Abstract
The Drosophila Toll receptor is activated by an endogenous cytokine ligand Spätzle. Active ligand is generated in response to positional cues in embryonic dorso-ventral patterning and microbial pathogens in the insect immune response. Spätzle is secreted as a pro-protein and is processed into an active form by the serine endoproteases Easter and Spätzle-processing enzyme during dorso-ventral patterning and infection, respectively. Here, we provide evidence for the molecular mechanism of this activation process. We show that the Spätzle prodomain masks a predominantly hydrophobic region of Spätzle and that proteolysis causes a conformational change that exposes determinants that are critical for binding to the Toll receptor. We also gather that a conserved sequence motif in the prodomain presents features of an amphipathic helix likely to bind a hydrophobic cleft in Spätzle thereby occluding the putative Toll binding region. This mechanism of activation has a striking similarity to that of coagulogen, a clotting factor of the horseshoe crab, an invertebrate that has changed little in 400 million years. Taken together, our findings demonstrate that an ancient passive defense system has been adapted during evolution and converted for use in a critical pathway of innate immune signaling and embryonic morphogenesis.
Keywords: Embryo, Neurotrophin, Protease, Signal Transduction, Toll Receptors
Introduction
The Toll pathway in Drosophila plays a crucial role in both embryonic dorso-ventral patterning and innate immunity to Gram-positive bacteria and fungi (1, 2). In both cases, the Toll receptor is activated by the endogenous protein ligand Spätzle, leading on the one hand to the formation of a morphogenetic gradient and on the other to the expression of genes involved in immune defense. Thus Spätzle acts as key regulator in two distinct but fundamental cellular processes in insects.
In both embryogenesis and immunity, the Spätzle ligand is synthesized as an inactive prepro-protein that is processed internally at the endoplasmic reticulum to remove the N-terminal signal peptide before being secreted from the cell as a homodimer (3, 4). The precursor is made up of two parts, a C-terminal domain (C-106)3 that contains the signaling activity and a regulatory N-terminal prodomain. Like the vertebrate homologue nerve growth factor, Spätzle is stabilized by an internal cysteine knot structure but unlike nerve growth factor also an intersubunit disulfide bond (5). By contrast, the N-terminal prodomain is natively unstructured but is necessary for biosynthesis, secretion, and stabilization, a property held in common with pro-nerve growth factor (6). Although largely unstructured, the prodomain may have short regions of secondary structure similar to so-called “loopy” proteins (7, 8).
The Spätzle pro-protein is unable to induce signaling through the Toll pathway but is activated by endoproteolysis. In dorso-ventral patterning, sulfated glycosaminoglycans present at ventral positions in the vitelline membrane activate a cascade of serine proteases leading to cleavage of Spätzle pro-protein located at ventral positions in the embryo (9, 10). By contrast, in immunity microbial carbohydrates such as lysine-containing peptidoglycan from Gram-positive bacteria are recognized by two proteins, peptidoglycan recognition protein and GNBP1 in the insect's hemolymph (11). This complex also initiates a proteolytic cascade with Spätzle being processed at the same trypsin-like cleavage site by a different serine protease, Spätzle-processing enzyme (12). The unprocessed Spätzle pro-protein is unable to interact with Toll, but after proteolysis it binds to Toll with a very high affinity (Kd = 0.4 nm) and induces signaling (4). Interestingly, the prodomain remains associated with C-106 after proteolysis but is displaced upon binding to the Toll receptor (8). On the basis of these results, we proposed that proteolysis causes a conformational change that exposes the Toll binding sites of Spätzle, but the structural basis for this molecular switch was unclear.
After activation Spätzle binds to the Toll receptor extracellular domain. This consists of two blocks of leucine-rich repeats, short sequence motifs that form an extended solenoidal structure, flanked by N- and C-terminal cysteine capping structures (13). Our previous work showed that one Spätzle dimer binds to the N terminus of the Toll ectodomain in an end-on configuration and that two Toll/Spätzle heterodimers then form into a heterotetrameric signaling complex (14).
In this paper, we now show experimentally that proteolytic activation of Spätzle exposes a surface in C-106 centered on a unique tryptophan residue. Spätzle with a mutation of this tryptophan is able to bind to Toll and to signal, but association with the prodomain is impaired. We also propose a mechanism in which a short motif in the prodomain forms an amphipathic helix that associates with C-106 and suppresses activity by analogy with coagulogen and zymogens.
EXPERIMENTAL PROCEDURES
Cell Culture
Sf9 cells were used for baculovirus generation and protein expression. The cells were grown at 28 °C in a suspension culture using Sf-900 II SFM (Invitrogen) supplemented with 0.1% pluronic acid (Sigma). A stable Drosophila cell line (648-1B6) expressing luciferase under the control of the drosomycin promoter was established from S2 cells (Invitrogen) and was a kind gift from Jean-Luc Imler (9). These cells were grown at 28 °C in Express Five SFM (Invitrogen), supplemented with 2 mm l-glutamine, 1% penicillin/streptomycin, and 0.5 μm puromycin. HEK293ET (human embryonic kidney 293 EBNA-T) cells were grown at 37 °C (5% CO2, 100% humidity) in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen) and 2 mm l-glutamine.
Generation of the Spätzle Expression Construct
The Spätzle “L” isoform expressed sequence tag clone HL01462 was modified previously (8) via several rounds of PCR to create the following construct: N-prodomain-His6-TEV-C106-C, containing the prodomain (Met1–Arg164), a His6 tag, a recombinant TEV protease cleavage site, and the C-106 domain (Val165–Gly270, referred to as Val1–Gly106 in keeping with C-106 numbering). A C-terminal FLAG tag was then introduced via PCR using this construct as a template to create a new dual-tagged construct: N-prodomain-His6-TEV-C106-FLAG-C. The following primers were used to generate this construct: 5′-ggaattccggatccatgatgacgcccatgtgg-3′ (BamHI site underlined) and 5′-tacatagcggccgcctacttgtcgtcatcgtctttgtagtccccagtcttcaacgcgcacttgcagcagg-3′ (NotI site and FLAG tag underlined). Following PCR, the fragment was purified, restricted with BamHI and NotI, then recloned into both the pcDNA3.1(+) and pFastBac backbone and sequenced.
Spätzle Mutagenesis
Site-directed mutagenesis was performed using the QuikChange II kit (Stratagene). The following primer (along with its reverse complement counterpart) was used for the W29F mutagenesis reaction (mutated bases underlined): 5′-cttgagggcggacgacaccttccagttaattgtcaataacgatg-3′. The mutagenized insert was sequenced and then recloned into both a fresh pcDNA3.1(+) and pFastBac backbone.
Protein Expression and Purification
For small scale expression of Spätzle, HEK293ET cells were cultured in 6-well plates and transfected using Lipofectamine 2000 (Invitrogen) with the relevant pcDNA3.1(+)-based Spätzle plasmid. Following a 24-h incubation period, the medium was replaced with Opti-MEM (Invitrogen) supplemented with 1% fetal calf serum and incubated overnight. Medium was then collected and concentrated using a 10,000 MWCO concentrator (Sartorius). His-tagged Spätzle was purified from the medium using the Ni-NTA Spin Kit (Qiagen) according to the manufacturer's instructions. Purified protein was then buffer-exchanged into PBS (pH 7.4), analyzed by SDS-PAGE for purity and quantified by a Bradford absorption assay (Bio-Rad) at 280 nm.
For large scale protein expression, recombinant His-tagged Toll ectodomain and His/FLAG-tagged Spätzle were produced in the baculovirus system and purified using Ni-NTA affinity chromatography as previously described in detail (4). In brief, constructs for the Toll ectodomain (Met1–Ala801, flanked by a C terminus His6 tag) and the dual-tagged Spätzle pro-protein were subcloned into pFastBac and then used to generate recombinant baculoviruses using the Bac-to-Bac system (Invitrogen). For protein expression, a 3-liter Sf9 culture at a density 1 × 106 cells/ml was infected at a multiplicity of infection of either 10 (Toll ectodomain) or 1 (Spätzle) with the virus. The supernatant was collected by centrifugation 2 days after infection, and filtered on a 0.45-μm Sartobran P capsule (Sartorius). The supernatant was then concentrated using the Centramate tangential flow filtration system (Pall Filtron) in 500 ml of buffer A (300 mm NaCl, 20 mm Tris-HCl, 20 mm imidazole (pH 7.5)), centrifuged at 10,000 × g for 60 min to remove residual debris, and then loaded onto a 5-ml Ni-NTA Superflow column (Qiagen) at 2 ml/min at 4 °C using an AKTA fast protein liquid chromatography (GE Healthcare). The resin was equilibrated in 10 column volumes of buffer A. Purified protein was eluted in buffer A supplemented with 250 mm imidazole. Peak fractions were pooled and purified by size exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare) at a flow rate of 1 ml/min in buffer C (150 mm NaCl, 20 mm Tris-HCl (pH 7.5)). Fractions were analyzed by Coomassie-stained SDS-PAGE, and protein concentration was calculated by Bradford absorption. Where necessary, protein was buffer exchanged into PBS (pH 7.4) using Vivaspin concentrators (Sartorius).
To obtain active C-106, Spätzle was either cleaved with AcTEV protease (Invitrogen), or the prodomain was proteolyzed using trypsin (Promega) as described previously (4). Where necessary, an additional purification step described previously (8) using high performance liquid chromatography (Varian), and reverse phase C8 chromatography was used to purify the C-106 domain following TEV protease cleavage.
Western Blot Analysis
Purified recombinant protein or domains thereof were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membrane (GE Healthcare) using the XCell II Blot Module kit (Invitrogen). Membranes were blocked in 3% milk powder and then probed with either the anti-His6 (BD Pharmingen) or anti-FLAG M2 (Sigma) primary antibodies at a 1:3,000 dilution. The membranes were then washed three times with PBS containing 0.1% Tween 20 and subsequently incubated with mouse TrueBlot ULTRA: HRP anti-mouse IgG (eBioscience). Chemiluminescent detection was performed using the SuperSignal West Pico substrate (Thermo) and captured using Hyperfilm ECL (GE Healthcare).
Fluorescence Spectroscopy
Fluorescence measurements were carried out on an LS 55 fluorescence spectrometer (PerkinElmer Life Sciences). Slit widths for excitation and emission were set at 5 nm and 2.5 nm, respectively. Purified protein was diluted to a concentration of 1 μm in PBS (pH 7.4), excited at a wavelength of 280 nm, and monitored between 290 nm and 400 nm for emission. For quenching experiments, acrylamide was used in 10 incremental steps (0.02 m) up to a final concentration of 0.2 m. Calculations were performed using Origin8 software (OriginLab).
Co-immunoprecipitation Assays
Binding of the Toll ectodomain to Spätzle was investigated using the ProFound Co-immunoprecipitation kit (Pierce). Up to 200 μg of anti-FLAG M2 antibody (Sigma) was covalently coupled to the resin according to the manufacturer's instructions and used to bind Spätzle (either the full pro-protein or the TEV protease-cleaved, active C-106 domain). Following three 5-min washes with Coupling Buffer, a 3-fold molar excess of Toll ectodomain was added to the mixture. After an hour, the complex was eluted and Western analysis performed.
Interaction Studies of Spätzle C-106 and Prodomain
Purified Spätzle was cleaved using TEV protease and then loaded onto a Ni-NTA spin column (Qiagen). Both flow-through and elution fractions were collected and analyzed by Western blotting to track either the C-106 (anti-FLAG) or prodomain (anti-His).
Interaction Studies of Spätzle C-106 and Toll Ectodomain
Purified Spätzle was cleaved with trypsin and separated by ion-exchange chromatography on a 5-ml HiTrap Q column (GE Healthcare) in a linear salt gradient from 50 mm to 1 m NaCl in 20 mm Tris-HCl (pH 8.0). Elution fractions were collected and analyzed by reducing SDS-PAGE to detect uncleaved C-106 dimer, C-106 with a single cut and C-106 with both protomers cleaved internally. N-terminal sequencing revealed that cleavage occurred in the Trp loop between Arg24 and Ala25. Cleaved C-106 was incubated for 1 h with Toll at 1:1 and 1:3 molar ratios and separated by gel filtration on a Superdex S200 column (GE Healthcare). Elution fractions were analyzed by reducing SDS-PAGE to reveal the formation of a Toll·C-106 complex.
Luciferase Assays
S2 cells were placed into 96-well plates and stimulated overnight by the addition of purified recombinant Spätzle (or mutant Spätzle) to the culture medium. Cells were lysed using Passive Lysis buffer (Promega) and the activity measured using a GloMax luminometer (Promega) immediately after the addition of the d-Luciferin substrate (Biosynth). All assays were performed three times in triplicate.
Bioinformatic Analysis and Molecular Modeling
Structural alignments were carried out using FATCAT software (15). Molecular visualization was performed using PyMOL and Chimera (Molecular Graphics System; DeLano Scientific, Palo Alto, CA) (16). Twenty three-dimensional models of the entire C-106 were generated using the crystal structure of the C-106 dimer as a template (16). Four missing loops were built simultaneously with MODELLER software version 9.7 (17). One protomer referred to as chain A in the crystal structure required the addition of 4 amino acids at the N terminus 1VGGS4 and a 14-amino acid-long Trp-containing loop (referred to as Trp loop) 22GLRADDTWQLIVNN35 to restore the intact domain. The following loops were added to chain B: 1VGG3 and 23LRADDTWQLIVNND36. The loop optimization step in MODELLER was performed in the context of the entire C-106 dimer using molecular dynamics with simulated annealing, which led to alternate conformations in adjacent regions. The quality of the models was assessed using the Structural Analysis and Verification server SAVES.
RESULTS
Molecular Dynamics Reveal How Areas of Flexibility Affect the Overall Structure of Spätzle
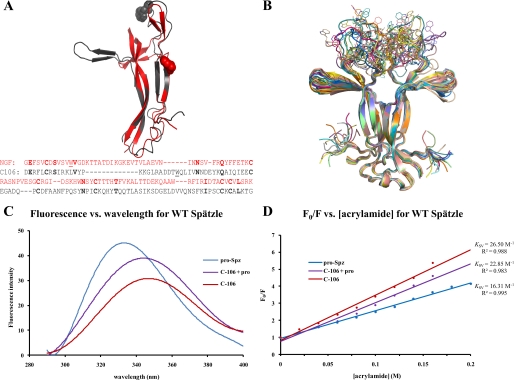
Unprocessed Spätzle pro-protein is unable to bind to the Toll receptor or induce signal transduction (4, 8). This suggests that the regions of C-106 required for receptor binding are sequestered by the prodomain and that proteolysis causes a conformational change that exposes the Toll binding surface. The Spätzle pro-protein has a single tryptophan residue (Trp29) located in the N terminus of C-106 (Fig. 1A). A crystal structure of C-106 (18) reveals that Trp29 is solvent-exposed and located in a disordered loop at the top of the molecule. We generated 20 models for the C-106 dimer containing the missing regions. We found that the Trp loops can adopt a variety of conformations in the absence of the prodomain (Fig. 1B). Surprisingly, modeling the Trp loops strongly affected neighboring structural elements. This can be explained by the energy-minimization step during modeling that was carried out on the entire domain. In particular, the β-sheet wings perpendicular to the dimer axis in the crystal structure fall back onto the flanks of Spätzle by up to 30°. As a result, residues at the tip of the wings are shifted by up to 10 Å away from their position in the crystal structure and closer to the N terminus of C-106. Prior to endoproteolytic activation, the presence of the prodomain and/or the acute angle between the wings and the flanks of Spätzle could prevent Toll receptor binding. Therefore, two areas of interest emerge from the modeling analysis as potential functional surfaces: the Trp loops and the flanks of C-106.
FIGURE 1.
Proteolytic activation of Spätzle induces a conformational change. A, structural and sequence alignment of nerve growth factor (residues 131–236, gray) and Spätzle C-106 (arbitrary sequence numbers because of N-terminal splice variant) (see “Experimental Procedures”). Tryptophan residues are illustrated as spheres, and the structures have a C-α backbone root means square deviation of 2.72 Å. In the sequence alignment, identical residues are in bold, and the tryptophans are underlined. B, C-106 dimer structure (18). The tryptophan loop is shown in 20 possible conformations selected on the basis of energy minimization. C, fluorescence spectroscopy of Spätzle (see “Experimental Procedures”). Red shifting of the spectra shows that the tryptophan residue is increasingly exposed as the prodomain is cleaved but remains noncovalently attached (C-106 + pro) and then most exposed once the prodomain is completely removed (C-106). D, quenching of the fluorescence signal using acrylamide. This allows for the calculation of the Stern–Volmer constant (Ksv), which gives a quantitative indication of the level of exposure of the tryptophan residue; the higher Ksv values indicate increased exposure of the residue.
Activation of Spätzle C-106 Exposes Tryptophan 29
We next decided to investigate the Trp loops using the technique of tryptophan fluorescence quenching. This technique provides information about the changes in the environment of the two tryptophan residues in the C-106 dimer when Spätzle protein is subjected to proteolytic activation.
Full-length Spätzle pro-protein, Spätzle activated by TEV protease, a treatment that maintains the noncovalent attachment of the prodomain to C-106, and Spätzle degraded with trypsin to produce intact C-106 lacking prodomain were purified for use in these fluorescence spectroscopy experiments (see “Experimental Procedures”). Using the Trp29 residue as the probe, a fluorescence spectrum was taken for each of the samples. The resulting spectra showed a red shift in the emission spectrum from 331 nm to 347 nm (Fig. 1C). This red shift implies that the tryptophan residue is increasingly exposed to solvent as the prodomain is removed from C-106. The exposure of the tryptophan residue can be quantified using the Stern–Volmer relationship (19).
This relates the changes in intensity of the fluorescence emission in the presence and absence of the quenching agent (Q), in this case acrylamide, which causes a decrease in intensity of the emission. F0 is the fluorescence intensity in the absence of quencher, and F is the intensity at a specific concentration of quencher. When F0/F is plotted as a function of [Q], a linear graph with intercept 1 and slope Ksv (Stern–Volmer constant) is obtained. This value reflects the level of exposure of the tryptophan residue to solvent; as the exposure of the residue to solvent increases, so does the Stern–Volmer constant (20).
In the case of Spätzle, Ksv values were shown to increase as the prodomain was removed, with the lowest value being obtained from full-length Spätzle, increasing as the prodomain is cleaved but still in noncovalent contact with C-106, and finally highest with C-106 alone (Fig. 1D). This result confirms that the Trp29 residue located in the Trp loop becomes exposed when the prodomain is removed, implying that the Toll-binding site is being unmasked.
Tryptophan 29 Is Required for Stable Association of the Prodomain and C-106
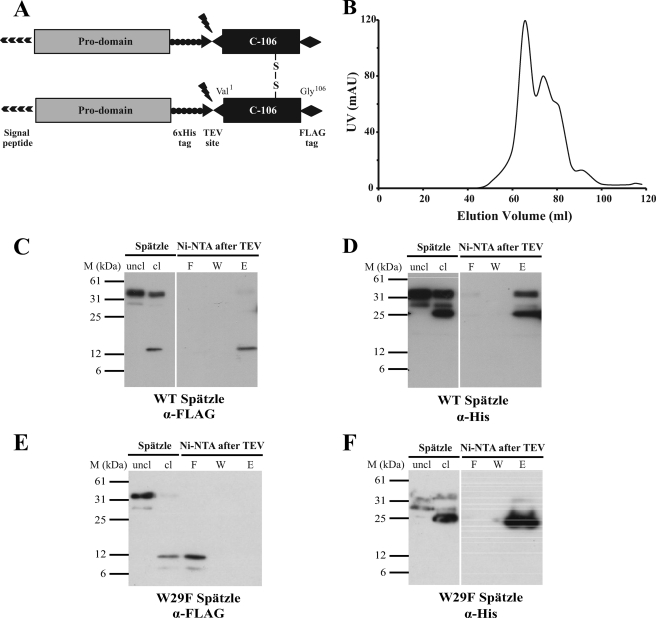
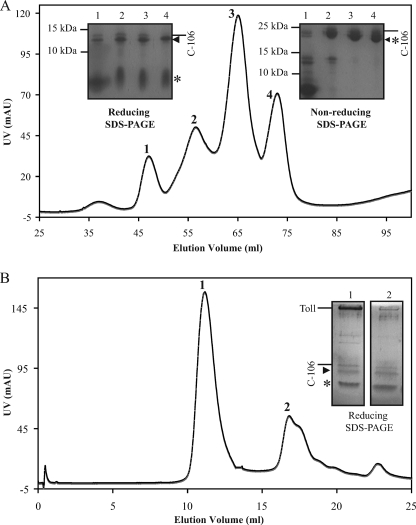
To test whether Trp29 is important for the noncovalent association of the prodomain and C-106, we expressed a mutant Spätzle pro-protein with a mutation of tryptophan 29 to phenylalanine (W29F). His6 and FLAG tags were incorporated into the prodomain and C-106, respectively (see Fig. 2A and “Experimental Procedures”). Wild-type and W29F Spätzle were expressed in Sf9 cells and purified by metal affinity chromatography. In contrast to the wild-type protein, W29F Spätzle had a marked tendency to aggregate as shown by the elution profile from a gel filtration column (Fig. 2B).
FIGURE 2.
Properties of W29F Spätzle. A, schematic of the Spätzle expression construct with TEV cleavage site, FLAG, and His epitope tags is shown. B, elution profile of W29F Spätzle separated by gel filtration on Superdex S-200 is shown. C and D, wild-type (WT) C-106 elutes along with the His-tagged prodomain following cleavage by TEV protease and subsequent Ni-NTA purification showing noncovalent interaction. E and F, W29F mutation causes C-106 to elute in the flow-through following Ni-NTA purification, implying a decreased interaction between the prodomain and C-106. The prodomain and C-106 are detected with anti-His and anti FLAG antibodies, respectively.
We then asked whether the prodomain of W29F Spätzle remains associated with C-106 after proteolysis. Approximately 120 μg of each protein was cleaved with TEV protease and loaded onto fresh Ni-NTA columns. As shown previously for the wild-type Spätzle protein (8), the prodomain and C-106 remain stably associated after proteolytic activation (Fig. 2, C and E) as C-106 can only be eluted by high concentrations of imidazole (Fig. 2C, lane E). By contrast, in the case of the W29F mutant, most of the C-106 protein is detected in the flow through fractions (Fig. 2E, lane F), indicating that it is not stably associated with the prodomain. On the other hand, W29F prodomain binds to the column and is eluted by a high concentration of imidazole.
W29F C-106 Binds to the Toll Ectodomain and Is Active in Signaling
To establish whether the W29F Spätzle can bind to Toll ectodomain, co-immunoprecipitation experiments were carried out using both Spätzle pro-protein and C-106. Anti-FLAG antibodies were covalently cross-linked to an amine-reactive gel, and then ∼30 μg of pro-Spätzle or C-106 was bound to the immobilized antibody. The samples were then washed three times with PBS to remove any residual TEV protease remaining from the cleavage of pro-Spätzle, and ∼100 μg of His-tagged Toll extracellular domain was added. Immunoprecipitated protein were then eluted and detected by Western blotting using anti-His antibodies.
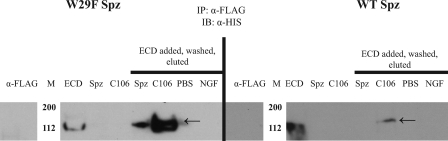
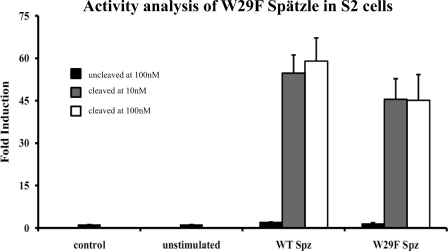
As expected, the uncleaved, full-length Spätzle does not bind to the Toll ectodomain, whereas C-106 does. Interestingly, the W29F mutation did not adversely affect the ability of C-106 to bind to Toll. However, uncleaved W29F Spätzle pro-protein did bind to Toll, implying that the Toll binding sites are at least partially exposed in the mutant protein (Fig. 4). We then tested whether W29F C-106 was active in signal transduction using a cell-based signaling assay. As shown in Fig. 3 the W29F mutant C-106 activated Toll signaling to about 80% of wild type. Furthermore, nearly maximum activation is induced with 10 nm W29F C-106, indicating that the binding affinity is similar to that of the wild-type protein (0.4 nm).
FIGURE 4.
Uncleaved W29F Spätzle pro-protein binds to the Toll ectodomain. Immunoprecipitation experiments were conducted by binding either uncleaved (Spz) or TEV protease-cleaved (C106) to the immunoprecipitation matrix using anti-FLAG antibody, then incubating with His-tagged Toll extracellular domain to test for interaction. The reducing gels were then probed with anti-His antibody. Left, W29F mutation in C-106 allows for Toll extracellular domain (ECD) binding of both uncleaved and cleaved Spätzle. Right, as expected, uncleaved Trp29 Spätzle does not bind to Toll, whereas wild-type (WT) C-106 does.
FIGURE 3.
W29F C-106 activates Toll signaling with characteristics similar to the wild-type protein. S2 cells expressing a drosomycin-luciferase reporter construct were cultured for 24 h in tissue culture medium supplemented with 10 nm or 100 nm Spätzle. After 24 h, cells were lysed, and luciferase activity was measured in triplicate (see “Experimental Procedures”). Data represent mean ± S.D. (error bars) and are shown as fold induction.
Trp Loop Has a Distal Effect on C-106 Activation
To test whether the Trp loop participates directly in Toll binding, we used overdigested C-106, which had been cleaved between residues Arg24 and Ala25 as revealed by N-terminal sequencing (see Fig. 1A). The N-terminal 31-residue-long fragment stays covalently bound to the 75-residue-long C terminus via the cystine-knot structure. It is also noteworthy that trypsin cleaves the prodomain first, which in turn is entirely degraded, before degrading C-106 at the tryptophan loop. Ion exchange chromatography was used to separate intact and cleaved forms of C-106. The intact C-106 dimer was separated from dimers with a single cut (one protomer cleaved, the other one left intact) and dimers with double cuts (both protomers cleaved) in a salt gradient. The latter eluted at higher salt concentrations (Fig. 5A). Overdigested C-106 was then mixed with Toll ectodomain at different molar ratios. Size exclusion revealed the presence of Toll·C-106 complex as described previously for uncleaved C-106 (Fig. 5) (14). It is therefore unlikely that the tryptophan loop is involved directly in Toll binding.
FIGURE 5.
An intact Trp loop is not required for Toll binding. A, Spätzle pro-protein was overdigested with trypsin, and the mixture was separated by ion-exchange chromatography (Mono Q). The four peaks were analyzed by reducing and nonreducing SDS-PAGE. Full-length C-106 is marked with a line (24 kDa nonreduced, 12 kDa reduced), whereas C-106 cleaved at Arg24 is indicated by an asterisk (8.9-kDa reduced fragment) and a triangle (3.1-kDa reduced fragment). B, Toll ectodomain was mixed with C-106 cleaved at Arg24. The Toll and cleaved C-106 co-elute at 11.1 ml as an apparent dimer on a Superdex S-200 (GE Healthcare) (see also Ref. 8).
DISCUSSION
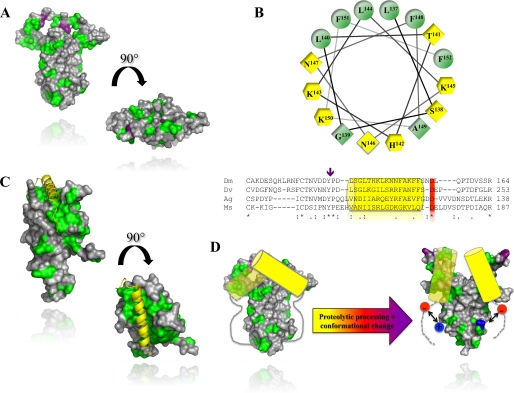
The activation of Spätzle pro-protein by endoproteases is critical for both embryonic dorso-ventral patterning and innate immune responses in invertebrates. It is thus of interest to understand how the prodomain acts as a molecular switch, exposing the high affinity binding sites that support Toll receptor signaling. In a previous study we showed that the prodomain of Spätzle remains associated with the cystine knot dimer after cleavage (8). We hypothesized that simply creating a new N terminus is not sufficient to release the binding activity of C-106 and that proteolysis would be accompanied by a conformational change that unmasks the Toll binding site of C-106. In this paper we provide direct biophysical evidence for this. Using fluorescence quenching experiments we found that a tryptophan residue, Trp29, in C-106 becomes more exposed to solvent upon endoproteolysis of Spätzle pro-protein and is further exposed by the complete removal of the prodomain by treatment with trypsin. A crystal structure of the C-106 dimer reveals that Trp29 is located in a disordered loop structure at the top of the cystine knot (Fig. 1A) (18). On the basis of our results it is likely that the Toll binding site is located in the region of C-106 in the vicinity Trp29, although the integrity of the Trp loop is not required for formation of an active complex with Toll. This is consistent with the “end-on” arrangement of Spätzle seen in low resolution structures of the Spätzle/Toll heterotetramer (14). This conclusion is also supported by mapping and mutagenesis of evolutionarily conserved epitopes in Spätzle. These are found in the same region of the molecule and are critical for signaling function (21). In Fig. 6A the surface of the C-106 dimer is depicted using the Kyte–Doolittle hydropathy index. This reveals that there are significant hydrophobic patches on the top surface and on the flanks of the molecule in the vicinity of Trp29 and suggests that specific binding to Toll may be driven by hydrophobic interactions.
FIGURE 6.
Conserved regulatory a-helix in Spätzle and horseshoe crab coagulogen. A, the molecular surface of C-106 with hydrophobic patches. Hydrophobic residues (alanine, glycine, valine, isoleucine, leucine, phenylalanine, methionine) are colored in green. The tryptophan residue Trp29 is shown in purple. B, lower, sequence alignment of the Spätzle prodomains from D. melanogaster (Dm), D. virilis (Dv), A. gambiae (Ag), and M. sexta (Ms). The position of the predicted α-helix is indicated by a yellow cylinder. A purple arrow indicates the position of the two Spätzle null mutations (Y134N, P135L). B, upper, helical-wheel projection of residues 132–157. In yellow hexagons are positively charged residues. Yellow diamonds are hydrophilic, and green positions are hydrophobic residues. C, crystal structure of coagulogen (PDB code 1AOC) with the same color scheme as in A. The regulatory helix is shown in yellow. D, schematic mechanism for Spätzle activation. Prior to activation, the conserved prodomain helix masks Trp29 and the Toll binding sites of Spätzle. Proteolysis induces a conformational change that partially exposes the Toll-binding determinants. Receptor binding displaces the prodomain otherwise tightly bound to C-106 via a charge clamp between the newly formed N terminus (in blue) and the conserved aspartate (in red) C terminus of the prodomain α-helix.
To gain further insights into the role of Trp29 in the regulation of Spätzle activity we have studied a mutant protein with a phenylalanine residue substituted for tryptophan 29. Although this is a relatively conservative change and a phenylalanine residue is found at this position in some other cystine knot proteins, the physico-chemical properties of W29F Spätzle are quite different from those of the wild-type protein. W29F pro-protein has a tendency to form soluble aggregates and is able to bind to the Toll receptor ectodomain prior to cleavage. By contrast, removal of the prodomain by proteolysis and purification of W29F C-106 yields a monodisperse, dimeric protein that can activate Toll signaling with kinetics similar to the wild-type protein.
These properties of the W29F Spätzle pro-protein are similar to those of two loss-of-function mutants, Spz2 and SpzU5, that we previously mapped to the prodomain sequence (8). Spz2 is a tyrosine to asparagine substitution at residue 134, 33 amino acids N-terminal to the cleavage site, whereas SpzU5 changes the adjacent residue from proline to leucine (Fig. 6B). Both of these mutants are defective in biosynthesis, and strikingly, Spz2, like W29F, is secreted as a soluble aggregate. As detailed above the prodomain is largely unstructured a finding that is confirmed by protein secondary structure predictions. Of particular note, the only part of the prodomain predicted to form secondary structure is a 15-amino acid region adjacent to tyrosine 134 (Fig. 6B). Significantly, this sequence would form into an amphipathic α-helix, as shown by a helical wheel plot (Fig. 6B). This putative α-helix is highly conserved in Spätzle homologues from distantly related species such as Drosophila virilis, Manduca sexta (22) and the mosquito Anopheles gambiae (Fig. 6B) but not in paralogues such as the Drosophila DNT1 (Spz2) (23, 24) or the vertebrate neurotrophins (25).
Taken together, these results suggest that during biosynthesis the amphipathic α-helix of the prodomain associates with C-106, burying Trp29 and occluding the Toll receptor binding sites. This arrangement has striking similarities to that of another cystine knot protein, coagulogen, a clotting factor from the horseshoe crab Tachypleus tridentatus. Tachypleus is an evolutionarily ancient invertebrate and is regarded as a living fossil because its form has remained largely unaltered for more than 500 million years (26). Coagulogen forms part of a primitive defense response and is secreted by Tachypleus hemocytes as an inactive, monomeric precursor with an amphipathic α-helix bound into a hydrophobic cleft at the top of the molecule (27) (Fig. 6C). Proteolysis of pro-coagulogen triggered by exposure to microbial products causes a conformational change that exposes the hydrophobic surface sequestered by the helix. This causes the coagulogen monomers to polymerize into a gel, passively trapping invading microorganisms.
Proteolysis of Spätzle creates new N and C termini at the site of cleavage but does not cause the prodomain to dissociate. Thus, a plausible mechanism for activation is that the amino and carboxylic groups formed by cleavage make new electrostatic contacts with C-106 or prodomain which directly or indirectly induce a change in the position of the regulatory helix (Fig. 6D). It may be significant that an acidic residue located in the linker between the putative α-helix and the processing site is highly conserved (Fig. 6C). This aspartate could make an ion pair with the newly generated N terminus and cause a shift in the position of the helix, exposing Trp29 and enabling binding to the Toll ectodomain (Fig. 6D). This mechanism has elements in common with the activation of the chymotrypsin zymogen. In that case, the N-terminal amino group released by tryptic proteolysis at arginine 15 forms an electrostatic interaction, and this leads to a series of discrete conformational changes that unblock the active site of the enzyme and allow access to substrate (28).
In conclusion, it appears that an ancient passive defense system of the horseshoe crab mediated by coagulogen has been adapted during evolution and converted for use in a critical pathway of innate immune signaling. It is also likely that later in the evolution of the insects it was further adapted for dorso-ventral pattern formation. Future studies will seek to define the precise molecular basis of the activating switch by crystallizing the prodomain·C-106 complex before and after proteolysis.
Footnotes
- C-106
- C-terminal 106 amino acids of Spätzle
- TEV
- tobacco etch virus
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Belvin M. P., Anderson K. V. (1996) Annu. Rev. Cell Dev. Biol. 12, 393–416 [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann J. A. (2003) Nature 426, 33–38 [DOI] [PubMed] [Google Scholar]
- 3.Schneider D. S., Jin Y., Morisato D., Anderson K. V. (1994) Development 120, 1243–1250 [DOI] [PubMed] [Google Scholar]
- 4.Weber A. N., Tauszig-Delamasure S., Hoffmann J. A., Lelièvre E., Gascan H., Ray K. P., Morse M. A., Imler J. L., Gay N. J. (2003) Nat. Immunol. 4, 794–800 [DOI] [PubMed] [Google Scholar]
- 5.Mizuguchi K., Parker J. S., Blundell T. L., Gay N. J. (1998) Trends Biochem. Sci. 23, 239–242 [DOI] [PubMed] [Google Scholar]
- 6.Rattenholl A., Ruoppolo M., Flagiello A., Monti M., Vinci F., Marino G., Lilie H., Schwarz E., Rudolph R. (2001) J. Mol. Biol. 305, 523–533 [DOI] [PubMed] [Google Scholar]
- 7.Liu J., Tan H., Rost B. (2002) J. Mol. Biol. 322, 53–64 [DOI] [PubMed] [Google Scholar]
- 8.Weber A. N., Gangloff M., Moncrieffe M. C., Hyvert Y., Imler J. L., Gay N. J. (2007) J. Biol. Chem. 282, 13522–13531 [DOI] [PubMed] [Google Scholar]
- 9.LeMosy E. K., Tan Y. Q., Hashimoto C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5055–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turcotte C. L., Hashimoto C. (2002) Dev. Dyn. 224, 51–57 [DOI] [PubMed] [Google Scholar]
- 11.Wang L., Weber A. N., Atilano M. L., Filipe S. R., Gay N. J., Ligoxygakis P. (2006) EMBO J. 25, 5005–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang I. H., Chosa N., Kim S. H., Nam H. J., Lemaitre B., Ochiai M., Kambris Z., Brun S., Hashimoto C., Ashida M., Brey P. T., Lee W. J. (2006) Dev. Cell 10, 45–55 [DOI] [PubMed] [Google Scholar]
- 13.Gay N. J., Gangloff M. (2007) Annu. Rev. Biochem. 76, 141–165 [DOI] [PubMed] [Google Scholar]
- 14.Gangloff M., Murali A., Xiong J., Arnot C. J., Weber A. N., Sandercock A. M., Robinson C. V., Sarisky R., Holzenburg A., Kao C., Gay N. J. (2008) J. Biol. Chem. 283, 14629–14635 [DOI] [PubMed] [Google Scholar]
- 15.Ye Y., Godzik A. (2003) Bioinformatics 19, Suppl. 2, 246–255 [DOI] [PubMed] [Google Scholar]
- 16.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 17.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann A., Funkner A., Neumann P., Juhnke S., Walther M., Schierhorn A., Weininger U., Balbach J., Reuter G., Stubbs M. T. (2008) J. Biol. Chem. 283, 32598–32609 [DOI] [PubMed] [Google Scholar]
- 19.Permyakov E. A. (1993) Luminescent Spectroscopy of Proteins, pp. 21–107, CRC Press, Boca Raton [Google Scholar]
- 20.Eftink M. R., Ionescu R., Ramsay G. D., Wong C. Y., Wu J. Q., Maki A. H. (1996) Biochemistry 35, 8084–8094 [DOI] [PubMed] [Google Scholar]
- 21.Wang Y., Zhu S. (2009) Cell Mol. Life Sci. 66, 1595–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An C., Jiang H., Kanost M. R. (2010) FEBS J. 277, 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu B., Pennack J. A., McQuilton P., Forero M. G., Mizuguchi K., Sutcliffe B., Gu C. J., Fenton J. C., Hidalgo A. (2008) PLoS Biol. 6, e284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker J. S., Mizuguchi K., Gay N. J. (2001) Proteins Struct. Funct. Genet. 45, 71–80 [DOI] [PubMed] [Google Scholar]
- 25.Paoletti F., Konarev P. V., Covaceuszach S., Schwarz E., Cattaneo A., Lamba D., Svergun D. I. (2006) Biochem. Soc. Trans. 34, 605–606 [DOI] [PubMed] [Google Scholar]
- 26.Iwanaga S. (2002) Curr. Opin. Immunol. 14, 87–95 [DOI] [PubMed] [Google Scholar]
- 27.Bergner A., Oganessyan V., Muta T., Iwanaga S., Typke D., Huber R., Bode W. (1996) EMBO J. 15, 6789–6797 [PMC free article] [PubMed] [Google Scholar]
- 28.Stroud R. M., Kossiakoff A. A., Chambers J. L. (1977) Annu. Rev. Biophys. Bioeng. 6, 177–193 [DOI] [PubMed] [Google Scholar]