Abstract
Increased expression of specific ATP-binding cassette (ABC) transporters is known to mediate the efflux of chemotherapeutic agents from cancer cells. Therefore, establishing how ABC transporter genes are controlled at their transcription level may help provide insight into the role of these multifaceted transporters in the malignant phenotype. We have investigated ABC transporter gene expression in a large neuroblastoma data set of 251 tumor samples. Clustering analysis demonstrated a strong association between differential ABC gene expression patterns in tumor samples and amplification of the MYCN oncogene, suggesting a correlation with MYCN function. Using expression profiling and chromatin immunoprecipitation studies, we show that MYCN oncoprotein coordinately regulates transcription of specific ABC transporter genes, by acting as either an activator or a repressor. Finally, we extend these notions to c-MYC showing that it can also regulate the same set of ABC transporter genes in other tumor cells through similar dynamics. Overall our findings provide insight into MYC-driven molecular mechanisms that contribute to coordinate transcriptional regulation of a large set of ABC transporter genes, thus affecting global drug efflux.
Keywords: Gene/Transcription, Transcription/Myc, Transcription/Regulation, ABC Transporter, Drug Transport, ABC Transporter Genes, MYCN, c-MYC, Multidrug Resistance, Neuroblastoma
Introduction
Chemoresistance can be due to several cellular mechanisms, including those related to the altered activity of specific ATP-binding cassette (ABC)6 drug transporters that shuttle hydrophobic lipophilic compounds across cell membranes in an ATP-dependent manner (1, 2). ABC transporter genes are highly conserved among evolutionarily distant species such as yeast and mammals. There are 48 functional ABC genes in Homo sapiens classified into seven subfamilies from A to G (3). ABCB1, also named MDR1, is the prototype of this gene superfamily, and its deregulation has been linked to a drug resistance phenotype in several types of cancers (4). Neuroblastoma is the most common solid cancer in childhood accounting for ∼10% of all pediatric cancers (5) and is characterized by a heterogeneous clinical behavior that ranges from spontaneous regression in a small proportion of cases to rapid progression and resistance to pharmacological treatment with fatal outcome. Patients affected by this tumor often show a multiple drug-resistant phenotype, and although many high risk neuroblastoma tumors initially respond to the first cycles of intensive chemotherapy, they frequently become refractory to treatment as the disease progresses. At present, patient risk classification includes MYCN gene amplification, deletions of chromosomal material on 1p and 11q23 (6), gain of 17q (7), tumor stage, and age of the child at diagnosis (8, 9). Amplification leading to overexpression of the MYCN oncogene is present in ∼25–30% of primary untreated neuroblastoma and is associated with advanced stage disease, rapid progression, and unfavorable prognosis (10, 11). Recent studies have shown that ABC transporters other than ABCB1 may be important for the chemoresistance phenotype of neuroblastoma. We have previously shown, both retrospectively and prospectively, that ABCC1 is a powerful independent prognostic marker in neuroblastoma (12, 13), with high levels of ABCC1 expression being strongly associated with reduced survival and event-free survival. ABCC4, another member of the ABCC subfamily, was also shown to be overexpressed in primary neuroblastoma tumors, and its overexpression has been significantly correlated with MYCN amplification and ABCC1 expression. Like ABCC1, high ABCC4 expression also correlated with poor clinical outcome in neuroblastoma (14). Consequently, the dissection of the molecular mechanisms underlying transcription regulation of ABC genes should enhance the understanding and prediction of drug response as also recently proposed by Gottesman and co-workers (15). However, most such mechanistic studies have focused on the ABCB1/MDR1 gene (16), and very few data have been collected on the transcriptional regulation of other ABC genes in normal and tumor tissues.
In this study we have investigated the role and mechanism of action of MYCN in transcriptional control of the entire set of human ABC transporter genes. Our findings show that MYCN can coordinate transcription of a large set of ABC genes. Moreover, coordinate regulation of ABC genes by MYCN also extends to an overlapping pattern of regulation of ABC family genes by c-MYC in a variety of c-MYC-driven tumor cell types. An increased level of MYC oncogene expression in tumor cells may trigger altered ABC transporter expression, hence contributing to the control of drug and physiological substrate efflux in cancer cells. Overall, in this study we identified a specific transcription signature of ABC transporter genes dictated by MYC oncoproteins that may provide additional value to the understanding of how a chemoresistance phenotype is established during cancer progression.
EXPERIMENTAL PROCEDURES
Cell Culture
Human neuroblastoma SHEP, human breast cancer MCF-7 cells, and human SJ-G2 glioblastoma cells were cultured in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum and 50 mg/ml gentamycin. Human neuroblastoma SK-N-BE(2)C, SH-SY5Y, LAN-1, and SK-N-SH cells were cultured in RPMI medium 1640 containing 20% fetal bovine serum and 50 mg/ml gentamycin. Human TET21/N cells were derived from SHEP neuroblastoma cell line and express MYCN under the control of tetracycline (tet-off).
Specifically, the human neuroblastoma cell line SHEP was transfected with two plasmids, pUHD15-1, that contains the tTA transactivator gene under the control of the human cytomegalovirus promoter, and pSV2neo, that confers neomycin resistance. Clones were selected in the presence of 200 μg/ml of G418. Independent clones were isolated and subsequently cotransfected with the plasmids pHUD10–3/MYCN, which contains a partial human MYCN cDNA and lacks the major portion of the noncoding exon I, and pHMR272, which provides resistance against hygromycin. Cell clones resistant to both G418 and hygromycin (90 μg/ml) were recovered and analyzed for tetracycline-dependent expression of MYCN. The TET21/N cells were grown in Dulbecco's modified Eagle's medium containing l-glutamine (2 mm), 50 mg/ml gentamycin, and 10% heat-inactivated tetracycline-free fetal bovine serum. The TET21/N cells were treated with tetracycline at a final concentration of 2 μg/ml for the indicated time (17, 18).
Human P493-6 cells were derived from the EREB2-5 lymphoblastoid cell line and express c-MYC under the control of tetracycline (tet-off). The P493-6 cells were grown in RPMI 1640 medium containing l-glutamine (2 mm), 50 mg/ml gentamycin, and 10% heat-inactivated tetracycline-free fetal bovine serum. The P493-6 cells were treated with tetracycline at a final concentration of 0.1 μg/ml for indicated time indicated (19).
Gene Expression Analysis
RNA samples were prepared using Tri-Reagent (Sigma) and treated with DNase (DNA-freeTM; Ambion). Reverse transcription and PCR were performed using a SuperScript reverse transcription-PCR kit (Invitrogen). Real time quantitative PCR (RQ-PCR) was performed using iQTM SYBR Green Supermix and the iQCycler thermocycler (Bio-Rad). Primers for RQ-PCR are listed in supplemental Table S1.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP and dual ChIP cross-linking were performed as previously described (20, 21). The antibodies employed in this study were: IgG (Santa Cruz sc-2027); MYCN monoclonal antibody B8.4.B (22), c-MYC (Santa Cruz N-262); MAX (Santa Cruz C-17), and Sp1 (Upstate 07–124). Specific pairs of primers used for quantitative ChIP are listed in supplemental Table S2.
Luciferase Assay
The pGL3-basic and Renilla-TK vectors were obtained from Promega. Promoters of selected ABC transporter genes were obtained using PCR and cloned into the pGL3-basic vector. The activity of firefly or Renilla luciferase was measured with a dual luciferase assay kit (Promega) according to the instructions.
Site-specific Mutagenesis
Mutagenesis of Sp1-binding sites on the ABCC3 promoter was performed with a QuikChange multisite-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The oligonucleotides used for site-directed mutagenesis are listed in supplemental Fig. S5.
Immunoblotting Analysis
Western blots were performed as previously described (12). The antibodies used were against MYCN (Oncogene OP13); β-actin (A2066; Sigma Aldrich); ABCC1 (Alexis Biochemicals), ABCC3 (M3II-21; Abcam ab3376), ABCC4 (Alexis Biochemicals), c-MYC (9E10; Santa Cruz), and α-tubulin (Sigma).
Analysis of Drug Sensitivity
Cell survival was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay as described previously (15).
Efflux Assay
The efflux assay was performed with a multidrug resistance direct dye efflux kit assay (Millipore) following the manufacturer's instructions.
Statistical Methods
Survival analyses for the 23 ABC family genes obtained from a publicly available microarray database (22) were performed according to the method of Kaplan and Meier. Expression data for each gene was defined as either “high” or “low” using the median, upper, and lower quartiles, and upper and lower deciles as cut points. Expression data for those genes showing prognostic significance (p < 0.05) were subjected to unsupervised k-means clustering using the Cluster 3 program, and the results were visualized using TreeView software. Student's t tests were used to compare data between groups, and p values less than 0.05 were considered statistically significant.
RESULTS
A Specific Transcription Signature of ABC Transporter Genes Correlates with MYCN Amplification and Reduced Survival in Neuroblastoma
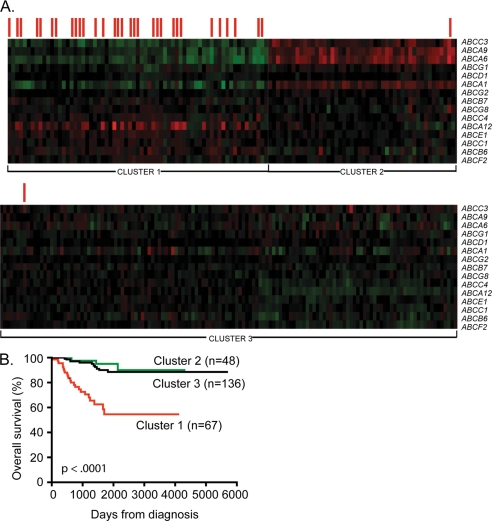
To understand whether the transcription profiles of multiple human ABC genes are relevant for predicting clinical outcome in neuroblastoma, we conducted a metanalysis of ABC gene expression in a publicly available data set containing clinical and gene expression data for 251 primary neuroblastoma samples described by Oberthuer et al. (23). The microarray contained 10,163 oligonucleotide probes, some of which matched 23 ABC drug transporter genes. Thus, the expression data for the 23 ABC genes were extracted and analyzed separately for risk stratification. A Kaplan-Meier plot was generated for each individual ABC gene and used to estimate survival for the 251 patients based on the expression level of the gene in tumor samples. Fifteen of the 23 ABC genes demonstrated prognostic significance following dichotomization of the expression levels around either the upper or the lower quartile with the exception of ABCF2, which became significant when analyzed at the upper decile (supplemental Fig. S1). For example, high levels of ABCA12, ABCB6, ABCB7, ABCC1, ABCC4, ABCE1, ABCF2, and ABCG8 were strongly associated with reduced survival. In contrast, low levels rather than high levels of ABCA1, ABCA6, ABCA9, ABCC3, ABCD1, ABCG1, and ABCG2 correlated with reduced survival (supplemental Fig. S1). This result remained significant even when the upper/lower decile and sometimes median were also used as cut points for dichotomization, with p values that ranged from 0.04 to <0.0001 (data not shown). The remaining eight ABC family genes analyzed showed no prognostic significance, regardless of the cut point used (data not shown). Expression data for the 15 genes demonstrating prognostic significance were analyzed using the Cluster 3 program (24). Tumors were grouped using unsupervised k-means clustering, based only on the similarity of expression patterns without taking into account any clinical variables such as MYCN amplification status or outcome. Essentially no tumors were identified that had high expression of all of the ABC transporter genes analyzed. However, the 251 tumors clearly partitioned into three groups as shown in Fig. 1A. Cluster 1 contained tumors that had high expression of approximately half of the ABC transporter genes and low expression of the remainder. Analysis of clinical parameters of the patients in this cluster indicated that Cluster 1 contained the majority of tumor samples from patients with amplified MYCN and with reduced survival (Fig. 1B), suggesting that the expression profile of this 15-ABC gene set may correlate with MYCN function. Thus, of the 67 patients in Cluster 1, 31 had amplification of the MYCN oncogene in their tumors, whereas only two of the remaining 184 patients in Clusters 2 and 3 had MYCN-amplified tumors. Cluster 2, which contained tumors with high expression of a small number of ABC genes that were expressed at low levels in Cluster 1 patients, as well as low levels of most other ABC genes, was found to include only one patient with MYCN amplification, and only three of the 48 patients in this cluster had died, by comparison with 23 of the 67 patients from Cluster 1 (Fig. 1B; p < 0.0001). Cluster 3 contained tumors that had uniformly low expression of all ABC transporter genes analyzed, and the majority of these patients also had good clinical outcome (Fig. 1B). Taken together these observations reveal a possible causal link between MYCN function and the expression of this specific set of ABC transporter genes.
FIGURE 1.
Combined expression of ABC transporter genes predicts cumulative survival in 251 patients with neuroblastoma. A, k-Means cluster analysis of ABC genes in primary neuroblastoma tumors. Expression profiles of 15 ABC transporter genes demonstrating prognostic significance following survival analysis in 251 primary neuroblastoma tumors were clustered into three groups using the Cluster 3 program and unsupervised k-means clustering. The rows represent the genes (indicated on the right side), whereas the columns represent patient samples. The levels of gene expression as log values are shown ranging from green (−1.0) to red (1.0). The red bars above the samples indicate those tumors displaying MYCN amplification. B, Kaplan-Meier survival analysis of these clusters revealed that the gene expression signature of the patients in Cluster 1 correlated with a significantly increased risk of death when compared with Cluster 2 or 3 (p < 0.0001).
MYCN Regulates mRNA Expression of a Large Set of ABC Transporter Genes
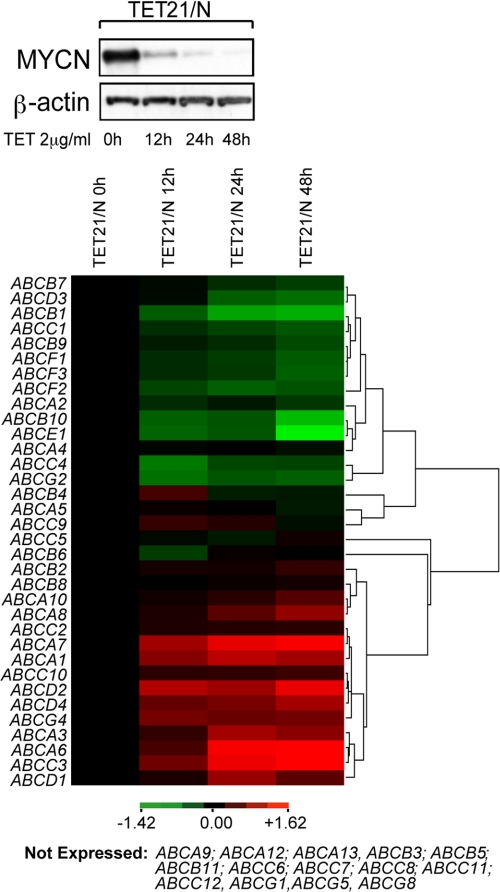
To determine whether the MYCN oncoprotein transcriptionally regulates specific ABC transporters, we examined the expression of all 48 ABC transporter genes in MYCN-inducible TET21/N human neuroblastoma cells (17). ABC transporter gene expression was measured by real time PCR and correlated with the level of MYCN (Fig. 2, upper panel and supplemental Table S3). The Cluster 3 program (24) was used to group transcription profiles of individual ABC genes as a function of their response to MYCN activity (Fig. 2, lower panel). Based on their transcription profile the ABC genes could be classified into four distinct groups: (i) genes positively regulated by MYCN; (ii) genes negatively regulated by MYCN; (iii) genes expressed but unresponsive to MYCN expression; and (iv) genes not expressed. With regard to the 15-gene signature identified in Fig. 1, four genes (ABCA9, ABCA12, ABCG1, and ABCG8) were found not to be expressed in the TET21/N cells; however, the pattern of expression of the remaining genes observed in MYCN amplified tumor samples (Cluster 1, Fig. 1A) essentially matched that seen following down-regulation of MYCN in the TET21/N cells (Table 1). Thus, genes that were seen to be down-regulated in most MYCN amplified tumors (Fig. 1) were up-regulated in TET21/N cells following suppression of MYCN expression and vice versa. Although ABCA12 was the most strongly expressed gene in the tumor samples of Cluster 1, it was not expressed in TET/21N cells. Thus, to verify whether ABCA12 transcription can be affected by MYCN, we analyzed its expression in SK-N-BE(2)C human neuroblastoma cells in which MYCN expression was silenced using MYCN-specific siRNA (supplemental Fig. S2A). The results show that ABCA12 expression correlated with that of MYCN, suggesting that ABCA12 may also be a target of MYCN transcriptional activity (supplemental Fig. S2B). With the exception of the ABCB6 and ABCG2 genes, our data show that a significant number of ABC transporter genes appear to respond to MYCN activity in both cell lines and tumors (Table 1). Furthermore, this study of ABC transporter gene expression in TET21/N cells revealed additional, potentially MYCN-regulated genes that had not been examined in the Oberthuer study, including ABCA2, ABCB9, ABCB10, ABCF1, and ABCF3, which were positively regulated by MYCN, and ABCA7, ABCD2, and ABCG4, which appeared to be suppressed by MYCN (Fig. 2, lower panel).
FIGURE 2.
MYCN regulates transcription of ABC transporter genes in TET21/N neuroblastoma cells. Modulation of MYCN expression by tetracycline (TET) in TET21/N cells was monitored by Western blot (upper panel). Cluster 3 was used to generate an image map showing expression levels of ABC genes as a function of tetracycline treatment time, relative to expression at time 0 and processed in log space. Hierarchical clustering on each axis used the average-linkage algorithm, and distance measures are based on Pearson correlation. Green indicates genes positively regulated by MYCN, whereas red indicates those negatively regulated (lower panel).
TABLE 1.
Concordance of the expression pattern of ABC transporter genes as a function of MYCN amplification or expression
H, high expression; L, low expression; NE, no expression; NS, no significant difference in expression; Y, concordance of expression profiles between Oberthuer's data set and TET21/N cells.
| ABC genes | Cluster1 MYCN amplified | Cluster2/3 single copy MYCN | Tet (+) MYCN | Tet (−) MYCN | Concordance of expression |
|---|---|---|---|---|---|
| A1 | L | H | L | H | Y |
| A6 | L | H | L | H | Y |
| A9 | L | H | NE | NE | |
| A12 | H | L | NE | NE | |
| B6 | H | L | NS | NS | |
| B7 | H | L | H | L | Y |
| C1 | H | L | H | L | Y |
| C3 | L | H | L | H | Y |
| C4 | H | L | H | L | Y |
| D1 | L | H | L | H | Y |
| E1 | H | L | H | L | Y |
| F2 | H | L | H | L | Y |
| G1 | L | H | NE | NE | |
| G2 | L | H | H | L | |
| G8 | H | L | NE | NE |
MYCN Is a Direct Transcriptional Regulator of Many ABC Transporter Genes
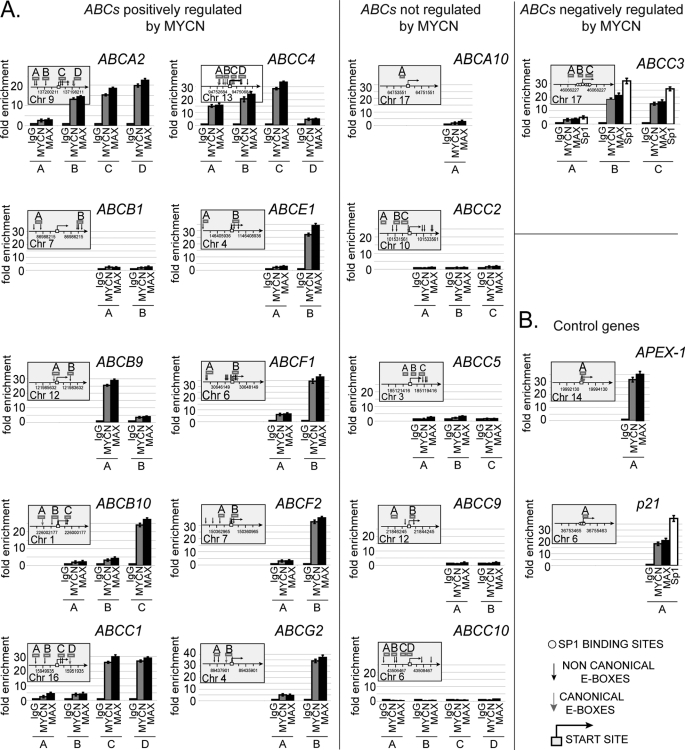
To investigate the role of MYCN in the regulation of ABC transporter genes, the promoter regions of those genes that responded to MYCN were bioinformatically scanned for putative MYC binding sites. MYC proteins can preferentially bind to a six-base pair canonical palindrome or E-box sequence, CACGTG, as well as other noncanonical sequences including CATGTG and CACGCG. MYC binding sites are often close to the transcription start site and in the context of CpG-rich unmethylated DNA regions of 1–2 kb in length (25, 26). Based on this knowledge, promoter regions from −2000 to +2000 base pairs with respect to the transcription start site were considered for analysis. As shown in Fig. 3A, the promoter regions of most of the ABC genes positively regulated by MYCN carry one or more E-box sites, the majority of which are located around the predicted transcription start site (Fig. 3A, left panel). No E-box sites were identified in the ABCB7, ABCD3, and ABCF3 gene promoters (data not shown), suggesting that MYCN may control their transcription through alternative mechanisms. For those genes carrying at least one potential MYC binding site, we examined physical association of MYCN with the gene promoters by ChIP assay in TET21/N cells. With the exception of the ABCB1 gene, the promoters of all such genes were directly bound by MYCN, as well as its dimerization partner, MAX (Fig. 3A, left panel). Because ABCA12 expression appeared to be regulated by MYCN in SK-N-BE(2)C cells, we tested promoter binding by MYCN in this cell line. No binding, however, was observed (supplemental Fig. S2C), suggesting that ABCA12 and ABCB1 may be secondary targets of MYCN activity. As expected, positive ChIP results were dependent on MYCN expression (supplemental Fig. S3). When ChIP was performed for several genes that did not respond to MYCN, no promoter association was observed, confirming that only a subset of ABC transporter genes are direct MYCN targets (Fig. 3A, middle panel). Importantly, for those ABC promoter regions bound by MYCN, the extent of fold enrichment was comparable with that of APEX-1, a well known MYC target gene (25) (Fig. 3B), indicating that the MYCN-MAX complex is strongly bound to these promoters. We also investigated the role of MYCN in transcriptional repression of ABC genes. We recently found that MYCN can repress transcription of the TG2 gene in neuroblastoma through interaction with Sp1 (27). Based on these findings, promoters from the five most strongly MYCN-repressed ABC genes (ABCA1, ABCA6, ABCA7, ABCC3, and ABCD2) were analyzed by dual cross-linking ChIP assay (20) to investigate whether MYCN and MAX proteins physically associate with these ABC promoters via Sp1. For each promoter we tested any regions bearing the predicted Sp1 binding sequences (28), which were usually proximal to or inclusive of the transcription start site, along with at least one other, selected 1–2 kb from the transcription start site, as a negative control of the ChIP reaction. Surprisingly, the results showed that MYCN exclusively associates with the ABCC3 core promoter region, suggesting that ABCC3 is the sole gene among those analyzed that may be directly repressed by MYCN (Fig. 3A, right panel). Sp1 was also bound to the ABCC3 promoter, thus reinforcing the notion that MYCN may associate with the ABCC3 promoter through interaction with the Sp1 transcription factor as previously described (27). The fact, however, that ABCA1, ABCA6, ABCA7, and ABCD2 were negatively regulated by MYCN, although their promoters were not bound by MYCN (supplemental Fig. S4), suggests that these genes may be secondary targets of MYCN transcriptional activity or repressed through MYC-driven transcriptional mechanisms not yet revealed.
FIGURE 3.
MYCN associates with the promoters of multiple ABC gene family members. A, quantitative ChIP was applied to TET21/N cells expressing MYCN. Fold enrichment is relative to the preimmune serum. The results represent the means ± S.E. of five independent ChIP experiments in which each region was amplified by RQ-PCR in triplicate ± S.E. Bent arrow, transcription start site; gray arrow, canonical E-box; black arrow, noncanonical E-box; open boxes, amplicons indicated with a capital letter. The chromosome and coordinates (bp) are also given (left and middle panel). Dual cross-linking ChIP for the ABCC3 gene promoter using MYCN, MAX, or Sp1 antibodies. The results represent the means ± S.E. of three independent ChIP experiments (right panel). B, quantitative ChIP and quantitative dual cross-linking ChIP was performed for APEX-1 and p21CIP/WAF, respectively.
MYCN Is Required to Activate or Repress ABC Gene Transcription
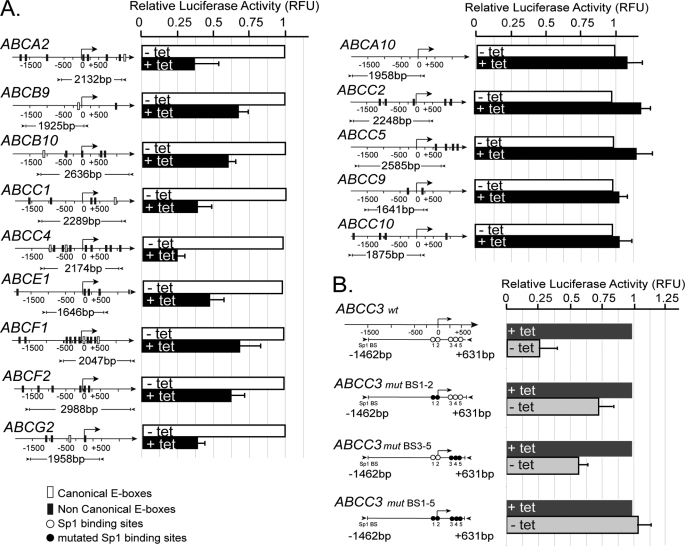
To further determine whether MYCN is indeed required to activate or repress transcription, individual ABC gene promoters found to be bound by MYCN were cloned upstream of a luciferase reporter gene, and activity was tested in TET21/N cells as a function of MYCN expression. For each of these gene promoters (ABCA2, ABCB9, ABCB10, ABCC1, ABCC4, ABCE1, ABCF1, ABCF2, and ABCG2), luciferase activity was dependent on MYCN expression (Fig. 4A, left panel). As expected, the promoters of transporter genes that showed no binding to MYCN in ChIP assays (ABCA10, ABCC2, ABCC5, ABCC9, and ABCC10) did not show MYCN-dependent reporter activity (Fig. 4A, right panel). By contrast, activity of the ABCC3 gene promoter was specifically repressed by MYCN (Fig. 4B). In this case, we also tested whether MYCN requires Sp1 sites to repress transcription by generating three variants of the ABCC3 promoter mutated in some or all of the Sp1 regulatory elements (supplemental Fig. S5 and Fig. 4B). As the results in Fig. 4B demonstrate, the partial or complete mutagenesis of the Sp1 binding sites was sufficient to reduce or ablate MYCN repressive function.
FIGURE 4.
MYCN regulates transcription from ABC promoters in a transient transfection assay. A, firefly luciferase activity was determined following transfection of reporter constructs into TET21/N cells, in the presence (+tet) or absence (−tet) of MYCN. Bent arrow, transcription start site. The cloned DNA region (bp) is indicated below the promoter map. B, luciferase assay of wt and mutated ABCC3 reporter vectors. The results are the means ± S.E. of three independent transfections. Fragment coordinates are expressed relative to the transcription start point.
ABC Transporter Gene Expression Levels Correlate with the Level of MYCN in Neuroblastoma Cell Lines
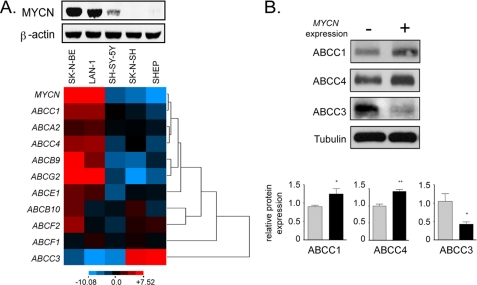
Transcription profiles of ABC transporter genes putatively regulated by MYCN were determined in a range of neuroblastoma cell lines and correlated with the level of MYCN expression. SK-N-BE(2)-C and LAN-1 cells express high levels of MYCN, whereas SH-SY5Y, SK-N-SH, and SHEP cells express relatively low levels of the MYCN protein (Fig. 5A, upper panel). Cluster analysis of gene expression across these five cell lines (supplemental Table S4) revealed that expression of many of the ABC transporters positively correlated with that of MYCN, with ABCA2, ABCB9, ABCC1, ABCC4, and ABCG2 showing particularly close correlation, whereas ABCC3 expression negatively correlated with that of MYCN (Fig. 5A, lower panel).
FIGURE 5.
Correlation of MYCN and ABC transporter gene expression in neuroblastoma cell lines. A, MYCN expression was evaluated in five different neuroblastoma cell line (upper panel). ABC gene expression was determined by RQ-PCR and data were clustered as described for Fig. 2 (lower panel). B, ABCC protein expression in cells overexpressing MYCN. MYCN gene expression was suppressed in TET21/N cells by the addition of 2 μg/ml tetracycline for at least 48 h (−) or induced by the removal of tetracycline (+). ABCC protein expression was measured by Western blotting of whole cell lysates (ABCC1 and ABCC4) or membrane preparations (ABCC3). Protein loading was normalized by tubulin Western blot (whole cell lysates) or by Ponceau staining (membrane preparations, not shown). Relative expression in triplicate samples was quantified using densitometry. *, p < 0.05; **, p < 0.005.
A similar correlation was observed when ABC protein expression in response to MYCN modulation was examined. For this purpose we chose to analyze two genes positively regulated by MYCN (ABCC1 and ABCC4) and one that was negatively regulated (ABCC3). As shown in Fig. 5B, ABCC1 and ABCC4 displayed significantly increased protein levels following induction of MYCN oncoprotein in TET21/N cells, whereas the negatively regulated ABCC3 gene exhibited significantly decreased levels following MYCN induction.
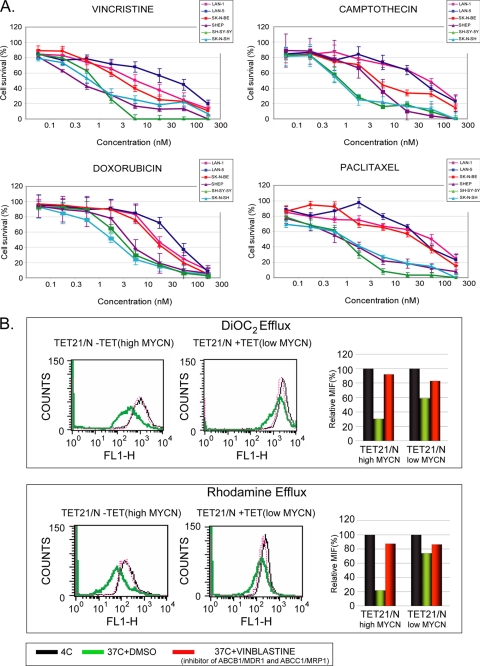
MYCN Overexpression Affects Drug Efflux Because of ABC Transporters in Neuroblastoma Cell Lines
To study the influence of MYCN on drug efflux and chemoresistance, we selected the same panel of neuroblastoma cell lines, which differ in terms of either expressing MYCN at a high level (LAN-1, LAN-5, and SK-N-BE(2)-C) or at a relatively low level (SH-SY5Y, SK-N-SH, and SHEP). We used the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay to test cell viability upon exposure to different chemotherapeutics, several of which are commonly used in the treatment of intermediate and high risk neuroblastoma. Neuroblastoma cell lines were grown in medium supplemented with increasing concentrations of vincristine, doxorubicin, camptothecin, and paclitaxel. Following 72 h of continuous exposure to each compound, the three cell lines with amplification/overexpression of MYCN displayed survival at an IC50 from five to ten times higher than the neuroblastoma cell lines expressing basal levels of MYCN (Fig. 6A). To determine whether the increased resistance observed in association with MYCN amplification/overexpression is associated with increased efflux activity of ABC transporters, the accumulation of two dyes, rhodamine and DiOC2, was evaluated in TET21/N as function of MYCN expression. DiOC2 is a substrate highly specific for ABCB1/MDR1, is weakly transported by ABCG2/BCRP, and is not transported by the related multidrug resistance protein MRP1. On the contrary, rhodamine 123 is effluxed by MDR1 and MRP1, representing a more broad indicator of total cell efflux activity. The functional activity of MDR1, MRP1, and BCRP transporters was measured as accumulation of fluorescence by flow cytometry. Consistent with the data obtained in the cytotoxicity assay, the accumulation of DiOC2 and rhodamine is highly reduced following MYCN induction (Fig. 6B), suggesting that MYCN overexpression resulted in a significant increase of cellular efflux activity. However, in the presence of vinblastine, a competitive inhibitor of MRP1 and MDR1, the induction of MYCN expression no longer affected the accumulation of the two dyes, indicating how MYCN can influence cellular efflux and drug response through altering the expression levels of specific ABC transporters.
FIGURE 6.
MYCN amplification/overexpression results in an increased IC50 of the different chemotherapeutic drugs in human neuroblastoma cells and affects cellular efflux activity in TET21/N. A, cell survival for the panel of chemotherapeutics shown in figures was evaluated in neuroblastoma cells with high levels (LAN-1, LAN-5, and SK-N-BE(2)-C) or low levels (SH-SY5Y, SK-N-SH, and SHEP) of MYCN. Each cell population was exposed to increasing concentrations of chemotherapeutic drug for 72 h. Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay for each cell line and plotted as percentages of cells that survived as compared with cells cultured in drug-free medium. The results represent the means ± S.D. from four independent experiments. B, accumulation of DiOC2 and rhodamine 123 was evaluated in TET21/N before and after MYCN induction. The efflux activity of ABC transporters is temperature-sensitive. ABC transporters function optimally near 37 °C but are effectively inactive at 4 °C. Hence, the cells incubated with fluorescent substrates retain the dye and consequently have high fluorescence when incubated at 4 °C (black lines and bars). Conversely, cells incubated at 37 °C (green lines and bars) more readily efflux the dye and show reduced fluorescence. In the presence of vinblastine (red lines and bars), MDR1 and MRP1 are inactive, and the cells reduce dye efflux. The data were obtained measuring the fluorescence of these two dyes by flow cytometry. For each condition, the mean fluorescence intensity was calculated and plotted below.
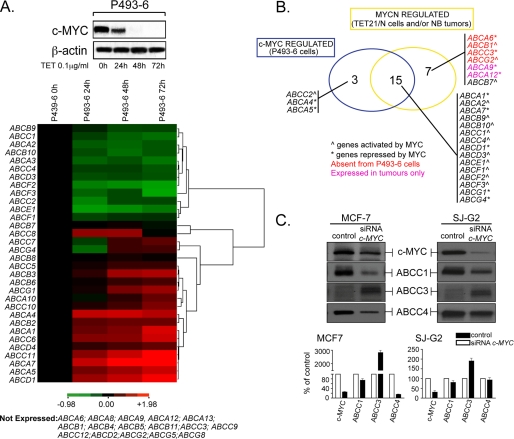
Transcriptional Regulation of ABC Transporter Genes by c-MYC Parallels That of MYCN
Although c-MYC and MYCN are expressed at different times during development and in distinct tissues, genetic evidence suggests considerable overlap in the spectrum of their target genes (29). To investigate whether c-MYC can regulate transcription of the same ABC genes as MYCN, we analyzed the transcription of ABC genes in P493-6 lymphoblastoid cells, as a function of c-MYC expression. Expression levels of the 48 ABC drug transporter genes were monitored in P493-6 cell lines (supplemental Table S5) where c-MYC expression can be silenced after treatment with tetracycline (19) (Fig. 7A, upper panel). ABC gene transcription profiles were determined by RQ-PCR and then clustered into different groups as a function of their response to c-MYC expression (Fig. 7A, lower panel). The results showed that c-MYC can regulate a large number of ABC transporter genes with dynamics that resemble those dictated by MYCN. Comparison of the ABC gene expression profiles of TET21/N and P493-6 cells revealed that 26 family members were expressed at detectable levels in both cell lines. Importantly, we found that all but one of the genes clearly up-regulated by c-MYC were also up-regulated, either directly or indirectly, by MYCN in TET21/N cells or associated with MYCN amplification in neuroblastoma tumors (Fig. 7B). Similarly, several MYCN repressed genes were also repressed by c-MYC in P493-6 cells. Furthermore, as shown by ChIP studies in P493-6 cells, we could demonstrate that in most cases such regulation occurred by direct binding of c-MYC to ABC gene promoters (supplemental Fig. S6), thus showing complete concordance between the c-MYC and MYCN regulated ABC transporter genes among tested cell lines and the Oberthuer tumor panel. In cases where discrepancies in gene expression and ChIP profiles among cell lines or the panel of tumors were observed, they may reflect differences in cellular background or particular epigenetic modifications occurring at gene promoters. For example, although ABCG1 was not expressed in TET21/N cells, this gene was down-regulated by c-MYC in P493-6 cells and was also inversely correlated with MYCN amplification (and poor prognosis) in neuroblastoma tumors (Figs. 7B and 1A). Furthermore ABCG2 was expressed in TET21/N but not in P493-6 cells. However, we found that the ABCG2 promoter is hypermethylated in P493-6 (data not shown). Considering that MYC activity is influenced by the methylation status of its cognate DNA binding sequences (26), this observation may explain some discrepancies about the ABCG2 transcription pattern in P493-6 cells and the panel of neuroblastomas.
FIGURE 7.
c-MYC can also regulate transcription of a large set of ABC transporter genes. A, modulation of c-MYC expression by tetracycline (TET) in P493-6 cells was monitored by Western blot (upper panel). RNA expression of the 48 ABC transporter genes was determined by quantitative reverse transcription-PCR as a function of c-MYC silencing. Cluster 3 was used to generate an image map showing expression levels of ABC genes as a function of tetracycline treatment time, relative to expression at time 0 and processed in log space. Hierarchical clustering was determined as described in Fig. 2. Green indicates genes positively regulated by c-MYC, whereas red indicates those negatively regulated (lower panel). B, Venn diagram representing the overlap of ABC transporter genes regulated by both MYCN and c-MYC transcription factors in TET21/N cells or neuroblastoma tumors and in P493-6 cells. C, protein (upper panel) and mRNA (lower panel) levels of ABCC1, ABCC3, and ABCC4 in MCF-7 and SJ-G2 cell lines after treatment with a specific c-MYC siRNA.
This analysis was extended to MCF-7 breast cancer and SJ-G2 glioblastoma cell lines, which express high endogenous levels of c-MYC. We examined ABC protein expression as a function of silencing of c-MYC using siRNA. We chose to analyze two genes positively regulated by c-MYC (ABCC1 and ABCC4) as well as ABCC3, which was negatively regulated by MYCN in TET21/N cells. As shown in Fig. 7C, reduction of c-MYC protein in MCF7 and SJ-G2 cells resulted in the down-regulation of ABCC1 and ABCC4 and the up-regulation of ABCC3. Overall, our findings demonstrate that c-MYC and MYCN can dictate almost identical ABC transcription signatures through similar mechanisms.
DISCUSSION
In this study we have shown that MYCN can coordinate transcription of a large set of ABC transporter genes, whose level of expression is strongly associated with clinical outcome in an aggressive childhood malignancy. The contribution of MYCN to the global transcription profile of the ABC gene family is quite complex in that it exerts its role as an activator on certain ABC genes and as a repressor on others. Regarding the activation of ABC genes, the association of MYCN with these promoters generally occurs close to the transcription start site, often binding E-box sites located after this site, a finding that is consistent with previously published work by Gallant and co-workers (30) in Drosophila. MYCN also associates with the core promoter region of ABCC3 to repress its transcription. Interestingly, ChIP data showed that MAX also co-occupies the same ABCC3 DNA region, indicating that not just MYCN alone but the heterodimer MYCN-MAX may be required for repression. In the context of transcription repression, MYCN does not recognize any detectable E-box sequence within the ABCC3 promoter but rather targets one region containing several Sp1 binding sites. These sites appear to be critical for MYCN-mediated function in that their deletion abolishes MYCN-mediated repression. Given that Sp1 has been shown to mediate c-MYC repression of p21CIP/WAF and p15Ink4b genes (31, 32), our data suggest functional equivalence of MYCN and c-MYC in repressing as well as in activating transcription. This idea is also fostered by our results showing that, in other tumor cell lines, c-MYC regulates the same set of ABC genes as that described for MYCN, apparently through similar dynamics.
Another important aspect of our study regards the fact that ABC gene transcription levels correlate well with MYC intracellular dosage. Because MYC expression is found to be dysregulated in more than 50% of human cancers, this suggests that knowing MYC expression levels in cancer may be important for prediction of ABC transporter gene transcription profiles and possibly for the choice of appropriate therapeutic protocols to treat affected patients. In addition, altered levels of expression of ABC genes are often detected at diagnosis prior to any medical treatment, suggesting that specific ABC genes may be relevant to tumor biology (33). Several recent studies support the idea that many transporters that we found to be regulated by MYCN may also play a more direct role in oncogenesis. For example, ABCE1 is an inhibitor of Ribonuclease L, an inherited susceptibility gene for familial prostate cancer (34, 35), and silencing of ABCE1 suppresses protein translation and tumor cell growth (36). A recent study identified ABCC4 as an androgen receptor-regulated gene and suggested that high levels of ABCC4 may also have a role to play in prostate cancer malignancy (37). Moreover, ABCC4 is involved in human dendritic cell migration (38), and its expression is affected during hematopoietic stem cell differentiation (39). The ABCA2 transporter has been associated not only with drug resistance but also with disease severity at diagnosis in acute myeloid leukemia (40), and recent evidence suggests that it may be an important molecular marker of oligodendroglioma (41). Likewise, ABCF2, which has been reported to be a candidate marker for clear cell adenocarcinomas of the ovary, may also have a direct role in tumor development (42, 43). Furthermore, when comparing the expression profile of ABC genes between melanocytes and melanoma cells, Schmitz and co-workers (44) found that the mRNA levels of ABCC4, ABCE1, and ABCF2 were significantly increased in melanoma, whereas ABCA7 and ABCD1 were down-regulated, raising the possibility of as yet unrevealed functions for these genes in melanoma tumorigenesis and progression. With respect to ABCC3, although a number of studies associated ABCC3 expression with outcome in different tumors, there is no clear evidence about its role in neuroblastoma tumorigenesis. However, interestingly, ABCC3 is one of the most expressed genes in the differentiated adrenal medulla, the tissue of origin for neuroblastoma (45). Finally, ABCC1, ABCC4, ABCA12, ABCE1, and ABCF1–3 were found to be highly expressed in retinoblastoma cells relative to normal tissues (46), and ABCG2/BCRP transporter expression is associated with the persistence of stem cells in normal and cancer tissues (47–49).
In conclusion, our findings provide strong support to a mechanistic model in which MYCN, and by analogy c-MYC, can contribute to the chemoresistance phenotype of cancer cells through the coordinate regulation of a specific set of ABC transporter genes. Furthermore expression analysis of ABC gene expression in primary neuroblastomas suggests that MYCN may drive the transcription of many ABC genes to influence outcome of patients with this disease. Indeed, combined expression of MYCN and ABC transporter genes identified a subset of neuroblastoma patients with particularly poor outcome, revealing how ABC gene transcriptional profiles may represent a novel risk estimation tool to improve outcome prediction and to adjust cancer treatment strategies.
Supplementary Material
Acknowledgments
We thank R. N. Eisenman and J. Reese for critical reading of the manuscript and D. Manzoni for excellent work with cell cultures.
This work was supported by Australian National Health and Medical Research Council Grants 350885 and 401129, Cancer Institute New South Wales Grant 05/RIG/1-30, Cancer Council New South Wales Grant PG/05/00, and funds from the German National Genome Research Network (NGNF plus), the Italian Association for Research on Cancer, The Italian Ministry of University and Research, and the University of Bologna.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S5 and Figs. S1–S6.
- ABC
- ATP-binding cassette
- RQ
- real time quantitative
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Dean M., Hamon Y., Chimini G. (2001) J. Lipid Res. 42, 1007–1017 [PubMed] [Google Scholar]
- 2.Higgins C. F. (1992) Annu. Rev. Cell Biol. 8, 67–113 [DOI] [PubMed] [Google Scholar]
- 3.Dean M., Rzhetsky A., Allikmets R. (2001) Genome Res. 11, 1156–1166 [DOI] [PubMed] [Google Scholar]
- 4.Gottesman M. M., Fojo T., Bates S. E. (2002) Nat. Rev. Cancer 2, 48–58 [DOI] [PubMed] [Google Scholar]
- 5.Westermann F., Schwab M. (2002) Cancer Lett. 184, 127–147 [DOI] [PubMed] [Google Scholar]
- 6.Attiyeh E. F., London W. B., Mossé Y. P., Wang Q., Winter C., Khazi D., McGrady P. W., Seeger R. C., Look A. T., Shimada H., Brodeur G. M., Cohn S. L., Matthay K. K., Maris J. M. (2005) N. Engl. J. Med. 353, 2243–2253 [DOI] [PubMed] [Google Scholar]
- 7.Bown N., Cotterill S., Lastowska M., O'Neill S., Pearson A. D., Plantaz D., Meddeb M., Danglot G., Brinkschmidt C., Christiansen H., Laureys G., Speleman F., Nicholson J., Bernheim A., Betts D. R., Vandesompele J., Van Roy N. (1999) N. Engl. J. Med. 340, 1954–1961 [DOI] [PubMed] [Google Scholar]
- 8.Brodeur G. M. (2003) Nat. Rev. Cancer 3, 203–216 [DOI] [PubMed] [Google Scholar]
- 9.Schwab M., Westermann F., Hero B., Berthold F. (2003) Lancet Oncol. 4, 472–480 [DOI] [PubMed] [Google Scholar]
- 10.Schwab M., Alitalo K., Klempnauer K. H., Varmus H. E., Bishop J. M., Gilbert F., Brodeur G., Goldstein M., Trent J. (1983) Nature 305, 245–248 [DOI] [PubMed] [Google Scholar]
- 11.Seeger R. C., Brodeur G. M., Sather H., Dalton A., Siegel S. E., Wong K. Y., Hammond D. (1985) N. Engl. J. Med. 313, 1111–1116 [DOI] [PubMed] [Google Scholar]
- 12.Haber M., Smith J., Bordow S. B., Flemming C., Cohn S. L., London W. B., Marshall G. M., Norris M. D. (2006) J. Clin. Oncol. 24, 1546–1553 [DOI] [PubMed] [Google Scholar]
- 13.Norris M. D., Bordow S. B., Marshall G. M., Haber P. S., Cohn S. L., Haber M. (1996) N. Engl. J. Med. 334, 231–238 [DOI] [PubMed] [Google Scholar]
- 14.Norris M. D., Smith J., Tanabe K., Tobin P., Flemming C., Scheffer G. L., Wielinga P., Cohn S. L., London W. B., Marshall G. M., Allen J. D., Haber M. (2005) Mol. Cancer Ther. 4, 547–553 [DOI] [PubMed] [Google Scholar]
- 15.Szakács G., Annereau J. P., Lababidi S., Shankavaram U., Arciello A., Bussey K. J., Reinhold W., Guo Y., Kruh G. D., Reimers M., Weinstein J. N., Gottesman M. M. (2004) Cancer Cell 6, 129–137 [DOI] [PubMed] [Google Scholar]
- 16.Scotto K. W. (2003) Oncogene 22, 7496–7511 [DOI] [PubMed] [Google Scholar]
- 17.Lutz W., Stöhr M., Schürmann J., Wenzel A., Löhr A., Schwab M. (1996) Oncogene 13, 803–812 [PubMed] [Google Scholar]
- 18.Manohar C. F., Bray J. A., Salwen H. R., Madafiglio J., Cheng A., Flemming C., Marshall G. M., Norris M. D., Haber M., Cohn S. L. (2004) Oncogene 23, 753–762 [DOI] [PubMed] [Google Scholar]
- 19.Schuhmacher M., Kohlhuber F., Hölzel M., Kaiser C., Burtscher H., Jarsch M., Bornkamm G. W., Laux G., Polack A., Weidle U. H., Eick D. (2001) Nucleic Acids Res. 29, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porro A., Perini G. (2007) The Epigenome-Network of Excellence: Epigenetics protocols database [Google Scholar]
- 21.Weinmann A. S., Farnham P. J. (2002) Methods 26, 37–47 [DOI] [PubMed] [Google Scholar]
- 22.Wenzel A., Cziepluch C., Hamann U., Schürmann J., Schwab M. (1991) EMBO J. 10, 3703–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberthuer A., Berthold F., Warnat P., Hero B., Kahlert Y., Spitz R., Ernestus K., König R., Haas S., Eils R., Schwab M., Brors B., Westermann F., Fischer M. (2006) J. Clin. Oncol. 24, 5070–5078 [DOI] [PubMed] [Google Scholar]
- 24.Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez P. C., Frank S. R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., Amati B. (2003) Genes Dev. 17, 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perini G., Diolaiti D., Porro A., Della Valle G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12117–12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu T., Tee A. E., Porro A., Smith S. A., Dwarte T., Liu P. Y., Iraci N., Sekyere E., Haber M., Norris M. D., Diolaiti D., Della Valle G., Perini G., Marshall G. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18682–18687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., Wingender E. (2006) Nucleic Acids Res. 34, D108–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malynn B. A., de Alboran I. M., O'Hagan R. C., Bronson R., Davidson L., DePinho R. A., Alt F. W. (2000) Genes Dev. 14, 1390–1399 [PMC free article] [PubMed] [Google Scholar]
- 30.Hulf T., Bellosta P., Furrer M., Steiger D., Svensson D., Barbour A., Gallant P. (2005) Mol. Cell Biol. 25, 3401–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu S., Cetinkaya C., Munoz-Alonso M. J., von der Lehr N., Bahram F., Beuger V., Eilers M., Leon J., Larsson L. G. (2003) Oncogene 22, 351–360 [DOI] [PubMed] [Google Scholar]
- 32.Staller P., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., Möröy T., Bartek J., Massagué J., Hänel F., Eilers M. (2001) Nat. Cell Biol. 3, 392–399 [DOI] [PubMed] [Google Scholar]
- 33.Fletcher J. I., Haber M., Henderson M. J., Norris M. D. (2010) Nat. Rev. Cancer 10, 147–156 [DOI] [PubMed] [Google Scholar]
- 34.Carpten J., Nupponen N., Isaacs S., Sood R., Robbins C., Xu J., Faruque M., Moses T., Ewing C., Gillanders E., Hu P., Bujnovszky P., Makalowska I., Baffoe-Bonnie A., Faith D., Smith J., Stephan D., Wiley K., Brownstein M., Gildea D., Kelly B., Jenkins R., Hostetter G., Matikainen M., Schleutker J., Klinger K., Connors T., Xiang Y., Wang Z., De Marzo A., Papadopoulos N., Kallioniemi O. P., Burk R., Meyers D., Grönberg H., Meltzer P., Silverman R., Bailey-Wilson J., Walsh P., Isaacs W., Trent J. (2002) Nat. Genet. 30, 181–184 [DOI] [PubMed] [Google Scholar]
- 35.Silverman R. H. (2003) Biochemistry 42, 1805–1812 [DOI] [PubMed] [Google Scholar]
- 36.Chen Z. Q., Dong J., Ishimura A., Daar I., Hinnebusch A. G., Dean M. (2006) J. Biol. Chem. 281, 7452–7457 [DOI] [PubMed] [Google Scholar]
- 37.Cai C., Omwancha J., Hsieh C. L., Shemshedini L. (2007) Prostate Cancer Prostatic. Dis. 10, 39–45 [DOI] [PubMed] [Google Scholar]
- 38.van de Ven R., Scheffer G. L., Reurs A. W., Lindenberg J. J., Oerlemans R., Jansen G., Gillet J. P., Glasgow J. N., Pereboev A., Curiel D. T., Scheper R. J., de Gruijl T. D. (2008) Blood 112, 2353–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oevermann L., Scheitz J., Starke K., Köck K., Kiefer T., Dölken G., Niessen J., Greinacher A., Siegmund W., Zygmunt M., Kroemer H. K., Jedlitschky G., Ritter C. A. (2009) Int. J. Cancer 124, 2303–2311 [DOI] [PubMed] [Google Scholar]
- 40.Mack J. T., Beljanski V., Tew K. D., Townsend D. M. (2006) Biomed. Pharmacother. 60, 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soichi O., Masanori N., Hideo T., Kazunori A., Nobuya I., Jun-ichi K. (2007) Neurosurgery 60, 707–714 [DOI] [PubMed] [Google Scholar]
- 42.Nishimura S., Tsuda H., Ito K., Jobo T., Yaegashi N., Inoue T., Sudo T., Berkowitz R. S., Mok S. C. (2007) Hum. Pathol. 38, 134–139 [DOI] [PubMed] [Google Scholar]
- 43.Tsuda H., Ito Y. M., Ohashi Y., Wong K. K., Hashiguchi Y., Welch W. R., Berkowitz R. S., Birrer M. J., Mok S. C. (2005) Clin. Cancer Res. 11, 6880–6888 [DOI] [PubMed] [Google Scholar]
- 44.Heimerl S., Bosserhoff A. K., Langmann T., Ecker J., Schmitz G. (2007) Melanoma Res. 17, 265–273 [DOI] [PubMed] [Google Scholar]
- 45.Borst P., de Wolf C., van de Wetering K. (2007) Pflugers Arch. 453, 661–673 [DOI] [PubMed] [Google Scholar]
- 46.Hendig D., Langmann T., Zarbock R., Schmitz G., Kleesiek K., Götting C. (2009) Mol. Cell Biochem. 328, 85–92 [DOI] [PubMed] [Google Scholar]
- 47.Bhattacharya S., Das A., Mallya K., Ahmad I. (2007) J. Cell Sci. 120, 2652–2662 [DOI] [PubMed] [Google Scholar]
- 48.Zhou S., Schuetz J. D., Bunting K. D., Colapietro A. M., Sampath J., Morris J. J., Lagutina I., Grosveld G. C., Osawa M., Nakauchi H., Sorrentino B. P. (2001) Nat. Med. 7, 1028–1034 [DOI] [PubMed] [Google Scholar]
- 49.Lin T., Islam O., Heese K. (2006) Cell Res. 16, 857–871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.