Abstract
Protein secretion and localization are crucial during eukaryotic development, establishing local cell environments as well as mediating cell interactions, signaling, and adhesion. In this study, we demonstrate that the glycosyltransferase, pgant3, specifically modulates integrin-mediated cell adhesion by influencing the secretion and localization of the integrin ligand, Tiggrin. We demonstrate that Tiggrin is normally O-glycosylated and localized to the basal matrix where the dorsal and ventral cell layers adhere in wild type Drosophila wings. In pgant3 mutants, Tiggrin is no longer O-glycosylated and fails to be properly secreted to this basal cell layer interface, resulting in disruption of integrin-mediated cell adhesion in the wing. pgant3-mediated effects are dependent on enzymatic activity, as mutations that form a stable protein yet abrogate O-glycosyltransferase activity result in Tiggrin accumulation within the dorsal and ventral cells comprising the wing. Our results provide the first in vivo evidence for the role of O-glycosylation in the secretion of specific extracellular matrix proteins, thus altering the composition of the cellular “microenvironment” and thereby modulating developmentally regulated cell adhesion events. As alterations in cell adhesion are a hallmark of cancer progression, this work provides insight into the long-standing association between aberrant O-glycosylation and tumorigenesis.
Keywords: Cell Adhesion, Development, Drosophila, Extracellular Matrix, Integrin, Protein Secretion, O-Glycosylation, Microenvironment, pgant3
Introduction
Integrin-mediated adhesion is a crucial process involved in many aspects of development, tissue maintenance, and disease. Integrin transmembrane receptors bind to extracellular matrix (ECM)2 ligands to coordinate cell-matrix and cell-cell adhesion as well as regulate cell morphology and signaling events (1–8). ECM ligands are a diverse group of molecules that are secreted by cells, creating local environments and matrices that modulate cell interactions. Mutations in integrins or their ECM ligands have been shown to disrupt cell adhesion, resulting in wing blistering (1, 3–5, 7, 9–12), muscle detachment (3, 13, 14), and defects in dorsal closure (15) in Drosophila, as well as the human disease epidermolysis bullosa, in which the epidermal and dermal skin layers are separated (16, 17). Loss of proper cell adhesive properties is also a hallmark of cancer progression and metastasis.
We have previously shown that an ECM protein, Tiggrin (3, 18, 19), is normally modified by the post-translational addition of the sugar GalNAc (20) through the action of a member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family of glycosyltranferases (EC 2.4.1.41). This type of evolutionarily conserved protein modification, known as mucin-type O-glycosylation (21–23), was shown to be required for proper cell adhesion between the epithelial cell layers comprising the Drosophila wing blade. Loss of the gene encoding the enzyme responsible for glycosylating Tiggrin (pgant3) resulted in the formation of wing blisters in adult flies (similar to those seen in homozygous tiggrin mutant escapers (3)), which could be rescued upon expression of wild type pgant3 (20). Additionally, pgant3 and tiggrin genetically interact to influence wing blistering frequency (20).
Although loss of mucin-type O-glycosylation is associated with a number of diseases and developmental defects (22, 24–30), the mechanistic role of mucin-type O-glycans in most of these instances remains unclear. Previous studies have suggested that O-glycans may influence protein stability, as is thought to be the case in the human disease, familial tumoral calcinosis (26, 31–32). Other studies in cell culture using chemical inhibitors of O-glycan extension have demonstrated that alterations in O-glycan formation can affect secretion and trafficking of certain proteins (33–36). However, these inhibitors also affect other glycosylation pathways, making specific conclusions about the role of mucin-type O-glycans difficult. In this study, we demonstrate that mucin-type O-glycosylation promotes a developmentally regulated cell adhesion event by influencing the proper secretion of an ECM component involved in integrin interactions. This study provides the first example of O-glycosylation influencing the composition of the extracellular matrix and suggests roles for this abundant protein modification in other developmental contexts as well as in disease states where matrix composition and cell adhesion are altered.
EXPERIMENTAL PROCEDURES
Fly Strains Used
The stocks used in this study are as follows: Bloomington stocks 3960 (if3); 4533 (w*; In(2LR)noc4LScorv9R, b1/CyO, P{ActGFP}JMR1); and 7748 (w1118; Df(2R)Exel6283, P{XP-U}Exel6263). Additionally, the following stocks from other sources were used: the w*; pgant3c01318/pgant3c01318 transposon insertion line (20, 37); the w*; pgant3hopout7/pgant3hopout7 transposon excision line (20); and finally, the w; Sco/SM6a and cn1 bw1 sp1 lines (the kind gifts of Dr. J. Kennison).
EMS Mutagenesis Screening and Sequencing
EMS mutagenesis was carried out as described previously (38). Briefly, cn1 bw1 sp1 males were fed EMS and crossed to the homozygous pgant3 transposon insertion line, (w*; pgant3c01318/pgant3c01318). From the F1 progeny, the individual males with blistered wings were collected and backcrossed to virgins from the homozygous pgant3 transposon insertion line. F2 progeny displaying blistered wings were then collected and crossed to w; Sco/SM6a to make a balanced stock of the putative pgant3 mutations.
Putative pgant3 mutation lines were then crossed to a deficiency line (Df(2R)Exel6283, P{XP-U}Exel6263) that uncovers pgant3. Heterozygous progeny from these crosses (pgant3mutant/Df(2R)Exel6283, P{XP-U}Exel6263) were used for sequencing to verify that the new EMS-generated mutations were in the pgant3 gene. Briefly, heterozygous adults were homogenized, and RNA was isolated using the FastRNA Pro Green kit (Qbiogene). cDNA synthesis was performed using iScript cDNA synthesis kit (Bio-Rad). PCR primers were designed to yield four products covering the pgant3 coding region: sense (GATCGGTTTGGATTGGATTG) and antisense (CGAGGCGGCACCACAACTG) to amplify the first exon and a portion of the second exon; sense (CCACTACATCGGCAAGGGAGAC) and antisense (ACCTTGCGTGATTCCTTAATGCG) to amplify part of the second, third, and fourth exon, and a portion of the fifth exon; sense (GGCGATGTGCTGACCTTCCTC) and antisense (TTCATGTGCTTGCTGTAGGC) to amplify the first portion of the fifth exon; and sense (GCCAAGGACAAGGTGAATGT) and antisense (ACCGGCATGACATGACATCCTACTC) to amplify the remainder of the fifth exon and the sixth exon. The resulting PCR fragments were purified using QIAquick gel extraction kit (Qiagen) and sequenced directly. From this analysis, two new pgant3 mutations were identified and designated pgant3m1 and pgant3m2. pgant3m1, and pgant3m2 stocks were then crossed to the previously described pgant3 transposon insertion line and deletion line to determine wing blistering frequencies.
Expression and Localization of PGANT3, PGANT3m1, and PGANT3m2 Enzymes in Drosophila Cells
cDNAs encoding pgant3, pgant3m1, and pgant3m2 were digested with EcoRI and NotI and subcloned into pIB/V5-His vector (Invitrogen) to form a C-terminal V5 fusion. S2R+ cells were transfected with plasmids using Effectene transfection reagent (Qiagen) according to the manufacturer's instructions. After 3 days, cells were fixed, stained with anti-V5 antibody (dilution 1:500) (Invitrogen), and the Drosophila Golgi marker GM130 (dilution, 1:100) (Abcam). After primary antibody staining, the cells were washed and incubated with Alexa 488-conjugated anti-mouse IgG secondary antibodies (dilution, 1:100) (Invitrogen) and Cy3-conjugated anti-rabbit IgG antibody (dilution, 1:100) (Jackson ImmunoResearch).
Expression of Secreted PGANT3, PGANT3m1, and PGANT3m2 Enzymes in COS7 Cells
The plasmids constructed above for localization studies of PGANT3 and PGANT3m2 were partially digested with AgeI to release fragments beginning just after the hydrophobic membrane spanning domain and ending after the V5 epitope at the C terminus. These fragments were then subcloned into the vector pF4-FlyB (39) to generate constructs encoding secreted enzymes with a C-terminal V5 tag (pF4-pgant3-V5 and pF4-pgant3m2-V5). For the pgant3m1 construct, sense (ATAACGCGTTCCAGGGCGGGGACGCGGAG) and antisense (GTGGCGCGGCGACCGGTTGATGAC) PCR primers were using to amplify a 435-bp pgant3m1 fragment beginning just after the hydrophobic region. The amplified PCR fragment was then digested with MluI and AgeI and cloned into the same sites of the vector pF4-pgant3-V5. All constructs were verified by DNA sequencing. Plasmids were transfected into COS7 cells using Lipofectamine reagent (Invitrogen). After 4 days, the media were harvested. To quantitate the relative amount of recombinant proteins secreted into the media, aliquots of media were electrophoresed under reducing conditions and transferred to nitrocellulose membranes. Membranes were incubated with anti-V5 antibody (dilution 1:2000) (Invitrogen) after blocking and developed with horseradish peroxidase-conjugated mouse IgG (dilution 1:2000) (Cell Signaling Technology).
Glycosyltransferase Assays
Assays for glycosyltransferase activity were performed as described previously (39). Briefly, media from COS7 cells expressing recombinant PGANT3 and PGANT3m1 were harvested, and recombinant proteins were quantitated by Western blotting as described above. Equal relative amounts of each recombinant protein were used in the in vitro reactions with sugar donor [14C]UDP-GalNAc and various concentrations (62.5–250 μm) of the EA2 acceptor substrate (PTTDSTTPAPTTK). All reactions were performed in triplicate at 37 °C for 1 h. Reaction products were purified by anion exchange chromatography, and [14C]GalNAc incorporation was measured. Reactions using media from cells expressing empty vector alone yielded background values that were subtracted from each experimental value. Adjusted experimental values were then averaged, and standard deviations were calculated. Glycosyltransferase activity is expressed as dpm/h.
Pupal Wing Disc Staining
Pupae were staged from pupariation (white prepupa). Staged pupae were immersed in 4% formaldehyde in PBS, and then an incision was made through the pupal case and the body wall. Fixation was continued on a shaker for at least 1 h at room temperature. Fixed pupae were then stored at 4 °C. Pupal cases were removed, and pupal wings were dissected. Samples were washed in PBST (PBS, 0.3% Triton X-100) and transferred to blocking buffer (4% goat serum/PBS, 0.3% Triton X-100) for 1 h on a shaker. Samples were then incubated with primary antibody overnight at 4 °C in blocking buffer. Primary antibodies used were mouse anti-β-PS-integrin (40) (Developmental Studies Hybridoma Bank, 1:100), rat anti-DE-cadherin (40) (Developmental Studies Hybridoma Bank, 1:100), mouse anti-Fasciclin III (40) (Developmental Studies Hybridoma Bank, 1:100), mouse anti-Tiggrin antibody (18) (the kind gift of Drs. L. and J. Fessler, 1:100), and mouse anti-Tn antibody (Ca3638) (41) (the kind gift of Dr. Richard Cummings, 1:50). Alexa 488-, 568-, and 647-conjugated secondary antibodies (Invitrogen, 1:100) and Cy3-conjugated donkey anti-mouse IgM antibody (Jackson ImmunoResearch Laboratories, 1:100) were used. Nuclei were stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (Sigma, 1:1000). Wings were mounted in aqueous mounting medium with anti-fading agents (Biomeda) and imaged on the Zeiss LSM 510 confocal laser scanning microscope. Optical cross-sections (X-Z images) of pupal wings in Figs. 2–4 were compiled from multiple X-Y images (1 μm thick) to form the 15–35-μm X-Z images shown. Fig. 6 shows representative 1-μm thick confocal images in the X-Y plane through the center of the dorsal and ventral cell layers of pupal wings. Images were processed using the LSM Imager Browser and Photoshop.
FIGURE 2.
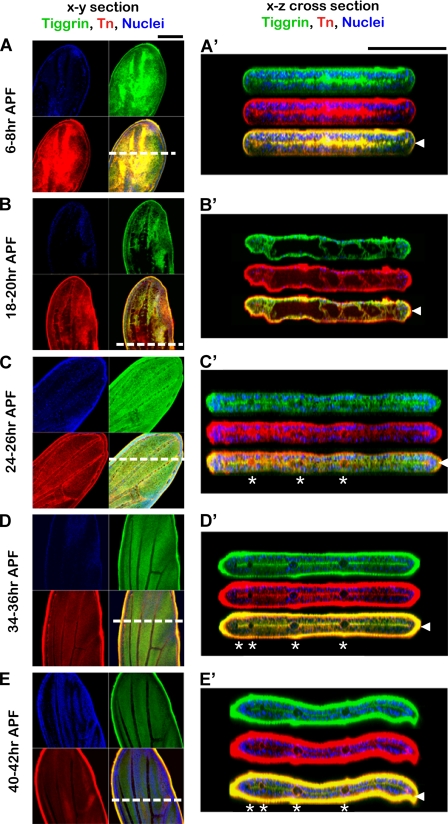
Tiggrin and O-glycans show dynamic and specific localization during pupal wing development. Shown are stages of wild type pupal wing development 6–8 h after puparium formation (6–8 h APF), 18–20 h after puparium formation (18–20 h APF), 24–26 h after puparium formation (24–26 h APF), 34–36 h after puparium formation (34–36 h APF), and 40–42 h after puparium formation (40–42 h APF). Wings were stained for Tiggrin (green), O-glycans (red), and DAPI (blue). Merged images show Tiggrin and O-glycan co-staining in yellow. Images shown are X-Y sections (A–E) or X-Z optical cross-sections (A′, B′, C′, D′, and E′). White dashed lines indicate regions used to produce X-Z optical cross-sections. Arrowheads at the right denote the basal dorsal/ventral cell layer interface, and asterisks denote the position of wing veins. Black bar, 100 μm.
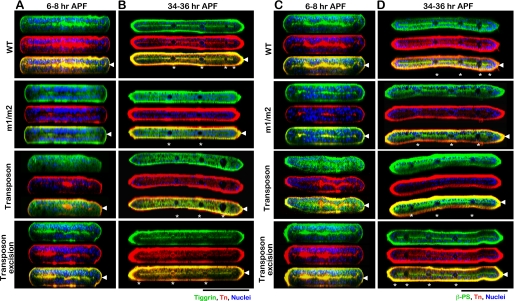
FIGURE 3.
Localization of Tiggrin, βPS integrin, and O-glycans in wild type and pgant3 mutant pupal wings. Pupal wings from either wild type (WT), pgant3m1/pgant3m2 homozygous point mutants (m1/m2), homozygous pgant3c01318/pgant3c01318 transposon insertion mutation (Transposon), or the pgant3hopout7/pgant3hopout7 transposon excision line (Transposon excision) were stained for Tiggrin (green) (A and B), βPS integrin (green) (C and D), and O-glycans (red) (A–D). Merged images show co-staining in yellow. X-Z optical cross-sections of confocal images of pupal wings at 6–8 h APF (A and C) or 34–36 h APF (B and D) are shown. Nuclei were detected with DAPI. Arrowheads at the right denote the basal dorsal/ventral cell layer interface, and asterisks denote the position of wing veins. Black bar, 100 μm.
FIGURE 4.
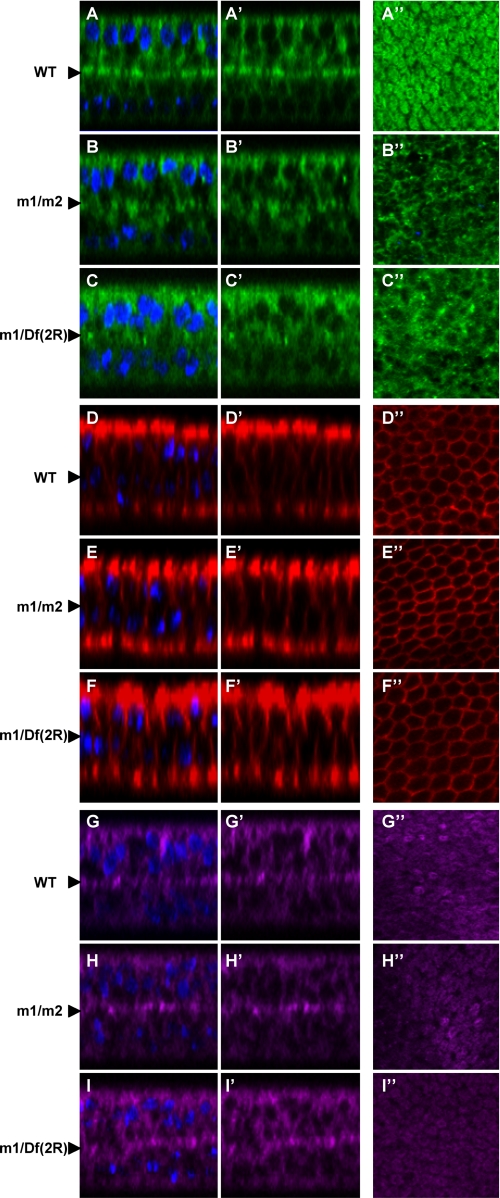
Localization of Tiggrin, but not other proteins, is affected in pgant3 mutant pupal wings. Wild type (WT) (A–A″, D–D″, and G–G″), pgant3m1/pgant3m2 (m1/m2) (B–B″, E–E″, and H–H″), and pgant3m1/Df(2R)Exel6283 (m1/Df(2R)) (C–C″, F–F″, and I–I″) pupal wings at 36 h APF were stained with Tiggrin (green) (A–C″), Fasciclin III (red) (D–F″), and DE-cadherin (purple) (G–H″). Shown are X-Z optical cross-sections of pupal wings with (A–I) or without (A′–I′) DAPI staining of nuclei. Images are oriented so that dorsal is at the top and ventral is at the bottom. Arrowheads at the left denote the basal dorsal/ventral cell layer interface. X-Y confocal images of the basal region comprising the dorsal and ventral boundary (A″–I″) are also shown. In wild type wings 36 h APF, Tiggrin localizes to the dorsal/ventral cell layer interface, where cell adhesion occurs. Mutations in pgant3 result in loss of Tiggrin localization at this interface, although DE-cadherin and Fasciclin III localizations are unaffected.
FIGURE 6.
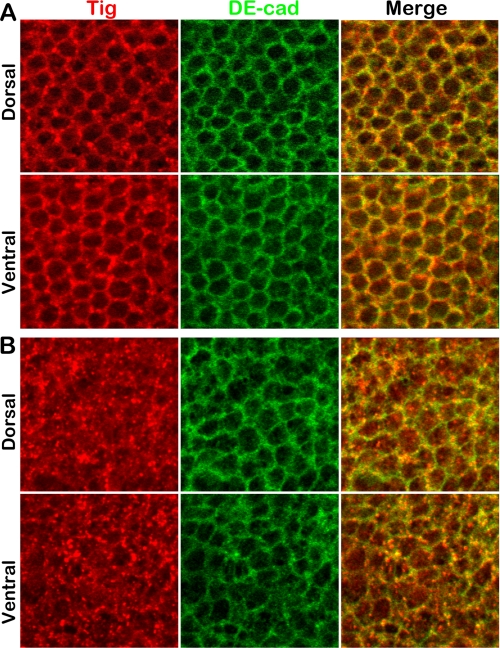
pgant3 mutations result in intracellular Tiggrin accumulation. Wild type (A) and pgant3m1/Df(2R)Exel6283 (B) pupal wings at 36 h APF were stained for Tiggrin (Tig) (red) and DE-cadherin (DE-cad) (green). Merged images are shown in yellow. Shown are representative X-Y confocal images (1 μm thick) of pupal wings in the center of the dorsal cell layer (Dorsal) and in the center of the ventral cell layer (Ventral). pgant3 mutants show loss of Tiggrin along cell membranes and accumulation within the cells of the dorsal and ventral cell layers.
RNAi in Drosophila Cell Culture
For the generation of dsRNA, regions from each gene of interest (YFP, mys, tiggrin, or pgant3) were amplified using primers containing T7 RNA polymerase-binding sites and gene-specific sequences to produce ∼500-bp fragments containing T7 promoters at the 5′ ends. We selected regions for dsRNA generation by using the off-target sequence search tool on the Drosophila RNAi Screening Center website, in an effort to minimize any potential off-target effects. The off-target size was set as low as 16 nucleotides for each analysis. Two regions for dsRNA generation were chosen for the pgant3 gene and for the tiggrin gene to verify knockdown and the cellular phenotypes observed. All primer sequences used for dsRNA generation are listed in supplemental Table 1.
PCR products from the above-mentioned primer pairs were purified and used as templates to produce RNA using the MEGASCRIPT T7 transcription kit (Ambion). RNA was LiCl-precipitated, resuspended in water, incubated at 65 °C for 30 min, and then slow-cooled to room temperature to allow annealing. dsRNA formed was then stored at −20 °C.
Drosophila cells were grown in Schneider's medium (Invitrogen) with 10% heat-inactivated fetal bovine serum (Invitrogen) at 25 °C in culture flasks. RNAi was performed as described on the Drosophila RNAi Screening Center website. Briefly, 2 × 105 S2R+ cells in 250 μl of serum-free media were added to each well of a 24-well plate. dsRNA (7 μg) was added to each well, and plates were mixed back and forth. The cells were then incubated for 30 min at room temperature before adding 750 μl of medium containing 10% heat-inactivated fetal bovine serum (Invitrogen). Cells were grown for 4–6 days at 25 °C. Each experiment was performed with two independent dsRNAs to pgant3 to verify gene-specific knockdown and observed phenotypes.
After dsRNA treatment, cells were fixed in 4% formaldehyde/PBS (Electron Microscopy Science) and then washed twice in PBS with 0.1% Triton X-100. Staining was then performed using TRITC-phalloidin (Sigma) and DAPI, followed by washes in PBS. Cells were examined using a Zeiss Axiophot microscope.
Cell Adhesion Assays
After incubating with dsRNA for 5 days, S2R+ cells were replated on glass slides and allowed to spread for 4 h before fixing. S2 cells treated with dsRNA were replated on glass slides coated with 0.5 mg/ml concanavalin A and allowed to spread for 2 h (42). After fixing, cells were stained with TRITC-phalloidin and DAPI.
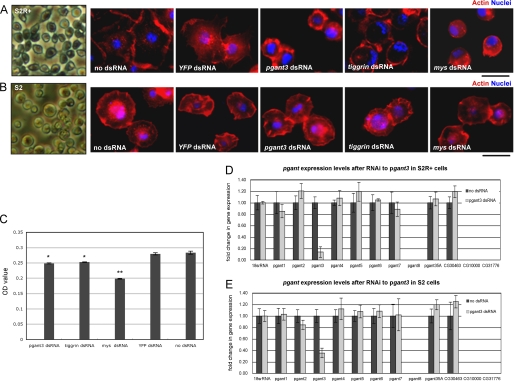
Quantitative cell adhesion assays were performed on S2R+ cells after incubation with dsRNA. S2R+ cells were harvested and resuspended in the medium at a density of 2 × 105 cells/ml. 100 μl of cell suspension was seeded into each well and then incubated for an additional 2 h at 25 °C. After removing the nonadherent cells, 150 μl of 0.2% crystal violet (Sigma) aqueous solution in 20% methanol was added to stain the cells that remained attached. Wells were washed twice with Milli-Q water and then air-dried. Finally, cells were dissolved in 150 μl of 1% SDS. The absorbance at 570 nm was measured using a Microplate Reader (TECAN). Cells adhesion assays were performed in triplicate and in multiple independent experiments. The data shown in Fig. 7C is representative of one experiment performed in triplicate.
FIGURE 7.
RNAi to pgant3 results in cell adhesion defects in cells that undergo integrin-mediated cell adherence. S2R+ (A) and S2 (B) cells were either untreated or treated with the dsRNA to pgant3, tiggrin, mys (βPS integrin), or YFP and then stained with phalloidin (Actin) and DAPI (Nuclei) to visualize cell spreading and adhesion. Decreases in S2R+-specific cell adhesion upon RNAi to pgant3, tiggrin, or mys were quantitated as described under “Experimental Procedures” (C). No cell adhesion alterations were seen in S2 cells plated on concanavalin A-coated surfaces following treatment with dsRNA to pgant3, tiggrin, or mys (B). Real time PCR analysis of the expression of all pgant family members in S2R+ cells (D) or S2 cells (E) confirmed that RNAi to pgant3 resulted in a specific reduction in pgant3 expression. RNA levels were normalized to 18 S rRNA. Scale bars shown below (A and B) = 20 μm. *, p < 0.05; **, p < 0.01. Error bars, standard deviation.
Western Blotting
Protein extracts were prepared from staged larval wing discs of wild type and pgant3m1/pgant3m2 mutants as described previously (43). Samples were electrophoresed under reducing conditions in a 4–12% SDS-polyacrylamide gradient gel and transferred to nitrocellulose membranes. Membranes were blocked with 1× blocking buffer (Sigma) and then incubated with an antibody to Tiggrin (dilution, 1:500) or the Tn antibody (dilution, 1:500) and developed with horseradish peroxidase-conjugated mouse IgG (dilution, 1:2000) (Cell Signaling Technology) or horseradish peroxidase-conjugated mouse IgM (dilution, 1:10,000) (StressGen Bioreagents) secondary antibodies, respectively. The upper band detected by the Tiggrin antibody is not specific to Tiggrin, as it is still present in tigx homozygous mutants.
Quantitative Real Time PCR
Cells were lysed, and RNA was extracted using the RNAqueous-4 PCR kit (Ambion). cDNA synthesis was performed using the iScript cDNA synthesis kit (Bio-Rad). Quantitative real time PCR primers for each pgant gene were described previously (20). Quantitative PCR was performed on a MyiQ real time PCR thermocycler (Bio-Rad) using the SYBR Green PCR master mix (Bio-Rad). Analyzed products were assayed in triplicate and in multiple independent experiments.
RESULTS
Point Mutations That Disrupt pgant3 Catalytic Activity Result in Wing Blistering and Abrogation of Tiggrin Glycosylation
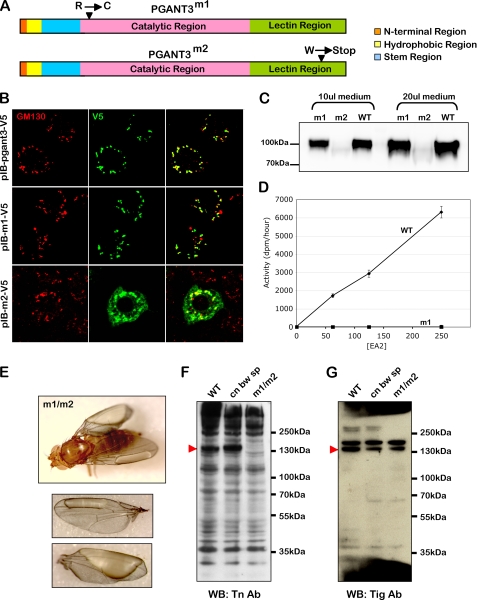
Previous work from our group demonstrated that a transposon insertion mutation in pgant3, which encodes an enzyme responsible for the initiation of mucin-type O-glycosylation, resulted in abrogation of pgant3 expression and subsequent disruption of cell adhesion in the adult wing (20). We set out to further examine the role of O-glycosylation in wing blade adhesion by generating additional pgant3 mutations that specifically affect catalytic activity. Through EMS mutagenesis, we obtained two novel point mutations in the coding region of pgant3 (Fig. 1A). One mutation, pgant3m1, is a C to T transition that changes a conserved arginine to a cysteine at amino acid position 130. pgant3m1, like wild type pgant3, encodes a protein that is localized properly to the Golgi apparatus and is stably expressed in cell culture (Fig. 1, B and C). However, the PGANT3m1 protein fails to glycosylate substrates in in vitro glycosyltransferase reactions, indicating that this mutation specifically compromises enzymatic activity (Fig. 1D). The second mutation, pgant3m2, is a G to A transition that creates a stop codon at amino acid 609, deleting the C-terminal 59 amino acids of the enzyme. This truncation results in an unstable protein that is not properly localized to the Golgi apparatus (Fig. 1B). Additionally, we were not able to stably express the pgant3m2 mutant as a secreted, recombinant protein to perform in vitro enzymatic assays (Fig. 1C).
FIGURE 1.
pgant3 point mutations cause wing blistering and loss of Tiggrin O-glycosylation. A, two point mutations in the coding region of pgant3 were obtained by EMS mutagenesis. The pgant3m1 mutation changes a conserved arginine to a cysteine at amino acid 130. The pgant3m2 mutation changes a tryptophan to a stop codon at amino acid 609 in the lectin-like region, deleting the C-terminal portion of the enzyme. B, expression of recombinant wild type pgant3 (pIB-pgant3-V5), pgant3m1 (pIB-m1-V5), and pgant3m2 (pIB-m2-V5) in Drosophila S2R+ cells. Cells were then stained for V5 and the Golgi marker, GM130, revealing Golgi localization of wild type pgant3 and pgant3m1. C, expression of secreted forms of pgant3 in COS7 cells demonstrates that wild type (WT) pgant3 and pgant3m1 are stably expressed as recombinant proteins, whereas pgant3m2 is not. D, pgant3m1 mutation results in loss of glycosyltransferase activity. Equal relative amounts of wild type pgant3 (circles) and pgant3m1 (m1, squares) proteins (as determined by Western blotting) produced and purified from COS7 cells were used with increasing concentrations of acceptor substrate (EA2) in enzyme assays as described under “Experimental Procedures.” All assays were performed in triplicate. Error bars indicate standard deviation. E, homozygous pgant3 mutants (pgant3m1/pgant3m2) have wing blisters, indicative of cell adhesion defects. Western blots (WB) of proteins from wing discs of wild type, the parental line used for mutagenesis (cn bw sp), and pgant3m1/pgant3m2 mutants (m1/m2) probed with the Tn antibody (F) or an antibody to Tiggrin (G) demonstrate that Tiggrin is present but not O-glycosylated in pgant3m1/pgant3m2 mutants. Protein size standards are shown to the right. Red arrowheads indicate the position of the glycosylated Tiggrin band.
Flies carrying the original transposon mutation (pgant3c01318), the pgant3m1 mutation, or pgant3m2 mutation were crossed to assess the effect of the new pgant3 point mutations on wing blistering frequency in adults (Table 1 and Fig. 1E). Although heterozygous mutants displayed no effect on wing integrity, all transheterozygous mutants (pgant3m1/pgant3m2, pgant3m1/pgant3c01318, or pgant3m2/pgant3c01318) as well as mutants over a deficiency for the region (pgant3m1/Df(2R)Exel6283, pgant3m2/Df(2R)Exel6283, or pgant3c01318/Df(2R)Exel6283) showed a high frequency of wing blistering (Table 1). Additionally, these point mutants also displayed a loss of Tiggrin glycosylation (Fig. 1, F and G), as was seen previously for the transposon mutant (20). Because the pgant3m1 mutation (which forms a stable protein yet is catalytically inactive) resulted in wing blistering when combined with other pgant3 mutations, we conclude that the loss of PGANT3 glycosyltransferase activity (and not simply loss of the PGANT3 protein) is responsible for the loss of wing integrity. This indicates that PGANT3 function in the developing wing is dependent upon its glycosyltransferase activity and not on a glycosylation-independent chaperone function seen previously for other glycosyltransferases (44, 45).
TABLE 1.
Frequency of wing blistering observed in pgant3 mutants
n = total number of flies scored.
| Genotype | Blistereda | n |
|---|---|---|
| % | ||
| pgant3m1/+ | 0 | 204 |
| pgant3m2/+ | 0 | 258 |
| pgant3c01318/+ | 0 | 107 |
| Def(2R) Exel6283, PXP-UExel6283/+ | 0 | 232 |
| pgant3m1/pgant3c01318 | 100 | 127 |
| pgant3m2/pgant3c01318 | 100 | 132 |
| w; pgant3m1/pgant3m2 | 90 | 109 |
| pgant3c01318/pgant3c01318 | 95 | 129 |
| pgant3m1/Def(2R) Exel6283, PXP-UExel6283 | 99 | 214 |
| pgant3m2/ Def(2R) Exel6283, PXP-UExel6283 | 100 | 198 |
a The following equation was used: (number of flies displaying blistered wings/total number of flies) × 100.
pgant3 Genetically Interacts with if (αPS2 Integrin) to Modulate Cell Adhesion
Prior studies from our group showed a genetic interaction between pgant3 and tiggrin (20), demonstrating a functional consequence of PGANT3 activity on Tiggrin-mediated cell adhesion. We next investigated whether a genetic interaction exists between pgant3 mutants and an αPS2 integrin mutant (if3). if3 mutants are hypomorphic and show a low level of wing blistering in a wild type background (9). Flies hemizygous for if3 in combination with the chromosome 2 used for mutagenesis (if3/Y; +/+) displayed 17% wing blistering (Table 2). As mentioned above, flies heterozygous for pgant3m1 or pgant3m2 displayed no wing blistering. However, flies that are both hemizygous for if3 and heterozygous for pgant3m1 (if3/Y; pgant3m1/+) or pgant3m2 (if3/Y; pgant3m2/+) displayed a dramatic increase in wing blistering (76 and 52%, respectively). Thus, mutations in pgant3 and αPS2 integrin interact genetically to exacerbate the same morphological phenotype, demonstrating that pgant3 modulates integrin-mediated cell adhesion in the developing wing.
TABLE 2.
Genetic interaction between pgant3 and if (αPS2 integrin)
n = total number of flies scored.
| Genotype | Blistereda | n |
|---|---|---|
| % | ||
| w/Y; pgant3m1/+ | 0 | 129 |
| w/Y; pgant3m2/+ | 76 | 176 |
| if3/Y; +/ + | 0 | 142 |
| if3/Y; pgant3m1/+ | 52 | 125 |
| if3/Y; pgant3m2/+ | 17 | 87 |
a The following equation was used: (number of flies displaying blistered wings/total number of flies) × 100.
Tiggrin and O-Glycans Are Specifically Localized during Pupal Wing Development
Pupal wing development undergoes a series of apposition, adhesion, expansion, and separation phases to ensure proper morphogenesis and establishment of cell contacts (46). To obtain insight into the role of O-glycans and Tiggrin in integrin-mediated cell adhesion in the wing, we examined pupal wings at multiple different developmental stages. Both Tiggrin and O-glycans were present in the developing wing and displayed dynamic localization throughout pupal wing development. Very specific localization of both was observed at the basal cell layer interface during two separate stages of adhesion (6–8 h after puparium formation (APF) and 34–42 h APF) (Fig. 2, A and A′ and D–E′), similar to what was previously seen for integrins (40, 46). Localization of both O-glycans and Tiggrin was diffuse during stages of separation of the epithelial cell layers (18–20 h APF) (Fig. 2, B and B′). Restoration of basal surface localization of both O-glycans and Tiggrin begins to occur again as the second apposition phase takes place (24–26 h APF) (Fig. 2, C and C′). Thus, the specific localization of Tiggrin and O-glycans to the basal region (where integrin is localized) during stages of cell adhesion further supports their involvement in integrin-mediated cell adhesion.
pgant3 Mutant Pupal Wing Discs Show Altered Glycosylation and Tiggrin Localization
To investigate the specific role of pgant3 in wing blade adhesion, we compared wild type and pgant3 mutant wings at the adhesive stages of pupal wing development. Pupal wings at either 6–8 h APF or 34–36 h APF were stained for Tiggrin, O-glycans, and βPS integrin (Fig. 3). Optical X-Z cross-sections of the wings are shown. In wild type wings, Tiggrin, O-glycans, and βPS integrin all localize as a diffuse line across the center of the wing where the two cell layers begin to appose at 6–8 h APF (Fig. 3, A and C). This localization becomes much more distinct at 34–36 h APF, when they are tightly localized at the basal surface of the dorsal/ventral cell layers (Fig. 3, B and D). However, pgant3m1/pgant3m2 mutants displayed a reduction in O-glycosylation and a loss of Tiggrin along the cell layer interface at both stages of wing development (Fig. 3), suggesting that either Tiggrin stability or localization is altered in these mutants. The same altered Tiggrin localization was also seen in pgant3c01318/pgant3c01318 transposon insertion mutants (Fig. 3, A and B). O-Glycosylation and Tiggrin localization were restored upon excision of the transposon (Fig. 3, A and B). Interestingly, βPS integrin was still present along the cell layer interface in pgant3 mutants, indicating that the presence of Tiggrin at this interface is not required for proper integrin localization and that the presence of βPS integrin is not sufficient to ensure Tiggrin localization (Fig. 3, C and D). However, βPS integrin staining was slightly more diffuse in the pgant3 mutants, suggesting that disruption of Tiggrin localization may influence integrin distribution.
To address whether other proteins are also mislocalized in pgant3 mutants, we examined the localization of Fasciclin III and DE-cadherin (Fig. 4). High magnification images of pupal wings at stage 36 h APF reveal that Tiggrin staining is still seen within the cells of the pgant3m1/pgant3m2 and pgant3m1/Df (2R)Exel6283 mutants (Fig. 4, B–C″), but it is not precisely localized to the basal cell layer interface as in wild type pupal wings (Fig. 4, A and A″). However, Fasciclin III and DE-cadherin, two membrane proteins with distinct localizations, showed no difference in staining patterns between wild type and pgant3 mutant wings (Fig. 4, D–I″). These results suggest that the loss of O-glycosylation specifically disrupts Tiggrin localization.
pgant3 Influences Tiggrin Secretion, Not Stability
The loss of Tiggrin from the basal cell adhesive region could be due to altered Tiggrin secretion, altered Tiggrin retention along this region, or an increase in Tiggrin degradation. To distinguish between these possibilities, we performed Western blots using extracts from pgant3 mutant and wild type pupal wings (Fig. 5). No significant difference in the amount or size of Tiggrin was seen between mutant and wild type wings at multiple developmental stages (Fig. 5, A–C), suggesting that loss of Tiggrin at the cell layer interface in pgant3 mutant wings is not due to degradation.
FIGURE 5.
Western blots of pupal wing extracts reveal no change in amount of Tiggrin present in pgant3 mutant wings relative to wild type. Equivalent amounts of protein extracts from wild type (WT) and pgant3m1/pgant3m2 homozygous point mutants (m1/m2) at 4 h APF (A), 8 h APF (B), and 48 h APF (C) were Western-blotted and probed with a Tiggrin antibody. No significant change in the amount or size of Tiggrin was seen when comparing wild type and pgant3 mutant wings at various times during wing development. Size markers are shown to the left of each blot.
To examine the effect of O-glycosylation on Tiggrin secretion, we stained wild type and pgant3 mutant pupal wings for Tiggrin and DE-cadherin (to outline the cells). Multiple confocal X-Y sections (1 μm) through the center of both the dorsal and ventral cell layers were taken to visualize whether Tiggrin is present at the cell boundaries defined by DE-cadherin staining. Confocal images revealed that although Tiggrin is present along the outer membrane of cells in wild type wings, it is largely absent from this region in pgant3 mutant wing cells (Fig. 6). Additionally, Tiggrin staining in pgant3 mutant wings is primarily seen in punctate structures inside the cell borders, indicating an accumulation of Tiggrin within the cells of pgant3 mutant wings. These results strongly support a role for O-glycosylation in proper Tiggrin secretion.
RNAi to pgant3 Affects Integrin-mediated Cell Adhesion in Cell Culture
To further investigate the role of pgant3 in cell adhesion, we employed a Drosophila cell line that has been used to examine integrin-mediated cell adhesion (11, 42). S2R+ cells, an adherent line of embryonic origin, were treated with a nonspecific dsRNA (YFP), dsRNA to a βPS integrin known to cause cell adhesion defects (mys) (47), dsRNA to tiggrin, or dsRNA to pgant3. Treated cells were fixed and stained with phalloidin to detect changes in cell spreading, shape, and adhesion. Untreated cells or cells treated with YFP dsRNA had highly developed actin-based lamellae and were well spread on cell culture dishes. As seen previously, cells treated with mys dsRNA became round and nonadherent, with a dramatic actin fibril reorganization (Fig. 7, A and C). Additionally, cells treated with dsRNA to tiggrin were also less adherent (Fig. 7, A and C). Interestingly, cells treated with pgant3 dsRNA displayed a similar nonadherent phenotype, with treated cells becoming round, detaching from the dish, and displaying altered actin organization (Fig. 7, A and C). The effects seen were verified with a second independent dsRNA to pgant3 (data not shown), and the degree and specificity of pgant3 knockdown were assessed by real time quantitative PCR (Fig. 7D). Additionally, we found that these morphological and cell adhesive changes were unique to pgant3, as RNAi to the other members of the pgant family did not result in similar changes (data not shown). Thus, as is the case in vivo, pgant3 appears to be required for proper integrin-mediated cell adhesion of S2R+ cells in culture.
To address the specificity of pgant3 in integrin-mediated cell adhesion, we compared the effects of pgant3 knockdown in a Drosophila cell line that utilizes different adhesion mechanisms. S2 cells, which are normally nonadherent, will adhere to concanavalin A-coated surfaces via an integrin-independent mechanism (42), in contrast to the integrin-dependent adherence of S2R+ cells. When βPS integrin (mys) or tiggrin gene expression in S2 cells was reduced by RNAi, S2 cells remained attached to the concanavalin A-coated surface, with no noticeable changes in cell morphology or spreading (Fig. 7B) (42). Similarly, RNAi to pgant3 in S2 cells significantly reduced pgant3 gene expression but had no effect on cell adhesion (Fig. 7, B and E). These data suggest that the cell adhesion defects caused by RNAi to pgant3 are unique to S2R+ cells and lend further support for a model where pgant3 is involved specifically in integrin-mediated cell adhesion. Taken together, our data support a model where pgant3 affects integrin-mediated cell adhesion in the developing wing by influencing the secretion of the integrin-ligand, Tiggrin.
DISCUSSION
In this study, we demonstrate that O-glycosylation influences the secretion and localization of a specific matrix protein, altering the microenvironment of the wing and resulting in direct biological consequences during development. Tiggrin, a secreted ECM protein known to bind integrin and mediate cell adhesion (3, 18), is normally O-glycosylated and located at the adhesive interface between the two epithelial cell layers of the developing wing. This area of the developing wing, where integrins are also localized, governs cell-cell contacts and proper wing blade formation. In pgant3 mutants, Tiggrin is not O-glycosylated and fails to be secreted to this cell layer interface, resulting in cell adhesion defects. Disruption of Tiggrin localization and cell adhesion are directly associated with the glycosylation status of Tiggrin, as restoration of pgant3 activity restores Tiggrin glycosylation, localization, and wing integrity. Additionally, a pgant3 mutant that is stably expressed, but lacks catalytic activity, demonstrates that these effects are due to the glycosyltransferase activity of PGANT3 and not due to a chaperone function in the absence of enzymatic activity. This distinguishes the effects of PGANT3 from those of another glycosyltransferase (O-fucosyltransferase 1) that influences Notch transport by functioning as a chaperone in the absence of glycosyltransferase activity (44, 45). Altogether, our studies provide the first evidence for the role of mucin-type O-glycans in proper secretion and localization of extracellular matrix proteins during eukaryotic development.
Our data further demonstrate that the influence of O-glycosylation on Tiggrin localization to the basal matrix is due to effects on Tiggrin secretion. Although other studies suggest that mucin-type O-glycans may influence protein stability, as is thought to be the case for FGF23 in familial tumoral calcinosis (26, 31, 32), we do not see evidence of altered Tiggrin stability in pupal wings. Rather, our analysis of Tiggrin localization in pgant3 mutant wings cells revealed dramatic intracellular accumulation, indicating defects in Tiggrin secretion. Prior studies examining proteins known to be glycosylated have suggested roles for O-glycans in secretion. For example, regions that are crucial for secretion or apical sorting of certain proteins have been shown to be rich in O-glycosylated residues (36, 48). Additionally, Muc4, a highly O-glycosylated protein, influences ErbB2 and ErbB3 receptor trafficking to the cell surface in cultured cells (49). Cell culture studies using competitive inhibitors of O-glycan extension resulted in reduced trafficking of certain proteins (33–36); however, these inhibitors also influence other forms of glycosylation. Studies examining protein distribution in subregions of the Golgi complex in larval imaginal discs found that those containing O-linked GalNAc were present predominantly in basally located Golgi units, suggesting that this polarized distribution may influence apicobasal polarity in these tissues (50). Here, we provide direct evidence for the role of mucin-type O-glycans in modulating proper secretion to this basal region in vivo by examining a specific substrate in a glycosyltransferase mutant background. Given that the multiple pgant genes responsible for initiating mucin-type O-glycosylation (22, 39) have unique tissue- and stage-specific patterns of expression (23, 51–53), it is likely that specific pgant family members may be playing unique roles in the secretion and localization of proteins within diverse cell types.
Our study further provides the first demonstration of a specific role for O-glycosylation in integrin-mediated cell adhesion events by demonstrating a genetic interaction between αPS2 integrin and pgant3. Mutations in pgant3 genetically interact with αPS2 integrin mutants to increase wing blistering, providing a functional connection between O-glycosylation and cell adhesion mediated by integrins. Additionally, Drosophila cell culture assays demonstrated that RNAi to pgant3 disrupted integrin-dependent cell adhesion but did not affect integrin-independent cell adhesion. Altogether, our studies demonstrate a specific role for O-glycosylation in integrin-mediated cell adhesion that is dictated by the influence of O-glycans on integrin-ligand secretion and localization. These results have implications for this protein modification in other developmental and pathological processes involving integrins.
pgant3 mutations did not appear to alter the localization of other proteins present in the developing wing. The specific localization of the cell membrane proteins DE-cadherin and Fasciclin III was unchanged in pgant3 mutants, suggesting that loss of O-glycosylation in this context is not leading to global changes in cell polarity as was seen previously for mutations in pgant35A, which disrupted tracheal cell polarity and diffusion barrier formation (28). The specific effect of pgant3 on cell adhesion is supported by the fact that adult wings in pgant3 mutants are, with the exception of blisters, normal in morphology. Additionally, β-integrin was still found at the cell layer interface, albeit in a more diffuse pattern, again indicating that pgant3 mutations do not result in an overall disruption of the basal surface. These data indicate that PGANT3 modulates cell adhesive functions by influencing the secretion of a specific ECM component. It will be interesting to see if other ECM components are influenced by PGANT3 or other UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family members in the future.
This study has implications for the role of O-glycosylation in other developing systems and in certain diseases (22). For example, our data could provide insight into the long-standing correlation between aberrant O-glycosylation and the progression of certain types of cancer (54–58). Indeed, recent work has demonstrated that germ line and somatic mutations in a member of this glycosyltransferase family (GALNT12) are associated with colon cancer in humans (59). Additionally, deletion of an extending glycosyltransferase in mice results in increased susceptibility to colorectal tumors (60). Although the molecular mechanisms underlying these associations are not known, our work supports a model where mutations in a glycosyltransferase could alter the composition of the secreted ECM or “microenvironment” surrounding cells, thus influencing adhesive, protective, or signaling properties. Such an alteration in the microenvironment may influence the dialogue occurring between a cell and its surrounding matrix, thereby altering the susceptibility of cells/tissues to other genetic or environmental changes. Indeed, many studies are focusing on the role of “tumor microenvironment” and the interplay between intra- and extracellular factors in cancer progression and metastasis (61). We have thus identified a factor that is capable of modifying the microenvironment of a cell, resulting in direct consequences on cell adhesion. We are currently investigating whether other UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family members are capable of modulating secretion and ECM composition, and what role this may play in both development and disease.
Supplementary Material
Acknowledgments
We sincerely thank our colleagues for many helpful discussions. We thank Drs. R. Cummings and T. Ju for the kind gift of the Tn antibody and Drs. J. Fessler and L. Fessler for the kind gift of the Tiggrin antibody. We thank Dr. J. Kennison for the kind gift of fly stocks. Finally, we also thank the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for providing fly stocks, antibodies, and other reagents.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program of the NIDCR.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- ECM
- extracellular matrix
- RNAi
- RNA interference
- YFP
- yellow fluorescent protein
- dsRNA
- double-stranded RNA
- APF
- after puparium formation
- DAPI
- 4′,6-diamidino-2-phenylindole, dihydrochloride
- EMS
- ethyl methanesulfonate
- PBS
- phosphate-buffered saline
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Brower D. L., Bunch T. A., Mukai L., Adamson T. E., Wehrli M., Lam S., Friedlander E., Roote C. E., Zusman S. (1995) Development 121, 1311–1320 [DOI] [PubMed] [Google Scholar]
- 2.Bloor J. W., Brown N. H. (1998) Genetics 148, 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunch T. A., Graner M. W., Fessler L. I., Fessler J. H., Schneider K. D., Kerschen A., Choy L. P., Burgess B. W., Brower D. L. (1998) Development 125, 1679–1689 [DOI] [PubMed] [Google Scholar]
- 4.Prokop A., Martín-Bermudo M. D., Bate M., Brown N. H. (1998) Dev. Biol. 196, 58–76 [DOI] [PubMed] [Google Scholar]
- 5.Walsh E. P., Brown N. H. (1998) Genetics 150, 791–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araujo H., Negreiros E., Bier E. (2003) Development 130, 3851–3864 [DOI] [PubMed] [Google Scholar]
- 7.Brower D. L. (2003) Curr. Opin. Cell Biol. 15, 607–613 [DOI] [PubMed] [Google Scholar]
- 8.Narasimha M., Brown N. H. (2004) Curr. Biol. 14, 381–385 [DOI] [PubMed] [Google Scholar]
- 9.Brabant M. C., Brower D. L. (1993) Dev. Biol. 157, 49–59 [DOI] [PubMed] [Google Scholar]
- 10.Prout M., Damania Z., Soong J., Fristrom D., Fristrom J. W. (1997) Genetics 146, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown N. H., Gregory S. L., Rickoll W. L., Fessler L. I., Prout M., White R. A., Fristrom J. W. (2002) Dev. Cell 3, 569–579 [DOI] [PubMed] [Google Scholar]
- 12.Domínguez-Giménez P., Brown N. H., Martín-Bermudo M. D. (2007) J. Cell Sci. 120, 1061–1071 [DOI] [PubMed] [Google Scholar]
- 13.Chanana B., Graf R., Koledachkina T., Pflanz R., Vorbrüggen G. (2007) Mech. Dev. 124, 463–475 [DOI] [PubMed] [Google Scholar]
- 14.Subramanian A., Wayburn B., Bunch T., Volk T. (2007) Development 134, 1269–1278 [DOI] [PubMed] [Google Scholar]
- 15.Schöck F., Perrimon N. (2003) Genes Dev. 17, 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bökel C., Brown N. H. (2002) Dev. Cell 3, 311–321 [DOI] [PubMed] [Google Scholar]
- 17.Devenport D., Bunch T. A., Bloor J. W., Brower D. L., Brown N. H. (2007) Dev. Biol. 308, 294–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fogerty F. J., Fessler L. I., Bunch T. A., Yaron Y., Parker C. G., Nelson R. E., Brower D. L., Gullberg D., Fessler J. H. (1994) Development 120, 1747–1758 [DOI] [PubMed] [Google Scholar]
- 19.Graner M. W., Bunch T. A., Baumgartner S., Kerschen A., Brower D. L. (1998) J. Biol. Chem. 273, 18235–18241 [DOI] [PubMed] [Google Scholar]
- 20.Zhang L., Zhang Y., Ten Hagen K. G. (2008) J. Biol. Chem. 283, 34076–34086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ten Hagen K. G., Fritz T. A., Tabak L. A. (2003) Glycobiology 13, 1R–16R [DOI] [PubMed] [Google Scholar]
- 22.Tian E., Ten Hagen K. G. (2009) Glycoconj. J. 26, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ten Hagen K. G., Zhang L., Tian E., Zhang Y. (2009) Glycobiology 19, 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ten Hagen K. G., Tran D. T. (2002) J. Biol. Chem. 277, 22616–22622 [DOI] [PubMed] [Google Scholar]
- 25.Xia L., Ju T., Westmuckett A., An G., Ivanciu L., McDaniel J. M., Lupu F., Cummings R. D., McEver R. P. (2004) J. Cell Biol. 164, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., Sprecher E. (2004) Nat. Genet. 36, 579–581 [DOI] [PubMed] [Google Scholar]
- 27.Ju T., Cummings R. D. (2005) Nature 437, 1252. [DOI] [PubMed] [Google Scholar]
- 28.Tian E., Ten Hagen K. G. (2007) J. Biol. Chem. 282, 606–614 [DOI] [PubMed] [Google Scholar]
- 29.Tenno M., Ohtsubo K., Hagen F. K., Ditto D., Zarbock A., Schaerli P., von Andrian U. H., Ley K., Le D., Tabak L. A., Marth J. D. (2007) Mol. Cell. Biol. 27, 8783–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu J., Gerhardt H., McDaniel J. M., Xia B., Liu X., Ivanciu L., Ny A., Hermans K., Silasi-Mansat R., McGee S., Nye E., Ju T., Ramirez M. I., Carmeliet P., Cummings R. D., Lupu F., Xia L. (2008) J. Clin. Invest. 118, 3725–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichikawa S., Guigonis V., Imel E. A., Courouble M., Heissat S., Henley J. D., Sorenson A. H., Petit B., Lienhardt A., Econs M. J. (2007) J. Clin. Endocrinol. Metab. 92, 1943–1947 [DOI] [PubMed] [Google Scholar]
- 32.Frishberg Y., Ito N., Rinat C., Yamazaki Y., Feinstein S., Urakawa I., Navon-Elkan P., Becker-Cohen R., Yamashita T., Araya K., Igarashi T., Fujita T., Fukumoto S. (2007) J. Bone Miner. Res. 22, 235–242 [DOI] [PubMed] [Google Scholar]
- 33.Delannoy P., Kim I., Emery N., De Bolos C., Verbert A., Degand P., Huet G. (1996) Glycoconj. J. 13, 717–726 [DOI] [PubMed] [Google Scholar]
- 34.Huet G., Hennebicq-Reig S., de Bolos C., Ulloa F., Lesuffleur T., Barbat A., Carrière V., Kim I., Real F. X., Delannoy P., Zweibaum A. (1998) J. Cell Biol. 141, 1311–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gouyer V., Leteurtre E., Delmotte P., Steelant W. F., Krzewinski-Recchi M. A., Zanetta J. P., Lesuffleur T., Trugnan G., Delannoy P., Huet G. (2001) J. Cell Sci. 114, 1455–1471 [DOI] [PubMed] [Google Scholar]
- 36.Potter B. A., Hughey R. P., Weisz O. A. (2006) Am. J. Physiol. Cell Physiol. 290, C1–C10 [DOI] [PubMed] [Google Scholar]
- 37.Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., Singh C. M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H. L., Ryner L., Cheung L. M., Chong A., Erickson C., Fisher W. W., Greer K., Hartouni S. R., Howie E., Jakkula L., Joo D., Killpack K., Laufer A., Mazzotta J., Smith R. D., Stevens L. M., Stuber C., Tan L. R., Ventura R., Woo A., Zakrajsek I., Zhao L., Chen F., Swimmer C., Kopczynski C., Duyk G., Winberg M. L., Margolis J. (2004) Nat. Genet. 36, 283–287 [DOI] [PubMed] [Google Scholar]
- 38.Lewis E. B., Bacher F. (1968) Drosophila Information Service 43, 193 [Google Scholar]
- 39.Ten Hagen K. G., Tran D. T., Gerken T. A., Stein D. S., Zhang Z. (2003) J. Biol. Chem. 278, 35039–35048 [DOI] [PubMed] [Google Scholar]
- 40.O'Keefe D. D., Prober D. A., Moyle P. S., Rickoll W. L., Edgar B. A. (2007) Dev. Biol. 311, 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avichezer D., Springer G. F., Schechter B., Arnon R. (1997) Int. J. Cancer 72, 119–127 [DOI] [PubMed] [Google Scholar]
- 42.Jani K., Schöck F. (2007) J. Cell Biol. 179, 1583–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickup A. T., Banerjee U. (1999) Dev. Biol. 205, 254–259 [DOI] [PubMed] [Google Scholar]
- 44.Okajima T., Xu A., Lei L., Irvine K. D. (2005) Science 307, 1599–1603 [DOI] [PubMed] [Google Scholar]
- 45.Okajima T., Reddy B., Matsuda T., Irvine K. D. (2008) BMC Biol. 6, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fristrom D., Wilcox M., Fristrom J. (1993) Development 117, 509–523 [DOI] [PubMed] [Google Scholar]
- 47.Kiger A. A., Baum B., Jones S., Jones M. R., Coulson A., Echeverri C., Perrimon N. (2003) J. Biol. 2, 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeaman C., Le Gall A. H., Baldwin A. N., Monlauzeur L., Le Bivic A., Rodriguez-Boulan E. (1997) J. Cell Biol. 139, 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Funes M., Miller J. K., Lai C., Carraway K. L., 3rd, Sweeney C. (2006) J. Biol. Chem. 281, 19310–19319 [DOI] [PubMed] [Google Scholar]
- 50.Yano H., Yamamoto-Hino M., Abe M., Kuwahara R., Haraguchi S., Kusaka I., Awano W., Kinoshita-Toyoda A., Toyoda H., Goto S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13467–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kingsley P. D., Ten Hagen K. G., Maltby K. M., Zara J., Tabak L. A. (2000) Glycobiology 10, 1317–1323 [DOI] [PubMed] [Google Scholar]
- 52.Tian E., Ten Hagen K. G. (2006) Glycobiology 16, 83–95 [DOI] [PubMed] [Google Scholar]
- 53.Tian E., Ten Hagen K. G. (2007) Glycobiology 17, 820–827 [DOI] [PubMed] [Google Scholar]
- 54.Kim Y. S., Gum J., Jr., Brockhausen I. (1996) Glycoconj. J. 13, 693–707 [DOI] [PubMed] [Google Scholar]
- 55.Kim Y. J., Varki A. (1997) Glycoconj. J. 14, 569–576 [DOI] [PubMed] [Google Scholar]
- 56.Springer G. F. (1997) J. Mol. Med. 75, 594–602 [DOI] [PubMed] [Google Scholar]
- 57.Ono M., Hakomori S. (2004) Glycoconj. J. 20, 71–78 [DOI] [PubMed] [Google Scholar]
- 58.Fuster M. M., Esko J. D. (2005) Nat. Rev. Cancer 5, 526–542 [DOI] [PubMed] [Google Scholar]
- 59.Guda K., Moinova H., He J., Jamison O., Ravi L., Natale L., Lutterbaugh J., Lawrence E., Lewis S., Willson J. K., Lowe J. B., Wiesner G. L., Parmigiani G., Barnholtz-Sloan J., Dawson D. W., Velculescu V. E., Kinzler K. W., Papadopoulos N., Vogelstein B., Willis J., Gerken T. A., Markowitz S. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12921–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.An G., Wei B., Xia B., McDaniel J. M., Ju T., Cummings R. D., Braun J., Xia L. (2007) J. Exp. Med. 204, 1417–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu R., Boudreau A., Bissell M. J. (2009) Cancer Metastasis Rev. 28, 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.