Abstract
Overexpression of the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), like exercise, increases mitochondrial content and inhibits muscle atrophy. To understand these actions, we tested whether PGC-1α or its close homolog, PGC-1β, influences muscle protein turnover. In myotubes, overexpression of either coactivator increased protein content by decreasing overall protein degradation without altering protein synthesis rates. Elevated PGC-1α or PGC-1β also prevented the acceleration of proteolysis induced by starvation or FoxO transcription factors and prevented the induction of autophagy and atrophy-specific ubiquitin ligases by a constitutively active FoxO3. In mouse muscles, overexpression of PGC-1β (like PGC-1α) inhibited denervation atrophy, ubiquitin ligase induction, and transcription by NFκB. However, increasing muscle PGC-1α levels pharmacologically by treatment of mice with 5-aminoimidazole-4-carboxamide 1-β-d-ribofuranoside failed to block loss of muscle mass or induction of ubiquitin ligases upon denervation atrophy, although it prevented loss of mitochondria. This capacity of PGC-1α and PGC-1β to inhibit FoxO3 and NFκB actions and proteolysis helps explain how exercise prevents muscle atrophy.
Keywords: Lysosomes, Mitochondria, Mouse, NF-κB, Proteasome, Protein Degradation, Protein Synthesis, Skeletal Muscle, Ubiquitin Ligase, AICAR
Introduction
The mass of a muscle and its functional capacity are determined by the balance between rates of protein synthesis and protein degradation. The rapid, debilitating loss of muscle that occurs upon inactivity, nerve damage, and in many systemic diseases (e.g. diabetes, cancer, sepsis, or renal failure) is characterized by an increased rate of protein degradation (1, 2) and coordinated changes in the expression of a set of atrophy-related genes, which have been termed “atrogenes” (3, 4). Many of these genes are induced by the FoxO family of transcription factors (5, 6), which is activated in atrophying muscles. In fact, activated FoxO3 alone stimulates overall protein degradation by both the ubiquitin-proteasome and the autophagic-lysosomal systems (7, 8) and induces fiber atrophy (5). Two FoxO-induced genes are particularly important in enhancing proteolysis, the muscle-specific ubiquitin ligases, Atrogin1/MAFBx and MuRF1 (5, 9, 10), and muscles that lack either of these ligases show reduced fiber atrophy upon denervation (9). Another transcription factor that plays an essential role in muscle atrophy is NFκB. Although activation of the NFκB pathway is sufficient to induce muscle wasting (11, 12), its precise role and the factors that control its activity in muscle are still poorly understood.
Despite appreciable recent progress in understanding the biochemical basis of atrophy, no pharmaceutical agents are available to inhibit this highly debilitating process. Exercise can protect against disuse atrophy as well as various systemic types of muscle wasting (13–15), but the mechanisms by which contractile activity reduces atrophy remain unclear. In principle, contractile activity may somehow enhance protein synthesis, suppress overall protein breakdown, and/or block the atrophy-related transcriptional program. Understanding exactly how exercise protects against atrophy may suggest novel means to inhibit the loss of muscle mass in bed-ridden individuals when exercise is impractical or impossible. Such information may lead to improved rehabilitation methods and allow restoration of muscle function in many disease states or in the frail (sarcopenic) elderly.
Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α)4 is a transcriptional coactivator that appears to be a link between muscle activity, energy metabolism, and the control of fiber size (16). The two homologous family members, PGC-1α and PGC-1β, are potent coactivators of gene transcription, although they lack DNA-binding activity. Instead, they dock on transcription factors such as ERRα, PPARα, or NRF-1 and influence their function. The expression of PGC-1α in muscle increases acutely and chronically with exercise (17–20), and its mRNA decreases during disuse atrophy and various other types of muscle wasting (4, 16). On the other hand, overexpression of either PGC-1α or its close homolog, PGC-1β, results in a substantial increase in muscle mitochondrial content (18, 21–23) and resistance to fatigue (22, 23), which are both characteristic responses to repeated exercise. Importantly, overexpression of PGC-1α in adult muscle fibers reduces the capacity of denervation, fasting, and even activated FoxO3 to cause rapid fiber atrophy (16). Recently, transgenic mice overexpressing PGC-1α in muscle was shown to both extend lifespan and prevent the muscle wasting characteristic of aged mice (24). Because protein degradation increases markedly during various types of rapid atrophy, and overexpression of PGC-1α can inhibit muscle wasting, it seemed likely that PGC-1α and perhaps PGC-1β can inhibit overall protein degradation. One major goal of this study was to test this hypothesis and to examine the alternative possibility that these coactivators might enhance rates of protein synthesis in muscle.
Because genetic overexpression of PGC-1α can inhibit atrophy (16, 24), it seems likely that one or more agents that raise cell levels of PGC-1α could be of therapeutic benefit to retard muscle wasting in bedridden patients, the frail elderly, and patients suffering from various catabolic conditions. One pharmacological agent that increases PGC-1α content is 5-aminoimidazole-4-carboxamide 1-β-d-ribofuranoside (AICAR), an activator of AMP-activated protein kinase and other AMP-sensitive enzymes. AICAR, which has been used in human clinical trials and in experimental studies (25), has been shown to elicit in sedentary animals some of the same adaptations as occur with exercise training. Mice treated with AICAR for many days or weeks have increased resistance to fatigue upon running (26), increased muscle levels of GLUT4 glucose transporter (27, 28), increased mitochondrial content (29–31), and, importantly, increased PGC-1α content (26, 28, 31), which presumably causes these effects.
On the other hand, AICAR may potentially exacerbate atrophy, because AICAR treatment of cultured myotubes for 1 day or less has been reported to induce Atrogin1 and MuRF1 expression, activate protein degradation, and induce loss of protein (32, 33). Such reports seem inconsistent with the ability of PGC-1α to retard atrophy in vivo (16). Therefore, we investigated whether treatment with AICAR, by raising levels of PGC-1α, might be a useful approach to inhibit muscle wasting in vivo. Such studies are also of interest because of the possibility that control of mitochondrial content and function are linked to the proper regulation of skeletal muscle mass (16, 24). In fact, loss of mitochondria and therefore endurance appears to be an important but poorly characterized component of muscle atrophy that would be valuable to block pharmacologically.
The primary goals of this study were to examine 1) whether PGC-1α and its much less studied homolog PGC-1β actually influence muscle mass through effects on rates of protein synthesis or breakdown in skeletal muscle and 2) whether overexpression of PGC-1β or AICAR treatment can protect against denervation (disuse) atrophy in vivo when muscles lose mitochondrial content. Using both myotubes in culture and adult mouse muscles, we demonstrate that overexpression of PGC-1α or PGC-1β can enhance growth of myotubes and inhibit atrophy of adult muscles by slowing protein degradation and blocking the activation of proteolysis by FoxO3. However, raising levels of PGC-1α with AICAR fails to prevent atrophy, though it still may yield the physiological benefits of increased mitochondrial content.
EXPERIMENTAL PROCEDURES
Cell Culture
C2C12 myoblasts (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and penicillin/streptomycin. Myoblasts were differentiated to myotubes and subsequently maintained in low serum media (DMEM plus 2% horse serum) as described previously (34). Infection with adenoviruses expressing PGC-1α, PGC-1β, or GFP was on the third or fourth day with differentiation medium, when myotubes were fully formed. For those cultures that were infected with a second adenovirus (caFoxO3 or GFP), the first virus was removed from the myotubes at 24 h, the cells were washed once, and the second virus was added in fresh differentiation medium. The adenoviruses expressing GFP, PGC-1α, PGC-1β, and caFoxO3 have been described previously (5, 21).
Protein Synthesis
Myotubes were incubated with l-[3,5-3H]tyrosine (4 μCi/ml, PerkinElmer Life Sciences) for 2 h. Then, cells were washed, and proteins were precipitated with 10% trichloroacetic acid and pelleted by centrifugation. The pellet was solubilized with 0.1 n NaOH, and the amount of radioactivity was measured by scintillation counting. The total protein was measured using Coomassie Plus (Pierce). Protein synthesis was expressed as [3H]tyrosine incorporated into trichloroacetic acid-insoluble proteins per microgram of protein.
Protein Degradation
As in our prior studies, long-lived cell proteins were labeled with l-[3,5-3H]tyrosine (4 μCi/ml, PerkinElmer Life Sciences) for 24–28 h (7, 34). The cells were washed and infected with adenoviruses in chase medium (DMEM plus 2% horse serum) plus high amounts of nonradioactive tyrosine (2 mm) to limit reincorporation of the [3H]tyrosine. This chase also allows degradation of short-lived proteins. Starting 48 h after infection, the cells were washed and fresh chase media were added. Aliquots of culture medium were taken at specified times for quantitation of [3H]tyrosine release. Proteins were precipitated with trichloroacetic acid (10% final concentration) and pelleted. Radioactivity in the trichloroacetic acid-soluble supernatant was measured using liquid scintillation counting. At the end of the chase period, cells were solubilized in 0.2 n NaOH, and the radioactivity was measured. Total radioactivity is the sum of the residual radioactivity in the cells and the trichloroacetic acid-soluble radioactivities at different time points. Protein breakdown was expressed as [3H]tyrosine released over time as a percentage of total [3H]tyrosine incorporated. These rates of breakdown of long-lived proteins were highly linear for at least 4 h.
Proteasomal and lysosomal proteolysis rates were determined precisely as done before (7) by treating cells with 1 μm bortezomib (kindly provided by Millennium Pharmaceuticals) and 0.1 μm concanamycin A (Wako), respectively. The inhibitors were added 1 h prior to measuring protein degradation, at the same time as HBSS (Cellgro) was added.
Luciferase Reporter Assays in Muscle Cell Cultures
Nearly confluent myoblasts were transfected using FuGENE 6 (Roche Applied Science) with a Renilla luciferase plasmid (pRL-TK, Promega, Madison, WI) and with either the 3.5-kb Atrogin1 promoter (5) or the 5-kb MuRF1 promoter cloned into pGL3 basic plasmids (Promega), which encode for firefly luciferase. The following day, the medium was shifted to differentiation medium to induce myotube formation. Three days later, myotubes were infected with GFP, PGC-1α, or PGC-1β adenovirus. One day later, the medium containing the first adenoviruses was removed, the cells were washed, and fresh media containing GFP or caFoxO3 adenoviruses was added. Twenty-four hours later, myotubes were lysed in 1× passive lysis buffer and analyzed using the Dual-Luciferase Reporter Assay System (Promega). To control for transfection efficiency, firefly luciferase activity was divided by Renilla luciferase activity.
Mitochondrial Enzyme Activities
Myotubes or muscles were homogenized on ice in a solution of 0.1% Triton X-100, 100 mm KH2PO4, 2 mm EDTA, pH 7.2. Then, total protein was measured using Coomassie Plus (Pierce). Citrate synthase activity was determined spectrophotometrically at 30 °C as done by Srere (35), and succinate dehydrogenase activity was measured at 37 °C as done by Green and Narahara (36).
Western Blotting
Muscles were homogenized with a Polytron in ice-cold lysis buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mm Tris (pH 7.5), 150 mm NaCl, and a protease inhibitor mixture (Roche Applied Science)). Total protein was measured using a BCA Protein or Coomassie Plus Assay Kit (Pierce). Then, equal amounts of total protein per lane were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and immunoblotted with specific primary antibodies. Primary antibodies were obtained from Abcam (β-actin, NRF-1, and succinate dehydrogenase), BD Pharmingen (cytochrome c), Santa Cruz Biotechnology (F1-ATPase and PGC-1α), Dr. Stewart Lecker (Atrogin1) (37), and Regeneron Pharmaceuticals (MuRF1) (9). Secondary antibodies were conjugated to alkaline phosphatase. Band intensities were analyzed using ImageJ software (National Institutes of Health).
Muscle Denervation
After anesthesia, hair was removed, and the skin disinfected from the lateral surface of the hind limb. A small (<5 mm) incision was made in the skin approximately mid-femur. With blunt dissection, the sciatic nerve was exposed, and a 2-mm section was removed. The skin was closed with surgical glue (Vet Bond).
DNA Transfection of Adult Muscle
Tibialis anterior muscles of adult male CD1 mice (28–32 g) were transfected by electroporation of plasmids precisely as described previously (5). In many experiments, denervation was performed at the same time as the electroporation. Muscles were analyzed 5 or 10 days later. No gross evidence for necrosis or inflammation as a result of the transfection procedure was noted.
To determine the effects on muscle fiber size, 30 μg of PGC-1β-IRES-GFP plasmid was injected. This plasmid was constructed by mutating the PGC-1β plasmid using PCR to add an XmaI restriction site just prior to the Kozak sequence and a NotI restriction site just prior to the STOP codon. After restriction digest, this PGC-1β sequence was inserted into the pIRES-hrGFPII plasmid (Stratagene). The coding region was sequenced to confirm proper insertion.
In reporter experiments, 10 μg of the expression vector (PGC-1α, PGC-1β, or GFP) with 10 μg of the 3.5-kb Atrogin1 promoter reporter construct (5) and 10 μg of pRL-TK (Promega) vectors were coinjected. Luminescence measurements in muscles transfected with reporter constructs were performed similarly to those done in myotubes, except that the harvested muscles were manually homogenized in lysis buffer.
Muscle Fiber Cross-sectional Area
Tibialis anterior muscles transfected with PGC-1β-IRES-GFP plasmids were cryosectioned (10 μm), and then the transverse sections were fixed with 4% paraformaldehyde. Fluorescent images were collected in the Nikon Imaging Center at Harvard Medical School. Using ImageJ, cross-sectional areas were measured in fibers expressing GFP and in an equal number of non-transfected fibers from the same muscle as we have done previously (5).
AICAR Treatment
AICAR (Toronto Research Chemical, Inc.) was prepared fresh daily in sterile phosphate-buffered saline (1 g of AICAR/20 ml). Mice were injected intraperitoneally with 500 mg of AICAR/kg of body weight/day for 2 weeks. Then, the muscles of one hind limb were denervated. AICAR treatment was continued for an additional 2 weeks until the muscles were collected. Therefore, muscles were treated with AICAR for a total of 4 weeks.
Statistics
Data are presented as mean ± S.E. Statistical significance was assessed using paired Student's t test (for comparisons of only denervated versus innervated muscles) or analysis of variance (for multiple comparisons) followed by the Tukey's honestly significant difference procedure, where appropriate. A p value of <0.05 was considered significantly different.
RESULTS
PGC-1α or PGC-1β Overexpression Inhibits Protein Degradation without Affecting Protein Synthesis in Myotubes
To test if PGC-1α or its close homolog, PGC1-β, can influence muscle growth, protein synthesis, or protein degradation, differentiated C2C12 myotubes were infected with adenoviruses expressing either coactivator. As expected (18, 21, 38, 39), mitochondrial content increased in the myotubes as illustrated by increased citrate synthase activity (Fig. 1A). By 62 h after infection with the adenoviruses, the levels of citrate synthase activity were nearly twice those in myotubes infected with a control (GFP-encoding) adenovirus (p < 0.001). Thus, infection of cultured muscle cells with either a PGC-1α- or PGC-1β-encoding adenovirus resulted in similar relative increases in mitochondrial content with similar time courses.
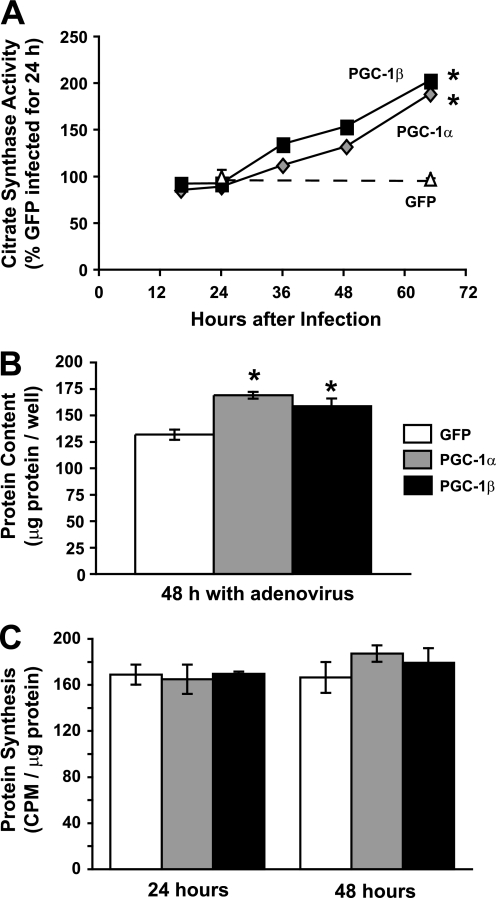
FIGURE 1.
Overexpression of PGC-1α or PGC-1β in C2C12 myotubes increases mitochondrial content and total protein content without altering protein synthetic rate. A, fully differentiated myotubes were infected at various times with adenoviruses expressing PGC-1α, PGC-1β, or GFP before all being collected at the same time after differentiation. Activity of citrate synthase, a mitochondrial enzyme, was determined spectrophotometrically from cell homogenates. *, p < 0.001 versus GFP. B, myotubes were infected with PGC-1α, PGC-1β, or GFP adenoviruses. Total protein per well of a 6-well plate was measured 48 h later. *, p < 0.001 versus GFP. C, protein synthetic rate was determined by measuring the incorporation of [3H]tyrosine for 2 h at the indicated times after adenoviral infection. No statistically significant differences were found.
We therefore compared the effects of the two coactivators on rates of cell growth and protein turnover. Two days after infection, the total protein content per well, which is proportional to cell size but can be measured with greater precision, was increased by ∼25% with either PGC-1-expressing adenovirus but not a control virus expressing GFP (Fig. 1B). However, to our surprise, the overexpression of either PGC-1α or PGC-1β had no measurable effect on the overall rate of protein synthesis in myotubes, measured as the incorporation of [3H]tyrosine for 2 h into protein at 24 or 48 h after infection (Fig. 1C). Alternatively, the increase in myotube protein content could have resulted from a fall in overall proteolysis. To test this possibility, we measured the overall rates of degradation of long-lived proteins, which comprise the bulk of cell proteins. After labeling cell proteins with [3H]tyrosine for 24 h, myotubes were infected with the PGC-1α-, PGC-1β-, or GFP-expressing adenoviruses. The rate of protein degradation measured 48 h later had decreased by 15–25% below levels in control cells (Fig. 2, A and C). Though relatively small, this reduction in proteolysis after PGC-1 overexpression was highly reproducible (p < 0.01) and sufficient to account fully for the increase in protein content (i.e. cell mass) in 2 days (see “Discussion”).
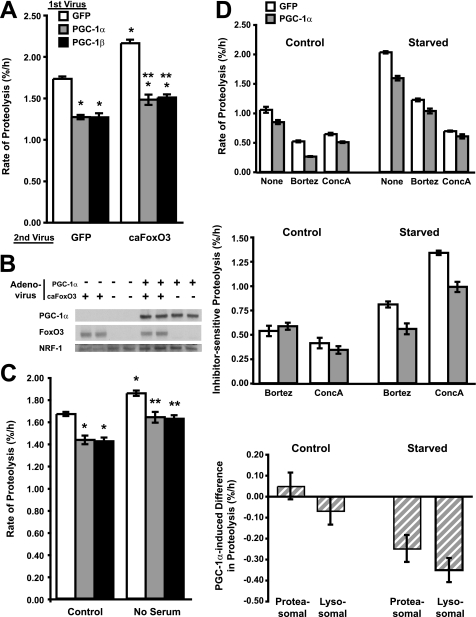
FIGURE 2.
PGC-1α or PGC-1β overexpression inhibits overall protein degradation by slowing both proteasomal and lysosomal degradation. A, after labeling with [3H]tyrosine, C2C12 myotubes were infected with adenoviruses expressing PGC-1α, PGC-1β, or GFP (1st Virus). 24 h later, the myotubes were given fresh media containing a second adenovirus (2nd Virus). *, p < 0.05 versus GFP-GFP; **, p < 0.05 versus GFP-caFoxO3. B, myotubes were infected with PGC-1α and caFoxO3 adenoviruses as in A. Protein levels were determined by Western blot. C, myotubes were incubated with [3H]tyrosine and 1st Virus as in A. 24 h later, fresh medium was added either with (control) or without serum. *, p < 0.05 versus GFP-control; **, p < 0.05 versus GFP-No serum. In D: Top panel, myotube proteins were labeled with [3H]tyrosine for 24 h and then infected with GFP or PGC-1α adenoviruses for 48 h. Fresh DMEM (control) or HBSS (starved) medium, both containing 2% horse serum, was then added with or without the proteasome (bortezomib = Bortz) or lysosomal (concanamycin A = ConcA) inhibitors. Rates of proteolysis were determined 1 h later as described previously. Middle panel, the amount of proteolysis sensitive to each inhibitor represented the amount of proteasome or lysosome-mediated degradation and was calculated from data in the top panel by subtracting the rates of proteolysis in cells treated with the inhibitors from those of untreated cells. Bottom panel, the PGC-1α-induced decrease in proteolysis was calculated by subtracting the amount of lysosomal or proteasomal protein degradation rate (i.e. the inhibitor-sensitive component) in the PGC-1α-overproducing cells from that in the controls (GFP-expressing).
Next, we examined whether either PGC-1α or PGC-1β could inhibit not only basal protein degradation but also the enhanced proteolysis characteristic of atrophying muscles. To stimulate protein degradation, either constitutively active FoxO3 (caFoxO3) was overexpressed for 24 h or serum was removed from the culture media. The FoxO3 transcription factor can increase the rate of protein degradation (7), and this effect accounts for its ability to cause rapid fiber atrophy (5). Both PGC-1α and PGC-1β overexpression blocked completely the increase in overall degradation rate by caFoxO3 (Fig. 2A) as well as the rapid increase in proteolysis induced by deprivation of serum from the culture media for 2 h (Fig. 2C). Interestingly, in these various conditions, PGC-1α and PGC-1β decreased protein degradation and increased myotube protein content to the same degree.
Importantly, these inhibitory effects of PGC-1 do not involve a down-regulation or phosphorylation of FoxO3 levels, because overexpression of PGC-1α did not prevent the accumulation of caFoxO3, which is mutated at the three AKT phosphorylation sites (Fig. 2B). Although both PGC-1s blocked the catabolic actions of FoxO3, caFoxO3 expression did not inhibit PGC-1α actions, such as its ability to induce the expression of nuclear respiratory factor-1 (NRF-1) content (Fig. 2B), a transcription factor that is necessary for the production of mitochondria by PGC-1α (24, 38).
PGC-1α and PGC-1β Inhibit Both Proteasomal and Autophagic/Lysosomal Degradation
The two main proteolytic systems in mammalian tissues are the ubiquitin/proteasome and the autophagic/lysosome. caFoxO3 stimulates both systems in skeletal muscle (7, 8), but most of the resulting increase in proteolysis in myotubes results from enhanced autophagy, as also occurs with serum deprivation (7). Therefore, to account for the inhibition of protein degradation by the PGC-1 coactivators (Fig. 2, A and C), some suppression of lysosomal proteolysis seems necessary, as has been suggested to occur in the muscles of aged PGC-1α transgenic mice (24).
To determine how PGC-1α and PGC-1β may influence the two proteolytic systems, myotubes in normal medium (DMEM) or in an autophagy-inducing “starvation” medium were treated with agents that block selectively and nearly completely proteasome function (bortezomib/Velcade) or lysosomal acidification (concanamycin A). Rates of proteolysis were then measured, and the contributions of proteasomes and lysosomes were determined by subtracting the absolute rates of proteolysis with either inhibitor present from that in untreated cultures as in our prior studies (7). The starvation medium used was Hanks' balanced salt solution (HBSS), which contains serum, the same inorganic salts, and 20% of the glucose as the standard medium, DMEM, but lacks pyruvate and amino acids. After incubation of the myotubes in HBSS, the rate of degradation of long-lived proteins was 93% faster than in DMEM (Fig. 2D). In the standard medium, where rates of proteolysis are low and the PGC-1α-induced inhibition is small, we were unable by this approach to definitively resolve which proteolytic system PGC-1α inhibits. However, in HBSS, where the absolute rates of proteolysis are much higher, PGC-1α overexpression clearly inhibits the lysosomal degradative process and also the proteasomal process.
PGC-1α and PGC-1β Suppress Induction of Ubiquitin Ligases by FoxO
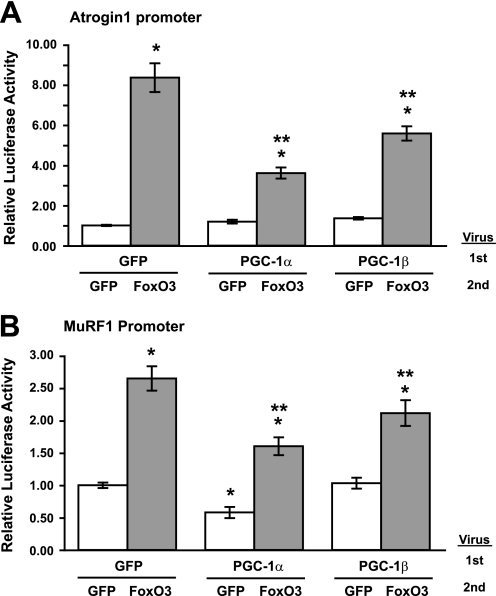
The ubiquitin ligases Atrogin1 and MuRF1 are expressed only in muscle and are highly induced during various forms of atrophy (9, 10, 40). Also, muscles lacking either ligase show reduced weight loss upon denervation or glucocorticoid treatment (9), and ones lacking MuRF1 are defective in the breakdown of myofibrillar components (41). To determine if PGC-1α or PGC-1β can inhibit the induction of these ligases, myoblasts were transfected with an Atrogin1 or a MuRF1 promoter luciferase plasmid construct. After differentiation, myotubes were infected with PGC-1α, PGC-1β, or control (GFP) adenoviruses and 24 h later with either caFoxO3 or control adenovirus. Infection with caFoxO3 increased the Atrogin1 promoter activity 8.3-fold (Fig. 3A) and the MuRF1 promoter activity 2.6-fold (Fig. 3B). However, the induction was strongly inhibited by overexpression of PGC-1α or PGC-1β (p < 0.05). Presumably, the ability of these coactivators to blunt the induction of these critical ubiquitin ligases accounts, in part, for the blockage of caFoxO3-induced proteasome-mediated degradation (cf. Fig. 2, A and C).
FIGURE 3.
PGC-1α or PGC-1β inhibits caFoxO3-induced Atrogin1 or MuRF1 promoter activity in myotubes. A, myoblasts were transfected with pRL-TK and an Atrogin1 promoter luciferase reporter plasmids and allowed to differentiate for 3 days. Myotubes were then infected with adenoviruses (1st) expressing GFP, PGC-1α, or PGC-1β. 24 h later, the myotubes were infected with a second adenovirus (2nd) expressing either GFP or caFoxO3. Extracts were collected 24 h later and analyzed for luciferase activity. *, p < 0.05 versus GFP-GFP; **, p < 0.05 versus GFP-caFoxO3. B, cells were prepared and data analyzed precisely as in A except that myoblasts were transfected with a MuRF1-promoter reporter in place of the Atrogin1 reporter.
Overexpression of PGC-1β Blocks Atrophy in Denervated Adult Muscle
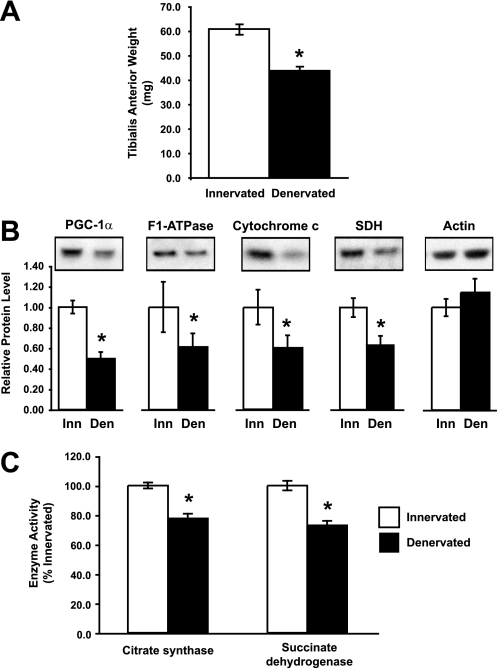
This ability to inhibit protein breakdown and atrogene induction in cultured myotubes implies that increased expression of either PGC-1α or PGC-1β should inhibit atrophy of adult muscle, as we previously reported for PGC-1α. To examine this possibility, we first determined how the endogenous levels of PGC-1α and the content of mitochondria change during denervation atrophy. Muscles of one hind limb in adult mice were denervated by removing a section of the sciatic nerve. As expected, 10 days later the mass of the tibialis anterior (TA) had decreased by 28% (Fig. 4A, p < 0.001) below that of the contralateral, innervated TA. Furthermore, the concentration of both PGC-1α protein and several mitochondrial proteins, F1-ATPase, cytochrome c, and succinate dehydrogenase, decreased by 40–50%, as measured by Western blot (Fig. 4B, p < 0.05). Accordingly, the enzymatic activity of citrate synthase decreased by 22% (p < 0.001) and that of succinate dehydrogenase decreased by 27% (p < 0.001). Because the Western blot and enzyme activity data are calculated per milligram of total protein, both PGC-1α and mitochondrial content decreased at a faster rate than the contractile proteins and overall muscle mass. In fact, there was no change in the concentration of actin at this time. Presumably, this loss of mitochondria contributes to the decrease in exercise capacity during disuse atrophy (42) and suggests additional benefits of maintaining high levels of PGC-1α and PGC-1β to counter these effects.
FIGURE 4.
PGC-1α and mitochondrial content decrease in TA muscle 10 days after denervation. A, weight of the TA decreased 10 days after cutting the sciatic nerve (denervated). *, p < 0.001, n = 6. B, PGC-1α and mitochondrial proteins were decreased in denervated TA. Protein levels were quantified by Western blot. Equal amounts of total protein were loaded per lane. *, p < 0.05. C, the activity of the mitochondrial enzymes, citrate synthase and succinate dehydrogenase, decreased in the denervated TA. *, p < 0.001.
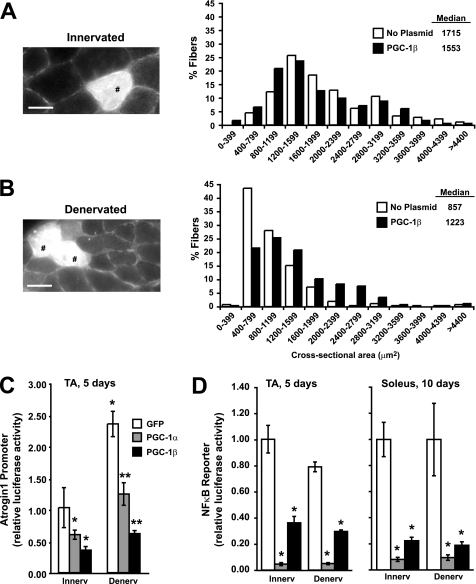
Because overexpression of PGC-1α or PGC-1β in cultured myotubes have a similar capacity to inhibit protein degradation and block FoxO actions, we tested whether in adult muscle overexpression of PGC-1β has the same ability to inhibit denervation atrophy as PGC-1α overexpression (16). An IRES bicistronic expression plasmid encoding both PGC-1β and GFP was electroporated bilaterally into TA muscles at the same time that the sciatic nerve of one hind limb was sectioned. Ten days later, the muscles were collected and cryosectioned, and the areas of fibers were determined by fluorescence microscopy. Interestingly, the median cross-sectional area of the innervated TA fibers overexpressing PGC-1β was 10% less than the area of those that were not transfected (Fig. 5A). This result agrees qualitatively with the observed smaller skeletal muscle fibers in transgenic animals with life-long overexpression of PGC-1β or PGC-1α and may reflect a switch to more oxidative fibers, which are typically smaller than glycolytic fibers (16, 23). On the denervated limb, muscle fibers overexpressing PGC-1β had a 43% greater median area than those that were not overexpressing PGC-1β (Fig. 5B). Consequently, the innervated and denervated muscle fibers that were transfected with PGC-1β had nearly the same median size. Thus, the coactivator protected muscle fibers from the loss of size typically seen with denervation atrophy.
FIGURE 5.
Electroporation of PGC-1β in mouse muscle inhibits fiber atrophy, and both PGC-1α and PGC-1β decrease Atrogin1 promoter activity and NFκB activity. Frequency histograms (right) showing the distribution of cross-sectional areas of innervated (A) or 10-day denervated (B) muscle fibers of the TA. Muscles were electroporated with PGC-1β-IRES-GFP plasmids at the same time as denervation. Transfected fibers were identified in transverse sections by GFP expression (left). Scale bar represents 30 μm. Tibialis anterior (TA) or soleus muscles of adult mice were co-electroporated with the pRL-TK and Atrogin1 promoter (C) or NFκB binding (D) luciferase reporter plasmids together with GFP (control), PGC-1α, or PGC-1β plasmids. Five or ten days later, muscles were collected and luciferase activity was measured. *, p < 0.05 versus innervated GFP-transfected fibers; **, p < 0.05 versus denervated GFP-transfected fibers.
To test whether PGC-1α or PGC-1β inhibits denervation atrophy in vivo by reducing the induction of atrophy genes like Atrogin1, as it does in cultured myotubes (Fig. 3A), we electroporated the Atrogin1 promoter luciferase construct into TA muscles together with either GFP, PGC-1α, or PGC-1β plasmids at the same time as these muscles were denervated. Both PGC-1α and PGC-1β prevented the increase in transcription off the Atrogin1 promoter that occurs following denervation (Fig. 5C, p < 0.05). In addition, both coactivators decreased the lower levels of Atrogin1 transcription in the contralateral innervated muscles. Presumably, these effects result from the ability of these coactivators to inhibit Atrogin1 transcription induced by activated FoxO3 in the denervated muscle (16).
There is growing evidence that disuse and other types of muscle atrophy, as well as the wasting in dystrophic muscles (43), is mediated in part by the NFκB pathway. Activation of the NFκB pathway (e.g. in muscles of transgenic mice) has been shown to induce muscle wasting, while inhibition of NFκB greatly slows disuse atrophy (11, 12, 44). Because PGC-1α and PGC-1β can inhibit many of the atrophy-inducing effects of FoxO in muscle, we tested whether these coactivators might also inhibit NFκB-dependent transcription. Muscles were electroporated with an NFκB luciferase reporter plasmid together with PGC-1α, PGC-1β, or GFP plasmids into the contralateral denervated and control TA muscles. Five days later in the innervated TA muscle, overexpression of PGC-1α had decreased the NFκB reporter activity by 95%, while overexpression of PGC-1β had decreased activity by 65% (Fig. 5D, p < 0.001). Similarly in the denervated muscles, NFκB activity also was decreased profoundly (p < 0.001) by both PGC-1α and PGC-1β.
One surprising observation was that denervation for 5 days did not increase the transcription of an NFκB reporter in TA muscles (Fig. 5D), as was expected due to the role of NFκB in atrophy (11, 44) as is found in the soleus during atrophy induced by cast immobilization (45). Because NFκB activity may play a more important role in muscle wasting at times later than 5 days denervation, as well as in the anti-gravity muscles containing more oxidative fibers (44, 45), we tested the effect of PGC-1α or PGC-1β overexpression on NFκB activity in the soleus 10 days after denervation. Unlike the TA, which is composed of ∼65% oxidative fibers and 35% glycolytic fibers, the soleus contains only oxidative fibers (46). In the soleus at 10 days, each coactivator dramatically inhibited NFκB activity in a similar way as at 5 days in the TA (Fig. 5D), and again denervation did not increase the muscle's NFκB activity. Therefore, the basal amount of NFκB activity appears to be sufficient for denervation atrophy for up to 10 days. In any case, the marked inhibition of NFκB actions by both PGC-1s seems to play a major role in their ability to retard muscle atrophy.
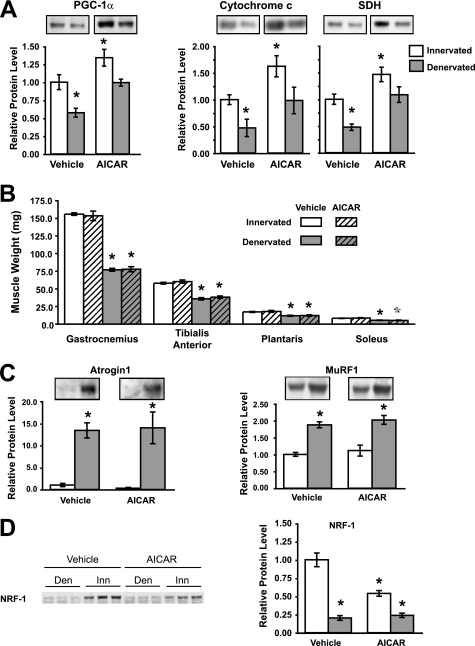
AICAR Treatment Maintains Normal PGC-1α and Mitochondrial Content in Denervated Muscles but Does Not Inhibit Muscle Weight Loss
Because overexpression of PGC-1α and PGC-1β can inhibit protein degradation and reduce atrophy, and because AICAR treatment of rodents has been shown repeatedly to increase PGC-1α content (26, 28, 31), we tested whether treatment with AICAR could inhibit denervation atrophy. Adult mice were injected daily with AICAR for 2 weeks. Then the muscles of one hind limb were denervated, and AICAR treatment continued for an additional 2 weeks (500 mg/kg). This dosage and duration were reported previously to enhance expression of genes for oxidative metabolism and to increase running endurance in mice (26). Accordingly, AICAR administration was effective at increasing the muscle's content of PGC-1α and mitochondrial proteins, cytochrome c, and succinate dehydrogenase by 35–60% in the innervated gastrocnemius (Fig. 6A, p < 0.05). Moreover, this treatment completely prevented the decrease in PGC-1α, cytochrome c, and succinate dehydrogenase proteins that is typically seen after denervation (cf. Fig. 4B).
FIGURE 6.
AICAR treatment for 4 weeks increases PGC-1α and mitochondrial content but has no effect on muscle mass or expression of ubiquitin ligases. A, levels in the gastrocnemius muscle of PGC-1α protein and the mitochondrial proteins, cytochrome c, and succinate dehydrogenase (SDH), increase with AICAR treatment, and decrease with denervation. *, p < 0.05 versus innervated vehicle-treated muscle, n = 6. B, 2 weeks after denervation, the weights of several muscles decreased similarly with or without AICAR treatment. *, p < 0.01 versus innervated vehicle-treated muscle. C, the levels of the ubiquitin ligases Atrogin1 and MuRF1 proteins in the gastrocnemius muscle increased after denervation similarly with or without AICAR treatment. *, p < 0.05 versus innervated vehicle-treated muscle. D, the content of NRF-1 protein decreased with AICAR treatment and with denervation. *, p < 0.01 versus innervated vehicle-treated muscle, in contrast to the induction of NRF-1 following PGC-1 overexpression (cf. Fig. 2B).
Sectioning the sciatic nerve on one side resulted in a 38–51% loss of mass of four muscles on the lower limb on that side (Fig. 6B, p < 0.01). These muscles (the gastrocnemius, TA, plantaris, and soleus) are composed of a broad range of fiber types with the gastrocnemius containing <40% oxidative fibers, whereas the soleus contains 100% oxidative fibers (46). AICAR treatment had no effect on the final body weight (AICAR 36.3 ± 0.8 g versus vehicle 37.6 ± 1.3 g) or the mass of any of the innervated or the denervated muscles (Fig. 6A). Furthermore, AICAR treatment failed, despite raising PGC1-α, to reduce the induction of Atrogin1 or MuRF1 protein in the denervated muscles just as it failed to influence the associated loss of muscle weight (Fig. 6C, p < 0.05).
This inability of AICAR treatment to block atrophy seems to be due to its inability to prevent atrogene induction in a similar fashion as PGC-1 overexpression. However, additional differences between these treatments were also found. Surprisingly, the levels of NRF-1, a transcription factor that enhances the expression of the vast majority of nuclear-encoded mitochondrial proteins and also the proteins encoded in the mitochondrial genome (47), decreased by 45% with AICAR treatment in the innervated muscle and even more (by 80%) following denervation (Fig. 6D, p < 0.05). This fall in NRF-1 with AICAR was unexpected given that NRF-1 expression is induced by PGC-1α (24, 38), as we confirmed in myotubes (Fig. 3C). It is noteworthy that, despite the decrease in NRF-1, the lack of contractile activity, and the enhanced autophagy, AICAR treatment could prevent the usual fall of PGC-1α content and mitochondria in the denervated muscle. Possibly, this inability of AICAR treatment to induce (or even maintain) NRF-1 may somehow contribute to its failure to block FoxO action and atrophy. In any case, the failure of AICAR to block atrophy in a similar fashion to PGC-1α and PGC-1β overexpression emphasizes that the effects of PGC-1α on mitochondrial content and cell mass involve distinct mechanisms that can be dissected.
DISCUSSION
PGC-1α or PGC-1β Overexpression Inhibits Muscle Protein Degradation by Multiple Mechanisms
PGC-1 coactivators have previously been shown to influence muscle fiber type, oxidative capacity, substrate utilization, and angiogenesis (48). To this list, we can now add control of muscle protein degradation, and in this way each coactivator may increase muscle protein content and size. The present findings reveal for the first time that overexpression of the transcriptional coactivators PGC-1α or PGC-1β slows protein degradation both in normal and atrophying muscle. In fact, in myotube cultures, PGC-1α and PGC-1β enhanced the growth of the culture by suppressing overall proteolysis, even without causing significant change in rates of protein synthesis. Importantly, these coactivators suppress both major proteolytic systems, the autophagic/lysosomal and the proteasomal pathways. The ubiquitin-proteasome system and autophagy seem to degrade distinct components of the muscle. Myofibrillar proteins are degraded primarily by the ubiquitin-proteasome pathway, and the ubiquitin ligase, MuRF1, is crucial in these responses (41), while autophagy can degrade organelles such as mitochondria. Therefore, inhibition of both major pathways would allow sparing of most muscle proteins. The effects on proteolysis demonstrated here can explain our previous findings that overexpression of PGC-1α inhibits muscle wasting induced by denervation, starvation, and even caFoxO3 expression (16).
Although the percent decrease in the overall protein degradation rates observed here with overexpression of either coactivator may seem small, these changes can completely explain the 25% increase in total cell protein content that we observed in 48 h (Fig. 1B). Thus, in Fig. 2A, PGC-1α or PGC-1β overexpression decreased the rate of degradation from 1.73%/h to 1.26%/h. Because the degradation of the bulk of cell proteins behaves as a simple first-order process (7), we can calculate that the difference in mean rates of protein degradation (0.47%/h) with either PGC-1 result in an apparent rate constant of 4.71 × 10−3/h. Over 48 h this would lead to a 25% greater amount of cell protein, without any change in overall protein synthesis. Because rapid muscle atrophy is characterized primarily by an increased rate of protein degradation (49, 50), our findings can explain one way that exercise, which increases PGC-1α content (17, 19), is effective at inhibiting atrophy.
This ability of PGC1s to suppress overall protein degradation, even in the face of activated FoxOs or NFκB, as may occur with disuse or low insulin or insulin-resistant states and also muscular dystrophies (43), while raising mitochondrial content and oxidative defenses (51), must have broad implications for maintenance of muscle function and metabolism. Recently, Wenz et al. (24) presented dramatic evidence that transgenic mice overexpressing PGC-1α in muscle lived significantly longer than control mice, and showed less age-related pathology, including a resistance of the loss of muscle mass (sarcopenia) characteristic of aged organisms. It seems likely that the present findings account at least in part for the maintenance of muscle mass in the aged mice. On the other hand, reduced autophagic and proteasomal proteolysis might actually be expected to contribute to the age-related accumulation of abnormal proteins, as has been reported to occur in liver of aged rodents (52), although such selective degradation of short-lived proteins may be regulated independently (53). An interesting related question for future research is whether these coactivators have similar effects on proteolysis in other cell types. Such studies would be of particular interest in liver, where PGC-1α functions and is regulated oppositely as in muscle; i.e. in fasting PGC-1α is induced in liver (54) (although its expression falls in muscle (4)), and PGC-1α functions synergistically with FoxO1 in promoting gluconeogenesis in the liver but inhibits FoxO actions in muscle (Figs. 2 and 3).
Multiple Actions of PGC-1α and PGC-1β in Muscles
As would be expected from their extensive structural similarities, PGC-1α and PGC-1β were found to have quite similar effects on the several metabolic properties of muscles studied here, including induction of mitochondrial components (Fig. 1), inhibition of protein degradation (Fig. 2), transcription of Atrogin1 and MuRF1 by FoxOs (Figs. 3 and 5), and reducing denervation atrophy (Fig. 5). However, these coactivators differ in their modes of regulation and have some distinct functions. For example, PGC-1α expression is highly responsive to exercise, whereas PGC-1β is not (18, 19). Mitochondria produced in myotubes upon overexpression of PGC-1β have less proton leakage and thus have more tightly coupled oxidative respiration than those induced by PGC-1α (21). Finally, transgenic PGC-1α overexpression promotes a shift toward type I muscle fibers (22), whereas overexpression of PGC-1β promotes a shift toward type IIx (23). Not surprisingly then, PGC-1α and PGC-1β do not target the exact same transcription factors (55), although they appear to bind similarly to several transcription factors such as with ERRα and NRF-1, which both control mitochondrial genes, or with PPARα, which regulates genes for fatty acid oxidation (55). Most likely, the ability of PGC-1α and PGC-1β to influence similarly muscle proteolysis and atrophy is through their similar interactions with FoxO and perhaps some other transcription factors.
Because FoxO transcription factors play such a central role in numerous types of atrophy (5–8), our results can most likely be explained by PGC-1α and PGC-1β directly inhibiting FoxO-induced transcription, as we had suggested previously (16). However, we show here that these coactivators also have the capacity to profoundly inhibit the functioning of NFκB, which is also necessary for rapid atrophy (11, 44). It was surprising therefore to find that transcription induced from NFκB binding sites, unlike FoxO-induced transcription, did not increase after denervation. Thus the basal NFκB activity is sufficient for it to trigger atrophy. At present, however, it is unclear what specific aspects of the atrophy program are activated by NFκB. Several recent studies have reported that in other tissues PGC-1α actually stimulates FoxO-induced transcription. Housley et al. demonstrated that PGC-1α stimulates the GlcNAcylation of FoxO1, which increases its transcriptional activity (56). Also, a previous study (57) showed in endothelial cells that PGC-1α can directly enhance FoxO3-dependent transcription. Thus, in both these cell types, PGC-1α activates transcription by FoxO, unlike in muscle where PGC-1α and FoxO3 have opposing actions, e.g. on protein degradation, myotube protein content, Atrogin1 transcription, and muscle atrophy. Together, these observations suggest that the ability of PGC-1s to inhibit the actions of FoxO requires a muscle-specific factor that is sensitive to both PGC-1α and PGC-1β.
Another noteworthy finding is that, during denervation atrophy, mitochondrial content decreases to a greater extent than overall cell protein and myofibrillar components. This selective destruction of mitochondria implies that the muscles are not just losing mass, but are undergoing remodeling to a different phenotype, apparently adapting to less contractile activity. Loss of mitochondria during disuse atrophy has been reported previously (42, 58, 59) and probably results from both reduced production of new mitochondria because of the lower PGC-1α content and enhanced degradation by autophagy (7, 8). Evidence is mounting for selective degradation of particular muscle components during atrophy. For example, the ubiquitin ligase MuRF1 is responsible for an ordered, progressive loss of the thick but not the thin filament during atrophy (41). Furthermore, mitochondria seem to be targeted directly for FoxO3-induced autophagy by BNIP3 (8, 60). Even though we have shown that both the proteasomal and the lysosomal degradative pathways can be inhibited by PGC-1α during atrophy, it remains to be determined whether PGC-1α overexpression protects all muscle components equally or whether it specifically reduces degradation of particular organelles or proteins.
Maintaining PGC-1α Content with AICAR Maintains Mitochondria but Does Not Inhibit Atrophy
The present findings imply that types of exercise or pharmacological agents that raise muscle PGC-1α or PGC-1β could have therapeutic benefit in blocking various types of muscle wasting and the associated loss of mitochondria. However, the mode of increasing PGC-1α or PGC-1β content is clearly critical. To increase PGC-1α and mitochondrial content, we treated mice with AICAR to test if this well characterized agent or related ones might be useful to inhibit disuse atrophy. The treatment regimen used here increased PGC-1α and mitochondrial enzymes to a similar extent as has been observed with chronic endurance exercise (19, 61). However, this treatment did not alter the loss of muscle weight or the induction of the atrophy-related genes, Atrogin1 or MuRF1, following denervation, despite its ability to prevent mitochondrial loss. Thus, two main deleterious effects of disuse appear to be dissociated by AICAR, even though PGC-1α or PGC-1β overexpression inhibits both processes, the loss of mitochondria and presumably endurance from the loss of myofibrillar mass and presumably strength.
This lack of effect with AICAR on muscle mass may have multiple explanations. First, because AICAR also alters the activity of proteins other than PGC-1α (62), these other proteins may counteract the inhibitory effects of PGC-1α on FoxOs. AICAR is an analog of AMP and functions principally as an allosteric activator of AMP-activated protein kinase (63). AMP-activated protein kinase increases PGC-1α content by phosphorylating PGC-1α, which leads then to positive feedback regulation of its transcription (28) possibly through the binding of the transcription factor upstream stimulatory factor-1 to the PGC-1α promoter region (64). However, AMP-activated protein kinase can also phosphorylate many other proteins, and AICAR can activate other AMP-sensitive kinases (62). In fact, in cultured myotubes AICAR treatment causes an up-regulation of Atrogin1 and MuRF1 mRNA, increases protein degradation, and causes a loss of protein content (32, 33), thus mimicking the effects of energy restriction and fasting. If AICAR had similar actions in adult muscle, they would clearly enhance atrophy and could counter any beneficial effects of PGC-1α. Thus, identification of molecules that are more selective inducers of PGC-1α or PGC-1β might still be beneficial in reducing wasting.
Secondly, the protection against muscle wasting may require a larger increase in PGC-1α (or PGC-1β) than is achievable with AICAR, which only prevented the atrophy-associated fall in PGC-1α. In other words, only high amounts of PGC-1α or PGC-1β as seen after electroporation of expression plasmids or use of adenoviruses may be protective against disuse atrophy, whereas normal or low levels of PGC-1 may be ineffective in influencing proteolysis. In support of this, 1 week of chronic contractile activity prior to 1 week of denervation is sufficient to prevent the loss of mitochondrial content (and presumably PGC-1α content) but does not prevent denervation atrophy (59). Furthermore, muscles without PGC-1α (as in muscle-specific PGC-1α knock-out mice) are similar in size and lose mass in response to denervation at the same rate as muscles from control mice (65). Therefore, activating PGC-1α or PGC-1β may still be an excellent approach to inhibit protein breakdown and atrophy, but possible therapeutic agents may have to increase PGC-1 levels significantly above normal levels to influence muscle size and FoxO function.
Another possible reason that AICAR treatment did not inhibit muscle atrophy is that this treatment did not actually mimic all the transcriptional effects of PGC-1α. In fact important differences seem likely, because we found that the transcription factor NRF-1 did not rise after AICAR treatment (Fig. 6D), although it did increase when PGC-1α was induced with adenovirus (Fig. 2B). NRF-1 is necessary for the transcription of the vast majority of mitochondrial genes, both in the nucleus and in the mitochondria (47). Up-regulating PGC-1α by genetic means (38) or by exercise (66, 67) results in a substantial increase in NRF-1 expression, whereas denervation, which lowers PGC-1α levels, was found to lower the content of NRF-1 protein (Fig. 6C). Perhaps lowering the amount of cellular NRF-1 may somehow limit the ability of PGC-1α to inhibit FoxO function and to reduce muscle atrophy. These findings thus support prior arguments that AICAR, while inducing some of the adaptations seen with exercise, is not an exercise mimetic (68, 69).
Thus, NRF-1 or another protein whose expression is regulated by PGC-1α or PGC-1β could be the link between PGC-1α and rates of protein degradation. Interestingly, NRF-1 binds to the promoter region of many genes, not just those necessary for mitochondrial production (70), and some of these same genes (such as some proteasome subunits, BNIP3, and cathepsin L) are highly up-regulated during muscle atrophy when protein degradation is accelerated (3). Thus, the ability of PGC-1 to increase NRF-1 levels could contribute to its effects on protein degradation.
Despite its inability to maintain muscle size during atrophy, AICAR or related agents may still be useful therapeutically during disuse or in other catabolic states to improve one aspect of muscle function. The loss of muscle mass during atrophy results in the loss of peak force production (42, 71). In addition, the atrophied muscles fatigue more readily than control muscles, probably due to the loss of mitochondrial content (42). Because AICAR treatment maintained mitochondrial content for 2 weeks after denervation, AICAR could still be an effective treatment to maintain muscle endurance in catabolic states or bed-ridden patients.
Acknowledgments
We thank Zoltan Arany (Dana-Farber Cancer Institute) for kindly providing the PGC-1β expression plasmid and adenovirus, Stewart Lecker (Beth Israel Deaconess Medical Center) for the Atrogin1 antibody, and Millennium, Inc., for bortezomib.
This work was supported, in whole or in part, by National Institutes of Health Grant AR055255 (to A. L. G.). This work was also supported by the Muscular Dystrophy Association (to A. L. G.).
- PGC-1α
- peroxisome proliferator-activated receptor γ coactivator 1α
- AICAR
- 5-aminoimidazole-4-carboxamide 1-β-d-ribofuranoside
- DMEM
- Dulbecco's modified Eagle's medium
- GFP
- green fluorescent protein
- HBSS
- Hanks' balanced salt solution
- IRES
- internal ribosomal entry site
- caFoxO3
- constitutively active FoxO3
- TA
- tibialis anterior
- NRF-1
- nuclear respiratory factor-1.
REFERENCES
- 1.Lecker S. H., Solomon V., Mitch W. E., Goldberg A. L. (1999) J. Nutr. 129, 227S–237S [DOI] [PubMed] [Google Scholar]
- 2.Tawa N. E., Jr., Odessey R., Goldberg A. L. (1997) J. Clin. Invest. 100, 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lecker S. H., Jagoe R. T., Gilbert A., Gomes M., Baracos V., Bailey J., Price S. R., Mitch W. E., Goldberg A. L. (2004) FASEB J. 18, 39–51 [DOI] [PubMed] [Google Scholar]
- 4.Sacheck J. M., Hyatt J. P., Raffaello A., Jagoe R. T., Roy R. R., Edgerton V. R., Lecker S. H., Goldberg A. L. (2007) FASEB J. 21, 140–155 [DOI] [PubMed] [Google Scholar]
- 5.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004) Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stitt T. N., Drujan D., Clarke B. A., Panaro F., Timofeyva Y., Kline W. O., Gonzalez M., Yancopoulos G. D., Glass D. J. (2004) Mol. Cell 14, 395–403 [DOI] [PubMed] [Google Scholar]
- 7.Zhao J., Brault J. J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S. H., Goldberg A. L. (2007) Cell Metab. 6, 472–483 [DOI] [PubMed] [Google Scholar]
- 8.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S. J., Di Lisi R., Sandri C., Zhao J., Goldberg A. L., Schiaffino S., Sandri M. (2007) Cell Metab. 6, 458–471 [DOI] [PubMed] [Google Scholar]
- 9.Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., Pan Z. Q., Valenzuela D. M., DeChiara T. M., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Science 294, 1704–1708 [DOI] [PubMed] [Google Scholar]
- 10.Gomes M. D., Lecker S. H., Jagoe R. T., Navon A., Goldberg A. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai D., Frantz J. D., Tawa N. E., Melendez P. A., Oh B. C., Lidov H. G., Hasselgren P. O., Frontera W. R., Lee J., Glass D. J., Shoelson S. E. (2004) Cell 119, 285–298 [DOI] [PubMed] [Google Scholar]
- 12.Van Gammeren D., Damrauer J. S., Jackman R. W., Kandarian S. C. (2009) FASEB J. 23, 362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wehling M., Cai B., Tidball J. G. (2000) FASEB J. 14, 103–110 [DOI] [PubMed] [Google Scholar]
- 14.Adams G. R., Haddad F., Bodell P. W., Tran P. D., Baldwin K. M. (2007) J. Appl. Physiol. 103, 1644–1654 [DOI] [PubMed] [Google Scholar]
- 15.Zinna E. M., Yarasheski K. E. (2003) Curr. Opin. Clin. Nutr. 6, 87–93 [DOI] [PubMed] [Google Scholar]
- 16.Sandri M., Lin J., Handschin C., Yang W., Arany Z. P., Lecker S. H., Goldberg A. L., Spiegelman B. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16260–16265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goto M., Terada S., Kato M., Katoh M., Yokozeki T., Tabata I., Shimokawa T. (2000) Biochem. Biophys. Res. Commun. 274, 350–354 [DOI] [PubMed] [Google Scholar]
- 18.Meirhaeghe A., Crowley V., Lenaghan C., Lelliott C., Green K., Stewart A., Hart K., Schinner S., Sethi J. K., Yeo G., Brand M. D., Cortright R. N., O'Rahilly S., Montague C., Vidal-Puig A. J. (2003) Biochem. J. 373, 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akimoto T., Pohnert S. C., Li P., Zhang M., Gumbs C., Rosenberg P. B., Williams R. S., Yan Z. (2005) J. Biol. Chem. 280, 19587–19593 [DOI] [PubMed] [Google Scholar]
- 20.Taylor E. B., Lamb J. D., Hurst R. W., Chesser D. G., Ellingson W. J., Greenwood L. J., Porter B. B., Herway S. T., Winder W. W. (2005) Am. J. Physiol. Endocrinol. Metab. 289, E960–968 [DOI] [PubMed] [Google Scholar]
- 21.St-Pierre J., Lin J., Krauss S., Tarr P. T., Yang R., Newgard C. B., Spiegelman B. M. (2003) J. Biol. Chem. 278, 26597–26603 [DOI] [PubMed] [Google Scholar]
- 22.Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., Spiegelman B. M. (2002) Nature 418, 797–801 [DOI] [PubMed] [Google Scholar]
- 23.Arany Z., Lebrasseur N., Morris C., Smith E., Yang W., Ma Y., Chin S., Spiegelman B. M. (2007) Cell Metab. 5, 35–46 [DOI] [PubMed] [Google Scholar]
- 24.Wenz T., Rossi S. G., Rotundo R. L., Spiegelman B. M., Moraes C. T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20405–20410 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Cuthbertson D. J., Babraj J. A., Mustard K. J., Towler M. C., Green K. A., Wackerhage H., Leese G. P., Baar K., Thomason-Hughes M., Sutherland C., Hardie D. G., Rennie M. J. (2007) Diabetes 56, 2078–2084 [DOI] [PubMed] [Google Scholar]
- 26.Narkar V. A., Downes M., Yu R. T., Embler E., Wang Y. X., Banayo E., Mihaylova M. M., Nelson M. C., Zou Y., Juguilon H., Kang H., Shaw R. J., Evans R. M. (2008) Cell 134, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes B. F., Kurth-Kraczek E. J., Winder W. W. (1999) J. Appl. Physiol. 87, 1990–1995 [DOI] [PubMed] [Google Scholar]
- 28.Jäger S., Handschin C., St-Pierre J., Spiegelman B. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jørgensen S. B., Treebak J. T., Viollet B., Schjerling P., Vaulont S., Wojtaszewski J. F., Richter E. A. (2007) Am. J. Physiol. Endocrinol. Metab. 292, E331–339 [DOI] [PubMed] [Google Scholar]
- 30.Winder W. W., Holmes B. F., Rubink D. S., Jensen E. B., Chen M., Holloszy J. O. (2000) J. Appl. Physiol. 88, 2219–2226 [DOI] [PubMed] [Google Scholar]
- 31.Suwa M., Nakano H., Kumagai S. (2003) J. Appl. Physiol. 95, 960–968 [DOI] [PubMed] [Google Scholar]
- 32.Nakashima K., Yakabe Y. (2007) Biosci. Biotechnol. Biochem. 71, 1650–1656 [DOI] [PubMed] [Google Scholar]
- 33.Krawiec B. J., Nystrom G. J., Frost R. A., Jefferson L. S., Lang C. H. (2007) Am. J. Physiol. Endocrinol. Metab. 292, E1555–E1567 [DOI] [PubMed] [Google Scholar]
- 34.Sacheck J. M., Ohtsuka A., McLary S. C., Goldberg A. L. (2004) Am. J. Physiol. Endocrinol. Metab. 287, E591–E601 [DOI] [PubMed] [Google Scholar]
- 35.Srere P. A. (1969) Methods Enzymol. 13, 3–11 [DOI] [PubMed] [Google Scholar]
- 36.Green J. D., Narahara H. T. (1980) J. Histochem. Cytochem. 28, 408–412 [DOI] [PubMed] [Google Scholar]
- 37.Bdolah Y., Segal A., Tanksale P., Karumanchi S. A., Lecker S. H. (2007) Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R971–R976 [DOI] [PubMed] [Google Scholar]
- 38.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 39.Rohas L. M., St-Pierre J., Uldry M., Jäger S., Handschin C., Spiegelman B. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7933–7938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanai J., Cao P., Tanksale P., Imamura S., Koshimizu E., Zhao J., Kishi S., Yamashita M., Phillips P. S., Sukhatme V. P., Lecker S. H. (2007) J. Clin. Invest. 117, 3940–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen S., Brault J. J., Gygi S. P., Glass D. J., Valenzuela D. M., Gartner C., Latres E., Goldberg A. L. (2009) J. Cell Biol. 185, 1083–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wicks K. L., Hood D. A. (1991) Am. J. Physiol. 260, C841–C850 [DOI] [PubMed] [Google Scholar]
- 43.Acharyya S., Villalta S. A., Bakkar N., Bupha-Intr T., Janssen P. M., Carathers M., Li Z. W., Beg A. A., Ghosh S., Sahenk Z., Weinstein M., Gardner K. L., Rafael-Fortney J. A., Karin M., Tidball J. G., Baldwin A. S., Guttridge D. C. (2007) J. Clin. Invest. 117, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mourkioti F., Kratsios P., Luedde T., Song Y. H., Delafontaine P., Adami R., Parente V., Bottinelli R., Pasparakis M., Rosenthal N. (2006) J. Clin. Invest. 116, 2945–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dodd S. L., Hain B., Senf S. M., Judge A. R. (2009) FASEB J. 23, 3415–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burkholder T. J., Fingado B., Baron S., Lieber R. L. (1994) J. Morphol. 221, 177–190 [DOI] [PubMed] [Google Scholar]
- 47.Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 48.Coussens L. M., Jacks T. (2008) Curr. Opin. Genet. Dev. 18, 1–2 [DOI] [PubMed] [Google Scholar]
- 49.Furuno K., Goodman M. N., Goldberg A. L. (1990) J. Biol. Chem. 265, 8550–8557 [PubMed] [Google Scholar]
- 50.Goldberg A. L. (1969) J. Biol. Chem. 244, 3223–3229 [PubMed] [Google Scholar]
- 51.St-Pierre J., Drori S., Uldry M., Silvaggi J. M., Rhee J., Jäger S., Handschin C., Zheng K., Lin J., Yang W., Simon D. K., Bachoo R., Spiegelman B. M. (2006) Cell 127, 397–408 [DOI] [PubMed] [Google Scholar]
- 52.Zhang C., Cuervo A. M. (2008) Nat. Med. 14, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Medicherla B., Goldberg A. L. (2008) J. Cell Biol. 182, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. (2003) Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- 55.Lin J., Handschin C., Spiegelman B. M. (2005) Cell Metab. 1, 361–370 [DOI] [PubMed] [Google Scholar]
- 56.Housley M. P., Udeshi N. D., Rodgers J. T., Shabanowitz J., Puigserver P., Hunt D. F., Hart G. W. (2009) J. Biol. Chem. 284, 5148–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olmos Y., Valle I., Borniquel S., Tierrez A., Soria E., Lamas S., Monsalve M. (2009) J. Biol. Chem. 284, 14476–14484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adhihetty P. J., O'leary M. F., Chabi B., Wicks K. L., Hood D. A. (2007) J. Appl. Physiol. 102, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 59.O'Leary M. F., Hood D. A. (2008) J. Appl. Physiol. 105, 114–120 [DOI] [PubMed] [Google Scholar]
- 60.Zhang J., Ney P. A. (2009) Cell Death Differ. 16, 939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Irrcher I., Adhihetty P. J., Sheehan T., Joseph A. M., Hood D. A. (2003) Am. J. Physiol. Cell Physiol. 284, C1669–C1677 [DOI] [PubMed] [Google Scholar]
- 62.Towler M. C., Hardie D. G. (2007) Circ. Res. 100, 328–341 [DOI] [PubMed] [Google Scholar]
- 63.Witczak C. A., Sharoff C. G., Goodyear L. J. (2008) Cell Mol. Life Sci. 65, 3737–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Irrcher I., Ljubicic V., Kirwan A. F., Hood D. A. (2008) PLoS. ONE 3, e3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Handschin C., Chin S., Li P., Liu F., Maratos-Flier E., Lebrasseur N. K., Yan Z., Spiegelman B. M. (2007) J. Biol. Chem. 282, 30014–30021 [DOI] [PubMed] [Google Scholar]
- 66.Murakami T., Shimomura Y., Yoshimura A., Sokabe M., Fujitsuka N. (1998) Biochim. Biophys. Acta 1381, 113–122 [DOI] [PubMed] [Google Scholar]
- 67.Baar K., Wende A. R., Jones T. E., Marison M., Nolte L. A., Chen M., Kelly D. P., Holloszy J. O. (2002) FASEB J. 16, 1879–1886 [DOI] [PubMed] [Google Scholar]
- 68.Goodyear L. J. (2008) N. Engl. J. Med. 359, 1842–1844 [DOI] [PubMed] [Google Scholar]
- 69.Richter E. A., Kiens B., Wojtaszewski J. F. P. (2008) Cell Metab. 8, 96–98 [DOI] [PubMed] [Google Scholar]
- 70.Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R. C., Young R., Kluger Y., Dynlacht B. D. (2004) Mol. Cell 16, 399–411 [DOI] [PubMed] [Google Scholar]
- 71.Patterson M. F., Stephenson G. M., Stephenson D. G. (2006) Am. J. Physiol. Cell Physiol. 291, C518–C528 [DOI] [PubMed] [Google Scholar]