Abstract
Low reprogramming efficiency and reduced pluripotency have been the two major obstacles in induced pluripotent stem (iPS) cell research. An effective and quick method to assess the pluripotency levels of iPS cells at early stages would significantly increase the success rate of iPS cell generation and promote its applications. We have identified a conserved imprinted region of the mouse genome, the Dlk1-Dio3 region, which was activated in fully pluripotent mouse stem cells but repressed in partially pluripotent cells. The degree of activation of this region was positively correlated with the pluripotency levels of stem cells. A mammalian conserved cluster of microRNAs encoded by this region exhibited significant expression differences between full and partial pluripotent stem cells. Several microRNAs from this cluster potentially target components of the polycomb repressive complex 2 (PRC2) and may form a feedback regulatory loop resulting in the expression of all genes and non-coding RNAs encoded by this region in full pluripotent stem cells. No other genomic regions were found to exhibit such clear expression changes between cell lines with different pluripotency levels; therefore, the Dlk1-Dio3 region may serve as a marker to identify fully pluripotent iPS or embryonic stem cells from partial pluripotent cells. These findings also provide a step forward toward understanding the operating mechanisms during reprogramming to produce iPS cells and can potentially promote the application of iPS cells in regenerative medicine and cancer therapy.
Keywords: Differentiation, Embryonic Stem Cell, Epigenetics, Induced Pluripotent Stem (iPS) Cell, MicroRNA, Dlk1-Dio3 Region, Pluripotency
Introduction
It is well known that embryonic stem (ES)4 cells have two defining properties: self-renewal and pluripotency (1, 2). Induced pluripotent stem (iPS) cells from mice have developmental potential identical to ES cells; recent studies confirmed that both have the ability to support full term development by tetraploid complementation (3). However, the success rate for obtaining iPS cells with full pluripotency is extremely low as compared with ES cells, which severely hinders basic iPS cell research and the use of this technology in regenerative medicine and other clinical applications.
Differences between ES and iPS cells in gene expression and differentiation ability have been identified (4, 5). Several reports indicated that iPS and ES cells express the same surface protein markers and are equally able to generate teratomas yet that they differ in their developmental potentials (6–8). As extensive chromatin remodeling and genome-wide epigenetic modifications occur during the reprogramming process, it is possible that the obtained iPS cells vary in their degrees of pluripotency due to different levels of reprogramming. Therefore, a fast and effective method to assess the extent of reprogramming and thereby predict the pluripotency of iPS cells would greatly improve iPS cell selection and applications.
The iPS process dramatically changes cell fate via a few transcription factors (6) and has sparked intense interest in molecular and cellular biology. However, its underlying mechanisms are still poorly understood. Multiple studies have aimed to identify the core molecular circuitry controlling cell pluripotency and self-renewal and identified several signaling pathways downstream of the key reprogramming factors (9–13). How these signaling networks invoked by the induced transcription factors convert cell fate still remains obscure.
By comparing gene and small RNA expression patterns in iPS cells with different degrees of pluripotency, we identified an imprinted genomic region, the Dlk1-Dio3 region, which was actively expressed in fully pluripotent stem cells but was repressed in the cells with partial pluripotency. The significant and consistent expression difference at this imprinted genomic region may serve as a marker to assess the pluripotency extent of cells. A large cluster of microRNAs (miRNAs) encoded by this imprinted region potentially form a feedback loop by regulating polycomb repressive complex 2 (PRC2) formation and are predicted to be involved in broad developmental regulatory processes, suggesting their master roles in gene expression regulatory networks. Protein-coding genes within the Dlk1-Dio3 region are highly conserved across species, whereas the miRNA cluster is conserved only in mammals. This indicated that it might have evolved early during selection for mechanisms that regulate stem properties, including those in human.
EXPERIMENTAL PROCEDURES
Generation of iPS Cell Lines and iPS Mice
Five iPS cell lines and their derived iPS mice used in this study were reported previously (3). Three other new iPS cell lines derived from adult tail tip fibroblasts and neural stem cells were induced by the “four Yamanaka factors” as previous described (3). iPS mice were produced by injecting 10–15 iPS cells into tetraploid blastocysts following the same procedure reported before (3). Two ES cell lines (ESC2 and R1) were also included. All pluripotent stem cells were cultured in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, 1,000 units of leukemia inhibitory factor (LIF), and 1× non-essential amino acids. All experiments requiring animal handling were conducted in accordance with Beijing animal protection laws.
Microarray Hybridization and Identification of Differentially Expressed Genes
Microarray hybridization data of all cell lines were obtained using identical procedures as described before (3). The microarray data were processed by R packages and BioConductor. The raw hybridization signals were normalized using the robust multichip average method and transformed to log 2 values. Differentially expressed genes were selected using Student's t test with multiple testing correction by false discovery rate cut off < 0.05. Expression heat maps of genes and small RNAs within the imprinted region were drawn using the heatmap.2 package of R.
Deep Sequencing Data Analysis, miRNA Identification, and miRNA Target Prediction
After removing adaptor sequences, the resulting Solexa sequencing data were mapped back to the mouse genome using BLAST; only sequences with perfect genomic matches were kept for future analysis. Known mouse miRNAs were downloaded from the miRBase (release 14) and mapped to the mouse genome using BLAST. Sequence reads overlapping with the genomic loci of known miRNAs with no more than 3 nucleotides of variation in length were identified. The 3′-untranslated region sequences of mouse genes were downloaded from the Ensemble database (NCBIM37). Genes whose 3′-untranslated region sequences contained at least two perfect complementary sequences to the 5′ 2–8-nucleotide seed region of a miRNA were selected as putative miRNA targets.
Gene Ontology and Pathway Analysis of Putative miRNA Targets
The enrichment of gene ontology terms among putative miRNA targets was analyzed using the GOEAST software with default parameters (14). Pathway analysis was performed using the DAVID Bioinformatics Resources with the BioCarta pathway database (15).
RESULTS
Mouse Stem Cells of Various Origins Exhibit Different Levels of Pluripotency
Previously we reported the production of live mice from iPS cells of different genetic backgrounds using the four Yamanaka factors (3, 6). In this study, in addition to the previously reported iPS cell lines, mouse embryonic fibroblasts (MEF), neural stem cells, and adult tail tip fibroblasts were collected from two mouse strains (B6 × D2 F1 and B6 × 129S2 F1). After transfection to introduce the four Yamanaka factors, the pluripotency of these iPS cells was assessed. Two ES cells lines (the R1 ES cells (16) and the ESC2 from the B6 × D2 F1 genetic background) were used as positive controls for pluripotency. All the iPS cell lines predominantly formed normal, diploid nuclei with 40 chromosomes, expressed pluripotency marker genes with expected patterns, and were able to form embryoid bodies in vitro. All lines generated teratomas with three germ layers in vivo and contributed to the generation of chimeric mice after injection into diploid blastocysts. However, two of the iPS cell lines could not produce full term mice through tetraploid complementation, and one of these lines even failed to support germline transmission in chimeras. These results indicated that the iPS cell lines varied in their pluripotency. Those with the ability to generate tetraploid blastocyst-complementated embryos (4n-iPS cells) exhibited full pluripotency equivalent to ES cells, whereas the two lines that were only able to produce diploid chimeric mice (2n-iPS cells) were classified as partially pluripotent. The pluripotency levels of all cell lines generated are summarized in Table 1.
TABLE 1.
Summary of ES and iPS cell lines
✓ indicates positive result from assay, and ✗ indicates negative result of assay. TTF, tail tip fibroblasts; NSC, neural stem cells.
| Pluripotent stem cell line | Nuclear donor | Genetic background | Immunohistochemistry (Oct4, Nanog, SSEA-1) | Karyotype | EB | Teratoma formation | Chimera formation (germline transmission) | Tetraploid complementation | Reference |
|---|---|---|---|---|---|---|---|---|---|
| IP36D-3 | MEF | B6 × D2 F1 | ✓ | Normal | ✓ | ✓ | ✓ (✗) | ✗ | (3) |
| IP20D-3 | MEF | B6 × D2 F1 | ✓ | Normal | ✓ | ✓ | ✓ (✓) | ✗ | (3) |
| ESC2 | Fertilized egg | B6 × D2 F1 | ✓ | Normal | ✓ | ✓ | ✓ (✓) | ✓ | Present work |
| R1 | Fertilized egg | 129X1/SvJ × 129S1 F1 | ✓ | Normal | ✓ | ✓ | ✓ (✓) | ✓ | (16) |
| IP14D-1 | MEF | B6 × D2 F1 | ✓ | Normal | ✓ | ✓ | ✓ (✓) | ✓ | (3) |
| IP14D-6 | MEF | B6 × D2 F1 | ✓ | Normal | ✓ | ✓ | ✓ (✓) | ✓ | (3) |
| IP14D-101 | MEF | B6 × 129S2 F1 | ✓ | Normal | ✓ | ✓ | ✓ (✓) | ✓ | (3) |
| IP26DT-115 | TTF | B6 × 129S2 F1 | ✓ | Normal | ✓ | ✓ | ✓ (✓) | ✓ | Present work |
| IP14DN-5 | NSC | B6 × D2 F1 | ✓ | Normal | ✓ | ✓ | ✓ (✓) | ✓ | Present work |
| IP14DN-7 | NSC | B6 × D2 F1 | ✓ | Normal | ✓ | ✓ | ✓ (✓) | ✓ | Present work |
Deep Sequencing Reveals MicroRNA Expression Differences between Full and Partial Pluripotent Stem Cells
To investigate whether miRNAs might contribute to the pluripotency differences among cell lines, we collected samples of RNA with size selection for 18–30 nucleotides from the 10 cell lines in Table 1. After Illumina library production for small RNA sequencing, the libraries were subjected to deep sequencing and quantitation using an Illumina genome analyzer. A total of 5–10 million small RNA reads were obtained from each sample, and read analysis indicated that the samples included 401–431 known miRNAs and an additional 77–97 miRNA star sequences).
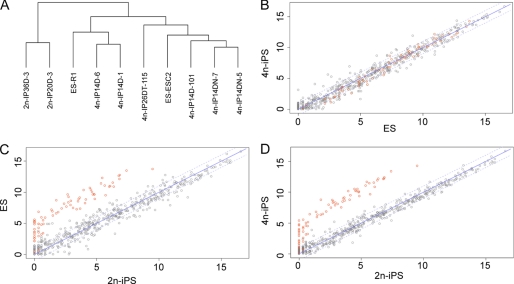
Hierarchical clustering analysis revealed that the miRNA expression patterns of the partially pluripotent stem cell lines (two 2n-iPS cell lines) differed significantly from those of the full pluripotent stem cell lines (two ES and six 4n-iPS cell lines), whereas those of the fully pluripotent stem cell lines were highly related (Fig. 1A). By examining the miRNA expression differences, we identified a group of miRNAs with significantly lower abundances in the partially pluripotent stem cells as compared with the fully pluripotent cells (Fig. 1, B–D). The normalized sequence counts for these miRNAs were almost all below 100 (and some close to zero) out of a total of 5–10 million reads in the two partially pluripotent stem cell lines but showed at least 10-fold (many over 100-fold) greater abundance in all eight fully pluripotent stem cell lines. To the contrary, little expression variance was observed for these miRNAs among the fully pluripotent stem cell lines (supplemental Table 1).
FIGURE 1.
miRNA expression patterns distinguish 2n-iPS cell lines from ES and 4n-iPS cell lines. A, clustering analysis of miRNA read counts in the 10 studied cell lines. B, scatter plot of miRNA expression in ES and 4n-iPS cell lines. C, scatter plot of miRNA expression in ES and 2n-iPS cell lines. D, scatter plot of miRNA expression in 4n-iPS and 2n-iPS cell lines. Normalized read counts (reads per million) were averaged within each cell line group (ES, 4n-iPS, and 2n-iPS) and log 2-transformed prior to drawing the plot.
Expression Levels from the Imprinted Dlk1-Dio3 Region Are Correlated with the Extent of Cell Pluripotency
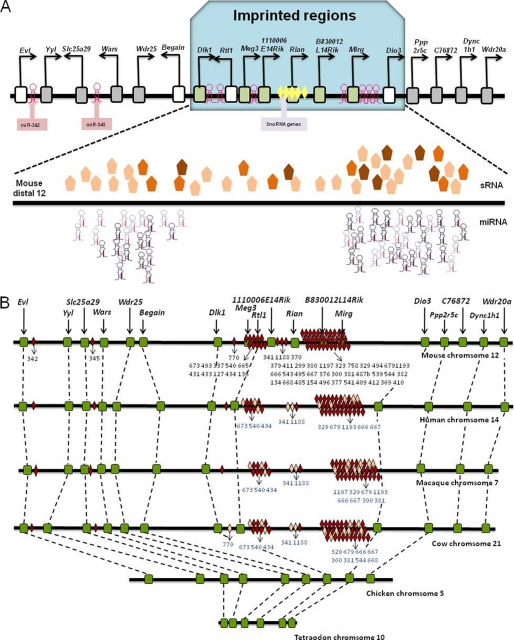
Interestingly, almost all of the miRNAs with reduced abundance in 2n-iPS cells are encoded by an imprinted genomic region (Fig. 1, red circles). This region of about 836 kb is located on the long arm of mouse chromosome 12 and includes the genes Dlk1 and Dio3 at either end. It encodes five protein-coding genes, Dlk1, Rtl1, 1110006E14Rik, B830012L14Rik, and Dio3; three long non-coding RNA genes, Meg3, Rian, and Mirg; one C/D box small nucleolar RNA gene cluster; and a cluster of 47 miRNAs (Fig. 2A). Protein-coding genes in this region were conserved during evolution (17), but other genes and the miRNA clusters within this region only arose in and are conserved among mammals (Fig. 2B). Previous studies showed that the expression of genes and miRNAs from this imprinted region is restricted to mouse embryos and adult brains (18, 19), but the functions of these genes and miRNAs are largely unknown.
FIGURE 2.
Expression and conservation analysis of the Dlk1-Dio3 imprinted region. A, expression difference of the Dlk1-Dio3 imprinted region between 4n-competent (ES and 4n-iPS) cell lines and 4n-incompetent (2n-iPS) cell lines. The protein-coding genes and long non-coding RNAs with higher expression in the 4n-competent cell lines are shown as green rectangles, the unaltered genes are shown as gray rectangles, and the non-detected genes are shown as empty boxes. Hairpin-shaped symbols represent miRNAs, and the darkness of the symbols represents the magnitude of expression difference between 4n-competent and 4n-incompetent cell lines. Pentagons represent other small RNAs, and the degree of color saturation represents the magnitude of expression difference between 4n-competent and 4n-incompetent cell lines. Genes, miRNAs, and other small RNAs (sRNA) encoded by the Dlk1-Dio3 imprinting region were activated in the 4n-competent cell lines but repressed in the 4n-incompetent cell lines. B, conservation analysis of the Dlk1-Dio3 region and neighboring genes. Conserved miRNAs are shown as red diamonds, non-conserved miRNAs are shown as pink diamonds, and other genes are shown as green boxes. Black numbers represent the IDs of conserved miRNAs, and blue numbers represent the IDs of non-conserved miRNAs.
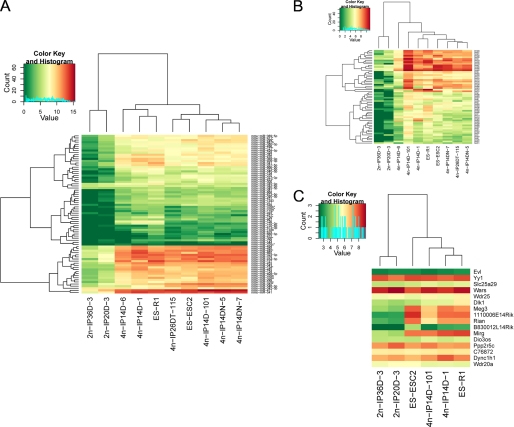
Except for 12 miRNAs that were almost undetectable in all cell lines, the remaining 35 Dlk1-Dio3 miRNAs all had comparable medium to high expression in the ES and 4n-iPS cells but were expressed at very low or undetectable levels in the 2n-iPS cells, whereas miR-342 and miR-345, which are encoded upstream of the imprinted region, had similar expression in all cell lines (supplemental Table 1). Besides miRNAs, a large number of small RNAs were also identified by sequencing and might resemble endogenous small interfering RNAs (Fig. 2). The expression levels of these small RNAs were also reduced in the partially pluripotent stem cell lines, similar to what was observed for miRNAs (Fig. 3B). Transcriptome analysis for 6 of the 10 cell lines using Affymetrix mouse genome 430 2.0 microarrays revealed that except for B830012L14Rik, the expression levels of other genes encoded by the Dlk1-Dio3 region were all significantly lower in the 2n-iPS cell lines as compared with ES and 4n-iPS cell lines, whereas genes outside of this region had no expression differences (Fig. 3C).
FIGURE 3.
Hierarchical clustering of expression levels of miRNAs (A), other small RNAs (B), and protein-coding genes (C) encoded by the Dlk1-Dio3 region. Neighboring genes surrounding the Dlk1-Dio3 region were also included in panel C. Log 2-transformed expression values detected either by deep sequencing or by microarray were used. Clusters were drawn using the hclust package of R.
Slight Expression Differences Are Observed between Partially Pluripotent Stem Cells Lines
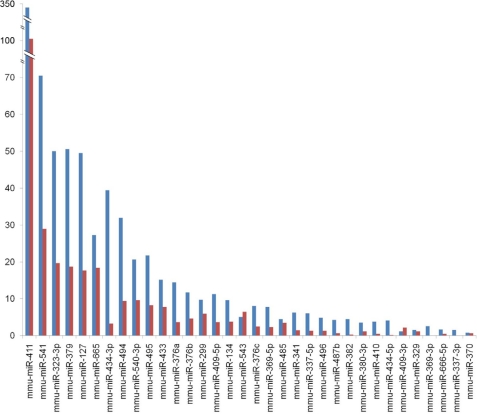
The two 2n-iPS cell lines used in this study exhibited different levels of pluripotency. The IP20D-3 cells were germline-transmittable, whereas the IP36D-3 cells were not. Although the Dlk1-Dio3 region encoded miRNAs all had significantly lower expression in the two 2n-iPS cell lines, their expression levels in the IP20D-3 cells were consistently higher than those in the IP36D-3 cells (Fig. 4).
FIGURE 4.
Expression differences of the Dlk1-Dio3 region encoded miRNAs between germline-competent and germline-incompetent 2n-iPS cell lines. Normalized counts of miRNA sequence reads from germline-competent and germline-incompetent 2n-iPS cell lines (IP20D-3 versus IP36D-3) are shown in blue and red, respectively.
Predicted Feedback miRNA Expression Regulatory Loop via Polycomb Silencing Complex 2
Microarray transcript profiling identified 1,639 and 3,467 genes with higher expression and 3,467 genes with lower expression levels in the ES and 4n-iPS cells compared to the 2n-iPS cells. As miRNAs and their target mRNAs usually exhibit reverse correlated expression patterns, the genes with reduced abundance in the ES and 4n-iPS cell lines may contain target recognition sites for the miRNAs that showed higher abundance. By searching these mRNAs for 3′-untranslated regions containing at least two perfect complementary binding sites to the 5′ 2–8-nucleotide seed regions of the Dlk1-Dio3 region-encoded miRNAs, 717 putative targets were identified for miRNAs with at least 50 clone counts in at least one ES or 4n iPS cell line. Gene ontology enrichment analysis revealed that genes related to multiple aspects of growth, differentiation, metabolism, and other developmental processes were significantly enriched among the putative miRNA target genes, indicating that activated miRNA expression may repress genes related to cell development.
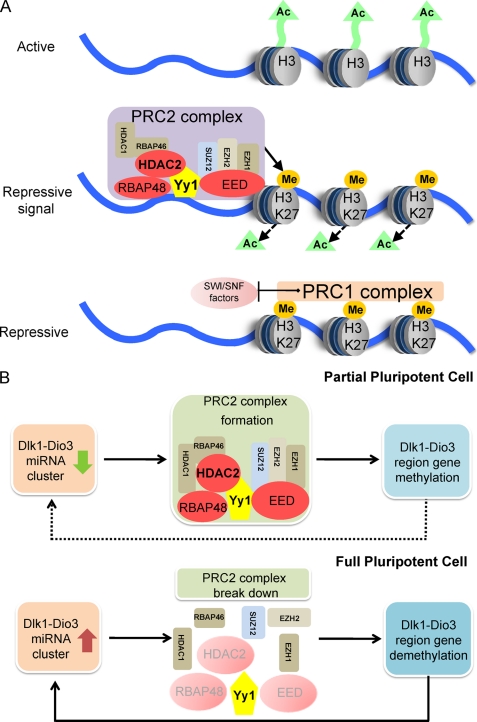
Pathway analysis revealed that three components of the polycomb repressive complex 2 (PRC2), namely HDAC2, RBAP48, and EED, were among the putative miRNA targets (supplemental Table 2). PRC2 induces gene silencing by trimethylating lysine 27 on histone 3 (3meH3K27) (20). Prior to the onset of PRC2-induced methylation, the action of histone deacetylase HDAC2 is required to remove the active acetylation marks on histone 3 (21). Our results indicated that in ES and 4n-iPS cells, the expression of the Dlk1-Dio3 region-encoded miRNAs was correlated with reduced HDAC2, RBAP48, and EED expression, which may prevent PRC2 formation. As the imprinting of the Dlk1-Dio3 region is regulated by methylation, the decreased PRC2 formation in ES and 4n-iPS cells might result in the maintenance of histone acetylation and failure to methylate the Dlk1-Dio3 region, which would be consistent with the increased gene and miRNA expression levels observed (Fig. 5).
FIGURE 5.
A working model for miRNA regulation of PRC2 complex. A, PRC2 induces gene silencing by methylating histone 3 at the Lys-27 sites. B, in partially pluripotent cells, the Dlk1-Dio3 region is methylated by PRC2 and the expression of the Dlk1-Dio3 region encoded miRNA cluster is repressed. In fully pluripotent cells, the Dlk1-Dio3 region is activated and the miRNA cluster is expressed. Consequently, transcripts encoding HDAC2, RBAP48, and EED are repressed by miRNAs, and methylation at the Dlk1-Dio3 region is blocked, resulting in a feedback loop to further increase miRNA and mRNA expression from this region.
DISCUSSION
The production of healthy cells with full pluripotency is essential for stem cell research and its application in regenerative medicine. However, the low efficiency of iPS cell generation and the reduced pluripotency of these cells have long been obstacles. Therefore, fast and effective methods to adjust the pluripotent properties of cells will greatly accelerate the exploration of reprogramming mechanisms and the use of iPS cells in regenerative medicine.
Here, we identified an imprinted genomic region in mouse that was actively expressed in fully pluripotent stem cell lines, such as ES and 4n-iPS cells, but was repressed in partially pluripotent cells as represented by two 2n-iPS cell lines. Protein-coding genes, a large cluster of miRNAs, and other small RNAs derived from this Dlk1-Dio3 imprinted region consistently exhibited significant expression repression or depletion in the 2n-iPS cells as compared with the ES and 4n-iPS cells. Regardless of the genetic background and origin of the ES and iPS cell lines, the expression differences between the fully and partially pluripotent stem cells were consistently observed. Furthermore, although genes and miRNAs from this region had low overall expression in the 2n-iPS cell lines, their expression was slightly higher in the germline-transmittable 2n-iPS cells than in those without germline transmission ability. These results indicated that the degree of activation of the Dlk1-Dio3 imprinted region is positively correlated with the pluripotency levels of stem cells. The coordinated expression changes of protein-coding genes and miRNAs encoded by this region, measured in independent assays, supports the conclusion that this region is affected by some regulatory mechanism that may be responsible for or respond to the pluripotency status of a cell.
Our analysis revealed that the miRNA cluster encoded by the Dlk1-Dio3 region only presented in mammalian genomes and is highly conserved among mammals, indicating its specific and crucial role in regulating mammalian development. We expect that this miRNA cluster should have conserved functions in human and other mammals. As some other genes within the Dlk1-Dio3 region are also conserved in non-mammal species, it is very likely that the pluripotency-correlated expression of the entire Dlk1-Dio3 region is indeed a functional reflection of the miRNA cluster.
After extensively screening the gene and small RNA expression data used in this study, we were not able to identify any other genomic region exhibiting similar or opposite expression patterns as that of the Dlk1-Dio3 imprinted region. These data indicated that the Dlk1-Dio3 region might be the only long genomic locus that exhibits a clear on-and-off switch in cells with full versus partial pluripotency; thus, the expression state of this region is a strong candidate for a marker of the quality of iPS and stem cells from other sources. Fully pluripotent human stem cells are very difficult to obtain via the iPS technique, so identification of such a marker site may greatly accelerate the selection process for good iPS or ES cells and pave the way for their application in regenerative medicine.
The putative targets of miRNAs from the Dlk1-Dio3 region include three genes in the PRC2 silencing complex. These genes are all expressed at lower levels in the ES and 4n-iPS cells as compared with the 2n-iPS cells, perhaps as a result of higher miRNA expression in the ES and 4n-iPS cells. We hypothesize that the repression of PRC2 complex formation contributes to the lack of methylation and silencing of the Dlk1-Dio3 region, resulting in the activated expression of its encoded genes as observed in our results. The activated miRNAs may act in a feedback loop to further prevent the formation of PRC2 and maintain the active state of the Dlk1-Dio3 region. A recent study showed that PRC2 is not necessary for stem cell pluripotency maintenance (22). Therefore, down-regulation of PRC2 in the ES and 4n-iPS cell lines should not affect their pluripotency. Many studies have been conducted to investigate the function of the Dlk1-Dio3 imprinted region, but no clear conclusions have been made. Here, we propose that some miRNAs from this region regulate cell pluripotency via gene silencing pathways. It is likely that the activated miRNA clusters are involved in the regulation of many developmentally related biological processes as revealed by gene ontology enrichment analysis.
The connection of the Dlk1-Dio3 region miRNAs with PRC2 also suggests potential applications of these miRNAs in cancer therapy. As one of the PRC2 components, the histone methyltransferase Ezh2 is frequently observed to be overexpressed in various types of cancers (23). Modulating the formation of PRC2 by manipulating the Dlk1-Dio3 region miRNAs might serve as an approach to prevent the uncontrolled proliferation of some types of cancer cells.
The discovery of iPS technology demonstrated that the developmental status of cells can be reverted by the activity of a few transcription factors, but its underlying mechanisms remain unknown. We hypothesize that changes in the methylation status of an imprinted genomic region might be involved in the pluripotency regulation of cells, providing a testable model toward understanding the mechanism of iPS cell formation.
Scientists have long attempted to identify key components that regulate complex traits but with very little success in protein-coding genes. Our results indicated that cell pluripotency levels might be in partially controlled by a group of miRNAs, suggesting the possibility of miRNAs as master regulators of gene expression networks. The synergetic effects of multiple miRNAs toward the same outcome revealed by our model also furthered our understanding of miRNA function mechanisms and shed light on future studies on the coordinated effects of miRNAs.
This work was in part supported by the National High Technology Research and Development Program of China Grant 2006AA02A101 (to Q. Z.), the Ministry of Science and Technology of China Grants 2006CB701501 (to Q. Z.) and 2007CB946901 to (X.-J. W.), and the National Natural Science Foundation of China Grants 90919060 (to Q. Z.) and 30725014 (to X.-J. W.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database under Accession Number GSE21515.
- ES
- embryonic stem
- iPS
- induced pluripotent stem
- MEF
- mouse embryonic fibroblast
- miRNA
- microRNA.
REFERENCES
- 1.Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T. W., Smith A. (2008) PLoS Biol. 6, e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva J., Smith A. (2008) Cell 132, 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao X. Y., Li W., Lv Z., Liu L., Tong M., Hai T., Hao J., Guo C. L., Ma Q. W., Wang L., Zeng F., Zhou Q. (2009) Nature 461, 86–90 [DOI] [PubMed] [Google Scholar]
- 4.Chin M. H., Mason M. J., Xie W., Volinia S., Singer M., Peterson C., Ambartsumyan G., Aimiuwu O., Richter L., Zhang J., Khvorostov I., Ott V., Grunstein M., Lavon N., Benvenisty N., Croce C. M., Clark A. T., Baxter T., Pyle A. D., Teitell M. A., Pelegrini M., Plath K., Lowry W. E. (2009) Cell Stem Cell 5, 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu B. Y., Weick J. P., Yu J., Ma L. X., Zhang X. Q., Thomson J. A., Zhang S. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4335–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 7.Meissner A., Wernig M., Jaenisch R. (2007) Nat. Biotechnol. 25, 1177–1181 [DOI] [PubMed] [Google Scholar]
- 8.Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007) Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- 9.Niwa H., Ogawa K., Shimosato D., Adachi K. (2009) Nature 460, 118–122 [DOI] [PubMed] [Google Scholar]
- 10.Yamanaka S. (2008) Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2079–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochedlinger K., Plath K. (2009) Development 136, 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sridharan R., Tchieu J., Mason M. J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. (2009) Cell 136, 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loh Y. H., Ng J. H., Ng H. H. (2008) Cell Cycle 7, 885–891 [DOI] [PubMed] [Google Scholar]
- 14.Zheng Q., Wang X. J. (2008) Nucleic Acids Res. 36, W358–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 16.Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards C. A., Mungall A. J., Matthews L., Ryder E., Gray D. J., Pask A. J., Shaw G., Graves J. A., Rogers J., Dunham I., Renfree M. B., Ferguson-Smith A. C. (2008) PLoS Biol. 6, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seitz H., Royo H., Bortolin M. L., Lin S. P., Ferguson-Smith A. C., Cavaillé J. (2004) Genome Res. 14, 1741–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tierling S., Dalbert S., Schoppenhorst S., Tsai C. E., Oliger S., Ferguson-Smith A. C., Paulsen M., Walter J. (2006) Genomics 87, 225–235 [DOI] [PubMed] [Google Scholar]
- 20.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002) Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 21.Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. (2002) Genes Dev. 16, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chamberlain S. J., Yee D., Magnuson T. (2008) Stem Cells 26, 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon J. A., Lange C. A. (2008) Mutat. Res. 647, 21–29 [DOI] [PubMed] [Google Scholar]