion protein-containing flanking sequences derived from the PQBP1 sequence and the expression vector. The GST tag was removed before using the bacterially expressed protein for biophysical studies.
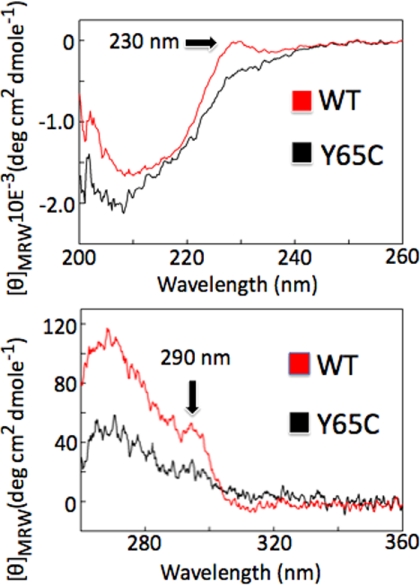
The far and near UV CD spectra of a protein provide information on the secondary and tertiary structure, respectively. The near UV spectrum in particular is a fingerprint of the tertiary structure, and even small structural perturbations can remarkably affect both its intensity and shape. The far-ultraviolet (UV) CD spectrum of both WT and mutant WW are typical of WW domains, having a positive band at 230 nm (44, 49). This band is, however, not equally intense in the spectrum of the mutant, suggesting a lower content of secondary structure in the former (Fig. 6, upper panel). The CD spectrum recorded in the near-UV region shows an intense signal in the 260–300-nm region with a maximum centered at ∼265 nm (Fig. 6, lower panel). The presence of a positive band at ∼290 nm is suggestive of Trp residues embedded in a rigid environment (50). The similarity of the near-UV CD spectra between the WT and mutant WW domain can be taken as an indication that these two proteins share a similar topology of aromatic residues within the three-dimensional protein structure, but the spectra intensities indicate a more packed tertiary structure for the WT WW domain than for the mutant domain. We cannot exclude the possibility that some of the differences we observed in the CD spectra between the two domains could reflect a strained β-sheet (51).
FIGURE 6.
CD spectra of recombinant WW-PQBP1 domains. Far UV (A) and near UV (B) CD spectra of WT (red) and Y65C mutant (black) WW-domains were recorded at 20 °C. MRW, mean residue weight.
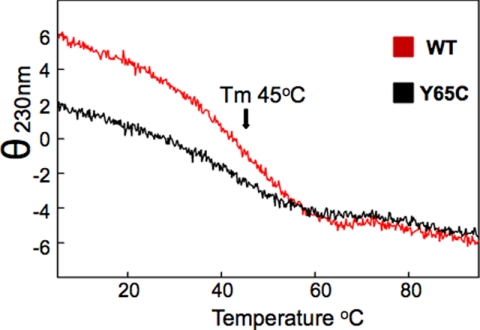
We also monitored the thermal denaturation curves of the WT and Y65C WW domain mutant by recording the CD signal at 230 nm. Both constructs have a melting temperature (Tm) at 45 °C. However, unfolding is clearly more cooperative for the WT than for the mutant domain (Fig. 7). Consistent results were obtained by fluorescence spectroscopy analysis performed at different temperatures. We observed reversible unfolding of the WT WW domain but not of the mutated domain (supplemental Fig. 4). In all the CD analyses reported here we are aware that the Y65C mutant has one less tyrosine and that this in part could explain the overall lower absorbance of the mutant compared with the WT domain.
FIGURE 7.
Thermal unfolding of recombinant WW-PQBP1 domains followed by CD and fluorescence techniques. Shown are CD curves of the WT (red) and mutant (black) proteins, obtained by detecting the signal at 230 nm as a function of temperature.
Finally, when we analyzed the domains by one-dimensional NMR, both the spectra of the WT and the mutant domains were in general typical of folded mono-dispersed proteins having well resolved, sharp, and widely spread resonances (supplemental Fig. 5). Particularly diagnostic were the high field resonances around 0.0–0.4 ppm, which are typically due to aliphatic groups shifted by spatially close aromatic groups. However, a more detailed analysis indicated a higher content of the tertiary structure for the WT domain compared with the mutant domain as indicated by the high field resonances. In the WT spectrum it is also possible to distinguish at least three resonances around frequency 10 ppm that should correspond to the indole resonances of the three tryptophan residues present in the sequence. Only one peak is visible at 10.2 ppm in the spectrum of the mutant. This is the typical position of tryptophan indole protons in a random coil conformation. Qualitatively similar results were obtained for the long and short versions, but they were clearer for the longer recombinant constructs.
The Y65C WW Domain Mutant Has a Tendency to Form Dimers under Non-reducing Conditions
When the PQBP1 WW domain polypeptides were analyzed on SDS gel electrophoresis under non-reducing conditions, we observed an aggregation and dimer formation for the Y65C mutant (supplemental Fig. 6). Although the normal cellular environment is generally reduced, the propensity of the mutant domain to form disulfide bridges in vitro should be noted.
Stability of the Mutated Protein Expressed in Human Cells
Because point mutations can destabilize mutant proteins (for review, see Ref. 49), we decided to express the full-length WT PQBP1 and its Y65C mutant in HEK293 cells and evaluate their relative levels as a function of time using cycloheximide treatment to inhibit de novo protein synthesis. The levels of expressed PQBP1 proteins were normalized to the level of actin, a “housekeeping” protein. The use of cycloheximide in HEK293 cells for 48 h was previously optimized and used successfully in our laboratory to show stabilization of p73 proapoptotic protein by YAP oncogene (24). As seen in supplemental Fig. 7, the full-length Y65C mutant did not show any significant change in the protein stability when compared with the WT PQBP1 protein. We cannot exclude the possibility that over longer periods of time there could be changes in the stability of the mutant protein expressed in the host neuronal cells. We also acknowledge a limitation of the assay that employs overexpression of tagged protein in an established cell line. Nevertheless, we conclude that the nature of the Y65C mutation seems subtle, and the Y65C PQBP1 mutant protein may not be recognized as overtly misfolded; otherwise, it would be targeted for fast degradation.
Three-dimensional Atomic Models Provide a Structural Basis for the Effect of the Y65C Mutation on the Structure and Fold Stability of the WW Domain of PQBP1
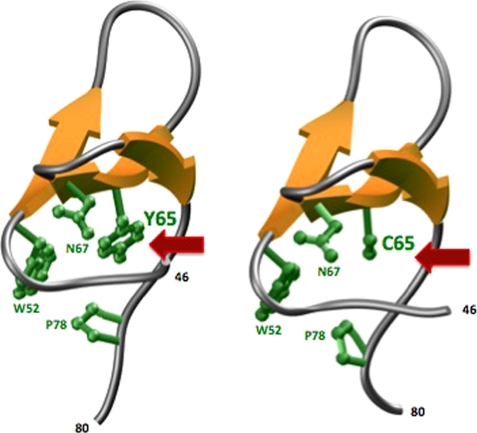
To further rationalize how the Y65C mutation might compromise the structure and stability of the WW domain of PQBP1 and, hence, its physiological function, we built three-dimensional atomic models of WT and Y65C mutant domains (Fig. 8). It is evident from our models that whereas the hydrophobic core of the WT WW domain is constituted by a highly conserved quartet of hydrophobic residues Trp-52, Tyr-65, Asn-67, and Pro-78 on the face opposite to that involved in ligand recognition, the substitution of Tyr-65 with cysteine in the mutant WW domain opens it up for serious structural consequences (46–48). Whereas Tyr-65 constitutes a key component of the hydrophobic core within the WT domain, the placement of cysteine at this position within the mutant WW domain leaves a gaping hole within the hydrophobic scaffold. This scenario could easily result in the collapse of the triple-stranded β-sheet roof and thereby compromise the binding potential of the mutant WW domain to cognate ligands of PQBP1 as observed in our ITC and SPR measurements. Additionally, the substitution of cysteine for tyrosine 65 could also render the mutant domain more vulnerable to oxidation relative to the WT domain, in agreement with our observations that the mutant domain exists in monomer-dimer equilibrium under non-reducing conditions (supplemental Fig. 6). Taken together, our three-dimensional atomic models provide key insights into structural consequences that might arise as a result of a naturally occurring mutant within the WW domain of PQBP1.
FIGURE 8.
Three-dimensional atomic models of WT (left) and Y65C-mutant (right) WW domains of PQBP1. The triple-stranded β-sheet of the WW domains is shown in yellow, and the intervening loops are in gray. The side chains of residues Trp-52, Tyr-65/Cys-65, Asn-67, and Pro-78 that constitute the hydrophobic core of the domains are colored green.
DISCUSSION
Using comparative screens of peptide repertoires, surface plasmon resonance binding assay, spectroscopic analyzes of proteins, transient expression of cDNAs in cells, and analysis of GIH syndrome-derived lymphoblasts, we have shown that the missense Y65C mutation that maps within the WW domain of the PQBP1 gene and causes the GIH X-linked mental retardation syndrome affects the folding of the WW domain and compromises its interaction with selected peptide ligands and their full-length proteins. One of the consequences of a compromised complex of the PQBP1-Y65C mutant with WBP11 splicing factor was a significant reduction of pre-mRNA splicing, as shown in lymphoblasts derived from a GIH patient. This decreased splicing efficiency was similar to that seen after small interfering RNA-mediated knockdown of PQBP1, indicating the PQBP1-Y65C is inactive in intact cells. Preliminary tests indicate that the Y65C mutation does not seem to cause any dramatic changes in the stability of the PQBP1 protein. We suggest that deregulated splicing is one of the hallmarks of GIH syndrome.
The following aspects of this work deserve further comment: (i) the sensitivity and variability of peptide arrays compared with the binding assay in solution; (ii) the misfolding of the PQBP1 protein and its relative stability; (iii) the information the Y65C mutation tells us about the molecular mechanism of the GIH syndrome and other XLMR conditions classified as Renpenning syndrome; (iv) the manipulation of the PQBP1 molecule to enhance the cognitive function of the brain; (v) cysteine 65 as a site of new regulatory modifications of the PQBP1 protein.
While appreciating the intrinsic variability of signals on the peptide arrays detected with tagged and untagged WW domains, we were surprised by differences observed between the array results and the preliminary results of the SPR and ITC assay. Atrophin 1 peptide SPOT1221 interacted strongly with the WT domain and moderately with the Y65C domain. However, in solution, we could not detect any interaction with the mutant (ITC assay data, not shown). Most likely, the covalent attachment of peptides to a solid support on the array via a linker of two β-alanines could, in part, explain the differences observed between the array and ITC results. Given that ITC analysis is solely based on the detection of heat change upon ligand binding, it is also conceivable that the SPOT1221 peptide does indeed bind to the Y65C mutant in solution, but the binding is accompanied by no net change in heat. Nevertheless, it is incumbent that more of the candidate peptides, which showed differential interactions between the WT and mutant PQBP1, be tested in solution to reinforce our observations and identify other potential signaling partners of PQBP1 for in vivo validation.
Single point mutations within proteins or within modular domains have been shown to affect the folding, stability, aggregation, or binding function of domains and, therefore, of the host proteins (19, 52–54). Several such examples of compromised protein stability and folding have been shown to directly underlie human diseases. Our data support a particular model in which a single amino acid-mutated protein is affected in terms of folding and binding activity of its protein-protein interaction domain. Most likely the mutation does not affect the protein stability. We suggest that these changes reflect a part of a repertoire of molecular changes underlying the GIH syndrome.
The PQBP1-Y65C mutation correlated with significant changes in splicing efficiency (Fig. 5). Our findings provide additional evidence for a role of PQBP1 in pre-mRNA splicing (55, 56). Alternative splicing is particularly important in the brain, and a switch in alternative splicing patterns of primary transcripts encoding neuron-specific proteins is known to accompany neuronal differentiation (57). Changes in alternative splice choices could, therefore, represent an important factor in the etiology of the GIH syndrome.
Other known mutations of the PQBP1 gene that cause XLMR syndromes result in significant deletions of the C-terminal region of the PQBP1 protein. Because clinically, the GIH syndrome differs only marginally from XLMR disorders that are associated with a reduction or complete loss of PQBP1 protein, this suggests that the WW domain is essential for the function of PQBP1. The diverse lesions in PQBP1 may lead to similar molecular changes in signaling. We envision two possibilities. The first is that for the PQBP1 to function properly it requires both the N-terminal-located WW domain and a putative motif in its C-terminal region. Mutation of either region will cause XLMR. The second possibility is that the only critical element for PQBP1 activity is the intact function of its WW domain, and even partially weakened binding activity of the domain will cause the disease. In the latter scenario we would presume that the deletion mutants of PQBP1 would be unstable as proteins, therefore reducing the level of total cellular PQBP1 protein. This process would mimic a reduced function of WW domain in cells. However, several recent reports question the simplicity of these models. The partial knock-down of PQBP1 in mice had a relatively mild effect. The mice showed an impairment of anxiety-related cognition (58). In general, this finding supports the idea that the loss of function of PQBP1 leads to mental retardation. In transgenic mice, elevated expression of PQBP1 caused motor neurodegeneration (59), whereas in Drosophila, the expression of human PQBP1 was shown to impair long-term memory and was correlated with abnormal social behavior (60). Although the reduced level of PQBP1 correlated with retarded phenotype, the enhanced level of PQBP1 did not seem to convey “super intelligence.” These observations suggest that the “right” level of intact PQBP1 protein is needed for normal development and normal cognitive functions. No matter which of the putative scenarios approximates the molecular changes that cause the syndrome, we support the proposal of Stevenson et al. (11), who suggested unifying the XLMR entities with PQBP1 mutations under one rubric of Renpenning syndrome.
Finally, it is important to point out that the WBP11 splicing factor, which we described here in detail, is one of the functional partners of PQBP1 from a potentially large repertoire of other signaling ligands. Therefore, we cannot exclude a possible “gain of function” with the introduction of Y65C mutation to PQBP1 protein, particularly given the fact that cysteine thiols in a certain local environment (pKa < 6) could rapidly form a thiolate anion. Such a reactive thiolate center may lead to several posttranslational modifications including phosphorylation, glutathionylation, or adduct formation with endogenous electrophilic molecules. These modifications could impart additional biochemical properties to PQBP1, as was described for selected protein-tyrosine kinases and phosphatases (61).
Supplementary Material
Acknowledgments
We thank E. Golemis, H. Okazawa, and S. Tsuji for cDNA constructs, J. Herrero, M. Becker, V. Alvarez, and K. Lokay of AxCell-Cytogen Co. for sharing unpublished data, D. H. Ledbetter, G. Gerhard, J. W. Kelly, M. A. Lemmon, K. S. Prabhu, and H. Okazawa for valuable advice, and W. Schwindinger and S. Knapp for constructive comments on the manuscript. We are grateful to the patient and to members of the family for their cooperation in our research. We also thank Dr. H. Lubs for originally contacting the family for our studies.
This work was supported, in whole or in part, by National Institutes of Health Grants T32-CA119929 (to C. B. M.), R01-GM083897 (to A. F.), and HD26202, NICHD (to C. E. S.). This work was also supported by Deutsche Forschungsgemeinschaft Grant SFB 449 (to V. T.), by the UMSylvester Braman Family Breast Cancer Institute (to A. F.), by Medical Research Council Grant U117584256 (to A. P.), by Concerted Research Action and the Fund for Scientific Research-Flanders Grants G.0670.09N and KAN1.5.101.07 (to M. B.), and by the Geisinger Clinic (to M. S.).
The recombinant plasmids reported in this paper have been deposited in the Addgene plasmid repository (www.addgene.org).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–7.
- PQBP1
- polyglutamine tract-binding protein 1
- CD
- circular dichroism
- CTD
- C-terminal domain
- GIH syndrome
- Golabi-Ito-Hall syndrome
- GST
- glutathione S-transferase
- HEK293
- human embryo kidney cell line 293
- ITC
- isothermal titration calorimetry
- SPR
- surface plasmon resonance
- WW domain
- tryptophan-tryptophan domain
- WBP11
- WW domain-binding protein 11
- XLMR
- X chromosome-linked mental retardation
- Fmoc
- 9-fluorenylmethoxycarbonyl
- TAMRA
- carboxytetramethylrhodamine
- TBS
- Tris-buffered saline
- WT
- wild type
- EGFP
- enhanced green fluorescent protein
- HA
- hemagglutinin
- SPR
- surface plasmon resonance
- HPLC
- high performance liquid chromatography.
REFERENCES
- 1.Komuro A., Saeki M., Kato S. (1999) Nucleic Acids Res. 27, 1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komuro A., Saeki M., Kato S. (1999) J. Biol. Chem. 274, 36513–36519 [DOI] [PubMed] [Google Scholar]
- 3.Waragai M., Lammers C. H., Takeuchi S., Imafuku I., Udagawa Y., Kanazawa I., Kawabata M., Mouradian M. M., Okazawa H. (1999) Hum. Mol. Genet. 8, 977–987 [DOI] [PubMed] [Google Scholar]
- 4.Waragai M., Junn E., Kajikawa M., Takeuchi S., Kanazawa I., Shibata M., Mouradian M. M., Okazawa H. (2000) Biochem. Biophys. Res. Commun. 273, 592–595 [DOI] [PubMed] [Google Scholar]
- 5.Enokido Y., Maruoka H., Hatanaka H., Kanazawa I., Okazawa H. (2002) Biochem. Biophys. Res. Commun. 294, 268–271 [DOI] [PubMed] [Google Scholar]
- 6.Okazawa H., Sudol M., Rich T. (2001) Brain Res. Bull. 56, 273–280 [DOI] [PubMed] [Google Scholar]
- 7.Okazawa H., Rich T., Chang A., Lin X., Waragai M., Kajikawa M., Enokido Y., Komuro A., Kato S., Shibata M., Hatanaka H., Mouradian M. M., Sudol M., Kanazawa I. (2002) Neuron 34, 701–713 [DOI] [PubMed] [Google Scholar]
- 8.Kalscheuer V. M., Freude K., Musante L., Jensen L. R., Yntema H. G., Gécz J., Sefiani A., Hoffmann K., Moser B., Haas S., Gurok U., Haesler S., Aranda B., Nshedjan A., Tzschach A., Hartmann N., Roloff T. C., Shoichet S., Hagens O., Tao J., Van Bokhoven H., Turner G., Chelly J., Moraine C., Fryns J. P., Nuber U., Hoeltzenbein M., Scharff C., Scherthan H., Lenzner S., Hamel B. C., Schweiger S., Ropers H. H. (2003) Nat. Genet. 35, 313–315 [DOI] [PubMed] [Google Scholar]
- 9.Kleefstra T., Franken C. E., Arens Y. H., Ramakers G. J., Yntema H. G., Sistermans E. A., Hulsmans C. F., Nillesen W. N., van Bokhoven H., de Vries B. B., Hamel B. C. (2004) Clin Genet 66, 318–326 [DOI] [PubMed] [Google Scholar]
- 10.Lenski C., Abidi F., Meindl A., Gibson A., Platzer M., Frank Kooy R., Lubs H. A., Stevenson R. E., Ramser J., Schwartz C. E. (2004) Am. J. Hum. Genet. 74, 777–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevenson R. E., Bennett C. W., Abidi F., Kleefstra T., Porteous M., Simensen R. J., Lubs H. A., Hamel B. C., Schwartz C. E. (2005) Am. J. Med. Genet A 134, 415–421 [DOI] [PubMed] [Google Scholar]
- 12.Lubs H., Abidi F. E., Echeverri R., Holloway L., Meindl A., Stevenson R. E., Schwartz C. E. (2006) J. Med. Genet 43, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cossée M., Demeer B., Blanchet P., Echenne B., Singh D., Hagens O., Antin M., Finck S., Vallee L., Dollfus H., Hegde S., Springell K., Thelma B. K., Woods G., Kalscheuer V., Mandel J. L. (2006) Eur. J. Hum. Genet. 14, 418–425 [DOI] [PubMed] [Google Scholar]
- 14.Martínez-Garay I., Tomás M., Oltra S., Ramser J., Moltó M. D., Prieto F., Meindl A., Kutsche K., Martínez F. (2007) Eur. J. Hum. Genet. 15, 29–34 [DOI] [PubMed] [Google Scholar]
- 15.Bork P., Sudol M. (1994) Trends Biochem. Sci. 19, 531–533 [DOI] [PubMed] [Google Scholar]
- 16.Chen H. I., Sudol M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7819–7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudol M., Hunter T. (2000) Cell 103, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 18.Sudol M. (2005) The WW Domain: Modular Protein Domains, pp. 59–72, Wiley VCH, Verlag GmbH & Co., KGaA, Weinheim, Germany [Google Scholar]
- 19.Lappalainen I., Thusberg J., Shen B., Vihinen M. (2008) Proteins 72, 779–792 [DOI] [PubMed] [Google Scholar]
- 20.Llorian M., Beullens M., Lesage B., Nicolaescu E., Beke L., Landuyt W., Ortiz J. M., Bollen M. (2005) J. Biol. Chem. 280, 38862–38869 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Lindblom T., Chang A., Sudol M., Sluder A. E., Golemis E. A. (2000) Gene 257, 33–43 [DOI] [PubMed] [Google Scholar]
- 22.Llorian M., Beullens M., Andrés I., Ortiz J. M., Bollen M. (2004) Biochem. J. 378, 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuji S. (1999) Adv. Neurol. 79, 399–409 [PubMed] [Google Scholar]
- 24.Oka T., Mazack V., Sudol M. (2008) J. Biol. Chem. 283, 27534–27546 [DOI] [PubMed] [Google Scholar]
- 25.Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 26.Frank R. (1992) Tetrahedron 48, 9217–9232 [Google Scholar]
- 27.Wenschuh H., Volkmer-Engert R., Schmidt M., Schulz M., Schneider-Mergener J., Reineke U. (2000) Biopolymers 55, 188–206 [DOI] [PubMed] [Google Scholar]
- 28.Kaelin W. G., Jr., Krek W., Sellers W. R., DeCaprio J. A., Ajchenbaum F., Fuchs C. S., Chittenden T., Li Y., Farnham P. J., Blanar M. A. (1992) Cell 70, 351–364 [DOI] [PubMed] [Google Scholar]
- 29.Chen H. I., Einbond A., Kwak S. J., Linn H., Koepf E., Peterson S., Kelly J. W., Sudol M. (1997) J. Biol. Chem. 272, 17070–17077 [DOI] [PubMed] [Google Scholar]
- 30.Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 31.Carson M. (1991) J. Appl Crystallogr. 24, 958–961 [Google Scholar]
- 32.Hu H., Columbus J., Zhang Y., Wu D., Lian L., Yang S., Goodwin J., Luczak C., Carter M., Chen L., James M., Davis R., Sudol M., Rodwell J., Herrero J. J. (2004) Proteomics 4, 643–655 [DOI] [PubMed] [Google Scholar]
- 33.Volkmer R. (2009) Chembiochem 10, 1431–1442 [DOI] [PubMed] [Google Scholar]
- 34.Gavva N. R., Gavva R., Ermekova K., Sudol M., Shen C. J. (1997) J. Biol. Chem. 272, 24105–24108 [DOI] [PubMed] [Google Scholar]
- 35.Chang A., Cheang S., Espanel X., Sudol M. (2000) J. Biol. Chem. 275, 20562–20571 [DOI] [PubMed] [Google Scholar]
- 36.Macias M. J., Wiesner S., Sudol M. (2002) FEBS Lett. 513, 30–37 [DOI] [PubMed] [Google Scholar]
- 37.Kato Y., Nagata K., Takahashi M., Lian L., Herrero J. J., Sudol M., Tanokura M. (2004) J. Biol. Chem. 279, 31833–31841 [DOI] [PubMed] [Google Scholar]
- 38.Schneider T. D., Stephens R. M. (1990) Nucleic Acids Res. 18, 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) Genome Res 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J., Ying M., Zhuang Y., Xu T., Han M., Wu X., Xu R. (2009) Dev Dyn. 238, 2471–2478 [DOI] [PubMed] [Google Scholar]
- 41.Guo F., Wang Y., Zhang Y. Z. (2007) Mol. Biotechnol 36, 38–43 [DOI] [PubMed] [Google Scholar]
- 42.Nicolaescu E., Beullens M., Lesage B., Keppens S., Himpens B., Bollen M. (2008) Eur. J. Cell Biol. 87, 817–829 [DOI] [PubMed] [Google Scholar]
- 43.Gibson T. J. (2009) Trends Biochem. Sci 34, 471–482 [DOI] [PubMed] [Google Scholar]
- 44.Macias M. J., Gervais V., Civera C., Oschkinat H. (2000) Nat. Struct. Biol. 7, 375–379 [DOI] [PubMed] [Google Scholar]
- 45.Nasim M. T., Chowdhury H. M., Eperon I. C. (2002) Nucleic Acids Res. 30, e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koepf E. K., Petrassi H. M., Ratnaswamy G., Huff M. E., Sudol M., Kelly J. W. (1999) Biochemistry 38, 14338–14351 [DOI] [PubMed] [Google Scholar]
- 47.Petrovich M., Jonsson A. L., Ferguson N., Daggett V., Fersht A. R. (2006) J. Mol. Biol. 360, 865–881 [DOI] [PubMed] [Google Scholar]
- 48.Jäger M., Dendle M., Kelly J. W. (2009) Protein Sci. 18, 1806–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernández-Escamilla A. M., Ventura S., Serrano L., Jiménez M. A. (2006) Protein Sci. 15, 2278–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strickland E. H. (1974) CRC Crit. Rev. Biochem. 2, 113–175 [DOI] [PubMed] [Google Scholar]
- 51.Bienkiewicz E. A., Moon, Woody A., Woody R. W. (2000) J. Mol. Biol. 297, 119–133 [DOI] [PubMed] [Google Scholar]
- 52.Gregersen N., Bross P., Jørgensen M. M., Corydon T. J., Andresen B. S. (2000) J. Inherit. Metab. Dis. 23, 441–447 [DOI] [PubMed] [Google Scholar]
- 53.Cohen F. E., Kelly J. W. (2003) Nature 426, 905–909 [DOI] [PubMed] [Google Scholar]
- 54.Jiang Y., Su J. T., Zhang J., Wei X., Yan Y. B., Zhou H. M. (2008) Int. J. Biochem. Cell Biol. 40, 776–788 [DOI] [PubMed] [Google Scholar]
- 55.Makarova O. V., Makarov E. M., Urlaub H., Will C. L., Gentzel M., Wilm M., Lührmann R. (2004) EMBO J. 23, 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Musante L., Kunde S. A., Sulistio T. O., Fischer U., Grimme A., Frints S. G., Schwartz C. E., Martínez F., Romano C., Ropers H. H., Kalscheuer V. M. (2010) Hum. Mutat. 31, 90–98 [DOI] [PubMed] [Google Scholar]
- 57.Fairbrother W. G., Lipscombe D. (2008) BioEssays 30, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ito H., Yoshimura N., Kurosawa M., Ishii S., Nukina N., Okazawa H. (2009) Hum. Mol. Genet. 18, 4239–4254 [DOI] [PubMed] [Google Scholar]
- 59.Okuda T., Hattori H., Takeuchi S., Shimizu J., Ueda H., Palvimo J. J., Kanazawa I., Kawano H., Nakagawa M., Okazawa H. (2003) Hum. Mol. Genet. 12, 711–725 [DOI] [PubMed] [Google Scholar]
- 60.Yoshimura N., Horiuchi D., Shibata M., Saitoe M., Qi M. L., Okazawa H. (2006) FEBS Lett. 580, 2335–2340 [DOI] [PubMed] [Google Scholar]
- 61.Xu D., Rovira I. I., Finkel T. (2002) Dev. Cell 2, 251–252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.