Abstract
Mtl1 is a member of the cell wall integrity (CWI) pathway of Saccharomyces cerevisiae, which functions as a cell wall sensor for oxidative stress. Genome-wide transcriptional analysis revealed a cluster of genes that were down-regulated in the absence of Mtl1. Many of these genes were potentially regulated by the general stress response factor Msn2/Msn4. In response to rapamycin, caffeine, glucose starvation and oxidative stress provoked by H2O2, mtl1 presents a significant loss of viability as well as a deficiency in the transcriptional response mediated by Msn2/Msn4. The Mtl1 function was required (i) to induce ribosomal gene repression, (ii) to induce the general stress response driven by the transcription factor Msn2/Msn4, and (iii) to activate the CWI pathway in response to both glucose starvation and oxidative stress. We also detected higher cAMP levels in the mtl1 mutant than in wild type cells indicative of up-regulated RAS2-PKA activity. Disruption of TOR1, disruption of RAS2, or hyperactivation of Rho1 restored both the viability and the transcriptional function (both ribosomal and Msn2/Msn4-dependent gene expression) in the mtl1 mutant to almost wild type levels when cells were starved of glucose or stressed with H2O2. Taking our results together, we propose an essential role for Mtl1 in signaling oxidative stress and quiescence to the CWI pathway and to the general stress response through Rho1 and the inhibition of either the TOR1 or RAS2 functions. These mechanisms would be required to allow cells to adapt to both oxidative and nutritional stresses.
Keywords: Cell Wall, MAP Kinases (MAPKs), Oxidative Stress, Signal Transduction, Yeast, Msn2/Msn4, Mtl1, Pkc1, TOR
Introduction
Organisms living in aerobic conditions are exposed to reactive oxygen species that provoke cellular damage and, as a consequence, various human diseases. The eukaryotic microorganism Saccharomyces cerevisiae serves as a model system in which to study the signal transduction pathways involved in the response to oxidative stress. In budding yeast the MAPK3 pathways are not well characterized for sensing and transmitting oxidative stress to the cytoplasm and nuclear elements. However, Mtl1 has already been characterized as a receptor for oxidative stress (1). Mtl1 was initially identified as a yeast homologue of Mid2 by Rajavel et al. (2) who observed that it performed a function in cell integrity signaling involved in vegetative growth. De Bettignies et al. (3) identified MTL1 as a suppressor of rgd1 mutants, with Rgd1 being a GTPase-activating protein of Rho3 and Rho4. MTL1 was also identified by Sekiya-Kawasaki et al. (4) as a multicopy suppressor of Rho1.
The cell integrity pathway in budding yeast involves a protein kinase (MAPK) cascade that participates in sensing and transmitting several extracellular signals and stresses that include cell-wall, osmotic, mating, and nutritional stress (5, 6) and, more recently, oxidative (1) and pH (7) stresses. The PKC1-MAPK pathway is integrated by several cell-wall proteins that are putative cell-membrane receptors of different stimuli; they are the Wsc1-Wsc4 family, Mid2, and Mtl1. They transmit signals to Rom2, which activates the G protein Rho1, which in turn activates the kinase Pkc1 (a protein that has high degree of homology with other isoforms of PKC in eukaryotic cells). Pkc1 activates a mitogen-activated protein kinase module: Bck1 (that is the MAPKKK) phosphorylates the redundant MAPK kinases Mkk1 and Mkk2, and together they activate Slt2, the last kinase member of the pathway. Two downstream events correlate with Slt2 activation: transcriptional activity driven by Rlm1 and Swi6 phosphorylation (5, 6). The upper elements of the CWI pathway are involved in the organization of the actin cytoskeleton under different conditions that include cell-wall and nutritional stresses (8–10), oxidative stress (1), and pH (11).
The Pkc1 pathway is also related to the TOR pathway. Budding yeasts have two different TOR genes, TOR1 and TOR2, that share 67% sequence identity and are partly redundant in function (12). Loewith et al. (13) have characterized two distinct TOR complexes, TORC1 and TORC2. TORC1 is sensitive to rapamycin, modulates translation initiation, inhibits protein turnover, and represses the transcription of genes related to nutrient starvation. TORC2 is closely related to the organization of the actin cytoskeleton and independent of rapamycin inhibition (13–16). Tor2 functions in both complexes, whereas Tor1 only participates in the TORC1 complex. The cellular function regulated by TOR contains a general mechanism; that is, the sequestration of the transcription factors Msn2/Msn4, Gln3 (17), and Rtg1/Rtg3 (18, 19) in its cytoplasm. The Tor function also regulates ribosomal protein expression in response to environmental conditions via PKA (20).
Ras signals via the cAMP-PKA pathway regulate the cellular metabolism in response to the carbon source (21, 22). Two Ras proteins are present in budding yeast and are encoded by the redundant small GTPases RAS1 and RAS2 (RAS) (23). The Ras-cAMP-PKA pathway also negatively regulates Msn2/Msn4 nuclear localization (24–26).
The Msn2/Msn4 transcription factor binds and activates genes containing the stress response element (CCCCT) in response to a wide variety of stresses, including nutritional, osmotic, acidic, and oxidative stress (17, 24, 27, 28).
In recent years several studies have been published that demonstrate the relationship between the TOR and cAMP-PKA pathways. Some authors have suggested that the RAS/cAMP pathway could be a novel TOR effector branch (29). Both the TOR and cAMP-PKA pathways regulate the expression of genes needed to overcome the diauxic and stationary phases (30, 31) and in whose regulation Msn2/Msn4 transcriptional activity has been reported to be essential (32). They also coregulate the expression of genes involved in fermentation and aerobic respiration (33). The CWI pathway is also required for viability in quiescence as Slt2 phosphorylation is necessary for cells to survive in stationary phase and upon rapamycin treatment (10, 34).
Here, we have employed a genomic approach to identify possible novel regulatory functions for the gene MTL1 and have demonstrated that Mtl1 signals toward Msn2/Msn4 transcriptional function in response to various stresses such as rapamycin, oxidative stress, and glucose starvation. We have also presented evidence to demonstrate that the Mtl1 function is required to down-regulate Tor1 and Ras2 activity upon glucose starvation and oxidative stress. This down-regulation is necessary to ensure cell viability in response to both types of stress. We show that Mtl1 is the member of the CWI pathway that acts as a sensor for oxidative stress and glucose starvation by activating the kinase Slt2. However, Slt2 activation is only visibly important for cell viability in conditions of quiescence.
EXPERIMENTAL PROCEDURES
Media and Growth Conditions
Yeasts were grown in SD medium (2% glucose, 0.67% yeast nitrogen base, and the required amino acids) (35). Diamide, hydrogen peroxide, caffeine, and rapamycin were purchased from Sigma. Diamide was dissolved in DMSO. Hydrogen peroxide was prepared in sterile distilled water.
Yeast Strains and Gene Disruptions
The yeast strains used in this study are listed in Table 1. MTL1 was disrupted either by the one-step disruption method using the kanMX4 module, as described in Vilella et al. (1), or by using the natMX4 module (36). RAS2 and TOR1 were disrupted by the same procedure but using the LEU marker.
TABLE 1.
Yeast strains used in this work
| Strain | Relevant genotype | Reference |
|---|---|---|
| CML128 | MATa leu2-3,112 ura3-52 trp1 his4 can1r | Gallego et al. (39) |
| GSL41 | MATa mtl1::kan MX4 | This worka |
| GSL34 | MATa tor1::kanMX4 | This worka |
| GSL48 | MATamtl1::kan MX4 tor1::LEU2MX5 | This worka |
| GSL53 | MATaras2::LEU2MX5 | This worka |
| GSL54 | MATamtl1::kan MX4 ras2::LEU2MX5 | This worka |
| W303-1A | MATa ade2-1, trp1-1, leu2-3,2-111, his3-11,75, ura3 | C. Gancedo |
| msn2msn4 | MATa ade2 can1 his3 leu2 trp1 ura3 msn2-delta3::HIS | F. Estruch |
| msn4-1::TRP1 |
a CML128 background.
DNA Manipulation and Plasmids
Plasmid pPkc1 contains Pkc1 under the tetO7 promoter and is also tagged with the hemagglutinin epitope, as initially described in Angeles de la Torre-Ruiz et al. (37) under the name of pMM126. Plasmid pSlt2 is a YEP352 derivative containing the Slt2 ORF under its own promoter and tagged with hemagglutinin in the C terminus; this plasmid was described in Vilella et al. (1). In this study, we used the pSlt2 plasmid for overexpression analysis and also to determine total Slt2 protein using the anti-hemagglutinin monoclonal antibody.
The plasmid pBCK1-20 is a pRS413 derivative bearing the constitutively activated BCK1–20 allele (38). The plasmid pRho1* contains the LEU marker and bears the Rho1his368 allele, which is constitutively activated, bears the hemagglutinin epitope in the C-terminal position, and is cloned under the Gal1 promoter. This plasmid was kindly provided by Dr. Tobias Schmelzle.
The plasmid pMsn2-GFP contains the LEU marker, cloned under the ADH1 promoter in a centromeric plasmid, as originally described in Görner et al. (44). This plasmid was kindly provided by Dr. Francisco Estruch. This plasmid was originally described as functional, but we checked its functionality in an msn2msn4 background. We demonstrated that it efficiently suppresses both the lack of viability and the deficiency in the transcription of HSP12 and CTT1 that were observed in the double mutant msn2msn4 in response to the stresses used in this study.
Microarray Experiments and Data Analysis
For microarray analysis, wild type and mtl1 cells were grown in SD minimum medium at 30 °C to a final A600 0.6. Total RNA from S. cerevisiae was extracted from 25 ml of each culture by mechanical disruption following the instructions for the RNeasy Midi kit manufacturer (Qiagen). RNA concentration was measured at 260 nm, and sample quality was checked using RNA Nano Labchips in a 2100B Bioanalyzer (Agilent Technologies, Palo Alto, CA).
For each experimental condition (wt and mtl1Δ strain), two microarray experiments corresponding to two biological replicates were processed and analyzed. Double-stranded cDNA was synthesized from 5 μg of total RNA using a “One-cycle cDNA Synthesis Kit” (Affymetrix, Santa Clara, CA). After cDNA purification using the “GeneChip Sample Cleanup Module” (Affymetrix), this DNA was used as a template for the in vitro transcription to obtain the biotin-labeled cRNA. The cRNA obtained was fragmented and hybridized to the Affymetrix GeneChip® Yeast Genome S98 array for 16 h at 45 °C. Hybridized microarrays were washed and stained with a streptavidin-phycoerythrin conjugate in a GeneChip® Fluidics Station 450. All these procedures were carried out as suggested by the manufacturer. Hybridized cRNA was finally identified by the fluorescence signal in a GeneChip® 3000 scanner.
After scanning, numerical data were obtained and processed with GCOS software (Affymetrix). We evaluated different parameters that accounted for the quality of the hybridization. The noise value was less than 2.51 (commonly between 1.5 and 3), the scaling factor was between 0.22 and 0.41, and the background was between 107 and 50.6. The arrays showed percentages of “Present” probe sets ranging between 72.3 and 79.5. The signals from the arrays were linearly scaled to an average signal value of 100.
Data from mutant and WT strains were compared in pairs, obtaining a total of four comparisons. For further analysis, a selection of significant results was carried for each comparison using the statistical criteria implemented by the GCOS software. Those genes labeled as “Absent” by the detection algorithm (using a p value of < 0.04) in both arrays of a comparison were discarded for the analysis. Those genes that gave “Increase” or “Decrease” values, according to the change algorithm (using a p value of <0.0025) in all four comparisons were used for further analysis. The signal log ratio was obtained as the mean of the signal log ratio obtained in each of the four comparisons; genes with signal ratios of >2 or <0.5 were selected for additional analysis. Probes on the yeast genome S98 chips that did not correspond to specific open reading frames were disregarded in this analysis.
RNA Preparation and Northern Blot Analyses
RNA purification, Northern blot, and probe labeling with digoxygenin were carried out according to Gallego et al. (39). Probes covering the entire open reading frame, without adjacent sequences, were generated by PCR from genomic DNA.
Yeast Extracts and Immunoblot Analyses
Both methods were performed as described in Angeles de la Torre-Ruiz et al. (37). The methodology for the use of both anti-phospho-p44/42 and anti-Swi6 antibodies was that previously described in Angeles de la Torre-Ruiz et al. (37). Horseradish peroxidase-linked anti-rabbit (NA931, Amersham Biosciences) was used at a 1:10,000 dilution in Tris-buffered saline-Tween buffer containing 1% milk fat. In all cases chemiluminescent detection was performed using the Supersignal substrate (Pierce) in a Lumi-Imager (Roche Applied Science).
cAMP Assay
The Amersham Biosciences cAMP Biotrak Enzyme-immunoassay system was used to determine intracellular levels of cAMP.
Strains wt, mtl1, ras2, ml1ras2, tor1, mtl1tor1 were grown in SD selective media at 30 °C. We performed the experiment using exponentially growing cultures (A600 0.6) and stationary cultures grown for up to 3 days. We basically followed the method described by Swiegers et al. (40) to obtain the samples. A total of 40 ml of each of the exponentially growing cultures (and the corresponding amount of cells for the stationary cultures) was harvested by centrifugation at 3000 rpm at room temperature for 4 min. After that, the wet weight was determined, the cells were subsequently resuspended in 300 μl of lysis buffer 1B (Amersham Biosciences), and 150 μl of glass beads were added. The suspension was vortexed for 30 min at 8 °C and then spun down for 10 min at 4 °C at 12,000 rpm. The supernatant (100 μl) was used for the protocol; that is, intracellular cAMP measurement using the non-acetylation enzyme-immunoassay procedure with the novel lysis reagents described in the kit.
RESULTS
The Mtl1 Function Is Required to Maintain the Basal Transcription of a Cluster of Genes Regulated by Msn2/Msn4
Our group previously demonstrated that Mtl1 functions as a cell-surface sensor for oxidative stress (1). To gain a further insight into the role that Mtl1 plays in cell integrity, we decided to analyze the genome-wide gene expression profile of the mtl1 mutant strain. The transcriptional profile of this mutant growing in SD medium was compared with the wild type strain CML128 under the same conditions using Affymetrix GeneChip® Yeast Genome S98 arrays, as detailed under “Experimental Procedures.” Microarray analysis of this profile revealed the presence of 102 genes that were repressed (-fold ≤0.5) (supplemental Table S1) and 13 that were induced genes (-fold ≥2) (supplemental Table S2) as a consequence of the MTL1 deletion. A functional classification of these genes was carried out according to MIPS and GO Databases using FUNSPEC. This analysis revealed the presence of several statistically significant functional groups (p value less than 0.01) within the repressed response. These included electron transport and energy uptake, stress response, and sugar and aromatic compound utilization among others (supplemental Table S3). Interestingly, and as deduced from the YEASTRACT data base, an important group of the repressed genes was potentially regulated by Msn2/Msn4 (34 of 102 repressed genes); most of these related in one way or another to the stress response (Table 2).
TABLE 2.
Genes down-regulated in mtl1 strain, which are potentially regulated by Msn2/Msn4
| Gene/open reading frame | Function | Biological process |
|---|---|---|
| ALD3 | Aldehyde dehydrogenase activity | Stress response |
| CTT1 | Catalase activity | |
| DAK1 | Glycerol kinase activity | |
| DDR2 | ||
| GAD1 | Glutamate decarboxylase activity | |
| GRE1 | ||
| HSP12 | ||
| HSP26 | Unfolded protein binding | |
| MCR1 | Cytochrome-b5 reductase activity | |
| SIP18 | Phospholipid binding | |
| SSA4 | Unfolded protein binding | |
| TRX3 | Thiol-disulfide exchange intermediate activity | |
| PHM7 | Unknown | |
| SOL4 | 6-Phosphogluconolactonase activity | |
| NQM1 | Transaldolase activity | |
| YHR033W | ||
| YKL151C | ||
| AIM33 | ||
| YMR090W | ||
| AGX1 | Alanine-glyoxylate transaminase activity | Amino acids |
| POT1 | Acetyl-CoA C-acyltransferase activity | Lipid metabolism |
| TES1 | Acyl-CoA thioesterase activity | |
| TKL2 | Transketolase activity | Carbohydrates |
| YJR096W | Aldehyde reductase activity | |
| TSA2 | Thioredoxin peroxidase activity | Homeostasis |
| ECM4 | Glutathione transferase activity | Cell wall |
| FMP45 | ||
| TFS1 | Lipid binding/protease inhibitor activity | Protein catabolism |
| VPS73 | Substrate specific transmembrane transporter activity | Transport |
| SPS100 | Sporulation | |
| HBT1 | Conjugation | |
| YLR294C | Hydrogen-transporting ATP synthase | Respiration |
| GTT1 | glutathione transferase activity | Others |
| YDL124W | α-Keto amide reductase activity, α-keto ester reductase activity |
In an initial characterization of Mtl1 we focused our efforts on studying the possible relationship between Mtl1 and Msn2/Msn4. First, and to validate our data, we performed Northern blot assays with samples taken from mtl1 and wt strains growing exponentially. We then tested HSP12, GRE1, TRX3, and DDR2 gene expression, as they are potentially regulated by the transcription factor Msn2/Msn4. In all cases we observed that the basal levels of these transcripts were lower in the mtl1 mutant than in the wt cells; this confirmed the results obtained in the genomic study described above and validated our genomic analysis (Fig. 1A).
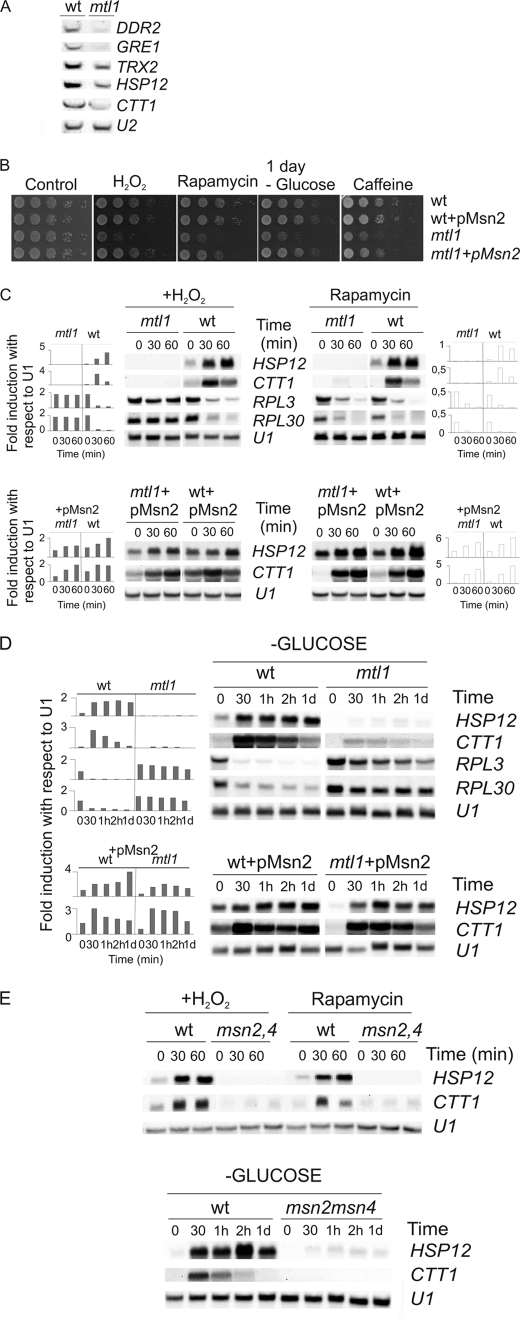
FIGURE 1.
Mtl1 is required for survival in response to various types of stress and it is essential for the regulation of the Msn2 transcription function. A, shown is Northern blot analysis of wt and mtl1 samples growing in SD medium at 30 °C to logarithmic phase using DDR2, GRE1, TRX2, HSP12, and CTT1 probes and U2 as a loading control. B, exponentially growing cultures of wt (CML128), mtl1, wt +pMsn2, and mtl1 + pMsn2 were serial-diluted and spotted onto plates containing 1 mm H2O2, 1 ng/ml rapamycin, and 5 mm caffeine. In parallel, aliquots were taken, washed four times with equivalent volumes of SD minus glucose, and incubated in fresh SD medium without glucose for 1 day before being serial diluted and spotted onto YPD plates. Growth on the plates was allowed to take place for 3 days at 30 °C. C, Northern blot analysis of wt, mtl1, wt +pMsn2, and mtl1 + pMsn2 samples growing in SD medium at 25 °C to logarithmic phase and then treated with either 1 mm H2O2 or 200 ng/ml rapamycin is shown. Samples were collected and processed at the indicated times using HSP12, CTT1, RPL3, and RPL30 probes and U1 as a loading control. Histograms represent the -fold induction normalized with respect to the corresponding U1 value for each of the genes analyzed. D, experiments were as in B, except that exponentially growing cells were washed four times with SD minus glucose and then transferred to fresh SD medium minus glucose. Samples were collected at the indicated times. Northern blot analyses of exponentially growing wt and mtl1 cultures. Histograms represent the -fold induction normalized with respect to the corresponding U1 value for each of the genes analyzed. E, Northern blot analyses of wt and msn2msn4 growing exponentially and then treated either with H2O2 or rapamycin or starved of glucose as in C and D. d, day.
Mtl1 Regulates the Msn2 Transcriptional Function in Response to Rapamycin, Glucose Starvation, and Oxidative Stress
To ascertain the physiological significance of the previously mentioned results, we proceeded to screen mtl1 survival under different conditions, inferred from mtl1 global transcription analysis. We hypothesized that a possible cause of the gene down-regulation observed in the mtl1 background was a defect in the Msn2/Msn4 function. As the Msn2/Msn4 is a transcription factor involved in the general stress response, we decided to screen the survival response of the mtl1 mutant to various stresses in which Msn2/Msn4 is involved: rapamycin, hydrogen peroxide, and glucose deprivation. In Fig. 1B we show that Mtl1 is needed for cell survival in response to rapamycin, hydrogen peroxide, caffeine, and carbon deprivation, whereas in stationary phase (not shown), cell survival is undistinguishable from wt cell. All these phenotypes are consistent with a defect affecting the general stress response; they also resemble those already described for msn2msn4 mutants (41, 42). We next analyzed the transcriptional regulation of CTT1 and HSP12 in wt and mtl1 mutant cells growing exponentially (time 0) or treated either with rapamycin or hydrogen peroxide or deprived of glucose (Figs. 1, C and D). We observed that the mtl1 mutant was deficient in the transcriptional induction of HSP12 and CTT1 upon treatment with rapamycin or peroxide or glucose deprivation. The regulation of these genes under these conditions was exerted by the Msn2 function because HSP12 and CTT1 genes were not induced in response to the three conditions in an msn2msn4 mutant (Fig. 1E). Moreover, we overexpressed Msn2 in both wt and mtl1 strains and observed that higher Msn2 levels were sufficient to complement the defects in HSP12 and CTT1 transcription (both regulated by Msn2/Msn4) in mtl1 cells in response to all the treatments to almost wild type levels (Fig. 1, C and D). Msn2 overexpression was also able to rescue the defect in mtl1 cell growth provoked by each of the previously mentioned stress types (Fig. 1B). Moreover, and in line with the previously mentioned results, Msn2 overexpression also restored the logarithmic levels of HSP12 and CTT1 transcription (Fig. 1, C and D). In view of these results and given that in the absence of Mtl1 function a wide cluster of genes regulated by Msn2 were down-regulated with respect to the wild type strain, we concluded that Mtl1 signals to Msn2 and regulates its function in response to the blockade of the TOR function, hydrogen peroxide, and glucose deprivation.
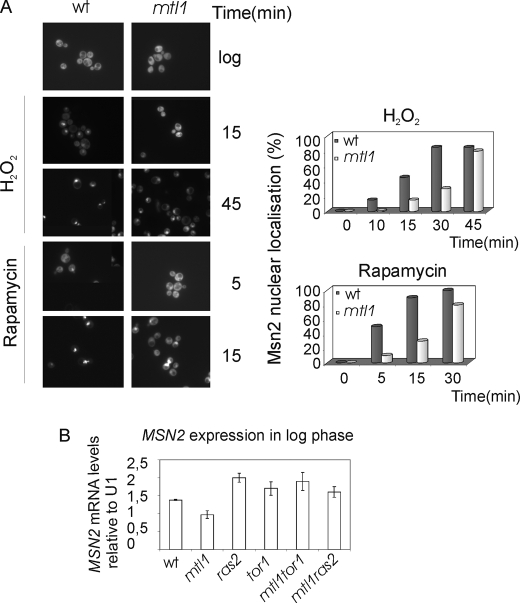
It is well known that Msn2 has a cytoplasmic localization when cells grow exponentially in rich medium and also that Msn2 translocates to the nucleus in response to several types of stress. We decided to investigate how the lack of Mtl1 function affected the subcellular localization of Msn2 to find an explanation for the transcriptional deficiency observed in the mtl1 mutant. To do this, we studied in vivo GFP-Msn2 localization in mtl1 mutant cells. In response to either hydrogen peroxide or rapamycin, Msn2 translocates to the nucleus in both wt and mtl1 cells. However, in mtl1 cells, we observed a significant delay in Msn2 translocation to the nucleus compared with wt cells (Fig. 2A). The timing of Msn2 translocation from the cytoplasm to the nucleus in response to stress has been attributed to RAS activity, whereas Msn2 translocation from the nucleus to the cytoplasm has been reported to be mainly dependent on TOR activity (30). PKA prevents the nuclear import of Ms2/Msn4, whereas the TOR function controls their nuclear export (43, 44). We also checked the mRNA levels of MSN2 in wild type, mtl1, ras2, mtl1ras2, tor1, and mtl1tor1 cells. In the mutant mtl1 we were able to observe a reduction in MSN2 basal transcription in exponentially growing cells with respect to wild type levels. However, deletion of either RAS2 or TOR1 in the mtl1 background restored the steady state transcription of MSN2 to wild type levels (Fig. 2B). These results together with the others described above led us to investigate the possible relationship between Mtl1 and the TOR and Ras pathways.
FIGURE 2.
Msn2 regulation in different mutants. A, shown is the Msn2 location in response to hydrogen peroxide and rapamycin. wt and mtl1 cells were transformed with a multicopy plasmid carrying Msn2 fused to GFP in the C terminus and under the control of the ADH1 promoter as described under “Experimental Procedures.” Cultures were grown in SD minimum medium at 30 °C to logarithmic phase and then treated with rapamycin and hydrogen peroxide for the times indicated in the figures. Histograms represent the percentage of cells with Msn2 localized in their nuclei counted from a total of 300 cells. We have chosen a representative experiment from three repetitions that were significantly similar. B, histograms represent the MSN2 mRNA levels calculated in wt, mtl1, ras2, mtl1ras2, tor1, and mtl1tor1 cells growing exponentially in logarithmic phase and normalized with respect to the values obtained using a U1 probe as a loading control. Error bars represent S.D. calculated from three repetitions.
Mtl1 Is Genetically Related to Tor1 and Ras2 in the Regulation of the Msn2 Function
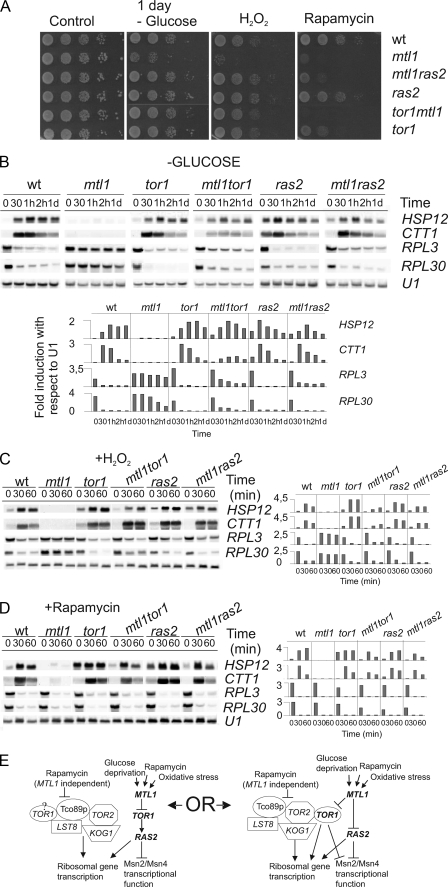
It has been reported that Tor signals to Ras regulating the Msn2 function (29). As the mtl1 mutant presented high sensitivity to rapamycin (a macrolide that specifically inhibits the TORC1 function) and as the induction of both HSP12 and CTT1 provoked by this drug was dependent on Mtl1, we next decided to investigate the relationship between the TORC1 complex and Mtl1. In a first approach we constructed an mtl1tor1 double mutant and observed that this strain exhibited greater sensitivity to rapamycin than either of the two single mutants: tor1 or mtl1 (Fig. 3A). However, when we tested for glucose starvation or hydrogen peroxide sensitivity, we observed that both the tor1 and mtl1tor1 strains presented similar levels of sensitivity, although both were less sensitive than the single mutant mtl1 (Fig. 3A). We next analyzed the transcriptional pattern in these strains and observed that tor1 deletion restored the induction of both HSP12 and CTT1 in mtl1 to almost wild type levels in response to oxidative stress and rapamycin and glucose starvation (Figs. 3, B–D). Because the TORC1 function also regulates ribosomal gene expression, we decided to analyze the transcription of two ribosomal genes: RPL3 and RPL30 in wt, mtl1, tor1, and tor1mtl1 cells under all the stress conditions used in the current study (Fig. 3, B–D). As expected and in accordance with previous studies, we observed clear ribosomal gene repression in wt, tor1, mtl1, and tor1mtl1 cells in response to the rapamycin treatment. Additionally, both the mtl1 and tor1mtl1 mutants presented similar responses to wild type cells; this indicated that Mtl1 was not required to repress ribosomal gene transcription when the TOR function was blocked by rapamycin (Fig. 3E). Our results suggest that both TOR1 and MTL1 regulate a common process in response to rapamycin and also that at least one of the two genes is required for cells to survive upon rapamycin treatment, although they are in different pathways. MTL1 and TOR1 signal to Msn2, suggesting that MTL1 could signal TOR1 inhibition for the regulation of the Msn2 function in response to the blockade of the TORC1 function (Fig. 3E). However, Mtl1 is not involved in the ribosomal gene repression mediated by rapamycin treatment.
FIGURE 3.
The absence of either TOR1 or RAS2 restores cell viability, ribosomal gene expression, and transcriptional regulation dependent on Msn2 in the mtl1 mutant to wild type levels. A, cell viability was as in Fig. 1 but using the following strains: wt, mtl1, ras2, ras2mtl1, tor1, and tor1mtl1. B, shown are Northern blot analyses of the above-mentioned strains growing exponentially and then starved of glucose or treated with either H2O2 (C) or rapamycin (D), as described in Fig. 1. Histograms represent the -fold induction normalized with respect to the corresponding U1 value for each of the genes analyzed. E, the schematic diagram shows two possible models, which could explain how Mtl1 signals to both TOR1 and RAS2 to regulate Msn2/Msn4 function in response to various stresses. d, day.
In response to the other two stresses, oxidative stress and glucose depletion, the mtl1 mutant was unable to induce ribosomal gene repression. Interestingly, in the double mutant tor1mtl1, the repression was restored to almost wild type levels (Fig. 3, B–C). These results, therefore, suggest that Mtl1 requires Tor1 repression to transmit the signal for glucose starvation and hydrogen peroxide stress to downstream effectors that regulate both ribosomal gene expression and the Msn2/Msn4 function (Fig. 3E).
Because it has been previously reported that TOR signals to Ras/cAMP for the regulation of ribosomal gene expression and the Msn2/Msn4 function (29), we further investigated the possible relationship between Mtl1 and Ras2. We constructed both ras2 and mtl1ras2 double mutants and obtained similar results to those already described above with respect to the interaction between Tor 1 and Mtl1. In the double mutant mtl1ras2 we observed (i) restoration of mtl1 viability after rapamycin treatment, oxidative stress or glucose deprivation, (ii) restoration of Msn2/Msn4 wild type activity (as deduced from the levels of expression of both transcripts, CTT1 and HSP12), and (iii) ribosomal gene repression similar to that associated with wild type and ras2 cells (Fig. 3, B–D). Thus, the deletion of Ras2 restored the majority of the phenotypes exhibited by the mtl1 cells to almost wild type levels in response to both oxidative stress and glucose depletion. Our results, therefore, suggest a role for Mtl1 in transmitting the signal for oxidative stress and glucose starvation to Msn2/Msn4 and also in the down-regulation of Ras2 functions (Fig. 3E).
The Absence of Either Tor1 or Ras2 Restores the Defects in the Accumulation of Glycogen Observed in the mtl1 Cells
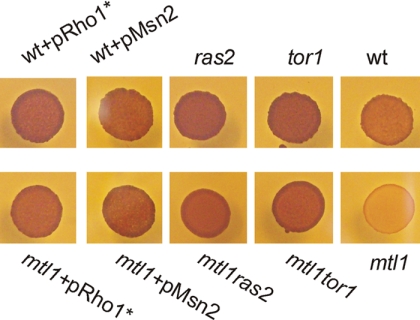
Accumulation of glycogen occurs when cells are starved of nutrients. It has been demonstrated that both the TOR and Ras-cAMP pathways control glycogen homeostasis in cells (45–47). Part of this effect is due to the fact that Msn2/Msn4 regulates the transcription of a subset of genes required for glycogen synthesis (48, 49). Because the absence of the Mtl1 function impaired the activity of Msn2/Msn4, we tested whether mtl1 cells presented any defects in the accumulation of glycogen. We confirmed our hypothesis and demonstrated that mtl1 was defective in the accumulation of the carbon storage source glycogen (Fig. 4). Interestingly, TOR1 and RAS2 deletion and also Msn2 overexpression restored the capacity to accumulate glycogen in mtl1 cells. This supports the hypothesis that Mtl1 signals the inhibition of Tor1 and Ras2 and consequently contributes to the activation of the Msn2 function in response to quiescence (Fig. 4).
FIGURE 4.
Glycogen accumulation in the different strains: wt, wt+pRHO1*, wt+pMsn2, ras2, tor1, mtl1+pRHO1*, mtl1+pMsn2, mtl1ras, and mtl1tor1. Cells were grown on SD medium to logarithmic phase then spotted onto SD medium plus amino acids to be grown for 5 days. A solution of 0.2% iodine/0.4% potassium iodide (Swiegers et al. (40)) was poured over the spots, and photographs were taken 3 min later. The darker the color, the more glycogen was accumulated, and the lighter the color, the less glycogen was present. Glycogen provides an indirect measurement of cAMP/PKA activity.
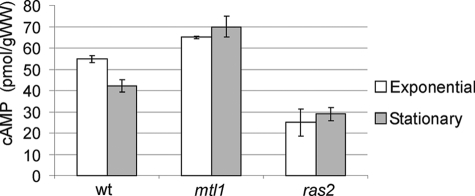
Mtl1 Affects Intracellular Levels of cAMP
Mtl1 is genetically related to Ras2 function. This is a logical consequence of what would be an additional relationship with the PKA pathway that should be reflected in cAMP intracellular levels. We decided to determine the effect of deleting Mtl1 on cellular cAMP levels. The levels of cAMP were determined in wild type, ras2, and mtl1 strains during exponential growth and in stationary phase. Under both conditions, mtl1 cells showed higher levels of cAMP than those determined in wild type cells (Fig. 5). Deletion of ras2 caused a significant decrease in cAMP levels compared with wild type cells. Taking all these results together we can conclude that Mtl1 regulates cAMP levels through the down-regulation of the RAS2 function.
FIGURE 5.
Intracellular cAMP in different cultures. wt, mtl1, and ras2 strains were grown either exponentially or to stationary phase (3 days of incubation at 30 °C) in SD medium plus amino acids and harvested for cAMP extraction and detection as described under “Experimental Procedures.” Values are the average of three independent experiments (error bars represent the S.D.). With respect to both conditions, exponential and stationary phases, the three strains (wt, mtl1, and ras2) were significantly different from one another, with a 0.01 < p < 0.02. These values were calculated upon performing two-tailed t tests.
Involvement of the Pkc1-MAPK Pathway in the Stress Response Mediated by Mtl1
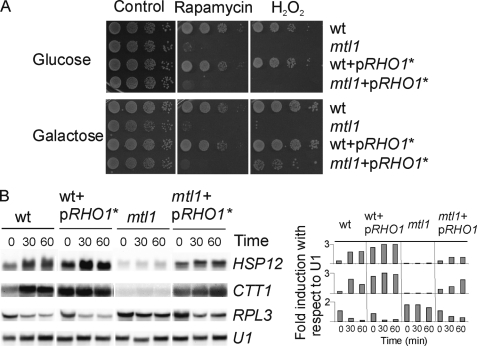
Mtl1 is a component of the cell integrity pathway and a sensor of oxidative stress. We wondered whether the effect that Mtl1 has on both Tor1 and Ras2 was also mediated by elements of the Pkc1-MAPK pathway. To ascertain this, we checked the functional relationship between Mtl1 and several members of the pathway. We first checked whether Rho1 was involved in this response. To do this we used a constitutively activated Rho1 allele in a centromeric plasmid under the control of the GAL1 promoter and transformed both wild type and mtl1 cells. As described in legend to Fig. 6, glucose repressed Rho1 expression (Fig. 6A), whereas galactose, the sole carbon source, induced Rho1 overexpression (Fig. 6A, middle panel corresponding to galactose). As shown in Fig. 6, expression of the hyperactive RHO1 allele in the mtl1 mutant strain led to (a) an increase in cell-survival in response to hydrogen peroxide (Fig. 6A) but not to rapamycin, (b) the partial restoration of the transcriptional induction of CTT1 and HSP12 (Fig. 6, B and C), and the repression of ribosomal genes under conditions of oxidative stress (Fig. 6B). We could not perform these experiments under conditions of glucose depletion because the expression of the RHO1 hyperactivated allele was regulated by the Gal1 promoter. However, we were able to detect that when wild type cells were transferred from glucose to galactose there was induction of the genes regulated by Msn2/Msn4 (Fig. 6B). Interestingly, when we transferred mtl1 cells from glucose to galactose (as the sole carbon source), we did not observe any significant induction of the genes used to report Msn2 activity (CTT1 and HSP12) with respect to the wild type cells, in which this shift induced the transcription of both genes. Importantly, overexpression of the RHO1 hyperactive allele in mtl1 cells not only rescued the transcriptional levels of Msn2-dependent genes to almost wild type levels but also the hyperactivation of the MAPK (Slt2) of the CWI pathway (Fig. 8A) when cells were transferred from glucose to galactose-containing medium and were subsequently treated with hydrogen peroxide. Furthermore, Rho1 also suppressed the deficiency in glycogen accumulation in mtl1 cells (Fig. 4). Taking these results together, we hypothesize that the signaling from Mtl1 to Tor1 and Ras2 that we have characterized occurred through Rho1. In addition we were able to observe that Rho1 presented stress specificity, as it did not rescue mtl1 cell viability in response to rapamycin treatment (Fig. 6A). We performed the same experiments by overexpressing Rom2 (a GAP for Rho1) and obtained equivalent results to those shown for Rho1 (data not shown).
FIGURE 6.
Hyperactivation of Rho1 restores mtl1 cell viability, transcriptional induction being dependent on Msn2 and the ribosomal gene transcription in mtl1 cells in response to hydrogen peroxide but not in response to rapamycin. A, serial dilutions of exponentially growing cultures of wild type, mtl1, wt+pRHO1*, and mtl1pRHO1* were grown in SD plus amino acids at 25 °C. Half of the cultures were serial-diluted and directly plated onto SD plates containing either hydrogen peroxide or rapamycin (first line in Fig. 5A). The other half was washed four times with two volumes of minimum medium containing galactose as the sole carbon source and then transferred to minimum medium containing galactose to induce the expression of RHO1* for 12 h of induction. After this period cells were serial-diluted and plated onto SD plates containing hydrogen peroxide or rapamycin (second line in Fig. 5B). B, shown is a Northern blot analysis of the strains growing logarithmically in SD minimum medium (wt and mtl1 strains) or in minimum medium containing galactose as a carbon source, as described in A (wt+pRHO1*, mtl1+pRHO1*) and then treated with 1 mm hydrogen peroxide. Samples were collected after the indicated times as in Fig. 2B. Histograms represent the -fold induction normalized with respect to the corresponding U1 value for each of the genes analyzed.
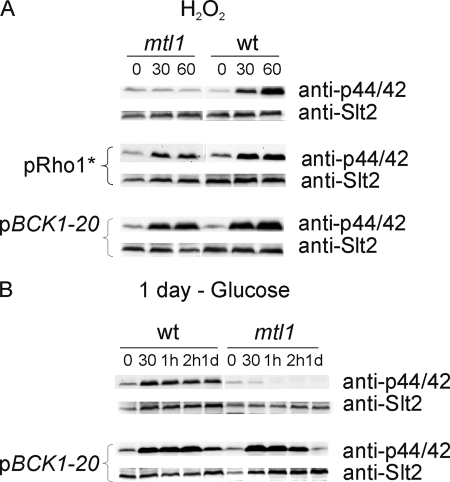
FIGURE 8.
Mtl1 is involved in signaling both oxidative stress caused by hydrogen peroxide and glucose starvation to the protein kinase Slt2 through elements of the Rho1 and Bck1 cell integrity pathways. A, Western blot analysis of Slt2 activity using the p44/42 polyclonal antibody in samples from wt, mtl1, wt+pRHO1*, mtl1+pRHO1*, wt+pBCK1-20, and mtl1+pBCK1-20 growing exponentially and treated with 1 mm hydrogen peroxide for the indicated times. For pRHO1* overexpression we followed the same procedure as described in the legend to Fig. 5A, the previous hydrogen peroxide treatment. B, procedures were the same as in A, but the strains were exponentially grown in SD, washed four times with minimum medium without glucose, and then transferred to SD minus glucose to be incubated at 25 °C. Samples were collected at the indicated times for Western blot analysis. Anti-GSTSlt2 antibody was used to detect total Slt2 protein (not shown).
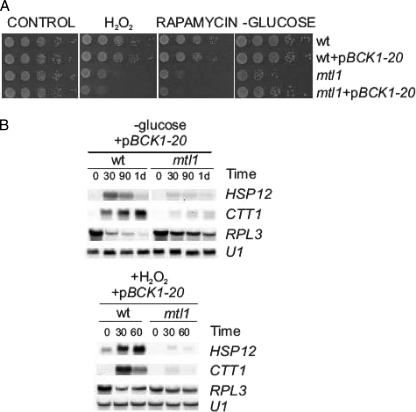
To further analyze the role of the downstream elements in the CWI pathway involved in this signaling process, we used a constitutively activated allele of the MAPKKK (MAPK kinase kinase) BCK1, the allele BCK1–20, and also checked for overexpression of the last kinase in the pathway, Slt2 (data not shown). The results shown in Fig. 7 indicate that upon treatment with hydrogen peroxide or upon glucose depletion, neither the expression of the BCK1–20 allele nor Slt2 overexpression (data not shown) was able to restore the following functions in the mutant mtl1; (i) cell viability (Fig. 7A), (ii) Msn2/Msn4 transcriptional function (Fig. 7B), and (iii) repression of ribosomal genes (Fig. 7B). Moreover, as in the case of the RHO1* allele, the BCK1–20 allele did not suppress the lethality caused by rapamycin in mtl1 cells. All these results indicate that there is cross-talk between Mtl1 and Tor1 and the Ras-cAMP pathways through the Rho1 protein. However, the other elements in the CWI pathway downstream of Rho1 did not seem to play any significant role in the signaling from Mtl1 to TOR and Ras/cAMP. However, the constitutive activation of the pathway at the level of the kinase BCK1 did restore cell viability in response to glucose depletion (Fig. 7A). This result stresses the role that the CWI pathway plays in quiescence.
FIGURE 7.
Constitutive activation of BCK1 only suppresses mtl1 cell lethality upon glucose deprivation, but it does not suppress the transcriptional regulation of ribosomal genes or of the Msn2 genes that are dependent on mtl1cells in response to hydrogen peroxide or glucose deprivation. A, shown are serial dilutions of exponentially growing cultures of wild type, mtl1, wt+pBCK1-20, and mtl1+pBCK1-20 plated onto plates containing either hydrogen peroxide or rapamycin as in Figs. 1, 2, and 5. As in Fig. 1B and 2A, aliquots were taken, washed four times with equivalent volumes of SD minus glucose, and then incubated in fresh SD medium without glucose for 1 day to be subsequently serial diluted and spotted onto YPD plates. B, Northern blots with samples treated with hydrogen peroxide or starved of glucose as in Figs. 1 and 2 are shown. Samples were harvested at the indicated incubation times.
Mtl1 Is Required to Activate the Pkc1-MAPk Pathway in Response to Hydrogen Peroxide and Glucose Starvation
As Mtl1 is a cell-wall sensor and a component of the CWI pathway, we decided to investigate the role of this protein in signaling through this pathway in response to hydrogen peroxide and glucose starvation. To check this we checked the phosphorylation of the MAPK Slt2 by Western blotting in wild type and mtl1 cells treated with hydrogen peroxide or starved of glucose. As detailed in Fig. 8, the MAPK Slt2 became notably activated in wt cells in response to both stresses. However, the hyperactivation of Slt2 was clearly abrogated in mtl1-stressed cells.
To further demonstrate that Mtl1 transmits the signal to Slt2 through the Pkc1 pathway, these experiments were carried out in mtl1 cells alternatively transformed with the RHO1 hyperactive allele and the constitutively active BCK1–20 allele. As shown in Fig. 8, A and B, hyperactivation of the pathway downstream from Mtl1 compensated for the absence of the receptor in the activation of Slt2 upon both oxidative and glucose depletion stresses. These results indicate that Mtl1 is involved in signaling the absence of glucose or oxidative stress caused by hydrogen peroxide and transmits it to Slt2 through the classical elements of the CWI pathway.
DISCUSSION
We have used a genomic approach to identify other functions in which the cell-surface protein Mtl1 could play a role. Having observed a large cluster of genes that were potentially regulated by Msn2 and down-regulated in the absence of Mtl1, we identified Msn2 as a possible target for Mtl1. The absence of Mtl1 makes cells sensitive to oxidizing agents, glucose starvation, and conditions in which the TORC1 function is compromised. This sensitivity is also associated with a notable defect in Slt2 activation. Msn2 overexpression was quite efficient at rescuing the loss of viability caused by the absence of the Mtl1 function in all the stress conditions studied here. It is very interesting to observe that a significant number of the Msn2-dependent genes that are down-regulated in the mtl1 mutant are related to the oxidative stress response. This is perhaps not surprising as Mtl1 has previously been characterized as an oxidative stress sensor (1). However, how Mtl1 senses these stimuli currently remains unknown. Some authors (28) have reported that Msn2 transcriptional control is important for the response to hydrogen peroxide. This paper helps to extend our existing knowledge of this field by attributing Mtl1, a sensor for oxidative stress, with a novel and essential role in the transduction of the oxidative signal to Msn2. One possible explanation for this is that Msn2 controls the transcription of the genes required for the oxidative stress response. This occurs, for example, with GAD1, a gene required for cellular resistance to hydrogen peroxide, and with other oxidant agents (50).
Msn2 is known to be regulated by PKA activity (26, 45) and the TOR pathway (17). It is also generally accepted that sensitivity to rapamycin is specific to mutants in the TORC1 complex but not in the TORC2 complex (13). The mutant mtl1 is sensitive to rapamycin, which is what connects it to the TORC1 complex. The observation that TOR1 deletion restores the Msn2/Msn4 function in mtl1 cells reinforces the hypothesis that Mtl1 acts as a possible negative regulator of TOR1. However, if this were the case, we should not expect any ribosomal gene repression to occur in the absence of Mtl1 as both TOR and Ras inhibition induce ribosomal gene repression (20, 34, 43, 51–53). The observation that rapamycin induced a marked repression of the ribosomal genes in mtl1 mutant cells was not unexpected as rapamycin blocks the TORC1 function and not only TOR1 activity. It has recently been reported that at least two independent branches of TORC1 are involved in the control of ribosomal protein genes (54). The presence of the other TORC1 components would be sufficient to signal to the ribosomal transcription independently of Mtl1. The synthetic lethality observed in the double mutant tor1mtl1 upon rapamycin treatment suggests that the two proteins act in different pathways but probably perform a common regulatory function (see Fig. 3E). However, more studies will be required to elucidate the meaning of this genetic interaction.
Upon oxidative treatment or glucose starvation, Tor1 deletion was required to restore ribosomal gene repression and the Msn2/Msn4 function in mtl1 cells. We consequently concluded that Mtl1 is a negative regulator of TOR1 and acts specifically in response to glucose starvation and oxidative stress. We wondered whether Mtl1 also negatively regulates other TOR readouts such as Gln3 and Rtg1/Rtg3 transcriptional functions (17, 55). We observed, however, that Mtl1 was not involved in such regulation in response to rapamycin (not shown).
It has been reported that RAS/cAMP may be a TOR effector branch (29). These authors reported that the activation of RAS causes 1) a deficiency in HSP12 transcription in response to rapamycin and 2) no ribosomal transcriptional repression in response to TOR inactivation. One of our initial assumptions was, therefore, that the RAS/cAMP pathway could be improperly up-regulated in the mtl1 mutant.
Deletion of Ras2 caused a significant decrease in cAMP levels, as expected, and as previously reported (by 56–58). We also obtained clear results indicating that cAMP levels were higher in mtl1 cells than in those determined in wild type cells. It has been demonstrated that high PKA levels negatively regulate MSN2 transcription. This could explain the lower transcription levels determined in mtl1 mutants in comparison with wild type cells, and this would also be in accordance with the increase in the transcription of MSN2 observed in both the mtl1tor1 and mtl1ras2 double mutants in the logarithmic phase. Taking all of these results together, we conclude that Mtl1 negatively affects cAMP levels through the regulation of the RAS2 function and more specifically in response to both oxidative stress and glucose deprivation (as depicted in Fig. 3E).
Glucose starvation induced a change from a fermentative to a respiratory metabolism in S. cerevisiae. This sudden shift to respiratory growth would lead to the accumulation of reactive oxidative species derived from the electronic chain in the mitochondria. It is not, therefore, surprising that both oxidative and nutritional stresses induce a common pathway that is sensed primarily by a common sensor, Mtl1, which would transmit the signal for both Tor1 and Ras2 inactivation, thus reducing the accumulation of cAMP. In mammal cells, amino acid depletion inhibits TORC1 (59). Unrestrained activity in mammals is associated with several diseases such as inflammation, cancer, and diabetes (60). Ras inhibition also increases resistance to oxidative stress in neurons (61). The inhibition of Tor1 and Ras must, therefore, be a common mechanism used by cells to survive nutrient depletion and other specific types of stress. In this context and because of the existence of either homologous TOR or RAS in unicellular organisms, they constitute a good cellular model for studying these signaling processes, stressing the interest of the current study.
It was recently (62) proposed that the TOR and PKA signals function independently of each other. However, other authors (29) have proposed that nutrient availability signals to the TOR function and that from there, the signal could diverge into two separate branches, one of which is directed to Ras/cAMP, whereas the other is related to ribosome biogenesis, stress response, and glycogen accumulation. In our studies we observed that in response to rapamycin, Mtl1 displayed an independent genetic relationship with TOR. However, in response to oxidative stress and glucose starvation, our data support the hypothesis that a signal is transmitted from Mtl1 and Rho1 to Tor1 and Ras/cAMP, which converges (at least) on the stress transcription factor Msn2/Msn4. Glycogen accumulation is also controlled by TOR, PKA, and Msn2/Msn4. The observation that Rho1 activation, Msn2 overexpression, or the deletion of either Tor1 or Ras2 suppressed the deficiency in glycogen accumulation in mtl1 mutants also supports our model.
The findings from our study were in line with a previous publication (63) that described a role for Rom2 in Ras2-cAMP down-regulation in response to several types of stress, including oxidative stress. These authors also indicate that this cross-talk between Ras-cAMP and the Pkc1-MAPK pathway could be mediated by Rho1. In the present study we have extended this information to Mtl1, as it is the cell-surface protein member of the cell integrity pathway that transmits the signal to Tor1 and Ras2 through Rom2 (data not shown) and Rho1. However, downstream elements of the cell integrity pathway do not participate in this cross-talk. This was deduced from the observation that the BCK1–20 allele did not restore the Msn2/Msn4 function in mtl1cells in response to oxidative stress and glucose starvation. Moreover, and in accordance with Park et al. (63), we also observed a certain specificity of stress in this signaling, as the RHO1* and BCK1–20 alleles did not suppress the loss of viability that the mtl1 cells experienced in response to other stress-inducing agents such as rapamycin.
In general, the cellular responses to oxidative stress or glucose starvation mediated by Mtl1 described in this paper were common ones. With only one exception, the constitutive activation of the Pkc1-MAPK pathway at the level of BCK1 clearly increased cell survival in mtl1 cells when they were starved of glucose. This result underlines the importance of cell integrity activity in quiescence (10, 34). The observation that Slt2 activation is not sufficient to recover cell viability in response to oxidative stress in an mtl1 mutant is in line with a previous publication (1). In it, we demonstrated that the elements of the MAPK module of the CWI pathway are dispensable for cell viability in response to oxidative stress. However, more studies are required to look more deeply into the biological significance of these divergences.
In conclusion, our results suggest that the essential functions mediated by Mtl1 are to inactivate the Tor1 and Ras2 function to repress ribosomal gene transcription, to induce Msn2/Msn4 gene transcription, and to reduce the cAMP cellular levels (see Fig. 3E). We believe that the Msn2 function directly contributes to the maintenance of cell viability in mtl1 mutants in response to oxidative stress, whereas Slt2 activity reflects an adaptive response. However, in response to glucose starvation, both Slt2 and Msn2 activity help to increase cell survival in the absence of Mtl1.
Supplementary Material
Acknowledgments
We acknowledge Dr. Luis Serrano Endolz for expert assistance in the statistical analyses. We sincerely thank Dr. F. Estruch for kindly providing the plasmid pGFPMsn2. We acknowledge the participation of Dr. Felipe Vilella and Gemma Ingles in the first steps of Mtl1 genomic characterization.
This work was supported by Ministerio de Educacion y Ciencia (Spanish government) Grants BFU2009-11215 and BIO2007-67821 and Comunidad de Madrid (Spain) Grant S-SAL-0246/2006.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3.
- MAPK
- mitogen-activated protein kinase
- PKC
- protein kinase C
- PKA
- protein kinase A
- CWI
- cell wall integrity
- wt
- wild type
- GFP
- green fluorescent protein.
REFERENCES
- 1.Vilella F., Herrero E., Torres J., de la Torre-Ruiz M. A. (2005) J. Biol. Chem. 280, 9149–9159 [DOI] [PubMed] [Google Scholar]
- 2.Rajavel M., Philip B., Buehrer B. M., Errede B., Levin D. E. (1999) Mol. Cell. Biol. 19, 3969–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Bettignies G., Thoraval D., Morel C., Peypouquet M. F., Crouzet M. (2001) Genetics 159, 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sekiya-Kawasaki M., Abe M., Saka A., Watanabe D., Kono K., Minemura-Asakawa M., Ishihara S., Watanabe T., Ohya Y. (2002) Genetics 162, 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heinisch J. J., Lorberg A., Schmitz H. P., Jacoby J. J. (1999) Mol. Microbiol. 32, 671–680 [DOI] [PubMed] [Google Scholar]
- 6.Levin D. E. (2005) Microbiol. Mol. Biol. Rev. 69, 262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serrano R., Martín H., Casamayor A., Ariño J. (2006) J. Biol. Chem. 281, 39785–39795 [DOI] [PubMed] [Google Scholar]
- 8.Helliwell S. B., Schmidt A., Ohya Y., Hall M. N. (1998) Curr. Biol. 8, 1211–1214 [DOI] [PubMed] [Google Scholar]
- 9.Delley P. A., Hall M. N. (1999) J. Cell Biol. 147, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres J., Di Como C. J., Herrero E., De La Torre-Ruiz M. A. (2002) J. Biol. Chem. 277, 43495–43504 [DOI] [PubMed] [Google Scholar]
- 11.Motizuki M., Yokota S., Tsurugi K. (2008) Biochim. Biophys. Acta 1780, 179–184 [DOI] [PubMed] [Google Scholar]
- 12.Helliwell S. B., Wagner P., Kunz J., Deuter-Reinhard M., Henriquez R., Hall M. N. (1994) Mol. Biol. Cell 5, 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 14.Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 15.Inoki K., Ouyang H., Li Y., Guan K. L. (2005) Microbiol. Mol. Biol. Rev. 69, 79–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoki K., Guan K. L. (2006) Trends Cell Biol. 16, 206–212 [DOI] [PubMed] [Google Scholar]
- 17.Beck T., Hall M. N. (1999) Nature 402, 689–692 [DOI] [PubMed] [Google Scholar]
- 18.Crespo J. L., Powers T., Fowler B., Hall M. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6784–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dilova I., Aronova S., Chen J. C., Powers T. (2004) J. Biol. Chem. 279, 46527–46535 [DOI] [PubMed] [Google Scholar]
- 20.Martin D. E., Soulard A., Hall M. N. (2004) Cell 119, 969–979 [DOI] [PubMed] [Google Scholar]
- 21.Broach J. R., Deschenes R. J. (1990) Adv. Cancer Res. 54, 79–139 [DOI] [PubMed] [Google Scholar]
- 22.Thevelein J. M. (1994) Yeast 10, 1753–1790 [DOI] [PubMed] [Google Scholar]
- 23.Broach J. R. (1991) Trends Genet. 7, 28–33 [DOI] [PubMed] [Google Scholar]
- 24.Martínez-Pastor M. T., Marchler G., Schüller C., Marchler-Bauer A., Ruis H., Estruch F. (1996) EMBO J. 15, 2227–2235 [PMC free article] [PubMed] [Google Scholar]
- 25.Boy-Marcotte E., Perrot M., Bussereau F., Boucherie H., Jacquet M. (1998) J. Bacteriol. 180, 1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Görner W., Durchschlag E., Wolf J., Brown E. L., Ammerer G., Ruis H., Schüller C. (2002) EMBO J. 21, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt A. P., McEntee K. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5777–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasan R., Leroy C., Isnard A. D., Labarre J., Boy-Marcotte E., Toledano M. B. (2002) Mol. Microbiol. 45, 233–241 [DOI] [PubMed] [Google Scholar]
- 29.Schmelzle T., Beck T., Martin D. E., Hall M. N. (2004) Mol. Cell. Biol. 24, 338–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardenas M. E., Cutler N. S., Lorenz M. C., Di Como C. J., Heitman J. (1999) Genes Dev. 13, 3271–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garreau H., Hasan R. N., Renault G., Estruch F., Boy-Marcotte E., Jacquet M. (2000) Microbiology 146, 2113–2120 [DOI] [PubMed] [Google Scholar]
- 32.Powers R. W., 3rd, Kaeberlein M., Caldwell S. D., Kennedy B. K., Fields S. (2006) Genes Dev. 20, 174–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J. C. Y., Powers T. (2006) Curr. Genet. 49, 281–293 [DOI] [PubMed] [Google Scholar]
- 34.Krause S. A., Gray J. V. (2002) Curr. Biol. 12, 588–593 [DOI] [PubMed] [Google Scholar]
- 35.Kaiser C., Michaelis S., Mitchell A. (1994) Methods in Yeast Genetics, Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 36.Goldstein A. L., McCusker J. H. (1999) Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 37.Angeles de la Torre-Ruiz M., Torres J., Arino J., Herrero E. (2002) J. Biol. Chem. 277, 33468–33476 [DOI] [PubMed] [Google Scholar]
- 38.Lee K. S., Levin D. E. (1992) Mol. Cell. Biol. 12, 172–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallego C., Garí E., Colomina N., Herrero E., Aldea M. (1997) EMBO J. 16, 7196–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swiegers J. H., Pretorius I. S., Bauer F. F. (2006) Curr. Genet. 50, 161–171 [DOI] [PubMed] [Google Scholar]
- 41.Estruch F. (2000) FEMS Microbiol. Rev. 24, 469–486 [DOI] [PubMed] [Google Scholar]
- 42.Estruch F., Carlson M. (1993) Mol. Cell. Biol. 13, 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Düvel K., Santhanam A., Garrett S., Schneper L., Broach J. R. (2003) Mol. Cell 11, 1467–1478 [DOI] [PubMed] [Google Scholar]
- 44.Görner W., Durchschlag E., Martinez-Pastor M. T., Estruch F., Ammerer G., Hamilton B., Ruis H., Schüller C. (1998) Genes Dev. 12, 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbet N. C., Schneider U., Helliwell S. B., Stansfield I., Tuite M. F., Hall M. N. (1996) Mol. Biol. Cell 7, 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.François J. M., Thompson-Jaeger S., Skroch J., Zellenka U., Spevak W., Tatchell K. (1992) EMBO J. 11, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hardy T. A., Huang D., Roach P. J. (1994) J. Biol. Chem. 269, 27907–27913 [PubMed] [Google Scholar]
- 48.Ruis H., Schüller C. (1995) BioEssays 17, 959–965 [DOI] [PubMed] [Google Scholar]
- 49.Smith A., Ward M. P., Garrett S. (1998) EMBO J. 17, 3556–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coleman S. T., Fang T. K., Rovinsky S. A., Turano F. J., Moye-Rowley W. S. (2001) J. Biol. Chem. 276, 244–250 [DOI] [PubMed] [Google Scholar]
- 51.Marion R. M., Regev A., Segal E., Barash Y., Koller D., Friedman N., O'Shea E. K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14315–14322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y., McIntosh K. B., Rudra D., Schawalder S., Shore D., Warner J. R. (2006) Mol. Cell. Biol. 26, 4853–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinboeck F., Krupanska L., Bogusch A., Kaufmann A., Heidenreich E. (2006) J. Biochem. 139, 741–751 [DOI] [PubMed] [Google Scholar]
- 54.Lee J., Moir R. D., Willis I. M. (2009) J. Biol. Chem. 284, 12604–12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohde J. R., Cardenas M. E. (2003) Mol. Cell. Biol. 23, 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsumoto K., Uno I., Kato K., Ishikawa T. (1985) Yeast 1, 25–38 [DOI] [PubMed] [Google Scholar]
- 57.Tatchell K., Robinson L. C., Breitenbach M. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. (1985) Cell 40, 27–36 [DOI] [PubMed] [Google Scholar]
- 59.Avruch J., Hara K., Lin Y., Liu M., Long X., Ortiz-Vega S., Yonezawa K. (2006) Oncogene 25, 6361–6372 [DOI] [PubMed] [Google Scholar]
- 60.Reiling J. H., Sabatini D. M. (2008) Mol. Cell 29, 533–535 [DOI] [PubMed] [Google Scholar]
- 61.Li Y., Xu W., McBurney M. W., Longo V..D. (2008) Cell Metab. 8, 38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zurita-Martinez S. A., Cardenas M. E. (2005) Eukaryotic Cell 4, 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park J. I., Collinson E. J., Grant C. M., Dawes I. W. (2005) J. Biol. Chem. 280, 2529–2535 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.