Abstract
The nucleobase-ascorbate transporter (NAT) signature motif is a conserved 11-amino acid sequence of the ubiquitous NAT/NCS2 family, essential for function and selectivity of both a bacterial (YgfO) and a fungal (UapA) purine-transporting homolog. We examined the role of NAT motif in more detail, using Cys-scanning and site-directed alkylation analysis of the YgfO xanthine permease of Escherichia coli. Analysis of single-Cys mutants in the sequence 315–339 for sensitivity to inactivation by 2-sulfonatoethyl methanethiosulfonate (MTSES−) and N-ethylmaleimide (NEM) showed a similar pattern: highly sensitive mutants clustering at the motif sequence (323–329) and a short α-helical face downstream (332, 333, 336). In the presence of substrate, N325C is protected from alkylation with either MTSES− or NEM, whereas sensitivity of A323C to inactivation by NEM is enhanced, shifting IC50 from 34 to 14 μm. Alkylation or sensitivity of the other mutants is unaffected by substrate; the lack of an effect on Q324C is attributed to gross inability of this mutant for high affinity binding. Site-directed mutants G333R and S336N at the α-helical face downstream the motif display specific changes in ligand recognition relative to wild type; G333R allows binding of 7-methyl and 8-methylxanthine, whereas S336N disrupts affinity for 6-thioxanthine. Finally, all assayable motif-mutants are highly accessible to MTSES− from the periplasmic side. The data suggest that the NAT motif region lines the solvent- and substrate-accessible inner cavity, Asn-325 is at the binding site, Ala-323 responds to binding with a specific conformational shift, and Gly-333 and Ser-336 form part of the purine permeation pathway.
Keywords: Bacteria, Evolution, Membrane Proteins, Nucleoside Nucleotide Transport, Purine, Site-directed Mutagenesis, Cys Scanning, Site-directed Alkylation
Introduction
The nucleobase-ascorbate transporter (NAT)2 or nucleobase-cation symporter-2 (NCS2) family is an evolutionarily ubiquitous family of purine, pyrimidine, and l-ascorbate transporters, with members specific for cellular uptake of uracil, xanthine, or uric acid (microbial and plant genomes) or vitamin C (mammalian genomes) (1, 2). Despite their importance for the recognition and uptake of several frontline purine-related drugs, NAT/NCS2 members have not been studied systematically at the molecular level, and high resolution structures or mechanistic models are missing. More than 1,000 sequence entries are known, but few are functionally characterized to date. Structure-function relationships have been studied extensively in two members of the family, the eukaryotic UapA, a high affinity uric acid/xanthine:H+ symporter from the ascomycote Aspergillus nidulans (3–6), and the prokaryotic YgfO, a specific, high affinity xanthine:H+ symporter from Escherichia coli (7–10). Mutagenesis data derived from both lines of study have shown that key NAT determinants are strikingly similar between the two transporters and that few residues conserved throughout the family may be invariably critical for function (10).
A prominent feature of the NAT/NCS2 family members is the NAT signature motif, a conserved 11-amino acid sequence between putative transmembrane helices (TMs) 9a and 9b (see Fig. 1), which appears to be essential for function and selectivity of the purine translocation pathway in both the bacterial (YgfO) (8) and the fungal (UapA) (4) NAT prototypes. The motif consensus sequence, (A/G/S)(Q/E/P)NXGXXXXT(R/K/G), was originally implicated with purine substrate selectivity based on in vivo complementation of a series of chimeric transporter constructs in the Aspergillus system (3); subsequently, site-directed mutagenesis of the conserved motif residues in UapA (4) showed specific alterations in substrate recognition and selectivity, and one of them (Gln-408) was suggested to bind directly to the imidazole moiety of purine substrates in mutant Q408E (4). In the E. coli system, Cys-scanning and site-directed mutagenesis of the NAT motif of YgfO (8) have shown that Gln-324 and Asn-325 are irreplaceable for active transport; Gln-324 is also essential for high affinity recognition of xanthine analogues, and an α-helical stripe of residues within and downstream of the motif (Ile-329, Thr-332, Gly-333, Ser-336, Val-339) is sensitive to inactivation by N-ethylmaleimide (NEM) (8). However, the substrate-interacting properties of the NAT motif mutants had not been addressed systematically in either the bacterial (8) or the fungal (4) transporter study.
FIGURE 1.
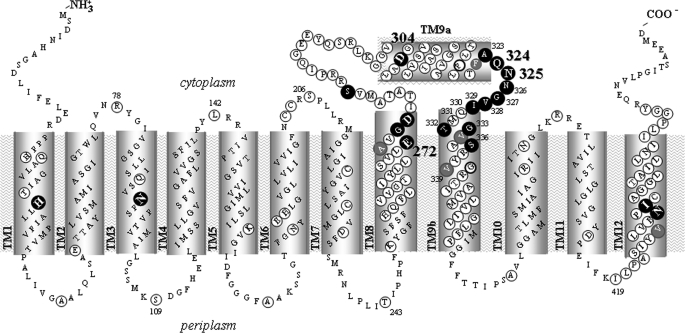
Topology model of YgfO permease. The model is based on program TMHMM, evidence that C terminus is cytoplasmic (7, 20), and our unpublished evidence (see Footnote 5) on the accessibility of loops to hydrophilic reagents. Irreplaceable residues of YgfO (10) are numbered and bolded. NEM-sensitive positions are shown in a dark background. Residues analyzed with site-directed mutagenesis are circled. Residues used in this study as targets for mutagenesis and/or mutant analysis are numbered. The ambiguous topology segment 299–323 upstream of the NAT motif is designated as TM9a, and the transmembrane segment 330–357 that follows is designated as TM9b.
In this report, we have studied the effect of substrate (xanthine) binding on the site-directed alkylation properties of a series of single-Cys mutants in the NAT motif sequence of YgfO. We have used two sulfhydryl-specific reagents, the hydrophilic, membrane-impermeable (11) 2-sulfonatoethyl methanethiosulfonate (MTSES−) and the small and relatively hydrophobic NEM (12). Our data show that the NAT motif region is highly accessible to solvent and the alkylating reagents from the periplasmic side, mutants sensitive to site-directed inactivation cluster at the motif and a short α-helical face immediately downstream, and Asn-325 is protected from alkylation with either reagent in the presence of substrate, whereas Ala-323 responds to substrate binding with enhanced sensitivity to NEM. In addition, site-directed replacements at the alkylation-sensitive α-helical face yield specific aberrations in the ligand recognition profile. In conjunction with the previous findings (4, 8), our results now establish that residues in the NAT signature motif region are proximal to or interact with the purine-binding site of YgfO.
EXPERIMENTAL PROCEDURES
Materials
[8-3H]xanthine (27.6 Ci mmol−1) and [8-14C]uric acid (51.5 mCi mmol−1) were purchased from Moravek Biochemicals. Non-radioactive nucleobases and analogs were from Sigma. Oligodeoxynucleotides were synthesized from BioSpring GmbH. High fidelity Taq polymerase (Phusion high fidelity PCR system) was from Finnzymes. Restriction endonucleases used were from Takara. ProBond resin was from Invitrogen. MTSES− was purchased from Toronto Research Chemicals. NEM was from Sigma. Oregon Green 488 maleimide (OGM) and polyclonal anti-OGM antibody were from Molecular Probes. Horseradish peroxidase (HRP)-conjugated penta-His antibody was from Qiagen. HRP-conjugated avidin and HRP-conjugated protein A were from Amersham Biosciences. All other materials were reagent grade and obtained from commercial sources.
Bacterial Strains and Plasmids
E. coli K-12 was transformed according to Inoue et al. (13). TOP10F′ (Invitrogen) was used for initial propagation of recombinant plasmids. T184 (14) harboring pT7-5/ygfO (7) with given replacements was used for isopropyl-1-thio-β-d-galactopyranoside-inducible expression from the lacZ promoter/operator.
DNA Manipulations
Construction of expression plasmids and His10-tagged or biotin-acceptor domain (BAD)-tagged versions of YgfO has been described (7). For construction of Cys-less YgfO (permease devoid of native Cys residues), the five native Cys codons were replaced simultaneously with Ser codons, using two-stage (multiple overlap/extension) PCR on the template of wild-type YgfO tagged at the C terminus with the BAD tag and transferred to the His10-tagged background by BamHI-HpaI restriction-fragment replacement (8). For construction of the NAT motif mutants, two-stage PCR was performed on the template of Cys-less or wild-type YgfO tagged at C terminus with either BAD or His10, as indicated. The entire coding sequence of all engineered constructs was verified by double-strand DNA sequencing in an automated DNA sequencer (MWG Biotech) (supplemental Table S1).
Growth of Bacteria
E. coli T184 harboring given plasmids was grown aerobically at 37 °C in Luria-Bertani medium containing streptomycin (0.01 mg/ml) and ampicillin (0.1 mg/ml). Fully grown cultures were diluted 10-fold, allowed to grow to mid-logarithmic phase, induced with isopropyl-1-thio-β-d-galactopyranoside (0.5 mm) for an additional 2 h at 37 °C, harvested, and washed with the appropriate buffers.
Preparation of Right-side-out Membrane Vesicles and Crude Membrane Fractions
Right-side-out (RSO) membrane vesicles were prepared from cultures of E. coli T184 (1 L) by lysozyme-EDTA treatment and osmotic lysis (15, 16). Vesicles were resuspended to a protein concentration of 4 mg/ml in potassium Pi buffer consisting of 100 mm potassium phosphate (potassium inorganic phosphate, pH 7.5) and 10 mm MgSO4, frozen in liquid nitrogen, and stored at −80 °C until use. Crude membrane fractions were prepared from 10-ml cultures of E. coli T184 by osmotic lysis and sonication as described (17) and used immediately after preparation.
Transport Assays and Kinetic Analysis
E. coli T184 or RSO membrane vesicles were assayed for active transport of [3H]xanthine (1 μm) by rapid filtration at 25 °C, as described (7, 18); active transport experiments in RSO vesicles were performed in the presence of phenazine methosulfate (0.2 mm) and potassium ascorbate (20 mm) (18, 19). For kinetic uptake measurements, initial rates were assayed in T184 cells, at 5–20 s, in the concentration range of 0.1–100 μm [3H]xanthine. Selected mutants were also assayed for transport of [14C]uric acid (0.04–2 mm), using the paralog YgfU as a positive control.3 For assaying the effect of NEM or MTSES− on xanthine uptake activity, T184 cells or RSO vesicles were preincubated with the reagent at the indicated conditions, the samples were diluted and washed three times with ice-cold potassium Pi buffer to remove excess reagents and ligands, and transport assays were performed in the presence of phenazine methosulfate (0.2 mm) and potassium ascorbate (20 mm) (8, 18). For ligand competition experiments, uptake of [3H]xanthine (1 μm) was assayed in the absence or presence of unlabeled analogs (1 mm) (7). For kinetic analysis, putative inhibitors were used in the concentration range of 0.1 μm to 1 mm, and data were fitted to the equation y = B + (T − B)/(1 + 10((log IC50 − log x) h)) for sigmoidal dose-response (variable slope), where x is the concentration variable, y (activity) values range from T (top) to B (bottom), and h is the Hill coefficient, using Prism4, to obtain IC50 values. In all cases, the Hill coefficient was close to −1, consistent with the presence of one binding site. Ki values for each analog were calculated from the Cheng and Prusoff equation Ki = IC50/(1+(L/Km)) (where L is the permeant concentration and Km is the value obtained for this permeant), assuming a simple model of competitive inhibition with the binding site of the transporter (8). Competitive inhibition has been shown with certain analogs (1-methyl, 2-thio, and 8-methylxanthine) by assaying their effect on Km and Vmax for wild type and selected mutants and showing that Vmax remains unaltered (8, 9).
Immunoblot Analysis
RSO membrane vesicles or membrane fractions of E. coli T184 harboring given plasmids were subjected to SDS-PAGE (12%), as described (7). Proteins were electroblotted to polyvinylidene difluoride membranes (Immobilon-PVDF; Pall Corp.). YgfO-BAD was probed with avidin-HRP. YgfO-His10 was probed with either anti-OGM antibody followed by protein A-HRP or penta-His antibody-HRP (7, 8). Signals were developed with enhanced chemiluminescence (ECL).
Sulfhydryl Alkylation Assay
RSO membrane vesicles prepared from E. coli T184 expressing His10-tagged versions of the indicated permeases (0.2 mg of total protein in 50 μl of potassium Pi buffer) were incubated for 10 min at 25 °C with NEM (1 μm−1 mm) or MTSES− (50 μm or 0.2 mm), in the absence or presence of xanthine (1 mm); reactions were stopped by the addition of a 10-fold excess of dithiothreitol followed by washing in potassium Pi buffer to remove excess reagents and ligands and extraction of the membrane proteins with n-dodecyl β-d-maltopyranoside (2% w/v) for 15 min, 25 °C. The YgfO-His10 permease was then purified with nickel-affinity chromatography using Ni2+-iminodiacetic acid beads (ProBond resin) and labeled with OGM, essentially as described (8). Briefly, after incubation with the protein extract, the beads were washed with equilibration buffer containing 100 mm imidazole and resuspended in 0.1 ml of denaturation buffer (containing 6 m urea and 0.5%, w/v, SDS); OGM was added at a final concentration of 0.2 mm, and the samples were incubated for 20 min at room temperature, with rotation. The OGM reaction was stopped by the addition of β-mercaptoethanol (5 mm), and the bound proteins were extracted from the beads in Laemmli buffer containing 0.07 m Na2EDTA at 30 °C for 10 min, prior to SDS-PAGE (12%) and immunoblotting. Protein concentrations were determined with the micro-BCA method (Pierce).
In Silico Analysis
Comparative sequence analysis of NAT/NCS2 homologs was based on BLAST-p search and ClustalW alignment; the most recent genome annotations were used for retrieving sequence data. Based on the functional (7) and phylogenetic data,4 the YgfO permease has been recently renamed also as XanQ (by the EcoGene Database of E. coli sequence and function). Initial analysis of transmembrane topology was performed using the program TMHMM (20). In the context of our work, extensive homology searches in conjunction with the functional data led us to extend the consensus sequence of the NAT motif, originally defined as (Q/E/P)NXGXXXXT(R/K/G) (3), by one amino acid residue (Ala-323 in YgfO); we thus refer to the NAT motif as an 11-amino acid sequence with consensus (A/G/S)(Q/E/P)NXGXXXXT(R/K/G).
RESULTS
NEM Sensitivity Profile of the NAT Motif Region
Our Cys-scanning analysis of the NAT motif of YgfO permease (8) has identified a stripe of residues, including Ala-323, Asn-326, Gly-327, Val-328, Ile-329, Thr-332, Gly-333, and Ser-336, which tolerate replacement with Cys but undergo strong inhibition upon further modification by NEM; some of these residues form a short α-helical face of TM9b (Fig. 1). Our previous work5 has also identified two irreplaceable residues of the motif, Gln-324 and Asn-325, where the sensitivity to NEM could not be tested with accuracy (8). Bioinformatic analysis (supplemental Tables S2 and S3) shows that all these inhibition-sensitive residues of YgfO correspond to positions of high side chain conservation among members of the NAT family (Fig. 2). As shown with other transporters (21–23), such inhibition-sensitive and conserved residues are expected to be at conformationally active faces or to line the substrate translocation pathway and be important for the transport mechanism. To address these possibilities further, we have analyzed the effect of substrate (xanthine) on the sensitivity (extent of inactivation) and reactivity (extent of site-directed alkylation) of single-Cys mutants with NEM and MTSES−.
FIGURE 2.
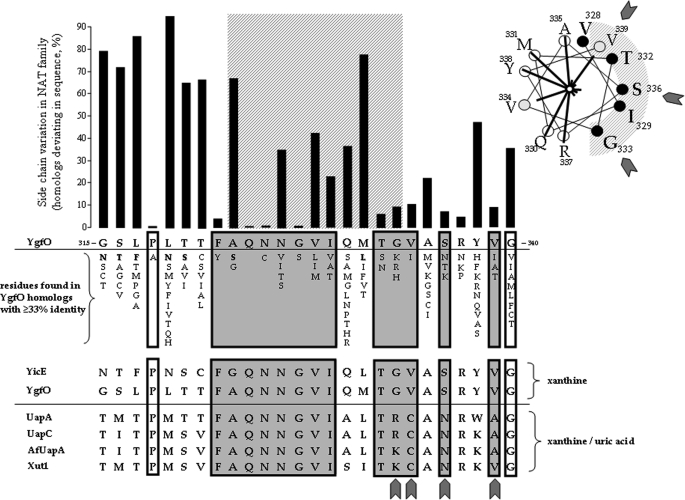
Correlation of sequence conservation with mutant sensitivity to NEM. The histogram shows side-chain variation among the 188 closest sequence homologs of YgfO (sequence identity ≥33%) as number of homologs deviating from YgfO at each position of sequence 315–340. The NAT motif (residues 323–333) is indicated as a cross-hatched orthogonal area. Also shown are residues of other homologs of this set that differ from the YgfO residue at the corresponding position and an alignment of the xanthine-specific YgfO (XanQ) (P67444) and YicE (XanP) (P0AGM9) from E. coli with the xanthine/urate dual specificity UapA (Q07307) and UapC (P487777) from A. nidulans, AfUapA (XP748919) from Aspergillus fumigatus, and Xut1 (AAX2221) from Candida albicans. Positions of YgfO residues important for expression in the membrane (8) are shown in white boxes. Positions of NEM-sensitive YgfO mutants (8) are shown in gray boxes; sequence deviations in the fungal transporters are indicated with filled cuneiform symbols. The inset on the upper right shows an α-helical wheel plot of residues 328–339 of YgfO with NEM-sensitive positions indicated as solid black circles and positions of intermediate sensitivity as gray circles; lines radiating form the center of the plot are proportional to the IC50 value of each mutant.
Substrate Effects on the NEM Sensitivity: Substrate Exposes A323C
Each active single-Cys mutant in the sequence region 321–339 of YgfO was assayed for the effect of NEM, a membrane-permeable sulfhydryl reagent, on the initial rate of xanthine transport after preincubation with NEM in the presence or absence of substrate (Fig. 3). Initial assays of sensitive mutants were performed with NEM concentrations yielding half-maximal inhibition of activity (IC50) in the absence of substrate (8), and the effect of substrate on the extent of inhibition was tested (Fig. 3A); then, a range of appropriate concentrations was applied to determine the effect of substrate on IC50 for each mutant (Table 1). Our data show that of the 15 single-Cys mutants, A323C responds to substrate by significant enhancement in sensitivity, yielding a shift of IC50 from 34 to 14 μm (Fig. 3B), and sensitivity of the other mutants is essentially unaffected by substrate (Fig. 3 and Table 1). For Phe-322, Asn-325, and Asn-326, where a single-Cys mutant is inactive, assays were performed with Cys replacement mutants in the wild-type background, F322C(wt) and N326C(wt), which are highly active (8), and N325C(wt), which has very low but detectable activity (8). These three mutants are sensitive to inactivation by NEM, whereas the wild-type YgfO, despite the presence of the five native Cys residues (7), is insensitive; in the presence of substrate, sensitivity of N325C(wt) decreases dramatically, and sensitivity of F322C(wt) or N326C(wt) remains unaffected (Fig. 3 and Table 1).
FIGURE 3.
Effect of xanthine on the NEM sensitivity of Cys replacement mutants. E. coli T184 harboring pT7-5/ygfO(C-less-BAD) or pT7-5/ygfO(wild-type-BAD) with given mutations were grown, induced, and assayed for transport of [3H]xanthine (1 μm) at 25 °C. Transport assays were performed in the presence of 20 mm potassium ascorbate and 0.2 mm phenazine methosulfate after preincubation of the cells with NEM for 10 min at 25 °C in the absence or presence of xanthine (1 mm) and removal of excess reagents. A, preincubations were performed at NEM concentrations of 10 μm (G327C), 20 μm (A323C), 40 μm (G333C), 50 μm (N326C(wt), I329C, T332C), 60 μm (S336C), 100 μm (N325C(wt), V328C), 0.2 mm (V334C), 0.4 mm (F322C(wt), V339C), 1 mm (T321C), or 2 mm (Q330C, M331C, A335C, R337C, Y338C), depending on the NEM sensitivity of each mutant (8). Uptake rates are presented as percentages of the rate measured in the absence of NEM with S.D. from three independent determinations shown. N.D., not determined due to negligible uptake rates of mutants Q324C and Q324C(wt) (8). B, NEM dose-response curves for mutant A323C in the absence or presence of substrate. IC50 values were deduced from non-linear regression fitting of the data to the sigmoidal dose-response equation using Prism4.
TABLE 1.
Effect of substrate on the sensitivity of mutants to N-ethylmaleimide
E. coli T184 expressing the corresponding constructs were assayed for initial rates of [3H]xanthine (1 μm) uptake at 5–20 s after preincubation with 0.001–2 mm NEM for 10 min in the absence or presence of unlabeled xanthine (1 mm), as described under “Experimental Procedures.” NEM concentrations inhibiting activity of each mutant by 50% (IC50 values) were deduced from non-linear regression fitting of the data to the sigmoidal dose-response equation using Prism4. Values were not determined for Q324C or Q324C(wt), which display negligible initial rates (8), nor determined using Cys-replacements in the wild-type background (wt) for mutants F322C(wt), N325C(wt), and N326C(wt). In control experiments, both Cys-less and wild type YgfO remained insensitive to NEM in any of the concentrations tested (≤2 mm). All mutants as well as the control permeases used in these experiments contained a carboxyl-terminal BAD. ND, assays were performed, but kinetic values were not determined due to very low uptake rates (≤0.05 nmol mg−1 min−1). Most significant differences from the wild-type profile are highlighted in bold.
| Permease | IC50 |
|
|---|---|---|
| Exposure to NEM in the absence of substrate | Exposure to NEM in the presence of 1 mm xanthine | |
| μm | ||
| YgfO(wt) | Insensitive | Insensitive |
| YgfO(Cys-less) | Insensitive | Insensitive |
| T321C | 1,000 | 1,500 |
| F322C(wt) | 400 | 450 |
| A323C | 34 | 14 |
| Q324C or Q324C(wt) | ND | ND |
| N325C(wt) | 89 | 1,000 |
| N326C(wt) | 50 | 50 |
| G327C | 12 | 10 |
| V328C | 96 | 80 |
| I329C | 50 | 50 |
| Q330C | Insensitive | Insensitive |
| M331C | Insensitive | Insensitive |
| T332C | 40 | 35 |
| G333C | 35 | 38 |
| V334C | 210 | 230 |
| A335C | Insensitive | Insensitive |
| S336C | 60 | 55 |
| R337C | Insensitive | Insensitive |
| Y338C | Insensitive | Insensitive |
| V339C | 400 | 410 |
Substrate Effects on the NEM Reactivity: Substrate Protects N325C
Single-Cys mutants Q324C, N325C, and N326C, which are inactive (8), and A323C, which responds to substrate by enhanced sensitivity to inactivation (Fig. 3), were subjected to site-directed alkylation analysis to determine the extent of modification with NEM in the presence or absence of substrate (Fig. 4). Our data show that the Cys residue in each one of the four mutants is reactive with NEM and that reactivity of single-Cys N325C is blocked in the presence of substrate, either completely (0.2–0.5 mm NEM) or partially (1 mm NEM) (Fig. 4A); using a range of NEM concentrations, we find that the xanthine substrate shifts the EC50 of N325C from 0.19 to 2.04 mm, as determined by Prism4 analysis (Fig. 4B). The reactivity of the other three mutants was not altered significantly by substrate, either at high (Fig. 4A) or at lower concentrations of NEM (10–200 μm) (data not shown). Finally, the relative reactivity of single-Cys mutants in the sequence region 323–334 was examined at a low NEM concentration (50 μm), in the presence or absence of xanthine substrate (Fig. 5); it is found that reactivity of mutants in this sequence region follows a pattern that is distinct from the NEM sensitivity profile (Fig. 5B) and that substrate exerts minor or no effect at these conditions.
FIGURE 4.
Effect of xanthine on the NEM reactivity of single-Cys mutants. RSO membrane vesicles were prepared from isopropyl-1-thio-β-d-galactopyranoside-induced cultures of E. coli T184 harboring pT7-5/ygfO(C-less-His10) with given mutations and subjected to sulfhydryl alkylation assay in the absence or presence of xanthine (1 mm). After termination of the NEM reaction, membrane proteins were extracted with n-dodecyl β-d-maltopyranoside and subjected to Ni2+-chelate chromatography to capture YgfO-His10. Samples were then denatured, incubated with OGM (0.2 mm), and subjected to SDS-PAGE (12%) and immunoblotting using anti-OGM and HRP-conjugated penta-His antibody, as indicated. A, representative blots for mutants A323C, Q324C, N325C, and N326C. Prestained molecular mass standards (Bio-Rad, low range) are shown on the left. B, NEM dose-response curves for mutant N325C in the absence or presence of substrate. EC50 values were deduced from non-linear regression fitting of the data to the sigmoidal dose-response equation using Prism4. The value of OGM signal retained, in each case, was deduced from the ratio (LENEM/LEOGM), where LEOGM and LENEM are the extent of OGM labeling in the absence and presence of NEM pretreatment, respectively. Quantitative estimation of the labeling, derived from the normalized density of each anti-OGM band divided by the density of the corresponding anti-penta-His band, was performed with the program Quantity One (Bio-Rad).
FIGURE 5.
Profile of the NEM reactivity of single-Cys mutants. RSO membrane vesicles were prepared from isopropyl-1-thio-β-d-galactopyranoside-induced cultures of E. coli T184 harboring pT7-5/ygfO(C-less-His10) with given mutations and subjected to sulfhydryl alkylation assay using NEM (50 μm) in the absence or presence of xanthine (1 mm). YgfO-His10 was extracted, affinity-purified, and labeled with OGM, exactly as described in the legend for Fig. 4; samples were analyzed with SDS-PAGE (12%) and immunoblotting using anti-OGM antibody (panel A, upper blots) and HRP-conjugated penta-His antibody (panel A, lower blots). A, representative blots for mutants A323C, Q324C, N325C, N326C, G327C, V328C, Q330C, M331C, G333C, and V334C. Prestained molecular mass standards are shown on the left. B, profile of the NEM reactivity of the NAT motif mutants in the absence or presence of substrate. The relative reactivity value of each mutant was deduced from the formula ((LEOGM − LENEM)/LEOGM), where LEOGM and LENEM are the extent of OGM labeling in the absence and presence of NEM pretreatment, respectively. Quantitative estimation of the labeling was performed as in Fig. 4. Also shown are values of the relative sensitivity of each mutant to inactivation (percentage of inactivation after incubation with 2 mm NEM), as taken from Karatza et al. (8).
The NAT Motif Region Is Accessible to Solvent
Single-Cys mutants of the NAT motif (sequence 323–333) were also examined in RSO membrane vesicles for reactivity with MTSES−, a hydrophilic and membrane-impermeable sulfhydryl reagent (supplemental Fig. S1). All assayable motif mutants are fully reactive with 0.2 mm MTSES− (data not shown), and most of them are highly reactive with 50 μm MTSES− (supplemental Fig. S1A); as a control, the internal loop mutants R78C, L142C, and S206C (Fig. 1) do not react with MTSES− at these conditions, whereas the external loop mutants S109C, T243C, and I419C react to near completion (supplemental Fig. S1B).
Substrate Effects on the Reactivity with MTSES−
In the presence of xanthine substrate, reactivity of N325C with MTSES− decreases dramatically, whereas reactivity of the other mutants does not change significantly (supplemental Fig. S1A). The pattern of reactivity and substrate responses of the motif mutants observed with MTSES− (supplemental Fig. S1C) is similar to the pattern of reactivity and substrate responses seen with NEM (Fig. 5B).
Sensitivity to MTSES− Recapitulates the NEM Sensitivity Profile
Each active His10-tagged single-Cys mutant in the sequence region 323–339 of YgfO was assayed for the effect of MTSES− on the initial rate of xanthine transport in energized RSO membrane vesicles (supplemental Fig. S2). When incubated with 0.2 mm MTSES−, A323C, G327C, V328C, G333C, and V334C are inactivated by 40–90%, whereas activity of Q330C, M331C, A335C, R337C, Y338C, or V339C is not altered significantly; this profile of sensitivity to the hydrophilic MTSES− is similar to the corresponding profile of sensitivity to NEM (8).
Site-directed Mutagenesis of Gly-333, Val-334, Ser-336, and Val-339
The observed correlation of side-chain conservation with the NEM sensitivity of each Cys mutant in the NAT motif region (Fig. 2) is clear in the pre-motif (315–322) and the core motif sequence (323–333) but less pronounced in the α-helical segment that follows (333–339). Interestingly, the NEM-sensitive motif residues of the xanthine-monospecific transporter YgfO (XanQ) are conserved nearly invariably in the uric acid/xanthine dual specificity UapA and the other fungal uric acid/xanthine transporters (Xut1, UapC, AfUapA), but the NEM-sensitive residues of the short α-helical face that follows differ significantly (Fig. 2). This observation prompted us to apply a rational site-directed mutagenesis approach, replacing the NEM-sensitive residues of this α-helical region, either one by one or in combination, with the corresponding amino acids found in UapA to test for modulation of the specificity profile. Thus, we constructed YgfO mutants V334C(wt), S336N(wt), V339A(wt), G333R/V334C/S336N/Y338W/V339A(wt), and G333R/S336N/V339A(wt) and analyzed them in comparison with wild type and G333R(wt), which had been studied by us previously (8). Of the six mutants, the single-replacement ones are highly active for xanthine transport and expressed in the membrane to levels comparable with wild type, and the multiple-replacement ones are inactive and expressed to low but significant levels (25–35% of wild type) (Fig. 6, A and B). Kinetic analysis shows that the active mutants display equal (V334C) or 2-fold higher affinity (lower Km) and 2.5-fold lower (G333R), 1.5-fold lower (S336N), 3-fold higher (V339A), or 4-fold higher (V334C) Vmax for xanthine uptake relative to wild type (Table 2). Ligand inhibition analysis of xanthine uptake revealed discrete changes in specificity, with G333R recognizing 7-methylxanthine and 8-methylxanthine, which are not sensed by wild type (8), S336N failing to recognize 6-thioxanthine, which is a high affinity ligand for wild type, and V334C and V339A deviating from the overall wild-type profile to a lesser extent (Fig. 6C and Table 3). With respect to the transport potential for other substrates (10), none of the six mutants displayed any detectable capacity for uric acid uptake at 0.04–2 mm (data not shown).
FIGURE 6.
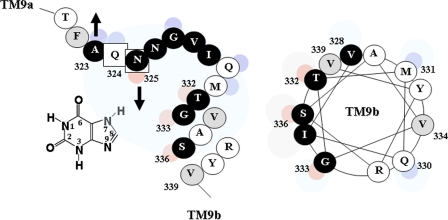
Functional properties of site-directed mutants at the α-helical face downstream of the motif. E. coli T184 harboring pT7-5/ygfO(wild-type-BAD) with given mutations were grown, induced, and subjected to immunoblot analysis of membrane fractions (A) or assayed for transport of [3H]xanthine (1 μm) at 25 °C in the absence (B) or presence of potential competitors (C). A, samples (100 μg of membrane protein) were subjected to SDS-PAGE (12%) and immunoblotting using HRP-conjugated avidin. Prestained molecular mass standards are shown on the left. B, open and closed histogram bars represent initial rates of xanthine uptake (measured at 5–20 s) and steady state levels of accumulation (reached at 1–10 min), respectively, expressed as percentages of the corresponding values of wild type (7), with S.D. shown from three independent determinations. triple mutant, G333R/S336N/V339A(wt); quintuple mutant, G333R/V334C/S336N/Y338W/V339A(wt). C, xanthine transport assays were performed in the presence of 0.001–1 mm competitor, and IC50 and Ki values were determined using Prism4. The purine bases or analogs shown are: 6-thioxanthine (6-SX); 2-thioxanthine (2-SX); 8-methylxanthine (8-MX); uric acid (8-OX); 7-methylxanthine (7-MX). Inset, bottom right, chemical formula of xanthine. Shaded circular areas denote numbered positions of the purine ring where a substitution leads to aberrant recognition by S336N or G333R, as indicated.
TABLE 2.
Functional characterization of YgfO mutants at positions Ser-336, Val-334, Val-339, and Gly-333: Km and Vmax values for xanthine uptake
E. coli T184 expressing the corresponding constructs were assayed for initial rates of xanthine uptake at 5–20 s in the concentration range of 0.1–100 μm; negative control values obtained from T184 harboring vector pT7–5 alone were subtracted from the sample measurements in all cases. Kinetic parameters were determined from non-linear regression fitting to the Michaelis-Menten equation using Prism4; values represent the means of three independent determinations with standard deviations shown. ND, assays were performed, but kinetic values were not determined due to very low uptake rates (≤0.05 nmol mg−1 min−1). All mutants and the wild-type YgfO version used contained a carboxyl-terminal biotin-acceptor domain (BAD); triple mutant, G333R/S336N/V339A(wt); quintuple mutant, G333R/V334C/S336N/Y338W/V339A(wt).
| Km | Vmax | Vmax/Km | |
|---|---|---|---|
| μm | nmol min−1mg−1protein | μl min−1mg−1 | |
| Permease | |||
| YgfO(wt) | 4.6 ± 0.3 | 6.4 ± 0.5 | 1391 |
| V334C(wt) | 4.6 ± 0.9 | 25.8 ± 1.5 | 5609 |
| G333R(wt) | 2.7 ± 0.2 | 2.8 ± 0.3 | 1037 |
| S336N(wt) | 2.0 ± 0.3 | 4.5 ± 0.2 | 2250 |
| V339A(wt) | 2.4 ± 0.3 | 19.6 ± 0.7 | 8167 |
| Triple mutant | ND | ND | |
| Quintuple mutant | ND | ND |
TABLE 3.
Functional characterization of YgfO mutants at positions Ser-336, Val-334, Val-339 and Gly-333: specificity profile of active mutants
E coli T184 expressing the corresponding constructs were assayed for initial rates of xanthine uptake at 5–20 s in the absence or presence of putative competitors. Upper part, values shown express % of xanthine (1 mm) uptake rate in the presence of 1,000-fold excess (1 mm) of unlabeled competitors. The uptake value obtained in the absence of competitor was taken as 100%. Values represent the means of three determinations, with standard deviations <20%. Lower part, xanthine (1 μm) transport assays were performed in the presence of 0.001–1 mm competitor, and IC50 and Ki values were determined as described under “Experimental Procedures.” Values shown for G333R have been taken from our previous study (8). Most significant differences from the wild-type profile are highlighted in bold.
| Competitor | [3H]Xanthine uptake rate retained (in %) |
||||
|---|---|---|---|---|---|
| YgfO(wt) | G333R(wt) | V334C(wt) | S336N(wt) | V339A(wt) | |
| Hypoxanthine | 91 | 80 | 87 | 97 | 84 |
| Adenine | 90 | 81 | 100 | 107 | 90 |
| Guanine | 75 | 90 | 67 | 91 | 67 |
| Uracil | 98 | 98 | 84 | 105 | 90 |
| 6-Thioxanthine | 12 | 11 | 8 | 61 | 17 |
| 2-Thioxanthine | 15 | 12 | 6 | 23 | 14 |
| 3-Methylxanthine | 15 | 1 | 21 | 8 | 17 |
| 8-Methylxanthine | 93 | 18 | 46 | 36 | 31 |
| Uric acid | 90 | 50 | 46 | 89 | 76 |
| 7-Methylxanthine | 100 | 20 | 79 | 69 | 64 |
| Oxypurinol | 30 | 10 | 8 | 7 | 8 |
| Allopurinol | 99 | 60 | 56 | 75 | 88 |
|
Ki |
|||||
|---|---|---|---|---|---|
| YgfO(wt) | G333R(wt) | V334C(wt) | S336N(wt) | V339A(wt) | |
| μm | |||||
| 6-Thioxanthine | 41 | 121 | 108 | >1000 | 111 |
| 2-Thioxanthine | 91 | 80 | 116 | 234 | 82 |
| 3-Methylxanthine | 72 | 14 | 32 | 50 | 56 |
| 8-Methylxanthine | >1000 | 409 | 832 | 685 | 843 |
| Uric acid | >1000 | 737 | 875 | >1000 | >1000 |
| 7-Methylxanthine | >1000 | 107 | >1000 | >1000 | >1000 |
DISCUSSION
Our previous Cys-scanning analyses of YgfO permease (8–10) have delineated the NAT motif sequence region as most important for the function and selectivity of the purine translocation pathway. Using site-directed alkylation analysis of Cys substitution mutants in the NAT signature motif of E. coli YgfO, we now provide evidence that residues in this sequence region are proximal to or interact with the purine-binding site. In brief, we propose that Asn-325, an invariably conserved residue of the NAT motif in NAT/NCS2 family, is at the binding site, accounting for the protective effect of substrate against site-directed alkylation of N325C (Fig. 4 and supplemental Fig. S1), whereas the neighboring Ala-323 responds to substrate binding with a specific conformational shift, detected as enhanced sensitivity of A323C to inactivation (Fig. 3). The intervening residue, Gln-324, which is invariably conserved in the purine-transporting members of the family, appears to be essential for high affinity binding (8) and might participate in binding substrate directly. Furthermore, a contiguous part of the motif (326–329) and a short α-helical face downstream (332, 333, 336) form a continuance of the purine permeation pathway, where alkylation of an engineered Cys does not hinder binding per se but leads to inactivation, presumably by blocking the release of substrate from the binding pocket.
A key finding is that substrate protects the position of Asn-325 from site-directed alkylation as it blocks both the NEM and the MTSES− reactivity of single-Cys N325C. Although one possibility might be that Asn-325 shifts to a less accessible environment due to a conformational effect of substrate, the most plausible interpretation is that Asn-325 lies at the binding site and substrate hinders alkylation at this position sterically. There are several lines of evidence that support the above contention. First, Asn-325 and the adjacent Gln-324 are functionally irreplaceable in YgfO. Replacement of Gln-324 impairs affinity (8), which accounts for the insensitivity of single-Cys Q324C alkylation to substrate (Figs. 4 and 5). Replacement of Asn-325 is compatible with high affinity binding, as evidenced by the finding that single-Cys N325C as well as N325C(wt) (Fig. 3) are subject to a substrate effect, but this binding does not elicit the proper changes of turnover, probably due to a local conformational aberration, leading to impairment of function. Importantly, the corresponding residues Gln-408 and Asn-409 of the fungal homolog UapA are also essential, and Gln-408 has been suggested to bind directly to the imidazole moiety of purine, based on the properties of mutant Q408E (4). Apart from the above, all mutant positions of the NAT motif of YgfO are highly accessible to solvent (supplemental Fig. S1), which is expected of a region forming part of the substrate-binding site in the alternately accessed hydrophilic cavity (11, 21–23). With the exception of N325C and A323C, the accessibility or reactivity of these positions with either MTSES− or NEM is practically unaltered by substrate, implying that they are close to the binding site and not at distal regions of the cavity, which are subject to more dynamic conformational changes (11). In any event, the alternative possibility that the motif is distal from the binding site and accessibility of Asn-325 is blocked by a conformational rather than a direct steric effect cannot be excluded with certainty. As seen with residues disposed on the cytoplasmic side of LacY and other transporters (11, 23), the conformational change induced by substrate might trap Asn-325 in a hydrophobic environment through closure of the cavity on the cytoplasmic side, limiting accessibility to NEM and MTSES−. On the other hand, our evidence that substrate protects Asn-325 from alkylation with NEM almost completely, whereas the sensitivity of the neighboring Ala-323 is enhanced (Fig. 3), and the aforementioned observations on the properties of the NAT motif mutants argue against the latter possibility.
Although all single-Cys mutants of the NAT motif region are highly reactive with both MTSES− and NEM, their sensitivity to inactivation varies greatly, and the sensitivity pattern is distinct from the reactivity pattern with either NEM (Fig. 5B) or MTSES− (supplemental Figs. S1 and S2). For example, mutants Q330C and M331C are highly accessible and reactive but display activity that is completely insensitive to either reagent, whereas mutants N326C and V328C display lower reactivity but are highly sensitive to inactivation by NEM. Thus, the sensitivity of mutants to inactivation does not reflect increased reactivity or accessibility of these positions to the reagent, but a more profound functional effect. Independently of the alkylating reagent used, the inhibition-sensitive positions cluster at the core motif sequence (323–329) and a short α-helical face downstream (332, 333, 336). This alkylation-sensitive subset of residues displays a high degree of evolutionary conservation of the side chain among members of the NAT family (Fig. 2), which is a recurring theme for NEM-sensitive residues of membrane transporters (12, 21, 23). All the above lines of evidence emphasize the importance of these residues for the mechanism of transport and, in conjunction with our data on Gln-324 (8) and Asn-325 (this study), imply that the inhibition-sensitive motif residues line the substrate translocation pathway in YgfO permease. Covalent attachment of the bulky maleimide or the 2-sulfonatoethyl (MTS) group at these positions might block turnover, leading to trapping of substrate in the binding pocket and impairment of active transport.
With the exception of N325C, saturating concentrations of xanthine substrate do not exert any major protective effect on the motif Cys replacement mutants, meaning that a direct steric hindrance on substrate binding should be excluded. On the other hand, accessibility or reactivity of most mutants in this region is too high to allow discrimination of subtle effects of substrate. For example, it is hard to detect any enhancing substrate effect for A323C and Q330C, which react completely and rapidly with either MTSES− or NEM even at low concentrations of reagent. However, a specific enhancing effect is detectable for A323C, based on the transport assay, showing a small but important increment in the sensitivity to inactivation by NEM (shift of IC50 from 34 to 14 μm). Such effects have also been described for residues of other transporters that are close to the binding site but not interfering sterically with it and are attributed to conformational shift of these residues to an environment that allows more efficient clogging of the substrate permeation pathway by the maleimidyl adduct (11, 18, 21).
Replacement of Gly-333 or Ser-336 at the short α-helical face downstream the motif with Arg or Asn, respectively, changes specific features in the ligand recognition profile of YgfO permease, implying that these two residues are implicated with substrate specificity. In particular, mutants G333R and S336N reverse in part the key specificity elements differentiating the xanthine-specific transporters YgfO and YicE (7) from the fungal, uric acid-transporting homologs UapA (4), Xut1 (24), and AfUapA (25), i.e. G333R and, to a lesser extent, S336N allow recognition of 8-methylxanthine, which is also recognized by the fungal homologs (Ki values from 50 to 100 μm), whereas S336N disrupts recognition of 6-thioxanthine, which is recognized with high affinity by wild-type YgfO (Ki 40 μm) but is a low affinity ligand for the fungal homologs (Ki values from 250 to 350 μm). However, the replacements have additional effects, which are unprecedented by either the bacterial or the fungal wild types, including the ability of G333R to bind 7-methylxanthine (Ki 107 μm) and the low affinity of S336N for 2-thioxanthine (Ki 234 μm). In any event, the pleiotropic effects seen with these mutants argue against a direct role on substrate binding but are consistent with the interpretation that the two YgfO residues lie in the vicinity of the binding site and contribute to the optimal specificity profile. Replacement of Thr-332, at the same α-helical face as Gly-333 and Ser-336, has also been shown to affect specificity in a similar manner, most prominently with mutant T332N conferring the ability to recognize 8-methylxanthine (Ki 196 μm) (8).
It is important to emphasize, at this point, that the predicted topology in the region of the NAT motif and flanking putative transmembrane helices TM9a and TM9b (Fig. 1) is unclear. Based on the comparative prediction analysis of known members of the NAT family with a series of different algorithms, the overall topology of bacterial (8), fungal (4), and mammalian (26) NAT transporters is expected to consist of 12 putative transmembrane α-helical segments, with the NAT motif sequence falling probably on the cytoplasmic side. However, analysis of the topology of YgfO permease with the TMHMM algorithm shows a striking ambiguity for the NAT motif region, predicting that sequence 300–325 may be transmembrane or cytoplasmic and sequence 325–340 may be periplasmic or cytoplasmic, respectively, with roughly equal probabilities.5 The ambiguous topology segment upstream of the motif (residues 301–318) conforms clearly to an amphipathic α-helix (10), which is not long enough to traverse the bilayer and has been designated by us as TM9a (10), whereas in silico analysis indicates that the NAT motif may form a short loop between two α-helical segments (4). In this respect, our current evidence that residues of the NAT motif are highly accessible to solvent from the periplasm and proximal to or interacting with the purine-binding site allows us to propose an alternative model, in which part of the NAT motif forms a re-entrant loop facing the central hydrophilic cavity (Fig. 7). This proposed new arrangement of the NAT motif allows positioning of important residues of TM9a (Asp-304) (10) and TM8 (Glu-272, Gly-275, Asp-276)6 in closer proximity with the putative binding site residues Gln-324 and Asn-325 (Fig. 7) and better accommodation of the specificity-related residues Thr-332, Gly-333, and Ser-336 in the binding pocket (Fig. 8). Another potential model differentiating from the one in Fig. 7 would entail TM9a crossing much of the membrane rather than being located in the cytosol and then the motif region crossing back to the other side of the membrane to connect to the cytosolic side of TM9b; this possibility is also consistent with the interpretation that the NAT motif may form part of the substrate permeation pathway. In any event, our ongoing Cys-scanning and substituted-Cys accessibility analysis of TM8, TM9a, and TM9b is expected to allow further refinement of the topology model in this permease region.6
FIGURE 7.
Revised model for the topology of the NAT motif and flanking putative transmembrane helices TM8, TM9a, and TM9b of YgfO permease. The NAT motif sequence is shown as a putative re-entrant loop with Gln-324 and Asn-325 pointing toward the solvent-accessible internal cavity. Irreplaceable residues of YgfO are numbered and bolded. NEM-sensitive positions are shown in a dark background. Residues analyzed with site-directed mutagenesis are circled. Cys-scanning data for TM8, loops 8–9, TM9a, and TM9b were taken from our unpublished evidence (see Footnote 6).
FIGURE 8.
Model of the NAT motif of YgfO permease with xanthine substrate and helical wheel plot of residues 328–339 in TM9b. NEM-sensitive positions are shown in a dark background. The small eccentric circles denote residues that are highly accessible to solvent (blue) or implicated with substrate interference or analog selectivity (red).
Given the key functional role of the NAT motif for purine substrate recognition in both a bacterial (this study) and a fungal (6) transporter and its sequence conservation across distantly related species, it would be interesting to examine whether such a role is also retained in the uracil-transporting NATs, including the quite recently characterized rSNBT1 from the rat small intestine (27), or in the mammalian homologs SVCT1 and SVCT2, which transport l-ascorbate and do not recognize nucleobases (1, 26). In this respect, it is important that residues found to be irreplaceable (7–10) or critical for function in the bacterial study model cluster at highly conserved positions of the motif (Ala-323, Gln-324, Asn-325, Gly-333) and neighboring sequence regions (Asp-304, Glu-272) (Fig. 7). Strikingly, as well, substrate selectivity differences of NAT transporters appear to be correlated with different side-chain occupation at specific conserved positions of the NAT motif. For example, the known purine-transporting NATs have a Gln at position of Gln-324 (10), the uracil-transporting NATs have a Glu, and the ascorbate-transporting mammalian NATs have a Pro at this position; the pattern is even retained for closely related pairs of paralogs, such as in the case of the recently recognized rSNBT1 (uracil transporter; Glu at position-324), which shares 50% sequence identity (67% similarity) with SVCT1 or SVCT2 (ascorbate transporter; Pro at position-324) (27). Rationalized site-directed mutagenesis approaches might help elucidate whether such side-chain differences are functionally relevant to specific shifts in substrate selectivity in the evolution of NAT transporters.
In conclusion, our data on the substrate-interacting properties and the reagent accessibility and reactivity of NAT motif mutants provide a basis to investigate the mechanism of substrate recognition and selectivity in the important, evolutionary ubiquitous family NAT/NCS2 of nucleobase-ascorbate transporters; ongoing work on the bacterial study model, in conjunction with potential future structural studies (28), is expected to provide valuable insight to this end.
Supplementary Material
Acknowledgments
We thank H. Ronald Kaback and George Diallinas for helpful discussions, Panayiotis Panos for mutants T321C and F322C(wt), and Panayiota Karatza for mutant G333R(wt).
This work was supported by the 03ED204 research project, implemented within the framework of the “Reinforcement Programme of Human Research Manpower” (PENED) and co-financed by National and Community Funds (25% from the Greek Ministry of Development, General Secretariat for Research and Technology, and 75% from the European Union European Social Fund).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1 and S2.
K. Papakostas and S. Frillingos, manuscript in preparation.
K. Rudd, personal communication.
G. Mermelekas and S. Frillingos, manuscript in preparation.
G. Mermelekas, E. Georgopoulou, A. Kallis, V. Vlantos, and S. Frillingos, manuscript in preparation.
- NAT
- nucleobase-ascorbate transporter
- TM
- transmembrane helix
- NEM
- N-ethylmaleimide
- MTSES−
- 2-sulfonatoethyl methanethiosulfonate
- OGM
- Oregon Green 488 maleimide
- HRP
- horseradish peroxidase
- BAD
- biotin-acceptor domain
- RSO
- right-side-out membrane vesicles
- Cys-less (C-less)
- permease devoid of native Cys residues
- wt
- wild-type.
REFERENCES
- 1.Tsukaguchi H., Tokui T., Mackenzie B., Berger U. V., Chen X. Z., Wang Y., Brubaker R. F., Hediger M. A. (1999) Nature 399, 70–75 [DOI] [PubMed] [Google Scholar]
- 2.Gournas C., Papageorgiou I., Diallinas G. (2008) Mol. Biosyst. 4, 404–416 [DOI] [PubMed] [Google Scholar]
- 3.Diallinas G., Valdez J., Sophianopoulou V., Rosa A., Scazzocchio C. (1998) EMBO J. 17, 3827–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koukaki M., Vlanti A., Goudela S., Pantazopoulou A., Gioule H., Tournaviti S., Diallinas G. (2005) J. Mol. Biol. 350, 499–513 [DOI] [PubMed] [Google Scholar]
- 5.Vlanti A., Amillis S., Koukaki M., Diallinas G. (2006) J. Mol. Biol. 357, 808–819 [DOI] [PubMed] [Google Scholar]
- 6.Papageorgiou I., Gournas C., Vlanti A., Amillis S., Pantazopoulou A., Diallinas G. (2008) J. Mol. Biol. 382, 1121–1135 [DOI] [PubMed] [Google Scholar]
- 7.Karatza P., Frillingos S. (2005) Mol. Membr. Biol. 22, 251–261 [DOI] [PubMed] [Google Scholar]
- 8.Karatza P., Panos P., Georgopoulou E., Frillingos S. (2006) J. Biol. Chem. 281, 39881–39890 [DOI] [PubMed] [Google Scholar]
- 9.Papakostas K., Georgopoulou E., Frillingos S. (2008) J. Biol. Chem. 283, 13666–13678 [DOI] [PubMed] [Google Scholar]
- 10.Karena E., Frillingos S. (2009) J. Biol. Chem. 284, 24257–24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaback H. R., Dunten R., Frillingos S., Venkatesan P., Kwaw I., Zhang W., Ermolova N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frillingos S., Sahin-Tóth M., Wu J., Kaback H. R. (1998) FASEB J. 12, 1281–1299 [DOI] [PubMed] [Google Scholar]
- 13.Inoue H., Nojima H., Okayama H. (1990) Gene 96, 23–28 [DOI] [PubMed] [Google Scholar]
- 14.Teather R. M., Bramhall J., Riede I., Wright J. K., Fürst M., Aichele G., Wilhelm U., Overath P. (1980) Eur. J. Biochem. 108, 223–231 [DOI] [PubMed] [Google Scholar]
- 15.Kaback H. R. (1971) Methods Enzymol. 22, 99–120 [Google Scholar]
- 16.Frillingos S., Wu J., Venkatesan P., Kaback H. R. (1997) Biochemistry 36, 6408–6414 [DOI] [PubMed] [Google Scholar]
- 17.Frillingos S., Sahin-Tóth M., Persson B., Kaback H. R. (1994) Biochemistry 33, 8074–8081 [DOI] [PubMed] [Google Scholar]
- 18.Nie Y., Zhou Y., Kaback H. R. (2009) Biochemistry 48, 738–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konings W. N., Barnes E. M., Jr., Kaback H. R. (1971). J. Biol. Chem. 246, 5857–5861 [PubMed] [Google Scholar]
- 20.Granseth E., Daley D. O., Rapp M., Melén K., von Heijne G. (2005) J. Mol. Biol. 352, 489–494 [DOI] [PubMed] [Google Scholar]
- 21.Frillingos S., Kaback H. R. (1997) Protein Sci. 6, 438–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura N., Konishi S., Iwaki S., Kimura-Someya T., Nada S., Yamaguchi A. (2001) J. Biol. Chem. 276, 20330–20339 [DOI] [PubMed] [Google Scholar]
- 23.Tavoulari S., Frillingos S. (2008) J. Mol. Biol. 376, 681–693 [DOI] [PubMed] [Google Scholar]
- 24.Goudela S., Karatza P., Koukaki M., Frillingos S., Diallinas G. (2005) Mol. Membr. Biol. 22, 263–275 [DOI] [PubMed] [Google Scholar]
- 25.Goudela S., Reichard U., Amillis S., Diallinas G. (2008) Fungal Genet. Biol. 45, 459–472 [DOI] [PubMed] [Google Scholar]
- 26.Velho A. M., Jarvis S. M. (2009) Exp. Cell Res. 315, 2312–2321 [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto S., Inoue K., Murata T., Kamigaso S., Yasujima T., Maeda J. Y., Yoshida Y., Ohta K. Y., Yuasa H. (2010) J. Biol. Chem. 285, 6522–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stroud R. M., Choe S., Holton J., Kaback H. R., Kwiatkowski W., Minor D. L., Riek R., Sali A., Stahlberg H., Harries W. (2009) J. Struct. Funct. Genomics 10, 193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.