Abstract
Bovine leukemia virus (BLV) proviral latency represents a viral strategy to escape the host immune system and allow tumor development. Besides the previously demonstrated role of histone deacetylation in the epigenetic repression of BLV expression, we showed here that BLV promoter activity was induced by several DNA methylation inhibitors (such as 5-aza-2′-deoxycytidine) and that overexpressed DNMT1 and DNMT3A, but not DNMT3B, down-regulated BLV promoter activity. Importantly, cytosine hypermethylation in the 5′-long terminal repeat (LTR) U3 and R regions was associated with true latency in the lymphoma-derived B-cell line L267 but not with defective latency in YR2 cells. Moreover, the virus-encoded transactivator TaxBLV decreased DNA methyltransferase expression levels, which could explain the lower level of cytosine methylation observed in the L267LTaxSN 5′-LTR compared with the L267 5′-LTR. Interestingly, DNA methylation inhibitors and TaxBLV synergistically activated BLV promoter transcriptional activity in a cAMP-responsive element (CRE)-dependent manner. Mechanistically, methylation at the −154 or −129 CpG position (relative to the transcription start site) impaired in vitro binding of CRE-binding protein (CREB) transcription factors to their respective CRE sites. Methylation at −129 CpG alone was sufficient to decrease BLV promoter-driven reporter gene expression by 2-fold. We demonstrated in vivo the recruitment of CREB/CRE modulator (CREM) and to a lesser extent activating transcription factor-1 (ATF-1) to the hypomethylated CRE region of the YR2 5′-LTR, whereas we detected no CREB/CREM/ATF recruitment to the hypermethylated corresponding region in the L267 cells. Altogether, these findings suggest that site-specific DNA methylation of the BLV promoter represses viral transcription by directly inhibiting transcription factor binding, thereby contributing to true proviral latency.
Keywords: CREB, DNA Methylation, Epigenetics, Transcription Regulation, Virus, Bovine Leukemia Virus, Latency
Introduction
Bovine leukemia virus (BLV)12 is a B-lymphotropic oncogenic retrovirus that infects cattle and is associated with enzootic bovine leucosis, a neoplastic proliferation of B cells (1–5). BLV is structurally and biologically closely related to human T-lymphotropic viruses HTLV-I and HTLV-II (4, 6). The majority of BLV-infected cattle are asymptomatic carriers of the virus. Only about 30% of BLV-infected animals develop a preneoplastic condition termed persistent lymphocytosis, with 2–5% developing B-cell leukemia and/or lymphoma after a long latency period (7). The virus can be inoculated experimentally into sheep, in which the disease appears earlier and with higher frequencies, providing a helpful model for understanding BLV and HTLV-induced leukemogenesis. BLV infection is characterized by viral latency in a large majority of infected cells and by the absence of viremia. These features are thought to be due to the transcriptional repression of viral expression in vivo (8–11). Latency is likely to be a viral strategy to escape the host immune response and allow tumor development.
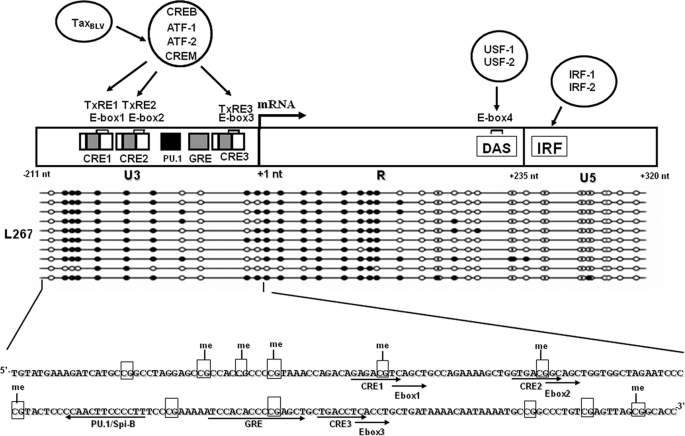
The BLV transcriptional promoter is located in the 5′-long terminal repeat (5′-LTR) and is composed of the U3, R, and U5 regions. Transcription initiates at the U3–R junction of the 5′-LTR. BLV gene expression is induced at the transcriptional level by the virus-encoded transactivator TaxBLV (12, 13). Transactivation by TaxBLV requires the presence of three 21-bp enhancer elements (called Tax-responsive elements (TxREs)) located in the U3 region of the 5′-LTR (12, 14). Each TxRE contains a core octanucleotide sequence corresponding to an imperfectly conserved cyclic AMP-responsive element (CRE), which binds cellular transcription factors: the CRE-binding protein (CREB), the CRE-modulator τ isoform (CREMτ), and activating transcription factor-1 and -2 (ATF-1 and ATF-2) (14–17). Besides the imperfect CRE consensus, each TxRE also contains an E-box sequence, which overlaps each of the three CRE motifs (4, 18). A PU.1/Spi-B site (19) and a glucocorticoid responsive-element (20–23) are also present in the U3 region. In addition, BLV expression is regulated by LTR sequences located downstream of the transcription initiation site: an upstream stimulatory factor-binding site in the R region (24) and an interferon regulatory factor-binding site in the U5 region (25).
During retroviral infection, the RNA viral genome is retrotranscribed into double-stranded DNA, which becomes integrated into the host genome and is organized into chromatin as all cellular genes. This chromatin environment is likely to be a key parameter for the control of viral gene expression because transcriptional activation by cellular or viral trans-acting factors is dependent on chromatin accessibility. BLV expression is controlled by chromatin structure. Indeed, our laboratory reported previously that histone acetylation is an important epigenetic modification for TaxBLV-dependent and -independent viral transcriptional regulation (26–29). BLV promoter activity is induced by deacetylase (HDAC) inhibitors, and this is correlated with an increased histone-4 acetylation at the viral promoter (27). Our laboratory has also demonstrated that HDAC inhibitors and TaxBLV synergistically activate BLV promoter transcriptional activity in a CRE- and CREB-dependent manner. This synergism is mediated by an HDAC inhibitor indirect action that requires de novo protein synthesis and that increases the level of CREB/ATF bound to the BLV promoter (28).
To further investigate the role of chromatin structure in the control of BLV gene expression, we studied in this report the involvement of another epigenetic modification, DNA methylation, in BLV gene expression regulation. The enzymes that establish and maintain specific DNA methylation patterns include the de novo DNA methyltransferases DNMT3A and DNMT3B and the maintenance DNA methyltransferase DNMT1 (30, 31). In mammalian cells, DNA methylation occurs predominantly at cytosines in CpG dinucleotides of transcriptional regulatory regions and is generally associated with gene silencing, either directly by inhibiting the binding of transcription factors to their recognition sequences or indirectly by preventing transcription factors from accessing their target sites through attachment of methyl-CpG-binding proteins, which “read” DNA methylation patterns. These proteins (MeCPs) recruit histone deacetylases and histone methyltransferases, thereby resulting in formation of a closed repressive chromatin structure (31, 32). Here, we have demonstrated that DNA cytosine hypermethylation is associated with BLV postintegration latency in a lymphoma-derived B-cell line, L267. Mechanistically, site-specific methylation in BLV promoter CRE elements inhibits CREB/CREM/ATF-1 in vitro binding and decreases by 2-fold BLV LTR-driven transcription. Importantly, by chromatin immunoprecipitation (ChIP) assays, we have demonstrated in vivo the recruitment of the transcription factors CREB and CREM, and to a lesser extent ATF-1, to the hypomethylated CRE region of the YR2 5′-LTR, whereas we detected no CREB/CREM/ATF recruitment to the hypermethylated corresponding region in the L267 cells. Interestingly, we show that TaxBLV protein decreases the expression levels of DNMT1, DNMT3A, and DNMT3B, which could, at least in part, explain the lower level of DNA methylation observed in the L267LTaxSN 5′-LTR compared with the L267 5′-LTR. These results indicate that DNA methylation contributes to proviral latency, a viral strategy to escape host immune response and allow tumor development.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
The Raji cell line, a human B-lymphoid Epstein-Barr virus-positive cell line derived from a Burkitt's lymphoma, and the Epstein-Barr virus-negative B-lymphoid cell line DG75 were maintained in RPMI 1640-Glutamax I medium (Invitrogen) supplemented with 10% fetal bovine serum, 50 μg of streptomycin/ml and 50 units of penicillin/ml. The human epithelial HeLa cell line is derived from a cervical carcinoma and is transformed by human papillomavirus type 18. HeLa cells were cultured in Dulbecco's modified Eagle's-Glutamax I medium (Invitrogen) supplemented with 5% fetal bovine serum, 50 μg of streptomycin/ml and 50 units of penicillin/ml. L267 is a clonal lymphoma-derived B-cell line established from a BLV-infected sheep (S267) injected with naked proviral DNA of an infectious BLV variant described previously (33–35). The tumor-derived L267 provirus displays a wild-type sequence that is identical to the infectious provirus used for sheep S267 inoculation (34, 35). The L267LTaxSN cell line results from the transduction of native L267 with the pLTaxSN retroviral vector expressing the tax cDNA following cocultivation with the PG13LTaxSN producer cell line and G418 selection of transduced cells (34, 36). L267 and L267LTaxSN were maintained in Opti-MEM medium (Invitrogen) supplemented with 10% fetal bovine serum, 1 mm sodium pyruvate, 2 mm glutamine, non-essential amino acids, and 100 μg of kanamycin. YR2 is a cloned B-lymphoid cell line established from peripheral blood lymphocytes isolated from a BLV-infected sheep (10, 36, 37). YR2 cells were maintained in Opti-MEM-Glutamax I medium (Invitrogen) supplemented with 10% fetal bovine serum, 50 μg of streptomycin/ml, and 50 units of penicillin/ml. All cells were grown at 37 °C in an atmosphere of 5% CO2.
Transient Transfection and Luciferase Assays
Raji and DG75 cells were transfected using the DEAE-dextran procedure as described previously (38). At 22 h after transfection, the cells were mock-treated or treated with increasing concentrations of procaine (Sigma), procainamide (Sigma), 5-aza-2′-deoxycytidine (Sigma), or zebularine (Calbiochem) as indicated. At 72 h after induction, the cells were lysed and assayed for luciferase activity (Promega). Luciferase activities were normalized with respect to protein concentrations using the detergent-compatible protein assay (Bio-Rad). HeLa cells were transfected using FuGENETM 6 (Roche Applied Science) as described previously (24). All transfection mixtures contained pRL-TK (50 ng), in which a cDNA encoding Renilla luciferase is under the control of the herpes simplex virus thymidine kinase promoter region and which is used as an internal control for transfection efficiency. At 48 h posttransfection, luciferase activities (firefly and Renilla) were measured in cell lysates by using the dual luciferase reporter assay system (Promega).
Western Blotting
Nuclear extracts prepared from transfected Raji cells were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes. The membranes were then blocked in Tris-buffered saline containing 5% nonfat dry milk and incubated with an anti-c-Myc (1/1000 dilution; Santa Cruz Biotechnology, catalog number sc-40) antibody or with an antibody from Imgenex directed against DNMT1 (IMG-261A), DNMT3A (IMG-268A), or DNMT3B (IMG-184A). A second antibody, a horseradish peroxidase-conjugated goat anti-rabbit IgG (1/3000 dilution; Santa Cruz Biotechnology, sc-2054), was used for enhanced chemiluminescence detection (Cell Signaling Technology).
Bisulfite Cytosine Methylation Analysis
Samples of total genomic DNA (700 ng) isolated with the DNeasy tissue kit (Qiagen) and digested overnight with EcoRI in a volume of 20 μl were prepared for methylation analysis as described previously by Olek et al. (39). Briefly, the digested DNA, which had been boiled for 5 min, chilled quickly on ice, and incubated in 0.3 m NaOH at 50 °C for 15 min, was mixed with 2.5 volumes of 2% low melting point agarose (SeaPlaque agarose, FMC Bioproducts). Ten-microliter volumes of agarose-DNA mixture were pipetted into 750 μl of chilled mineral oil to form agarose beads. Aliquots of 1 ml of 2.5 m sodium metabisulfite (Sigma) and 125 mm hydroquinone (Sigma) (pH 5.0) were then added to mineral oil containing up to four agarose beads. The reaction mixtures were kept on ice for 30 min and then incubated in the dark at 50 °C for 3.5 h. The agarose beads were equilibrated four times (15 min each time) with 1 ml of Tris-EDTA (pH 8.0) and DNA desulfonated in 500 μl of 0.2 m NaOH (twice for 15 min each time) at room temperature. Finally, the beads were washed with 1 ml of Tris-EDTA (pH 8.0) (3 times for 10 min each time) and then with 1 ml of sterile water (twice for 15 min each time) and kept at 4 °C.
Bisulfite-treated DNA in melted agarose was amplified by PCR in a 100-μl reaction mixture containing 1× PCR buffer (GoTaq polymerase buffer, Promega, Madison, WI), 1 mm MgCl2, each dNTP at 400 μm, with 1 unit of GoTaq polymerase, and 50 pmol of each primer: NA (5′-TGTATGAAAGATTATGT-3′, nucleotides (nt) −211 to −195, sense) and NB (5′-CATAATATAATTTAAAAAAAAAACCCAAAA-3′, nt 391–420, antisense), primers complementary to the BLV 5′-LTR and Gag-coding region, respectively (Fig. 2C); NE (5′-TAGAAAATGAATGGTTTTTT-3′, nt 7930–7949, sense) and NF (5′- TATTTACCAATCTCTCCTAA-3′, nt 8484–8503, antisense), primers complementary to the X region and the 3′-LTR, respectively (Fig. 2C). The amplification products were subjected to a second round of PCR with nested primers: NC (5′-GATTATGTTGATTTAGGAGT-3′, nt −202 to −183, sense) and ND (5′-AAAATAAATCCAAAAAATTATTAAAATTCC-3′, nt 351–380, antisense), primers complementary to the BLV 5′-LTR and Gag coding regions, respectively (Fig. 2C); NG (5′-GAGGGGGAGTTATTTGTATG-3′, nt 8060–8079, sense) and NH (5′-TATTTACCAATCTCTCCTAACC-3′, nt 8482–8503, antisense), primers complementary to BLV 3′-LTR and X regions, respectively (Fig. 2C). The sense primers contained T and the antisense primers contained A instead of C in positions complementary to nonmethylable C (i.e. C out of CpG dinucleotides). PCR was performed with about 50 ng of genomic DNA (of 140 ng of DNA/1 agarose bead) for 40 cycles at 95 °C for 1 min, 50 °C (45 °C for the 3′-LTR) for 2 min, and 72 °C for 1 min 30 s for the first round of PCR. The second round of PCR was performed with 5 μl from the first round of PCR for 50 cycles at 95 °C for 1 min, 56 °C for 2 min (52 °C for the 3′-LTR), and 72 °C for 1 min 30 s. Amplification products were cloned in the TOPO TA cloning vector system (Invitrogen) and sequenced. The frequency of conversion of C to T following the sodium bisulfite treatment and PCR in our studies was greater than 99% at non-CpG sites, indicating the adequacy of the approach.
FIGURE 2.
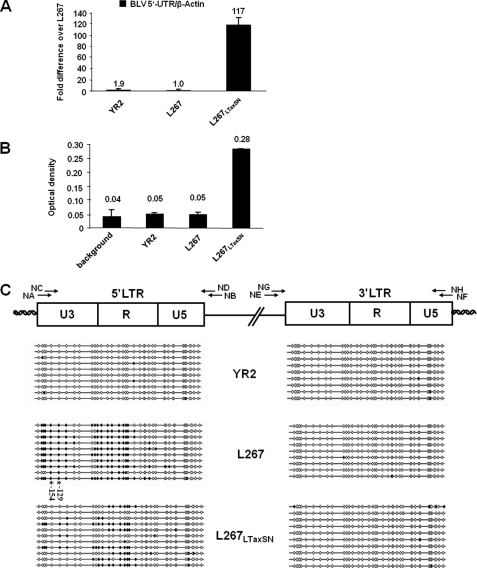
CpG methylation status in the 5′ and 3′-LTR regions of BLV proviruses integrated in different infected cell lines. A, mRNA expression levels in the BLV-infected YR2, L267, and L267LTaxSN cell lines. Total RNA samples were extracted and digested by DNase I. First strand cDNA was synthesized by reverse transcription, and quantitative PCR was performed with oligonucleotide primers amplifying the 5′-UTR region of BLV or the β-actin gene. Results are presented as the ratio of 5′-UTR to β-actin and are the mean values of triplicate samples. The results from a representative experiment of three independent experiments are shown. B, ex vivo expression of BLV in infected YR2, L267, and L267LTaxSN cell lines. The BLV p24 major gag antigen levels were titrated in the culture supernatants by ELISA. Data are the mean values of triplicate samples. The results from a representative experiment of three independent ELISA experiments are shown. C, bisulfite sequencing analysis of the CpG methylation status in the 5′- and 3′-LTRs of the YR2, L267, and L267LTaxSN cell lines. Genomic DNA from the three cell lines was extracted and treated by sodium bisulfite/hydroquinone. The two LTRs were amplified by nested PCR, amplified PCR products were subcloned in a TA cloning vector, and 10–11 clones were sequenced for each cell line. The methylation status of these clones is presented for each cell line. The positions and orientations of PCR primers (NA, NB, NC, ND, NE, NF, NG, and NH) used to amplify bisulfite-treated BLV DNA by nested PCR are indicated by arrows. Open circles represent unmethylated CpG dinucleotides, and filled circles represent methylated CpG dinucleotides. The −154 CpG and −129 CpG (located within the CRE1 and CRE2 sites, respectively) are hypermethylated in the L267 cell line and are indicated by asterisks.
ChIP Assays
ChIP assays were performed using the ChIP assay kit (EZ ChIP technology, Upstate). YR2 and L267 cells were cross-linked for 10 min at room temperature with 1% formaldehyde. To detect chromosomal flanking regions, pellets were sonicated (Bioruptor sonicator) to obtain DNA fragments at an average size of 400 bp. Chromatin immunoprecipitations were performed with an antibody directed against CREB (Santa Cruz Biotechnology, catalog number sc-186), CREM (sc-440), ATF-1 (sc-243), and ATF-2 (sc-242). To test aspecific binding to the beads, a purified IgG was used as a control for immunoprecipitation (Vector Laboratories, catalog number I-1000). Quantitative PCR reactions were performed with the MESA GREEN qPCR MasterMix (Eurogentec). Relative quantification using standard curve method was performed for each primer pair, and 96-well optical reaction plates were read in an Applied Biosystems ABI Prism 7300 real-time PCR instrument (absolute quantification method). Fold enrichments were calculated as percentages of input values following this formula: Immunoprecipitated DNA/total DNA. Primer sequences used for quantification in a region overlapping the three CRE sites of the BLV LTR (Fw, 5′-GCCAGAAAAGCTGGTGACG-3′ and Rv, 5′-GGGTGTGGATTTTTCGGG-3′) and in the U5 region of the BLV LTR, where no CRE binding site has been reported (Fw, 5′-CCTCTGACCGTCTCCACGT-3′ and Rv, 5′-AAGTAAGACAGGAAACAAGCGC-3′), were designed using the software Primer Express 2.0 (Applied Biosystems).
Electrophoretic Mobility Shift Assays
Nuclear extracts were prepared from cells using a rapid protocol described by Osborn et al. (40). All buffers contained the following protease inhibitors: antipain (10 μg/ml), aprotinin (2 μg/ml), chymostatin (10 μg/ml), leupeptin (1 μg/ml), and pepstatin (1 μg/ml). Protein concentrations were determined according to the Bradford methodology (41) with bovine serum albumin as a standard. Electrophoretic mobility shift assays (EMSAs) were performed as described previously (24). The DNA sequences of the coding strand of TxRE1 and TxRE2 probes (nt −164 to −144 and nt −139 to −119, respectively) were as follows: 5′-CAGACAGAGACGTCAGCTGCC-3′ and 5′-AGCTGGTGACGGCAGCTGGT-3′ (the CRE sites are underlined, and the CpG dinucleotides are highlighted in boldface type). Methylated versions of these oligonucleotides (where CpGs at positions −154 (located within the CRE1 site) or −129 (located within the CRE2 site) were methylated) were synthesized chemically by Eurogentec (Liege, Belgium) and named TxRE1-154me (Fw, 5′-CAGACAGAGACMeGTCAGCTGCC-3′; Rv, 5′-GGCAGCTGACMeGTCTCTGTCTG-3′) and TxRE2–129me (Fw, 5′-AAGCTGGTGACMeGGCAGCTGGT-3′; Rv, 5′-ACCAGCTGCCMeGTCACCAGCTT-3′).
Plasmid Constructs and Generation of Constructs with Site-specific Methylation
The eukaryotic expression vector pSG-TaxBLV was a kind gift from Drs. Richard Kettmann and Luc Willems (Faculty of Agronomy, Gembloux, Belgium). The eukaryotic pcDNA3.1/Myc-His-derived expression vectors for DNMT1, DNMT3A, and DNMT3B (pCMV-Myc-Dnmt1, pDNMT3aMyc, and pDNMT3bMyc, respectively) were kind gifts from Drs. François Fuks and Tony Kouzarides (42, 43). The non-episomal plasmid pLTRwt-luc and the episomally replicating plasmid pLTR*wt(direct)-luc were described previously (in Refs. 24 and 27, respectively) and contain the luciferase gene under the control of the complete 5′-LTR of the 344 BLV provirus. Strain 344 is an infectious and pathogenic molecular clone (33). The vector pLTRwt-luc was used as a substrate for mutagenesis with the QuikChange site-directed mutagenesis method (Stratagene) using 50 ng of pLTRwt-luc DNA/reaction. Following PCR, the samples were treated with DpnI for 2 h to remove the DNA template. The products were purified using the QIAquick method (Qiagen), reannealed, and precipitated. DNA concentration was determined by UV absorbance before transfection. Mutations in the CRE1 and CRE2 sites were generated with the following pairs of mutagenic oligonucleotides: CRE1 (Fw, 5′-CAGACAGAGTGGTCAGCTGCC-3′; Rv, 5′-GGCAGCTGACCACTCTGTCTG-3′) and CRE2 (Fw, 5′- AAGCTGGTGTGGGCAG CTGGT-3′; Rv, 5′-ACCAGCTGCCCACACC AGCTT-3′) (mutations are highlighted in boldface type, and the CRE motif is underlined). These mutated plasmids were designated pLTRmutCRE1 and pLTRmutCRE2, respectively. All mutated constructs were fully sequenced after identification (Genome Express).
The pLTRwt-luc vector with methylation either at the −154 CpG, or the −129 CpG or at both CpGs in combination (−154 CpG and −129 CpG) was generated using site-specific methylated oligonucleotides (purchased from Eurogentec) and the QuikChange site-directed mutagenesis kit (Stratagene) following a protocol adapted from Martinowich et al. (44). In brief, two oligonucleotide primers, each complementary to opposite strands of the parental vector and methylated at the −154 CpG (Fw, 5′-CAGACAGAGACMeGTCAGCTGCC-3′; Rv, 5′-GGCAGCTGACMeGTCTCTGTCTG-3′) or the −129 CpG (Fw, 5′-AAGCTGGTGACMeGGCAGCTGGT-3′; Rv, 5′-ACCAGCTGCCMeGTCACCAGCTT-3′), were extended by the PfuTurbo DNA polymerase. The product of this reaction was then treated with the endonuclease DpnI, which is specific for methylated and hemimethylated DNA (target sequence 5′-Gm6ATC-3′), and digested the parental DNA template to select for mutation-containing synthesized DNA. This experimental strategy led to the production of circular closed plasmids with site-directed methylation on both strands. These CpG methylated plasmids were designated pLTR-154me, pLTR-129me, and pLTR-154me/-129me, respectively, and transfected directly after the mutagenesis reaction without amplification in bacteria. As an additional control, we generated pLTRwt-luc and pLTR*wt(direct)-luc derivatives, which were hypermethylated only in the LTR (i.e. in which every CpG of the BLV LTR, but no CpG elsewhere in the pLTRwt-luc or pLTR*wt(direct)-luc plasmid, were methylated in vitro by the SssI methyltransferase). To this end, we methylated in vitro the whole pLTRwt-luc (or pLTR*wt(direct)-luc) plasmid with SssI and then cloned back the purified fragment corresponding to the LTR in the parental reporter vector pGL3basic (or pREP10), thereby generating pLTRmSssIme (or pLTR*mSssIme).
RNA Purification and Analyses of Transcripts
Total RNA samples were extracted from 1 × 106 cells using the RNeasy Plus mini kit (Qiagen) according to the manufacturer's instructions and digested with TURBO DNase I (TURBO DNA-freeTM kit, Ambion) to ensure the removal of genomic DNA. First strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen). Quantitative real-time PCR (qPCR) reactions were performed using MESA GREEN qPCR MasterMix (Eurogentec), and 96-well optical reaction plates were read in an Applied Biosystems StepOnePlus real-time PCR system (comparative Ct (ΔΔCt) quantification method). The following sets of primers were used for amplification of the BLV 5′-LTR and the β-actin genes, respectively: Fw, 5′-GAGACCCTCGTGCTCAGCT-3′, and Rv 5′-CAGAAGGTCTCGGGAGCAA-3′; and Fw 5′-TCCACTGCTCCTGTCTTCGA-3′, and Rv 5′-GATCTTTTCCCGTCCCAAAGT-3. Primer sequences used to quantify DNMT1, DNMT3A, and DNMT3B mRNAs were as follows: DNMT1 (Fw, 5′-ACGCTAACGAATCTGGCTTTG-3′; Rv, 5′-CGCGTTTACAGTACACGCTG-3′); DNMT3A (Fw, 5′-CTGGGTCATGTGGTTCGG-3′; Rv, 5′-GCACTGCAAAACGAGCTCAG-3′); and DNMT3B (Fw, 5′-TTTGGCGATGGCAAGT TCT-3′; Rv, 5′-GGCCAAATTAAAGTGCTGGC-3′).
ELISA
After culture, the cells were harvested by centrifugation, and the supernatants were removed. The BLV p24 protein in the supernatants was titrated by an immunoenzymatic capture technique as described by Portetelle et al. (45).
RESULTS
Activation of BLV Promoter Transcriptional Activity by Inhibitors of DNA Methylation
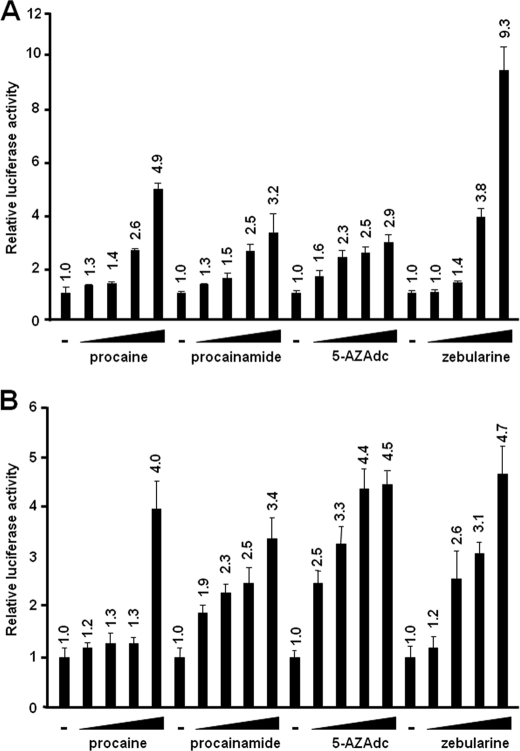
Our laboratory had previously demonstrated that histone acetylation is an important chromatin modification for TaxBLV-dependent and -independent transcriptional regulation of BLV (26–28). To further study the molecular mechanisms underlying the epigenetic control of BLV expression, we examined the potential role of DNA methylation in BLV transcriptional activity. To this end, we tested the effects of DNA methylation inhibitors on transcriptional activity of the BLV promoter (the 5′-LTR) in transient transfection experiments (Fig. 1). A pLTRmSssIme reporter plasmid containing the firefly luciferase (luc) gene under the control of the complete BLV 5′-LTR (nucleotides −211 to +320) (24), which had been methylated previously in vitro by the Sss1 methyltransferase (see “Experimental Procedures”), was transiently transfected into the human B-lymphoid Raji cell line. Transfected cells were mock-treated or treated with increasing concentrations of procaine (0–10 mm), procainamide (0–10 mm), 5-aza-2′-deoxycytidine (5-AZAdc) (0–200 nm), or zebularine (0–1 mm) for 72 h and assayed for luciferase activity. As shown in Fig. 1A, the BLV LTR was activated in a dose-dependent manner by each of the four DNA methylation inhibitors used. pLTRmSssIme presented procaine inductions varying from 1.3- to 4.9-fold, procainamide inductions varying from 1.3- to 3.2-fold, 5-AZAdc inductions varying from 1.6- to 2.9-fold, and zebularine inductions varying from 1.0- to 9.3-fold. Similar results were observed using the human B-lymphoid DG75 cell line (data not shown). Bisulfite sequencing analyses revealed that the increased transcriptional activity of the BLV LTR following treatment of the cells with each of the four DNA methylation inhibitors used was correlated with a partial CpG demethylation in the U3 and R regions of the BLV LTR (data not shown).
FIGURE 1.
Activation of BLV LTR activity by DNA methylation inhibitors. A, Raji cells were transiently transfected using the DEAE-dextran procedure with 500 ng of pLTRmSssIme, where the BLV LTR had previously been methylated in vitro by the SssI methyltransferase. At 22 h after transfection, cells were mock-treated or treated with increasing concentrations of procaine (0, 2.5, 5, 7.5, and 10 mm), procainamide (0, 2.5, 5, 7.5, and 10 mm), 5-AZAdc (0, 50, 100, 150, and 200 nm), or zebularine (0, 0.05, 0.1, 0.25, and 0.5 mm). Luciferase activities were measured in cell lysates 96 h after transfection and normalized to protein concentrations. Results are presented as histograms indicating the procaine, procainamide, 5-AZAdc, and zebularine inductions of pLTRmSssIme with respect to its basal activity, which was assigned a value of 1. Means ± S.E. are shown. Results from a representative experiment of three independent transfections are shown. B, Raji cells were transiently transfected using the DEAE-dextran procedure with 500 ng of the episomal vector pLTR*mSssIme, where the BLV LRT had previously been methylated in vitro by the SssI methyltransferase. At 22 h posttransfection, cells were mock-treated or treated with increasing concentrations of procaine (0, 0.1, 0.5, 1, and 10 mm), procainamide (0, 0.1, 0.5, 1, and 10 mm), 5-AZAdc (0, 250, 500, 1000, and 2500 nm), or zebularine (0, 0.05, 0.075, 0.1, and 1 mm). Luciferase activities were measured in cell lysates 96 h after transfection and normalized to protein concentrations. Results are presented as histograms indicating the procaine, procainamide, 5-AZAdc and zebularine inductions of pLTR*mSssIme with respect to its basal activity, which was assigned a value of 1. Means ± S.E. are shown. The results from a representative experiment of three independent transfections are shown.
Because transiently transfected DNA does not always form proper chromatin structure, we next wanted to confirm the above described transient transfection results in a nucleosomal context. To this end, we used an episomally replicating BLV LTR-luc reporter plasmid (pLTR*wt(direct)-luc), described previously by our laboratory from a modified pREP10 episomal vector that contained the Epstein-Barr virus replication origin and encoded nuclear antigen EBNA-1 (27). Several studies have reported that pREP-based episomal constructs display the hallmarks of proper chromatin structure when transiently transfected into cells (46–49). We transiently transfected into Raji cells the episomally replicating pLTR*mSssIme plasmid, where the BLV promoter had been methylated previously in vitro by the Sss1 methyltransferase. Transfected cells were mock-treated or treated with increasing concentrations of the four precited DNA methylation inhibitors and assayed for luciferase activity. As shown in Fig. 1B, the BLV promoter was activated in a dose-dependent manner by procaine (up to 4.0-fold), procainamide (up to 3.4-fold), 5-AZAdc (up to 4.5-fold), and zebularine (up to 4.7-fold). Moreover, a cellular model in which a BLV 5′-LTR-luciferase cassette was stably integrated into the B-lymphoid DG75 cell line (construction named clone 27; its characterization is part of a separate, unpublished study from our laboratory) was also used to address the effect of DNA methylation inhibition. Using clone 27, we observed that increasing concentrations of DNA methylation inhibitors enhanced BLV 5′-LTR-directed luciferase expression in a dose-dependent manner (data not shown). These results demonstrate that the BLV LTR-directed gene reporter expression is activated in a dose-dependent manner in response to selective DNA methylation inhibitors in the context of non-episomal and episomal LTR methylated constructs, thereby suggesting a role of DNA methylation in BLV transcriptional regulation.
Hypermethylation of the U3 and R Regions in the BLV 5′-LTR in L267 Lymphoma-derived B-cell Line
To further examine the potential role of DNA methylation in transformation-associated BLV silencing, we investigated the DNA methylation status of the 5′-LTR in the context of integrated BLV proviruses. To this end, we used three different BLV-infected cell lines representing two distinct viral transcriptional states: the YR2 and L267 cell lines, which represent the silent/nonproductive state, and the L267LTaxSN cell line, which represents the productive state. On the one hand, the YR2 cell line is an ovine B-cell line, which was cloned from leukemic B cells of a BLV-infected sheep (10, 36, 37, 50). This cell line contains a single monoclonally integrated silent provirus in which two Glu-to-Lys amino acid substitutions in the TaxBLV protein impair the infectious potential of the integrated provirus (33). The L267 cell line is a B-cell line isolated from sheep S267, a BLV-infected sheep injected with naked proviral DNA of an infectious BLV variant (33–35). Provirus expression is completely suppressed in L267 lymphoma B cells, as demonstrated conclusively by their incapacity to generate infection in the in vivo sheep model, and the tumor-derived L267 provirus displays a wild-type sequence that is identical to the infectious provirus used for sheep S267 inoculation (34, 35). In contrast to the YR2 cell line, which represents a defective latency state, the L267 cell line represents a true latency state, i.e. a reversible transcriptionally silent but replication-competent provirus. On the other hand, the L267LTaxSN cell line is a virus-expressing cell line resulting from transduction of the native L267 cell line with a retroviral vector pLTaxSN expressing a wild-type TaxBLV protein. Transduction of this TaxBLV coding vector restores viral expression from the silent integrated provirus (34).
To verify the productive versus nonproductive state of our cell lines, we first measured BLV transcripts by reverse transcription qPCR in each of the three cell lines. We observed very low levels of viral transcripts in the YR2 and L267 cell lines, as compared with the L267LTaxSN cell line, which abundantly produces BLV mRNAs (Fig. 2A). Similarly, measurement of p24 major capsid antigen by ELISA in the supernatants of all cultured cell lines showed a dramatic increase in the production of this structural antigen in the L267LTaxSN cell line compared with the silent L267 and YR2 cell lines (Fig. 2B). These results indicate that exogenous TaxBLV is capable of restoring viral expression from silent integrated proviruses, in agreement with previous studies (34).
To determine the methylation status of each CpG dinucleotide in the 5′-LTR of integrated BLV proviruses, we used the sodium bisulfite sequencing technique to analyze genomic DNA from the YR2, L267, and L267LTaxSN cell lines. To distinguish between the 5′- and 3′-LTRs, which have the same nucleotide sequence, we used reverse primers located in the gag gene (Fig. 2C, primers NB and ND) or forward primers located in the X region (primers NE and NG) for the nested PCR, respectively. As shown in Fig. 2C, the U3 region and the 5′-half of the R region of the 5′-LTR in L267 cells were densely methylated. Indeed, six consecutive CpG dinucleotides in the U3 region and nine consecutive CpG dinucleotides in the R region, overlapping the transcription initiation site, were strongly methylated. Among these methylated CpGs, the −154 CpG and the −129 CpG (located in the distal CRE1 and middle CRE2 sites, respectively) were methylated in 100% of the cloned sequences tested for the L267 cell line. These two methylated −154 and −129 CpGs are indicated by asterisks in Fig. 2C. In the L267LTaxSN cells, only the 5′-half of the R region was methylated although to a lesser extent than in the L267 cells. Remarkably, the 5′-LTR in the YR2 cell line was completely unmethylated.
In all retroviruses, LTRs localized at the 5′ and 3′ extremities of the proviral genome have identical nucleotide sequences but display different functions; the 5′-LTR functions as a transcriptional promoter, whereas the 3′-LTR has a polyadenylation function. Therefore, we addressed the question of differential CpG methylation patterns in the sequentially identical but functionally different BLV LTRs. Our bisulfite sequencing analysis of the 3′-LTR revealed a lack of CpG methylation in the three tested cell lines: YR2, L267, and L267LTaxSN (Fig. 2C).
In conclusion, our results indicate that the BLV true latency state correlates with CpG hypermethylation in the 5′-LTR U3 and R regions of the integrated proviral DNA, directly demonstrating for the first time the association of this epigenetic modification with a specific viral expression state. This could be a viral strategy to escape host immune response and allow tumor onset and propagation. Importantly, these methylation marks were not observed in the YR2 proviral system, representative of a defective latency state.
Ectopically Expressed DNMT1 and DNMT3A, but Not DNMT3B, Down-regulate BLV Promoter Activity
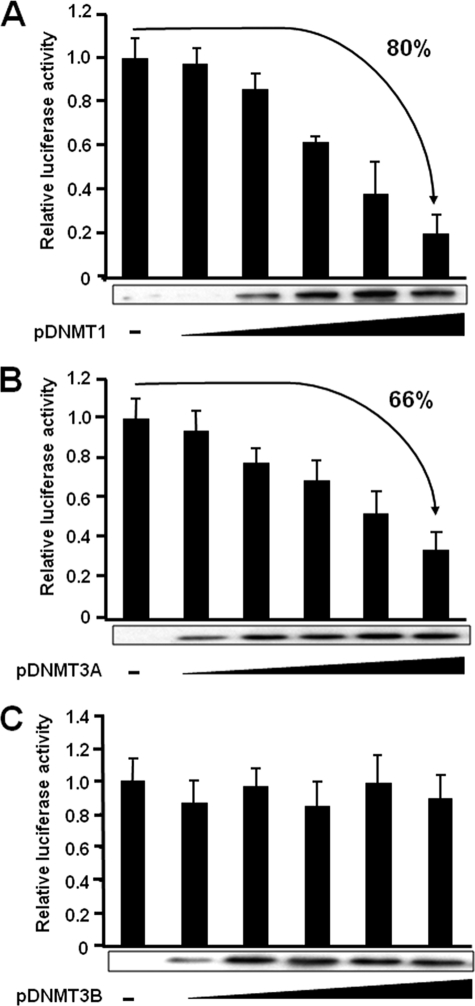
To address the role of DNMTs in transcriptional regulation of the BLV promoter, transient cotransfections of Raji cells were performed with the pLTRwt-luc reporter construct and increasing amounts of expression vectors coding for DNMT1, DNMT3A, or DNMT3B. As shown in Fig. 3A and B, DNMT1 and DNMT3A strongly repressed basal LTR-directed gene expression, suggesting that these DNMTs could negatively regulate BLV promoter activity. In contrast, DNMT3B had no effect on reporter gene expression (Fig. 3C), suggesting a certain functional specificity of DNMTs in the transcriptional regulation of the BLV promoter. Western blot experiments allowed us to verify the efficiency of each DNMT overexpression in correlation with the level of BLV 5′-LTR promoter activity (Fig. 3). These results confirmed that the inability of DNMT3B to repress BLV 5′-LTR activity was not due to a lack of DNMT3B protein expression (Fig. 3C).
FIGURE 3.
Response of the BLV promoter to ectopically expressed DNA methyltransferases. Raji cells were transiently cotransfected using the DEAE-dextran procedure with 500 ng of pLTRwt-luc and increasing amounts (0, 50, 100, 250, 500, and 1000 ng of plasmid DNA) of the DNMT1, DNMT3A, and DNMT3B expression vectors (A, B, and C, respectively). To maintain the same amount of transfected DNA and avoid squelching artifacts, the different amounts of DNMT expression vector cotransfected were complemented to 1000 ng of DNA by using the empty vector. Luciferase activities measured in cell lysates 44 h after transfection were normalized to protein concentrations. The results are presented as histograms indicating the induction by DNMT1, DNMT3A, or DNMT3B with respect to the activity of the reporter construct in the absence of DNMT, which was assigned a value of 1. Means ± S.E. are indicated. A representative experiment of four independent transfections is shown. Nuclear extracts were prepared and analyzed by Western blot with an anti-c-Myc antibody to detect DNMT1, DNMT3A, and DNMT3B expression levels. These results are presented in parallel with the transfection results for each DNMT.
In addition, we performed ChIP experiments to evaluate in vivo the recruitment of DNMTs to the BLV promoter using chromatin samples prepared from the transiently transfected cells. We immunoprecipitated the chromatin either with an antibody directed against the c-Myc epitope fused to the different DNMTs present in the recombinant expression vectors we had used in the overexpression experiments or with antibodies directly targeting DNMT1, DNMT3A, or DNMT3B. However, we did not succeed in detecting any statistically relevant ChIP signal compared with the negative control (consisting of a purified IgG antibody). In additional ChIP experiments, we used chromatin samples from YR2 and L267 cells to study DNMT recruitment in vivo to the hypomethylated and hypermethylated BLV promoters, respectively. Despite many attempts, we could not detect in vivo the recruitment of these DNMTs to the BLV 5′-LTR region in our ChIP assays neither with the chromatin extracted from the transiently transfected cells nor with the chromatin prepared from the BLV-infected cell lines YR2 and L267 (data not shown).
Moreover, we evaluated the DNA methylation profile of the BLV LTR in the LTR-reporter constructs following overexpression of each DNMT. Methylation-sensitive restriction analyses revealed that overexpression of DNMT1 and DNMT3A (and to a lesser extent DNMT3B) increased CpG methylation in the BLV LTR. However, the results we obtained by the bisulfite sequencing technique revealed in each case a very weakly methylated profile (data not shown), which did not correspond to the methylation pattern we observed in the infected cell lines (see Fig. 2C), thereby suggesting that the establishment of the DNA methylation profile during or after natural integration into the host cell genome can not be recapitulated using LTR-reporter constructs.
Taken together, our results suggest that the activity of the BLV promoter is negatively regulated by the DNA methyltransferases DNMT1 and DNMT3A but not by DNMT3B. However, we could not demonstrate the recruitment of these enzymes in vivo in the BLV 5′-LTR region.
Synergistic Activation of BLV Promoter by TaxBLV and DNA Methylation Inhibitors
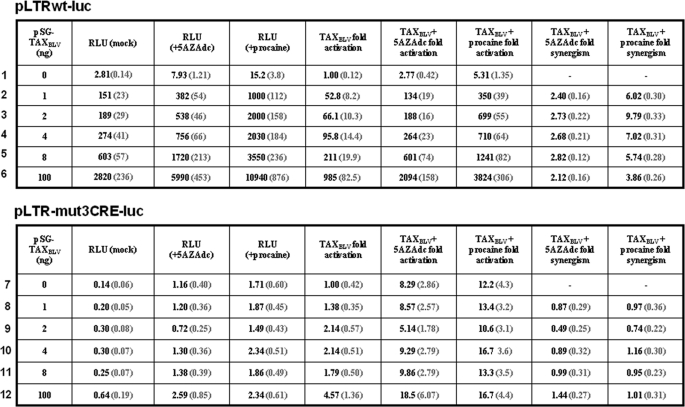
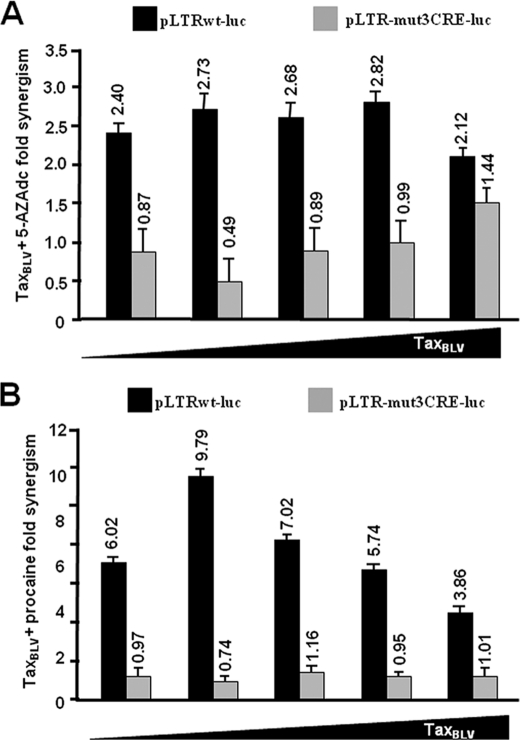
To examine the potential link between DNA methylation and TaxBLV transactivation, we tested the effect of two DNA methylation inhibitors (5-AZAdc and procaine) on basal and TaxBLV-induced BLV LTR activity. To this end, human B-lymphoid Raji cells were transiently cotransfected with the pLTR-luc reporter construct (methylated in vitro by Sss1 before transfection) and with increasing amounts of a TaxBLV expression vector, pSG-TaxBLV. Twenty-two hours posttransfection, transfected cells were mock-treated or treated with either 5-AZAdc (250 μm) or procaine (7.5 mm) for 72 h and assayed for luciferase activity (Table 1). As expected, in the absence of 5-AZAdc or procaine, TaxBLV transactivated the BLV promoter in a dose-dependent manner up to 985-fold (Table 1, TaxBLV fold activation, lines 2–6). Treatment of transfected cells with 5-AZAdc or procaine alone resulted in a 2.77- or 5.31-fold activation of luciferase expression, respectively (Table 1, line 1). Remarkably, when cells were both cotransfected with increasing amounts of the TaxBLV expression vector and treated with 5-AZAdc (or procaine), a strong synergism was observed between TaxBLV and these DNA methylation inhibitors, resulting in transactivations ranging from 134- to 2094-fold in the presence of 5-AZAdc (Table 1, lines 2–6, TaxBLV + 5-AZAdc fold activation) and from 350- to 3824- fold in the presence of procaine (Table 1, lines 2–6, TaxBLV + procaine fold activation). Transcriptional activators synergize when their combination produces a transcriptional rate that is greater than the sum of the effects produced by individual activators (51). Transfection of 1 ng of pSG-TaxBLV led to a 52.8-fold stimulation of transcription in the absence of DNA methylation inhibitors, whereas in the presence of 5-AZAdc or procaine, it led to a 134- or 350-fold stimulation, respectively (Table 1, line 2). This amount of transcription is 2.40- or 6.02-fold greater than the sum of the effect produced by each activator separately (2.77 + 52.8 or 5.31 + 52.8) (Table 1, line 2). Likewise, transfection of 8 ng of pSG-TaxBLV in the presence of 5-AZAdc (or procaine) stimulated transcription 601 (or 1241)-fold, corresponding to a 2.82 (or 5.74)-fold synergism (Table 1, line 5). The data shown in Table 1 are presented in Fig. 4 as histograms indicating the synergistic activation (-fold) by TaxBLV and 5-AZAdc and by TaxBLV and procaine (Fig. 4, A and B, respectively). This synergism between TaxBLV and 5-AZAdc (or procaine) persisted even at saturating amounts of the TaxBLV transactivator (data not shown), indicating that the observed effect was not the consequence of increased TaxBLV expression because of activation of simian virus 40 (SV40) promoter by 5-AZAdc or procaine. Moreover, we confirmed by Western blot experiments that 5-AZAdc or procaine did not increase the amount of TaxBLV protein by activating the SV40 promoter (data not shown). Similar results were observed using another B-lymphoid cell line (the DG75 cell line; data not shown).
TABLE 1.
Synergistic activation of BLV promoter activity by TaxBLV and inhibitors of DNA methylation
Raji cells were transiently cotransfected using the DEAE-dextran procedure with 500 ng of either the pLTRwt-luc (lines 1–6) or pLTR-mut3CRE-luc (lines 7–12) (both methylated in vitro by the SssI methyltransferase before transfection) and with increasing amounts of pSG-TaxBLV (from 0 to 100 ng of plasmid DNA). To maintain the same amount of transfected DNA and to avoid squelching artifacts, the different amounts of pSG-TaxBLV cotransfected were complemented to 100 ng of DNA by using the pSG5 empty vector. Twenty-two hours posttransfection, cells were mock-treated or treated with either 5-AZAdc (250 μm) or procaine (7.5 mm) for 72 h. Luciferase activities were measured in cell lysates 96 h after transfection and normalized with respect to protein concentrations. Results are presented as relative light units (RLU), TaxBLV fold activation, TaxBLV + 5-AZAdc fold activation, and TaxBLV + procaine fold activation of the reporter constructs (pLTRwt-luc and pLTR-mut3CRE-luc) with regard to their respective basal activity level, which was arbitrarily set at a value of 1. The TaxBLV + 5-AZAdc (or TaxBLV + procaine) fold synergism was determined as described previously (51) using the following formula: [Fold activation by TaxBLV + 5-AZAdc (or TaxBLV + procaine)]/{[fold activation by TaxBLV alone] + [fold activation by 5-AZAdc (or procaine) alone]}. Values represent the means of triplicate samples with S.E. indicated in parentheses. An experiment representative of four independent transfections is shown.
FIGURE 4.
-Fold activation of the BLV promoter activity by TaxBLV and inhibitors of DNMTs. The histograms represent -fold inductions of BLV promoter activity by TaxBLV and 5-AZAdc (A) or procaine (B) as detailed in Table 1. Values represent the means of triplicate samples ± S.E. are indicated. An experiment representative of four independent transfections is shown.
Synergistic activation by ectopically expressed TaxBLV and inhibitors of DNA methylation required intact CRE binding sites in the three BLV TxREs, because point mutations in these three CRE motifs abrogated the synergistic effect (pLTR-mut3CRE-luc; Table 1, lines 7–12, and Fig. 4). This implied that the synergistic effect was mediated by interactions at the CRE motifs and not at the otherwise intact LTR BLV DNA sequences. In conclusion, these results demonstrate that the DNA methylation inhibitors 5-AZAdc and procaine synergistically enhance the transcriptional activity of the BLV promoter mediated by TaxBLV in a CRE-dependent manner.
Methylation at −154 CpG and −129 CpG in BLV 5′-LTR CRE1 and CRE2 Sites, Respectively, Impairs CREB, CREM, and ATF-1 Binding
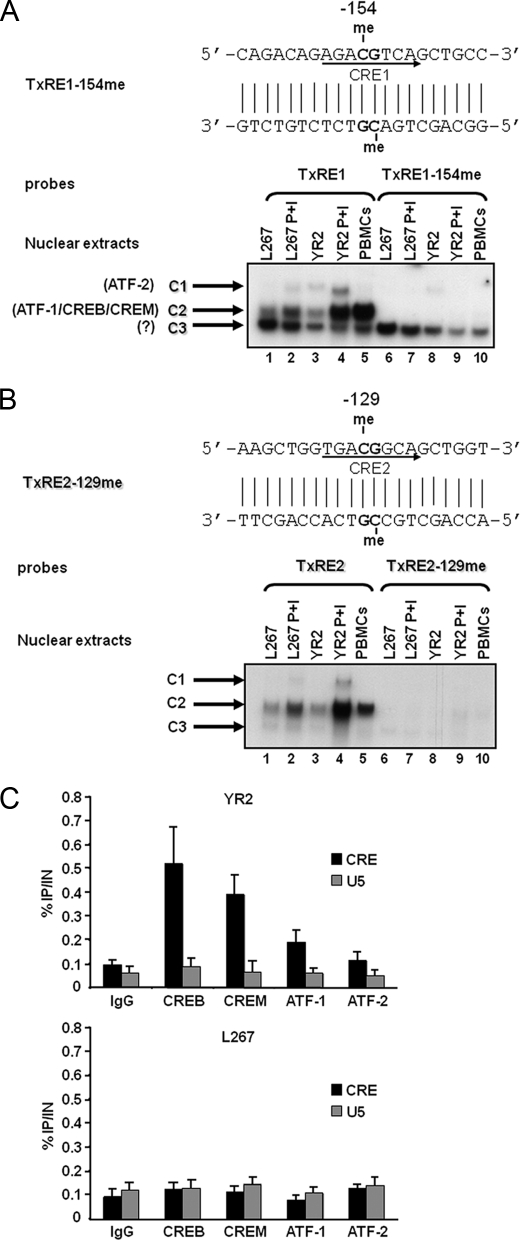
Our bisulfite sequencing results indicate that the central CpGs located in the 5′-LTR CRE1 and CRE2 sites (i.e. −154 CpG and −129 CpG) are hypermethylated in the integrated BLV provirus of L267 cells (Figs. 2C and 5). This hypermethylation correlates with the transcriptionally silent viral state in these cells. We investigated the in vitro effect of methylation at the −154 CpG (distal CRE1 site) and at the −129 CpG (middle CRE2 site) on transcription factor binding by EMSAs. For these studies, we used oligonucleotide probes corresponding to the TxRE1 (nucleotides −164 to −144) or to the TxRE2 (nucleotides −139 to −119) sites of the BLV 5′-LTR, which were either unmethylated (referred to as TxRE1 or TxRE2) or methylated at the −154 or −129 CpG (referred to as TxRE1–154me or TxRE2–129me, respectively). These probes were incubated with nuclear extracts from the latently BLV-infected B-cell line L267 treated (Fig. 6, A and B, lanes 2 and 7) or not (lanes 1 and 6) with a combination of PMA + ionomycin (P+I), two known activators of BLV expression (26, 52, 53); from the defective cell line YR2 treated (lanes 4 and 9) or not (lanes 3 and 8) with PMA + ionomycin; or from PBMCs derived from a BLV-infected sheep (BLV-infected sheep M298 presenting a persistent lymphocytosis) (lanes 5 and 10). When the EMSAs were carried out with the unmethylated oligonucleotide TxRE1 and TxRE2 probes, we observed, as reported previously by our laboratory (17), a minor and two major retarded bands (called C1, C2, and C3, respectively) (Fig. 6, A and B, lanes 1–5). Our laboratory had previously reported that the C1 complex contains the ATF-2 transcription factor and the C2 complex contains ATF-1, CREB, and CREM proteins (14, 15, 17, 27). Supershift experiments did not identify the nature of the proteins present in complex C3 (17). Treatment of YR2 or L267 cells with PMA + ionomycin increased the intensity of the retarded DNA-protein complex C2 as compared with mock-treated cells (Fig. 6, A and B, compare lanes 1 and 2 and lanes 3 and 4). Moreover, we observed a strong binding of CREB/CREM/ATF-1 proteins to the TxRE1 and TxRE2 probes when these probes were incubated with nuclear extracts prepared from BLV-infected PBMCs (Fig. 6, A and B, lanes 5). Remarkably, when the methylated TxRE1–154me probe or the methylated TxRE2–129me probe was used, we observed the complete disappearance of complex C2, even with nuclear extracts from infected PBMCs or from PMA + ionomycin-treated YR2 or L267 cells (Fig. 6, A and B, compare lanes 1–5 with lanes 6–10). Following PMA + ionomycin treatment of YR2 cells, complex C1 seemed to migrate slower than in mock-treated cells (Fig. 6A, compare lanes 3 and 4), but we could not identify by supershift experiments any potential additional (co)factor(s) associated with complex C1 following PMA + ionomycin treatment (data not shown). However, complex C1 was not observed when PBMC nuclear extracts were used (Fig. 6, A and B, lanes 5), suggesting a minor function of this complex.
FIGURE 5.
Schematic representation of the BLV 5′-LTR aligned with the CpG methylation pattern of the L267 cell line. The transcription initiation site at the U3–R junction of the 5′-LTR (mRNA start site is at +1 nt) is indicated by an arrow. The TxREs are three major transcriptional enhancer sequences of 21 bp, which interact with the cellular transcription factors CREB, CREM, ATF-1, and ATF-2 and which are required for the transcriptional activation of the BLV LTR by the virus-encoded TaxBLV transactivator. Each of the 21-bp enhancers contains a sequence homologous to the consensus E-box binding motif (E-box1, E-box2, and E-box3) overlapping an imperfect CRE (CRE1, CRE2, and CRE3). The U3 region also contains a glucocorticoid-responsive element (GRE) and a PU.1/Spi-B site. A USF-1/USF-2 binding site (E-box 4) and an interferon regulatory factor (IRF-1/IRF-2)-binding site are located in the R region and the U5 region, respectively. The results of bisulfite genomic sequencing that we obtained in the L267 cell line are aligned with the schematic representation of the 5′-LTR. The complete nucleotide sequence of the U3 region is shown with the transcription factor binding sites indicated by arrows and all of the CpG dinucleotides shown in boxes. me, methylated CpGs. DAS, downstream activating sequence.
FIGURE 6.
Inhibition of CREB/CREM/ATF-1 transcription factor binding by methylation at the −154 and −129 CpGs. A, EMSA analysis of nuclear factors interacting with the methylated or unmethylated TxRE1 probes. The nucleotide sequence of the methylated TxRE1–154me double-stranded oligonucleotide is shown with the CRE1 site indicated by an arrow on the coding strand and with the methylated cytosine residues indicated by the abbreviation me. The unmethylated TxRE1 double-stranded oligonucleotide (lanes 1–5) or its methylated version, TxRE1–154me (lanes 6–10), was used as probe and incubated with nuclear extracts from the BLV latently infected B-cell line L267 (30 μg) treated (lanes 2 and 7) or not (lanes 1 and 6) with PMA + ionomycin (P+I) for 30 min, from the defective cell line YR2 (15 μg) treated (lanes 4 and 9) or not (lanes 3 and 8) with PMA + ionomycin, or from BLV-infected ovine PBMCs (10 μg) (lanes 5 and 10). Sequence-specific retarded bands of interest are shown. A minor and two major protein-DNA complexes were observed and named C1, C2, and C3, respectively (indicated by arrows). The presence of ATF-2 in the minor protein-DNA complex C1 and the presence of ATF-1, CREB, and CREM in the major protein-DNA complex C2 were reported previously by our laboratory (17). B, EMSA analysis of nuclear factors interacting with the methylated or unmethylated TxRE2 probes. The nucleotide sequence of the methylated TxRE2–129me double-stranded oligonucleotide is shown with the CRE2 site indicated by an arrow on the coding strand and with the methylated cytosine residues indicated by the abbreviation me. The unmethylated TxRE2 double-stranded oligonucleotide (lanes 1–5) or its methylated version, TxRE2–129me (lanes 6–10), was used as probe and incubated with nuclear extracts from the BLV latently infected B-cell line L267 (30 μg) treated (lanes 2 and 7) or not (lanes 1 and 6) with PMA + ionomycin for 30 min; from the defective cell line YR2 (15 μg) treated (lanes 4 and 9) or not (lanes 3 and 8) with PMA + ionomycin; or from BLV-infected ovine PBMCs (10 μg) (lanes 5 and 10). Sequence-specific retarded bands of interest are shown. A minor and two major protein-DNA complexes were observed (C1, C2, and C3, respectively; indicated by arrows). The presence of ATF-2 in the minor protein-DNA complex C1 and the presence of ATF-1, CREB, and CREM in the major protein-DNA complex C2 were reported previously by our laboratory (17). C, CREB/CREM/ATF-1 are recruited in vivo to the BLV 5′-LTR CRE region. YR2 and L267 cells were cross-linked for 10 min at room temperature with 1% formaldehyde. To detect chromosomal flanking regions, pellets were sonicated to obtain DNA fragments of an average size of 400 bp. Chromatin immunoprecipitations were performed with an antibody directed against CREB, CREM, ATF-1, or ATF-2. To test aspecific binding to the beads, a purified IgG was used as a control for immunoprecipitation. Quantitative PCR reactions were performed with oligonucleotide primers hybridizing in a region overlapping the three CRE sites of the BLV LTR or in the U5 region of the BLV LTR, where no CRE binding site has been reported previously. -Fold enrichments were calculated as the percentage of input values following the formula: Immunoprecipitated DNA (IP)/total DNA (IN). Values represent the means of triplicate samples ± S.E. An experiment representative of three independent ChIP assays is shown.
To confirm these results, unlabeled double-stranded oligonucleotides were prepared and used as competitors in the EMSAs with the unmethylated TxRE1 or TxRE2 probe and nuclear extracts from BLV-infected PBMCs (supplemental Fig. S1, A or B, respectively). As shown previously by our laboratory (17), formation of complex C2 was competed out by molar excesses of the TxRE1 or TxRE2 unlabeled homologous oligonucleotides but not by the same molar excesses of a heterologous oligonucleotide of unrelated sequence. Moreover, the C2 complex was not or was weakly competed out by the same molar excesses of methylated TxRE1–154me (supplemental Fig. S1B) or TxRE2–129me (supplemental Fig. S1A) unlabeled oligonucleotides, respectively, thereby confirming that formation of complex C2 was dependent on the presence of an unmethylated TxRE1 or TxRE2 sequence (supplemental Fig. S1). Altogether, these results indicate that methylation at the −154 CpG or at the −129 CpG abolish in vitro binding of the CREB/CREM/ATF-1 transcription factors to the CRE1 or CRE2 site, respectively.
To confirm these results in vivo, we conducted ChIP experiments using the YR2 and L267 cell lines, representative of defective and true latency, respectively. These two cell lines allowed us to compare CREB/CREM/ATF recruitment to the hypomethylated YR2 BLV 5′-LTR with their recruitment to the hypermethylated L267 BLV 5′-LTR. We used specific antibodies directed against CREB, CREM, ATF-1, and ATF-2 proteins and a purified IgG antibody as a control. Two different probes were designed in the BLV 5′-LTR to compare the recruitment of these proteins to the TxRE site region with their recruitment to the U5 region in which no CRE sites had been reported previously. As shown in Fig. 6C, we observed the recruitment of transcription factors CREB and CREM, and to a lesser extent ATF-1, to the hypomethylated CRE region of YR2 5′-LTR. In contrast, no CREB/CREM/ATF recruitment was observed to the hypermethylated corresponding region in the L267 cell line (Fig. 6C). Moreover, in both cell lines, the ATF-2 protein, which is part of complex C1 (see the EMSAs in Fig. 6, A and B, and as reported previously by our laboratory (17)), did not seem to be recruited in vivo to the 5′-LTR region (Fig. 6C). This latter result is in agreement with our in vitro binding studies showing that complex C1, which contains the ATF-2 transcription factor, is not the major retarded band observed in gel shift assays using the TxRE probes (Fig. 6, A and B), suggesting a less important function of ATF-2 compared with that of CREB/CREM/ATF-1. Altogether, these results indicate that CpG methylation impairs in vivo CREB/CREM/ATF-1 binding to the CRE binding sites of the BLV 5′-LTR.
Methylation at the −129 CpG in the CRE2 Site Down-regulates BLV 5′-LTR Transcriptional Activity
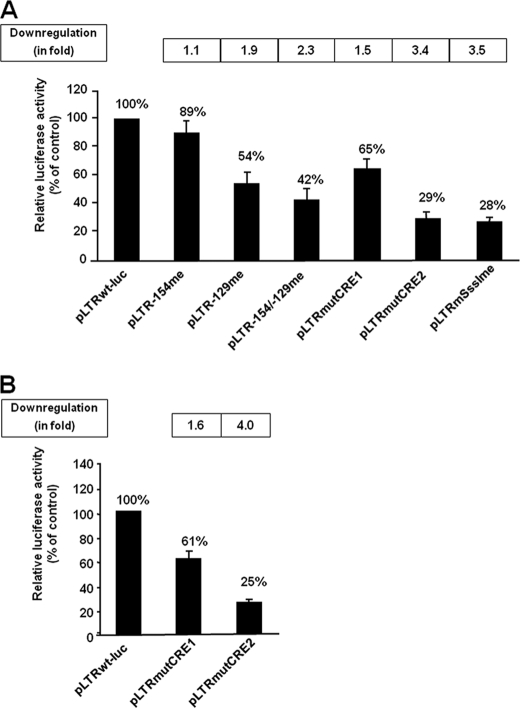
To assess the transcriptional regulatory function of methylation at the −154 and −129 CpGs, we modified the pLTRwt-luc vector so that the −154 CpG and/or the −129 CpG were methylated by a PCR-mediated mutagenesis protocol (see “Experimental Procedures” and Ref. 44). As controls, we also generated, by the same technique, an unmethylated pLTRwt-luc as well as unmethylated mutant reporter constructs named pLTRmutCRE1 or pLTRmutCRE2, which contained point mutations abolishing CREB/CREM/ATF-1 transcription factor binding to the CRE1 or CRE2 site, respectively. Directly after purification, the PCR products from these site-directed mutagenesis experiments were transiently transfected into the human epithelial HeLa cell line (which presents high transfection efficiency). At 44 h posttransfection, cells were lysed and assayed for luciferase activity. As shown in Fig. 7A, methylation at the −129 CpG caused a 46% decrease in luciferase activity. Methylation at the −154 CpG did not affect basal BLV LTR-directed gene expression. Moreover, when combined with methylation at the −129 CpG, methylation at the −154 CpG did not further decrease BLV promoter activity compared with what we observed with the plasmid methylated only at the −129 CpG.
FIGURE 7.
Down-regulation of BLV promoter activity by site-specific methylation. A, using site-directed mutagenesis we generated BLV promoter luciferase constructs with site-specific methylation at the −154 CpG or/and −129 CpG (pLTR-154me, pLTR-129me, and pLTR-154/-129me, respectively) as well as its unmethylated counterpart, pLTRwt-luc. By the same technique, we generated mutated BLV promoter luciferase constructs containing point mutation abolishing transcription factor binding to the CRE1 or CRE2 motif (pLTRmutCRE1 or pLTRmutCRE2, respectively). The products from these mutagenesis reactions (500 ng of DNA) were transfected directly into HeLa cells using the FuGENE procedure. Each transfection included 50 ng of the internal control plasmid, pRL-TK, in which the herpes simplex virus thymidine kinase promoter drives Renilla luciferase gene expression. Luciferase activities (firefly and Renilla) were measured in cell lysates 44 h after transfection. The results are expressed as luciferasefirefly/luciferaseRenilla. As an additional control, we generated a pLTRwt-luc derivative that was hypermethylated only in the LTR (i.e. in which every CpG of the BLV LTR, but no CpG elsewhere in the pLTRwt-luc plasmid, was methylated in vitro by SssI methyltransferase) and named pLTRmSssIme. The results are presented as histograms indicating the luciferase activity of each reporter construct relative to that measured with the pLTRwt-luc vector, which was arbitrarily assigned an activity value of 100%. The down-regulation of BLV promoter activity by methylation(s) or by mutation(s) is also indicated (-fold) at the top of the histogram representation. Means ± S.E. from five independent transfections performed with different DNA products are shown. B, HeLa cells were transiently cotransfected using the FuGENE procedure with 500 ng of pLTRwt-luc, pLTRmutCRE1, or pLTRmutCRE2 and 50 ng of the internal control plasmid, pRL-TK. Here, in contrast to the plasmids used in A, the reporter plasmids were amplified in bacteria prior to transfection. Luciferase activities (firefly and Renilla) were measured in cell lysates 44 h after transfection. The results are expressed as Luciferasefirefly/luciferaseRenilla. The results are presented as histograms indicating the luciferase activity of each reporter construct relative to that measured with the pLTRwt-luc vector, which was arbitrarily assigned a value of 100% of activity. The down-regulation of BLV promoter activity by mutations is indicated (in -fold) at the top of the histogram representation. Means ± S.E. from triplicate samples are represented. An experiment representative of two independent transfections is shown.
Point mutation in the CRE1 site (pLTRmutCRE1) or in the CRE2 site (pLTRmutCRE2) caused a 35 or 71% decrease, respectively, of basal LTR-directed gene expression. As an additional control, we generated a pLTRwt-luc derivative that was hypermethylated only in the LTR (i.e. in which every CpG of the 5′-LTR, but no CpG elsewhere in the pLTRwt-luc plasmid, was methylated in vitro by SssI methyltransferase). This construction was named pLTRmSssIme (see “Experimental Procedures”). As shown in Fig. 7A, when transiently transfected the pLTRmSssIme exhibited a 72% decrease in luciferase activity compared with that observed with the unmethylated pLTRwt-luc construct.
To validate our method, we performed additional independent transfection experiments using the pLTRwt-luc, pLTRmutCRE1, and pLTRmutCRE2 constructs resulting from the same PCR mutagenesis technique but followed by a classical amplification step in bacteria and not directly transfected after purification of the mutagenized PCR products. These constructs were transiently transfected into HeLa cells. At 44 h posttransfection, cells were lysed and assayed for luciferase activity. As shown in Fig. 7B, mutation in the CRE1 or the CRE2 site reduced BLV promoter activity by 39 or 75%, respectively. Remarkably, these reductions were almost identical to those observed by directly transfecting the same plasmids after mutagenesis (compare pLTRmutCRE1 and pLTRmutCRE2 in Fig. 7), thereby validating the mutagenesis technique we used in Fig. 7A.
Altogether, these results demonstrate that methylation at the −129 CpG significantly reduces BLV promoter-driven reporter gene expression, whereas methylation at the −154 CpG has no effect on the BLV promoter activity. These data suggest that methylation at the −154 CpG position in the BLV promoter, even if it impairs CREB/CREM/ATF-1 transcription factor binding, does not interfere with basal transcriptional activity of this promoter ex vivo.
Transactivator Protein TaxBLV Down-regulates DNMT Expression
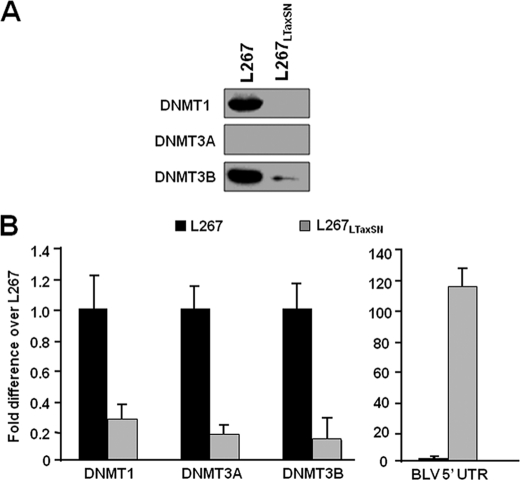
In our bisulfite sequencing experiments, we observed a partial loss of DNA methylation of the BLV 5′-LTR in L267LTaxSN cells compared with L267 cells (Fig. 2C). A possible explanation is that the transactivating TaxBLV protein could decrease the expression level of DNA methyltransferases. In order to address this issue, we quantified DNMT1, DNMT3A, and DNMT3B protein expression levels in the L267LTaxSN cell line compared with the L267 cell line by Western blot experiments. Remarkably, DNMT1 and DNMT3B protein expression was strongly down-regulated in L267LTaxSN cells compared with L267 cells (Fig. 8A). However, we could not assess the protein level of DNMT3A because no ovine antibody against DNMT3A was available commercially and the murine anti-DNMT3A antibody did not reveal any specific band in either L267 or L267LTaxSN cellular extracts. We next measured DNMT mRNA expression levels in these two cell lines by reverse transcription qPCR. As shown in Fig. 8B, we observed significant decreases in DNMT1, DNMT3A and DNMT3B mRNA levels in L267LtaxSN cells compared with L267 cells, whereas BLV transcripts were more abundant in the productive L267LtaxSN cells than in the silent L267 cells. These results suggest that TaxBLV down-regulates DNMT expression at the transcriptional level, which could, at least in part, explain the lower level of DNA methylation we observed in the L267LTaxSN 5′-LTR compared with that observed in the L267 5′-LTR.
FIGURE 8.
Down-regulation of DNMT expression levels by TaxBLV. A, nuclear extracts from the two BLV-infected cell lines, L267 and L267LTaxSN, were analyzed by Western blotting with an antibody directed against DNMT1, DNMT3B, or DNMT3A. We could not assess the protein level of DNMT3A because no ovine antibody against DNMT3A was available commercially, and the murine anti-DNMT3A antibody did not reveal any specific band in either the L267 or the L267LTaxSN cellular extracts. B, total RNA samples were extracted from L267 or L267LTaxSN cells and digested by DNase I. First strand cDNA was synthesized by reverse transcription, and qPCR reactions were performed with oligonucleotide primers amplifying DNMT1, DNMT3A, or DNMT3B transcripts and β-actin transcripts. The results are presented as the ratio of DNMT to β-actin. Means ± S.E. from triplicate samples are represented. An experiment representative of three independent experiments is shown. As a control, we performed quantitative PCR with oligonucleotide primers amplifying the 5′-UTR region of the BLV as presented in Fig. 2A.
On the basis of our results, we concluded that TaxBLV-dependent down-regulation of DNMT expression could lead to a lower level of DNA methylation in the BLV promoter region, notably in the CRE site region. The transcription factors CREB/CREM/ATF-1 could subsequently bind to their cognate sites and allow TaxBLV-mediated transactivation of the BLV 5′-LTR, thereby facilitating virus escape from latency.
DISCUSSION
BLV proviral latency appears to represent a viral strategy to escape host immune system and allow tumor development. Our laboratory had previously demonstrated the role of histone deacetylation in BLV transcriptional repression (26–29). In this report, we further studied the epigenetic control of BLV expression and showed that BLV promoter activity was induced by several DNA methylation inhibitors (such as 5-AZAdc). Moreover, ectopically expressed DNMT1 and DNMT3A, but not DNMT3B, down-regulated BLV promoter activity. However, we failed to detect any recruitment in vivo of DNMTs to the BLV promoter neither in the context of transiently transfected LTR-luc reporter constructs nor in the latently infected cell lines YR2 and L267. It should be stressed that several viral promoters have been shown to be regulated by CpG methylation (54–57), but in vivo DNMT recruitment to these viral regions has never been reported so far. This could be explained by a rapid kinetics of DNMT action or by a transient and/or weak recruitment of these enzymes to the viral promoters.
Importantly, using the sodium bisulfite sequencing technique, we showed that DNA cytosine hypermethylation in the 5′-LTR U3 and R regions of the integrated proviral DNA was associated with a BLV true latency state in the lymphoma-derived B-cell line L267 but not with a defective latency state in the YR2 cell line. Indeed, the YR2 cell line is a B-cell line characterized by a silent defective TaxBLV-mutated provirus (33, 36), whereas the L267 cell line is a transformed B-cell clone isolated from a lymphomatous BLV-infected sheep, which carries a single full-length, nondefective, transcriptionally silent provirus (36, 37). This latter cell line represents a true latency state, i.e. a reversible, transcriptionally silent, but replication-competent provirus. These results suggest that BLV gene expression is inactivated in tumoral cells by epigenetic mechanisms such as DNA methylation or by genetic alterations such as inactivating mutations in the tax gene.
Efficient BLV transcription requires the virus-encoded TaxBLV protein, which acts through three TxREs located in the 5′-LTR. The binding of CREB, CREM, and ATF transcription factors to the three imperfect CRE sites located in each TxRE has been proposed to allow TaxBLV-mediated transcriptional activation. In this report, we showed that DNA methylation inhibitors synergistically enhanced the transcriptional activation of the BLV promoter by TaxBLV in a CRE-dependent manner. Mechanistically, methylation of the CpG located at the −154 position or at the −129 position relative to the transcription start site impairs in vitro CREB/CREM/ATF-1 transcription factor binding to the distal CRE1 or middle CRE2 site, respectively. Moreover, methylation at the −129 CpG alone was sufficient to decrease by 2-fold BLV promoter-driven reporter gene expression. By ChIP experiments, we demonstrated in vivo the recruitment of the transcription factors CREB and CREM, and to a lesser extent ATF-1, to the hypomethylated CRE region of the YR2 5′-LTR, whereas we detected no CREB/CREM/ATF recruitment to the hypermethylated corresponding region in the L267 cell line, thereby indicating an inverse correlation between hypermethylation of, and in vivo CREM/CREM/ATF-1 binding to, the BLV promoter. As TaxBLV recruitment to the BLV LTR requires CREB/CREM/ATF binding to the TxREs, CpG methylation also impairs TaxBLV-mediated transactivation (see above). These findings suggest that site-specific DNA methylation of the BLV promoter represses viral transcription by directly inhibiting transcription factor binding and that CpG methylation contributes to true proviral latency, together with histone deacetylation.
The methylation status of the 5′-LTR of BLV has been controversial so far. Indeed, an earlier study using methylation-sensitive restriction enzymes has shown that the proviral DNA is hypermethylated in BLV-induced tumors (58). Later on, another study reported by Tajima et al. (59) examined the extent of DNA methylation of the U3 region and part of the R region of the 5′-LTR in BLV-infected cattle and in experimentally BLV-infected sheep at various stages of the disease; and they detected no or minimal CpG methylation of the U3/R region by the bisulfite genomic sequencing method even in tumor cells. The first study (58) is in agreement with our results in the true latent L267 cell line, whereas the second study (59) seems more compatible with what we observed in the defective latent YR2 cell line. Therefore, a possible explanation for these discrepancies could be that the tumor cells examined in Tajima's study carried proviruses silenced by genetic alteration(s), such as deletions in the BLV provirus or mutations into functionally important domains of the TaxBLV protein, that are well established mechanisms of transformation-associated virus silencing in ovine B-cell tumors (10, 36, 60).
We also studied the BLV 5′-LTR methylation status in the L267LTaxSN cell line in which expression of TaxBLV via the pLTaxSN vector relieves the repressive state of the L267 provirus and allows viral expression, as shown by viral RNA expression and by virus release in the naive host (34). In this productive L267LTaxSN cell line, the U3 region is almost completely devoid of methylated CpGs, and the 5′-half of the R region is methylated but to a lesser extent compared with the L267 cell line. These results reinforce the inverse correlation between the degree of methylation of cytosine residues at CpG dinucleotides in proviral DNA and the transcriptional activity of the BLV promoter.
Importantly, our results indicate that TaxBLV expression decreases DNMT1, DNMT3A, and DNMT3B expression levels, which could explain, at least in part, the lower level of DNA methylation observed in the L267LTaxSN 5′-LTR compared with the L267 5′-LTR. In addition to playing a direct tumorigenic function, TaxBLV and its more extensively studied HTLV-1 counterpart TaxHTLV-1 modulate the expression of several genes, either up- or down-regulating their expression through mechanisms including Tax interaction with cellular transcription factors, chromatin-modifying enzymes, and proteins involved in the posttranscriptional control of mRNA expression (61). Selective down-regulation of gene expression by TaxBLV may result from its interaction with methyl-CpG-binding domain 2 demethylase (MBD2), with histone methyltransferase Suv39h1, and/or with HDACs, interactions already reported for TaxHTLV-1 (Ref. 62–64 and our laboratory).13 Proteins from other oncogenic viruses have been shown to disturb the host-DNA methylation system by up-regulating DNMT expression, thereby maintaining hypermethylation of tumor suppressor genes (64–66). Here, we show that TaxBLV down-regulates DNMT expression, which leads to a lower level of methylation in the BLV promoter region and contributes to virus escape from latency.
Despite their similar sequences, the 5′-LTR contains the transcriptional promoter, whereas the 3′-LTR has a polyadenylation function. In this report, we also have shown the hypomethylated status of the 3′-LTR by bisulfite genomic sequencing analysis of the proviruses integrated in the three BLV-infected cell lines (YR2, L267, and L267LTaxSN), revealing a different methylation pattern of the 3′-LTR compared with the 5′-LTR. Such a differential level of methylation between the two LTRs has already been reported for the HTLV-I virus, where the 5′-LTR and the 3′-LTR are hypermethylated and not methylated at all, respectively, in the same population of adult T-cell leukemia cells (67–70). The unmethylated state of the HTLV-I 3′-LTR is consistent with its transcriptionally active role in promoting the expression of the HTLV-I bZIP (HBZ) gene, which is important for the proliferation of adult T-cell leukemia- and HTLV-I-infected cells and is encoded by the minus strand of the provirus (71).
Mechanistically, we have shown that CpG methylation in the BLV 5′-LTR impairs in vitro and in vivo CREB/CREM/ATF-1 binding to their cognate sites in the U3 region. Furthermore, CpG methylation at the −129 position alone was sufficient to reduce by 2-fold the BLV LTR-driven luciferase expression in our transient transfection experiments. The literature has reported a few examples of such transcriptional repression by direct inhibition of factor binding by site-specific methylation, such as for the mouse Bdnf (brain-derived neurotrophic factor) gene (44) or the interferon-γ gene (72). In addition to directly prevent the binding of transcription factors to their cognate binding sites, CpG methylation could also act through attachment of methyl-CpG-binding proteins, which in turn recruit repressive chromatin modification enzymatic complexes.
Moreover, inhibition of CREB/CREM/ATF-1 binding impairs TaxBLV-mediated transcriptional activation of the BLV promoter (as strongly suggested by the fact that DNA methylation inhibitors and TaxBLV synergistically activated BLV gene expression in a CRE-dependent manner). This binding inhibition could also impair the recruitment of the histone acetyltransferase coactivators CREB-binding protein (CBP)/p300; their role in BLV transcriptional regulation, recently demonstrated by our laboratory (17), leads to a reduction in the level of histone acetylation, reinforcing the transcriptionally silenced state of integrated BLV proviruses.
In conclusion, our results constitute direct evidence for an interplay between the DNA methylation status of, and CREB/CREM/ATF protein binding to, the BLV promoter, a mechanism that could contribute to true proviral latency. Together with previous data on the role of chromatin modifications in BLV expression, the present work contributes to a deeper understanding of the complex epigenetic control of BLV transcriptional latency.
Supplementary Material
Acknowledgments
We thank Drs. Tony Kouzarides (Cambridge, United Kingdom) and François Fuks (University of Brussels, Belgium) for DNA methyltransferase expression vectors used in this study and for helpful discussions and Dr. Anne Van den Broeke (University of Brussels, J. Bordet Institute, Brussels, Belgium) for the BLV-infected cell lines.
This work was supported by grants from the Belgian Fund for Scientific Research (FRS-FNRS, Belgium), the Télévie Program, the “Action de Recherche Concertée du Ministère de la Communauté Française” (Université Libre de Bruxelles, ARC Program 04/09-309), the Internationale Brachet Stiftung, the CGRI-INSERM Cooperation, the Région Wallonne-Commission Européenne FEDER (Fonds Européen de Développment Régionale), the “Fortis Banque Assurance,” ANRS, Sidaction, the French Ministry of Research, and the Theyskens-Mineur Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
C. Van Lint, unpublished results.
- BLV
- bovine leukemia virus
- 5-AZAdc
- 5-aza-2′-deoxycytidine
- ATF
- activating transcription factor
- ChIP
- chromatin immunoprecipitation
- CRE
- cAMP-responsive element
- CREB
- CRE-binding protein
- CREM
- cAMP-responsive element modulator
- DNMT
- DNA methyltransferase
- ELISA
- enzyme-linked immunosorbent assay
- EMSA
- electrophoretic mobility shift assay
- Fw
- forward
- HDAC
- histone deacetylase
- HTLV
- human T-lymphotropic virus
- LTR
- long terminal repeat
- nt
- nucleotide
- PBMC
- peripheral blood mononuclear cell
- PMA
- phorbol 12-myristate 13-acetate
- qPCR
- quantitative PCR
- Rv
- reverse
- TxRE
- Tax-responsive element.
REFERENCES
- 1.Burny A., Cleuter Y., Kettmann R., Mammerickx M., Marbaix G., Portetelle D., Van den Broeke A., Willems L., Thomas R. (1987) Cancer Surv. 6, 139–159 [PubMed] [Google Scholar]
- 2.Burny A., Bruck C., Chantrenne H., Cleuter Y., Dekegel D., Ghysdael J., Kettmann R., Leclercq M., Leunen J., Mammerickx M., Portetelle D. (1980) in Viral Oncology (Klein G. ed) pp. 231–289, Raven Press, New York [Google Scholar]
- 3.Burny A., Willems L., Callebaut I., Adam E., Cludts I., Dequiedt F., Droogmans L., Grimonpont C., Kerkhofs P., Mammerickx M., Portetelle D., Van den Broeke A., Kettmann R. (1994) Bovine Leukemia Virus: Biology and Mode of Transformation, pp. 213–234, Cambridge University Press, Cambridge, UK [Google Scholar]
- 4.Willems L., Burny A., Dangoisse O., Colette D., Dequiedt F., Gatot J. S., Kerkhofs P., Lefèbre L., Merezak C. P., Portetelle D., Twizere J. C., Kettmann R. (1999) Curr. Topics Virol. 1, 139–167 [Google Scholar]
- 5.Willems L., Burny A., Collete D., Dangoisse O., Dequiedt F., Gatot J. S., Kerkhofs P., Lefèbvre L., Merezak C., Peremans T., Portetelle D., Twizere J. C., Kettmann R. (2000) AIDS Res. Hum. Retroviruses 16, 1787–1795 [DOI] [PubMed] [Google Scholar]
- 6.Gillet N., Florins A., Boxus M., Burteau C., Nigro A., Vandermeers F., Balon H., Bouzar A. B., Defoiche J., Burny A., Reichert M., Kettmann R., Willems L. (2007) Retrovirology 4, 18. [DOI] [PubMed] [Google Scholar]
- 7.Ferrer J. F. (1979) J. Am. Vet. Med. Assoc. 175, 1281–1286 [PubMed] [Google Scholar]
- 8.Lagarias D. M., Radke K. (1989) J. Virol. 63, 2099–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kettmann R., Cleuter Y., Mammerickx M., Meunier-Rotival M., Bernardi G., Burny A., Chantrenne H. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 2577–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van den Broeke A., Cleuter Y., Chen G., Portetelle D., Mammerickx M., Zagury D., Fouchard M., Coulombel L., Kettmann R., Burny A. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 9263–9267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta P., Ferrer J. F. (1982) Science 215, 405–407 [DOI] [PubMed] [Google Scholar]
- 12.Willems L., Kettmann R., Portetelle D., Burny A. (1987) Haematol. Blood Transfus. 31, 482–487 [DOI] [PubMed] [Google Scholar]
- 13.Derse D. (1987) J. Virol. 61, 2462–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adam E., Kerkhofs P., Mammerickx M., Kettmann R., Burny A., Droogmans L., Willems L. (1994) J. Virol. 68, 5845–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adam E., Kerkhofs P., Mammerickx M., Burny A., Kettmann R., Willems L. (1996) J. Virol. 70, 1990–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willems L., Kettmann R., Chen G., Portetelle D., Burny A., Derse D. (1992) J. Virol. 66, 766–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyên T. L., de Walque S., Veithen E., Dekoninck A., Martinelli V., de Launoit Y., Burny A., Harrod R., Van Lint C. (2007) J. Biol. Chem. 282, 20854–20867 [DOI] [PubMed] [Google Scholar]
- 18.Unk I., Kiss-Toth E., Boros I. (1994) Nucleic Acids Res. 22, 4872–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekoninck A., Calomme C., Nizet S., de Launoit Y., Burny A., Ghysdael J., Van Lint C. (2003) Oncogene 22, 2882–2896 [DOI] [PubMed] [Google Scholar]
- 20.Bloom J. C., Ganjam V. K., Gabuzda T. G. (1980) Cancer Res. 40, 2240–2244 [PubMed] [Google Scholar]
- 21.Bloom J. C., Kenyon S. J., Gabuzda T. G. (1979) Blood 53, 899–912 [PubMed] [Google Scholar]
- 22.Niermann G. L., Buehring G. C. (1997) Virology 239, 249–258 [DOI] [PubMed] [Google Scholar]
- 23.Xiao J., Buehring G. C. (1998) J. Virol. 72, 5994–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calomme C., Nguyen T. L., de Launoit Y., Kiermer V., Droogmans L., Burny A., Van Lint C. (2002) J. Biol. Chem. 277, 8775–8789 [DOI] [PubMed] [Google Scholar]
- 25.Kiermer V., Van Lint C., Briclet D., Vanhulle C., Kettmann R., Verdin E., Burny A., Droogmans L. (1998) J. Virol. 72, 5526–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merezak C., Reichert M., Van Lint C., Kerkhofs P., Portetelle D., Willems L., Kettmann R. (2002) J. Virol. 76, 5034–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calomme C., Dekoninck A., Nizet S., Adam E., Nguyên T. L., Van Den Broeke A., Willems L., Kettmann R., Burny A., Van Lint C. (2004) J. Virol. 78, 13848–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyên T. L., Calomme C., Wijmeersch G., Nizet S., Veithen E., Portetelle D., de Launoit Y., Burny A., Van Lint C. (2004) J. Biol. Chem. 279, 35025–35036 [DOI] [PubMed] [Google Scholar]
- 29.Achachi A., Florins A., Gillet N., Debacq C., Urbain P., Foutsop G. M., Vandermeers F., Jasik A., Reichert M., Kerkhofs P., Lagneaux L., Burny A., Kettmann R., Willems L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10309–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermann A., Gowher H., Jeltsch A. (2004) Cell. Mol. Life Sci. 61, 2571–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenner C., Fuks F. (2006) Curr. Top. Microbiol. Immunol. 301, 45–66 [DOI] [PubMed] [Google Scholar]
- 32.Latham T., Gilbert N., Ramsahoye B. (2008) Cell Tissue Res. 331, 31–55 [DOI] [PubMed] [Google Scholar]
- 33.Willems L., Kettmann R., Dequiedt F., Portetelle D., Vonèche V., Cornil I., Kerkhofs P., Burny A., Mammerickx M. (1993) J. Virol. 67, 4078–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merimi M., Klener P., Szynal M., Cleuter Y., Kerkhofs P., Burny A., Martiat P., Van den Broeke A. (2007) J. Virol. 81, 5929–5939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merimi M., Klener P., Szynal M., Cleuter Y., Bagnis C., Kerkhofs P., Burny A., Martiat P., Van den Broeke A. (2007) Retrovirology 4, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Den Broeke A., Bagnis C., Ciesiolka M., Cleuter Y., Gelderblom H., Kerkhofs P., Griebel P., Mannoni P., Burny A. (1999) J. Virol. 73, 1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kettmann R., Cleuter Y., Gregoire D., Burny A. (1985) J. Virol. 54, 899–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Lint C., Ghysdael J., Paras P., Jr., Burny A., Verdin E. (1994) J. Virol. 68, 2632–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olek A., Oswald J., Walter J. (1996) Nucleic Acids Res. 24, 5064–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborn L., Kunkel S., Nabel G. J. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 2336–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 42.Fuks F., Burgers W. A., Brehm A., Hughes-Davies L., Kouzarides T. (2000) Nat. Genet. 24, 88–91 [DOI] [PubMed] [Google Scholar]
- 43.Hsieh C. L. (1999) Mol. Cell. Biol. 19, 8211–8218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinowich K., Hattori D., Wu H., Fouse S., He F., Hu Y., Fan G., Sun Y. E. (2003) Science 302, 890–893 [DOI] [PubMed] [Google Scholar]
- 45.Portetelle D., Mammerickx M., Burny A. (1989) J. Virol. Methods 23, 211–222 [DOI] [PubMed] [Google Scholar]
- 46.Liu R., Liu H., Chen X., Kirby M., Brown P. O., Zhao K. (2001) Cell 106, 309–318 [DOI] [PubMed] [Google Scholar]
- 47.van der Vlag J., den Blaauwen J. L., Sewalt R. G., van Driel R., Otte A. P. (2000) J. Biol. Chem. 275, 697–704 [DOI] [PubMed] [Google Scholar]
- 48.Marban C., Suzanne S., Dequiedt F., de Walque S., Redel L., Van Lint C., Aunis D., Rohr O. (2007) EMBO J. 26, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanfield-Oakley S. A., Griffith J. D. (1996) J. Mol. Biol. 256, 503–516 [DOI] [PubMed] [Google Scholar]
- 50.Van den Broeke A., Cleuter Y., Droogmans L., Burny A., Kettmann R. (1997) in Immunology Methods Manual, pp. 2127–2132, Academic Press, London [Google Scholar]
- 51.Herschlag D., Johnson F. B. (1993) Genes Dev. 7, 173–179 [DOI] [PubMed] [Google Scholar]
- 52.Jensen W. A., Wicks-Beard B. J., Cockerell G. L. (1992) J. Virol. 66, 4427–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerkhofs P., Adam E., Droogmans L., Portetelle D., Mammerickx M., Burny A., Kettmann R., Willems L. (1996) J. Virol. 70, 2170–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kauder S. E., Bosque A., Lindqvist A., Planelles V., Verdin E. (2009) PLoS Pathog. 5, e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blazkova J., Trejbalova K., Gondois-Rey F., Halfon P., Philibert P., Guiguen A., Verdin E., Olive D., Van Lint C., Hejnar J., Hirsch I. (2009) PLoS Pathog. 5, e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dickerson S. J., Xing Y., Robinson A. R., Seaman W. T., Gruffat H., Kenney S. C. (2009) PLoS Pathog. 5, e1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhende P. M., Seaman W. T., Delecluse H. J., Kenney S. C. (2005) J. Virol., 79, 7338–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kashmiri S. V., Mehdi R., Gupta P., Ferrer J. F. (1985) Biochem. Biophys. Res. Commun. 129, 126–133 [DOI] [PubMed] [Google Scholar]
- 59.Tajima S., Tsukamoto M., Aida Y. (2003) J. Virol. 77, 4423–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kettmann R., Deschamps J., Cleuter Y., Couez D., Burny A., Marbaix G. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 2465–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boxus M., Twizere J. C., Legros S., Dewulf J. F., Kettmann R., Willems L. (2008) Retrovirology 5, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ego T., Tanaka Y., Shimotohno K. (2005) Oncogene 24, 1914–1923 [DOI] [PubMed] [Google Scholar]
- 63.Kamoi K., Yamamoto K., Misawa A., Miyake A., Ishida T., Tanaka Y., Mochizuki M., Watanabe T. (2006) Retrovirology 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ego T., Ariumi Y., Shimotohno K. (2002) Oncogene 21, 7241–7246 [DOI] [PubMed] [Google Scholar]
- 65.Park I. Y., Sohn B. H., Yu E., Suh D. J., Chung Y. H., Lee J. H., Surzycki S. J., Lee Y. I. (2007) Gastroenterology. 132, 1476–1494 [DOI] [PubMed] [Google Scholar]
- 66.Villanueva R., Iglesias A. H., Camelo S., Sanin L. C., Gray S. G., Dangond F. (2006) Oncol. Rep. 16, 581–585 [PubMed] [Google Scholar]
- 67.Koiwa T., Hamano-Usami A., Ishida T., Okayama A., Yamaguchi K., Kamihira S., Watanabe T. (2002) J. Virol. 76, 9389–9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taniguchi Y., Nosaka K., Yasunaga J., Maeda M., Mueller N., Okayama A., Matsuoka M. (2005) Retrovirology 2, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takeda S., Maeda M., Morikawa S., Taniguchi Y., Yasunaga J., Nosaka K., Tanaka Y., Matsuoka M. (2004) Int. J. Cancer 109, 559–567 [DOI] [PubMed] [Google Scholar]
- 70.Saggioro D., Panozzo M., Chieco-Bianchi L. (1990) Cancer Res. 50, 4968–4973 [PubMed] [Google Scholar]
- 71.Matsuoka M., Green P. L. (2009) Retrovirology 6, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones B., Chen J. (2006) EMBO J. 25, 2443–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.