Abstract
A retro-inverso peptide is made up of d-amino acids in a reversed sequence and, when extended, assumes a side chain topology similar to that of its parent molecule but with inverted amide peptide bonds. Despite their limited success as antigenic mimicry, retro-inverso isomers generally fail to emulate the protein-binding activities of their parent peptides of an α-helical nature. In studying the interaction between the tumor suppressor protein p53 and its negative regulator MDM2, Sakurai et al. (Sakurai, K., Chung, H. S., and Kahne, D. (2004) J. Am. Chem. Soc. 126, 16288–16289) made a surprising finding that the retro-inverso isomer of p53(15–29) retained the same binding activity as the wild type peptide as determined by inhibition enzyme-linked immunosorbent assay. The authors attributed the unusual outcome to the ability of the d-peptide to adopt a right-handed helical conformation upon MDM2 binding. Using a battery of biochemical and biophysical tools, we found that retro-inverso isomerization diminished p53 (15–29) binding to MDM2 or MDMX by 3.2–3.3 kcal/mol. Similar results were replicated with the C-terminal domain of HIV-1 capsid protein (3.0 kcal/mol) and the Src homology 3 domain of Abl tyrosine kinase (3.4 kcal/mol). CD and NMR spectroscopic as well as x-ray crystallographic studies showed that d-peptide ligands of MDM2 invariably adopted left-handed helical conformations in both free and bound states. Our findings reinforce that the retro-inverso strategy works poorly in molecular mimicry of biologically active helical peptides, due to inherent differences at the secondary and tertiary structure levels between an l-peptide and its retro-inverso isomer despite their similar side chain topologies at the primary structure level.
Keywords: p53, Peptide Conformation, Peptide Interactions, Ubiquitin Ligase, X-ray Crystallography, MDM2, Native Chemical Ligation, Retro-inverso Peptide
Introduction
Protein-protein interactions govern a great variety of biological processes and present important targets for therapeutic intervention (1, 2). Small peptides emulating the activity of one binding partner to antagonize the other play instrumental roles in drug screening and design. Despite their ability to bind proteins with high affinity and unsurpassed specificity, peptides themselves are rarely used as therapeutic agents due primarily to their poor in vivo stability. Even for in vitro applications, efficacy often necessitates peptide resistance to proteolytic degradation. To tackle peptide susceptibility to proteolysis, various peptidomimetic chemistries have been developed, involving the use of d-amino acids, unnatural amino acids, peptide backbone modifications, cyclizations, and secondary structure-inducing templates, among others (3). Peptide retro-inverso isomerization, pioneered by Chorev and Goodman (4), represents an elegant solution to functional peptides stable under physiological conditions.
It is postulated that a retro-inverso peptide (a peptide of the reversed sequence made up of d-amino acids, also known as a retro-all-d- or retro-enantio-peptide) assumes a side chain topology, in its extended conformation, similar to that of its native l-sequence, thus emulating biological activities of the parent molecule while fully resistant to proteolytic degradation (4). Some success has been achieved immunologically in using retro-inverso peptides toward antigenic mimicry of their parent l-peptides (5). Failures, however, have also been noted for retro-inverso isomers to elicit antibodies that cross-react with native immune epitopes (6). In fact, retro-inverso peptides are not isofunctional to their parent l-peptide molecules with respect to binding energetics even in some successful immunological applications of antigenic mimicry (7). This mixed outcome comes as no surprise, because antibody-antigen recognition is notoriously lenient at the structural level to tolerate conformational plasticity (8).
What surprises, however, is different outcomes arising from well characterized peptide-protein interacting systems where conformational rigidity or stability of a helical peptide ligand is key to its high affinity binding to the preformed cavity of a folded protein (9–11). Limited digestion of ribonuclease A by subtilisin generates S peptide (residues 1–20) and S protein (residues 21–124), which can reassociate with high affinity to form enzymatically active ribonuclease S (12). Two recent reports on the extensively studied S peptide-S protein interacting system have shown that it is structurally impossible for the retro-inverso isomer of the α-helical S peptide to functionally mimic S protein binding (9, 10). Moreover, the exquisite specificity for recognition of the native S peptide is strictly maintained in antibody recognition and T cell stimulation (9). In studying the interaction between the tumor suppressor protein p53 and its negative regulator MDM2 (13, 14), however, Sakurai et al. (11) found that p53(15–29) and its retro-inverso isomer displayed nearly identical binding affinities for the p53-binding domain of MDM2. To rationalize their unexpected finding, Sakura et al. (11) suggested that the retro-all-d-peptide isomer of p53(15–29), like its parent l-peptide, adopted a right-handed helical conformation in the complex.
These conceptually conflicting reports motivated us to carry out a comprehensive study of functional effects of peptide retro-inverso isomerization on the p53-MDM2 interaction, using a combination of biochemical, biophysical, and structural tools. The interactions of p53(15–29) with MDMX (a MDM2 homolog) and of PMI (a high affinity, dual specific peptide ligand of MDM2 and MDMX) with MDM2 were also subjected to investigation. To determine whether or not conclusions from the p53/PMI-MDM2/MDMX interactions are applicable to others, we expanded our study to include two additional, well characterized peptide-protein interacting systems: the C-terminal domain of HIV-12 capsid protein (CCA) and the Src homology 3 (SH3) domain of Abl tyrosine kinase with their respective peptide ligands.
MATERIALS AND METHODS
Chemical Synthesis of Peptides and Proteins
Total chemical synthesis of highly pure and correctly folded p53-binding domains of MDM2 (residues 25–109, referred to hereafter as synMDM2) and MDMX (residues 24–108 or synMDMX) has been described elsewhere (15). Abl SH3 domain was prepared as described (16). Folding of Abl SH3 domain was initiated by dissolving the polypeptide at 3 mg/ml in 0.2 m phosphate buffer containing 6 m guanidine HCl and 1 mm dithiothreitol, pH 7.5, followed by extensive dialysis (molecular weight cut-off 3000) in water at 4 °C. HIV CCA (S146PTSILDIRQG156PKEPFRDYVD166RFYKTLRAEQ176ASQEVKNWMT186ETLLVQNANP196DCKTILKALG206PGATLEEMMT216ACQGVGGPGH226KARVL) was made using native chemical ligation (17, 18). The two peptide fragments of CCA, H2N-CA(146–197)αCOSR (where R represents CH2CO-Leu-OH) and H2N-CA(198–231)COOH, were individually synthesized on t-butoxycarbonyl-Leu-OCH2-PAM resin using an optimized HBTU activation/DIEA in situ neutralization protocol developed by Kent and co-workers (19). Crude peptides, after hydrogen fluoride cleavage and deprotection in the presence of 5% p-cresol at 0 °C, were precipitated with cold ether and purified by preparative C18 reversed-phase HPLC, and their molecular masses were ascertained by electrospray ionization mass spectrometry. The determined molecular mass of 9518.7 Da of the full-length ligation product agreed with the expected value of 9519.0 Da calculated on the basis of the average isotopic compositions of CCA. Spontaneous folding of CCA and formation of the disulfide bond between Cys198 and Cys218 was achieved through air oxidation in aqueous buffer. The determined molecular mass of 9517.1 ± 0.6 Da of oxidized CCA, differing by 2 mass units from the calculated value of 9519.0 Da of reduced CCA, confirmed the disulfide formation.
End-capped p53(15–29) (Ac-SQETFSDLWKLLPEN-NH2) and its retro-inverso isomer RI-p53(15–29) (Ac-DNDEDPDLDLDKDWDLDDDSDFDTDEDQDS-NH2) were synthesized on 4-methyl-benzhydrylamine resin using the above described t-butoxycarbonyl chemistry. Both peptides were N-terminally acetylated on resin using acetic anhydride and DIEA (95:5). All other peptides were only C-terminally amidated: PMI (TSFAEYWNLLSP-NH2) and RI-PMI (DPDSDLDLDNDWDYDEDADFDSDT-NH2), CAI (ITFEDLLDYYGP-NH2) and RI-CAI (DPDGDYDYDDDLDLDDDEDFDTDI-NH2), Y4W-P40 (APTWSPPPPP-NH2) and RI-Y4W-P40 (DPDPDPDPDPDSDWDTDPDA-NH2), DPMI-α (DTDNDWDYDADNDLDEDKDLDLDR-NH2) and DPMI-β (DTDADWDYDADNDFDEDKDLDLDR-NH2), and RI-DPMI-β (RLLKEFNAYWAT-NH2). All peptides were purified to homogeneity by preparative C18 reversed-phase HPLC, and their molecular masses were ascertained by electrospray ionization mass spectrometry. Peptide and protein quantification was achieved by UV measurements at 280 nm using molar extinction coefficients calculated according to a published algorithm (20).
CD and Fluorescence Spectroscopy
Far-UV CD spectra were obtained on a Jasco J-810 spectropolarimeter at room temperature using a 0.1-cm path length. p53(15–29) and RI-p53(15–29) were thoroughly dialyzed (2000 molecular weight cut-off dialysis cassette, Pierce) against 10 mm phosphate buffer, pH 7.2, to remove trace amounts of trifluoroacetic acid that interferes with CD measurements. Trp fluorescence spectra of p53(15–29) and RI-p53(15–29) in the presence and absence of synMDM2 were recorded at room temperature in phosphate-buffered saline (PBS) containing 0.5 mm tris(2-carboxyethyl)phosphine (TCEP) on a Varian (Cary) Eclipse fluorometer. TCEP was added to prevent oxidative dimerization of synMDM2 via a free Cys residue in the protein. The excitation wavelength was 295 nm, and the width of both slits was set to 5 nm.
The binding affinity of Y4W-P40 and RI-Y4W-P40 for Abl SH3 domain was determined essentially as described (16). Briefly, different concentrations of Y4W-P40 (0–20 μm) or RI-Y4W-P40 (0–200 μm) were incubated with SH3 (5 μm for Y4W-P40 and 15 μm for RI-Y4W-P40) for 15 min in 5 mm phosphate buffer, pH 7.0, and ligand-induced changes of Trp fluorescence of SH3 were measured at 350 nm in a cuvette. The Kd values were obtained using a four-parameter non-linear regression analysis as previously described (16).
Surface Plasmon Resonance (SPR)
Competition binding kinetics was carried out at 25 °C on a Biacore T100 SPR instrument using an N-terminally uncapped p53(15–29) peptide immobilized on a CM5 sensor chip (17 resonance units (RUs)). The buffer was 10 mm HEPES, 150 mm NaCl, 0.005% surfactant P20, pH 7.4. 50 nm synMDM2 or 100 nm synMDMX was incubated at room temperature for 30 min with varying concentrations of peptide and injected at a flow rate of 20 μl/min for 2 min, followed by a 4-min dissociation. The concentration of unbound synMDM2 or synMDMX in solution was deduced, based on p53 association RU values, from a calibration curve established by RU measurements of different concentrations of synMDM2 or synMDMX injected alone. Non-linear regression analysis was performed using GraphPad Prism 4 to give rise to Kd values using the equation, Kd = [peptide][MDM2]/[complex]. For Kd measurements of CAI and RI-CAI with CCA, steady state binding kinetics of CAI (0.78–100 μm) and RI-CAI (6.25–800 μm) on oxidized CCA (500 RUs) immobilized on a CM5 biosensor chip were obtained.
Isothermal Titration Calorimetry (ITC)
Direct protein-peptide interactions were quantified at 25 °C using a MicroCal VP-ITC microcalorimeter. A typical experiment involved injection of 30 aliquots (8 μl each) of 200 μm peptide solution (prepared in PBS containing 0.1 mm TCEP and 0.01% NaN3) into a 1.4-ml ITC cell containing 15 μm protein solution in the same buffer. For background subtraction, a reference set of injections of peptide was made in a separate experiment into the buffer alone. The integrated interaction heat values were analyzed using the Origin 7.0-based ITC fitting software provided by MicroCal, yielding the binding affinity, stoichiometry, and other thermodynamic parameters.
Inhibition ELISA
GST-MDM2(1–150) and His6-p53 were expressed in Escherichia coli and purified by binding to glutathione-agarose and Ni2+-nitrilotriacetic acid beads under non-denaturing conditions. ELISA plates were incubated with 2.5 μg/ml His6-p53 in PBS for 16 h. After washing with PBS plus 0.1% Tween 20 (PBST), the plates were blocked with PBS plus 5% nonfat dry milk plus 0.1% Tween 20 (PBSMT) for 30 min. GST-MDM2(1–150) (5 μg/ml) was mixed with compounds in PBSMT plus 10% glycerol plus 10 mm dithiothreitol and added to the wells. The plates were washed with PBST after incubation for 1 h at room temperature and incubation with MDM2 antibody 5B10 in PBSMT for 1 h, followed by washing and incubation with horseradish peroxidase-rabbit anti-mouse Ig antibody for 1 h. The plates were developed by incubation with TMB peroxidase substrate (KPL) and measured by absorbance at 450 nm.
NMR Spectroscopy
All NMR experiments were performed at 293 K on a Varian INOVA 500 spectrometer operating at a 1H resonance frequency of 499.754 MHz. The peptides were dissolved at 5 mm in d3-trifluoroethanol/H2O co-solvent (1:1) with 10% D2O, pH 4.4. The homonuclear DQF-COSY, TOCSY, and NOESY spectra were collected using standard protocols (21). TOCSY spectra were recorded with a mixing time of 80 ms, whereas mixing times of 80 and 200 ms were used for NOESY experiments and evaluation of spin diffusion effects. All data were processed with the program NMRPipe (22). The spin systems of all residues were identified using the Wüthrich strategy (23), aided by the CARA software (24). Structure calculation was carried out using a standalone ATNOS/CANDID program (25), combined with the molecular modeling software XPLOR-NIH (26). A total of 234 (RI-p53(15–29)) or 318 (p53(15–29)) meaningful nuclear Overhauser effect upper distance constraints, extracted from a total of 525 (RI-p53(15–29)) or 642 (p53(15–29)) assigned NOESY cross-peaks, were used as input for the final structure calculation by XPLOR. 20 conformers with the lowest residual target function values from energy-refined cycles were superimposed in the figure.
Crystal Structure Determination of Oxidized CCA
Crystals were grown in 24-well plates at room temperature using the hanging drop, vapor diffusion method. 1 μl of 10 mg/ml CCA in water was mixed with an equal volume of crystallization solution and equilibrated against 800 μl of mother liquor consisting of 0.1 m HEPES, pH 7.5, 0.8 m sodium phosphate monobasic monohydrate, and 0.8 m potassium phosphate monobasic. Crystals were soaked briefly in reservoir solution plus 30% (v/v) glycerol as cryoprotectant and subsequently flash-frozen in liquid nitrogen.
X-ray diffraction data were collected using a rotating anode x-ray generator Rigaku-MSC Micromax 7 and a Raxis-4++ image plate detector (at the X-ray Crystallography Core Facility, University of Maryland, Baltimore, MD) and were integrated and scaled with the HKL2000 package (27). The structure was solved by the molecular replacement method with the program Phaser from the CCP4 suite (28), based on the Protein Data Bank entry 1A43 model (29), and refined with Refmac and coupled with manual refitting and rebuilding with COOT (30, 31). Data collection and refinement statistics are summarized in supplemental Table S1. Molecular graphics were generated using PyMOL (DeLano Scientific LLC, San Carlos, CA).
RESULTS
15–29p53 and RI-15–29p53 Differ Structurally in Solution
The N-terminal transactivation domain of p53 encompasses the sequence F19S20D21L22W23K24L25L26 (critical residues highlighted in boldface type) minimally required for effective MDM2 binding (14, 32, 33). The p53 transactivation domain itself is unstructured in solution (34). Upon binding to the N-terminal domain of MDM2, however, it acquires a right-handed amphipathic α-helical structure where Phe19, Trp23, and Leu26 bury their hydrophobic side chains inside the p53-binding cavity of MDM2 (14). We recently identified PMI (TSFAEYWNLLSP) by phage display, a potent, dual specific peptide inhibitor with low nanomolar binding affinity for both MDM2 and MDMX (15). Shown in Fig. 1A is the synMDM2-PMI complex superimposed with the co-crystal structure of recombinant MDM2(17–125) and p53(15–29) (14). The synthetic and recombinant MDM2 proteins are nearly identical, as evidenced by a root mean square deviation of 0.7 Å between their equivalent Cα atoms. The functionally critical residues, Phe3, Trp7, and Leu10 of PMI, are topologically equivalent to Phe19, Trp23, and Leu26 of p53 in bound states. Notably, we crystallized synMDM2 in complex with p53(15–29) but failed to crystallize the same protein with RI-p53(15–29) (data not shown).
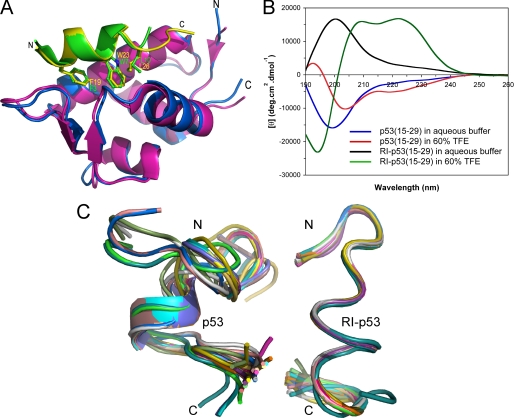
FIGURE 1.
p53(15–29) and RI-p53(15–29) are structurally different in solution. A, superposition of the crystal structures of synMDM2-PMI in magenta and green and recombinant MDM2(17–125)-p53(15–29) in blue and yellow. The image was created from Protein Data Bank entries 3EQS (15) and 1YCR (14) by PyMOL (DeLano Scientific LLC). Note that only residues 25–109 of MDM2 and 17–29 of p53 are visible in the complex structure due to disordered termini. B, CD spectra of 100 μm p53(15–29) and RI-p53(15–29) in 10 mm phosphate buffer, pH 7.2, with or without 60% (v/v) TFE. The double maxima at 208 and 222 nm and a strong negative peak at 195 nm shown by RI-p53(15–29) (green) are characteristic of left-handed α-helical secondary structure. C, 20 superimposed structures of p53(15–29) (left) and of RI-p53(15–29) (right) determined at 20 °C in 50% TFE by NMR spectroscopy.
Shown in Fig. 1B are the CD spectra of 100 μm p53(15–29) and RI-p53(15–29) obtained at room temperature. Both peptides were unstructured in 10 mm phosphate buffer, pH 7.2. However, in the presence of the α-helix-inducing agent trifluoroethanol (TFE) (60%, v/v), although p53(15–29) became only partially ordered, the retro-inverso isomer readily adopted a distinct left-handed α-helix, as indicated by double maxima at 208 and 222 nm and a strong negative peak at 195 nm. We further characterized both peptides in 50% TFE by NMR spectroscopy at 20 °C. The solution structures of p53(15–29) and RI-p53(15–29) were determined by the standard two-dimensional homonuclear methods using TOCSY, NOESY, and DQ-COSY spectra (23). As shown in Fig. 1C, except for a less-than-a-full-turn α-helical segment, p53(15–29) is largely disordered, particularly at its N terminus. By contrast, the retro-inverso isomer is well structured, adopting a somewhat irregular left-handed helix expected for d-peptides, fully consistent with the CD spectroscopic data. Clearly, p53(15–29) and RI-p53(15–29) differ not only in the handedness of helix but also in helical propensity; the d-peptide has a significantly stronger propensity to form a left-handed helix than the l-peptide to form a right-handed one in solution.
Retro-inverso Isomerization of 15–29p53 Diminishes Its Binding to MDM2 and MDMX by 3.2–3.3 kcal/mol
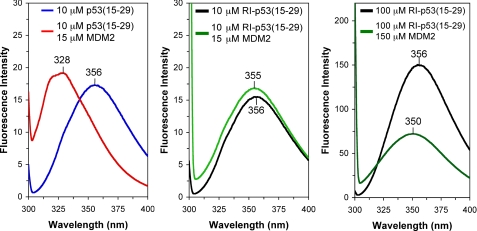
One of the hallmarks of the p53-MDM2 interaction is the blue shift of Trp fluorescence, resulting from the burial of Trp23 of p53 in the hydrophobic cavity of MDM2 (35). As shown in Fig. 2, Trp fluorescence of p53(15–29) and RI-p53(15–29) emitted maximally at 356 nm in phosphate buffer, suggesting that Trp23 in both peptides was fully solvent-exposed (36). The addition of 15 μm synMDM2 to 10 μm p53(15–29) accentuated a marked shift of Trp fluorescence maximum by 28 nm, indicative of a complex formed between p53(15–29) and synMDM2. (MDM2 itself lacks Trp fluorescence.) By contrast, when 15 μm synMDM2 was added to 10 μm RI-p53(15–29), the Trp fluorescence maximum shifted by only 1 nm. A modest shift of 6 nm (from 356 to 350 nm) was observed only after the concentrations of both synMDM2 and RI-p53(15–29) were increased by 10-fold to favor complex formation. These data strongly suggest that RI-p53(15–29) binds to synMDM2 at a significantly lower affinity than that of the wild type p53 peptide.
FIGURE 2.
Trp fluorescence spectra of p53(15–29) and RI-p53(15–29) in the presence and absence of excess synMDM2 in PBS containing 0.5 mm TCEP: 10 μm p53(15–29) and 15 μmsynMDM2 (left), 10 μm RI-p53(15–29) and 15 μmsynMDM2 (middle), and 100 μm RI-p53(15–29) and 150 μmsynMDM2 (right). A decrease in Trp fluorescence intensity with 100 μm RI-p53(15–29) and 150 μm synMDM2 (right) was due to minor precipitation at high peptide/protein concentrations.
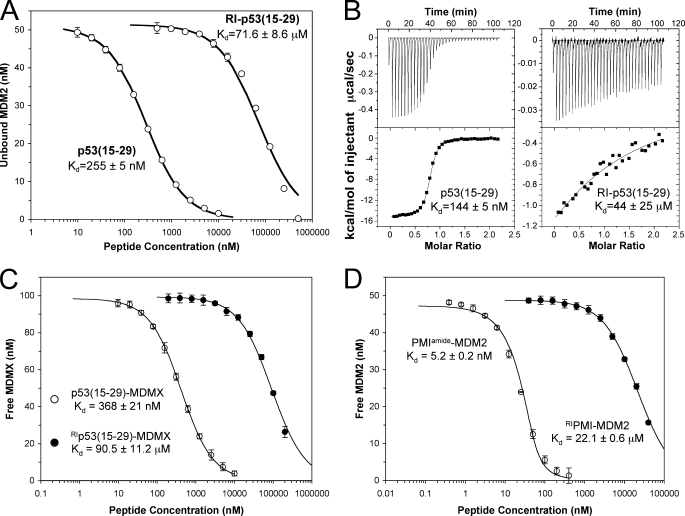
To quantify these effects, we used a previously established, SPR-based competition binding assay (15, 35, 37), in which an N-terminally uncapped p53(15–29) peptide was immobilized on a CM5 biosensor chip for kinetic analysis of 50 nm synMDM2 preincubated in solution with varying concentrations of peptide. As shown in Fig. 3A, solution peptides competed with immobilized p53(15–29) for synMDM2 binding in a dose-dependent manner, giving rise to a Kd value of 255 ± 5 nm. In sharp contrast, the retro-inverso p53 isomer showed a greatly reduced binding affinity for synMDM2 with a Kd value of 71.6 ± 8.6 μm. The 280-fold difference in Kd between p53(15–29) and RI-p53(15–29) was independently verified by ITC measurements, which yielded Kd values of 144 ± 5 nm and 44.1 ± 25.4 μm for p53(15–29) and RI-p53(15–29), respectively (Fig. 3B and Table 1). Importantly, weakened binding of the retro-inverso p53 isomer to MDM2 was due entirely to a huge loss in enthalpy, suggesting that the binding modes or conformations of p53(15–29) and RI-p53(15–29) are significantly different in the complexes. In addition, the negative entropic change for RI-p53(15–29) binding to MDM2 (−7.5 cal/mol/K) was substantially smaller than that for p53(15–29) (−19.8 cal/mol/K).
FIGURE 3.
p53(15–29) and RI-p53(15–29) are functionally different. A, quantification of the interaction of synMDM2 (50 nm) with varying concentrations of p53(15–29) and RI-p53(15–29) by SPR-based competition assays. Each curve is the mean of three independent measurements at 25 °C in 10 mm HEPES, 150 mm NaCl, 0.005% surfactant P20, pH 7.4. B, isothermal titration calorimetric measurements of the interaction between synMDM2 and p53(15–29) (left) and RI-p53(15–29) (right) at 25 °C in PBS containing 0.1 mm TCEP and 0.01% NaN3. C, SPR-based quantification of p53(15–29) and RI-p53(15–29) interacting with synMDMX. Each curve is the mean of three independent measurements. D, determination of the Kd values of PMI and RI-PMI with synMDM2 by competition SPR in three separate experiments.
TABLE 1.
Thermodynamic parameters measured by isothermal titration calorimetry at 25 °C in PBS containing 0.1 mm TCEP and 0.01% NaN3
| Protein | Peptide | na | Ka | Kd | ΔH | ΔS |
|---|---|---|---|---|---|---|
| m−1 | nm | cal/mol | cal/mol/K | |||
| synMDM2 | p53(15–29) | 0.78 ± 0.01 | (6.96 ± 0.22) × 106 | 144 | 15,230 ± 40 | −19.8 |
| synMDM2 | RI-p53(15–29) | 0.96 ± 0.59 | (2.27 ± 1.31) × 104 | 44,100 | −3717 ± 3108 | −7.5 |
a Stoichiometry of binding.
We further compared p53(15–29) and RI-p53(15–29) in a well established inhibition ELISA using recombinant GST-MDM2(1–150) and His6-tagged p53 (38–40). In this assay, PMI (acid form) inhibited the p53-MDM2 interaction with an IC50 value of 20 nm (40), 6-fold higher than its Kd value (3.2 nm) measured by SPR (37). The IC50 value of p53(15–29) was determined by ELISA to be ∼2 μm (supplemental Fig. S1), ∼8-fold higher than its Kd value. These similar IC50/Kd ratios demonstrate an internal consistency between the two different assay methods. Not surprisingly, RI-p53(15–29) was barely inhibitory at the highest concentration of 50 μm tested. The IC50 values of p53(15–29) and RI-p53(15–29) probably differed by orders of magnitude, as inferred by the inhibition curves (supplemental Fig. S1), corroborating the findings from the SPR and ITC measurements as well as the fluorescence spectroscopic studies.
The p53-binding domains of MDM2 and MDMX share over 50% sequence identity and are highly similar both structurally and functionally (15, 41). We also quantified via SPR the binding of p53(15–29) and RI-p53(15–29) to synMDMX. p53(15–29) bound synMDMX at an affinity of 368 nm, 246-fold stronger than the retro-inverso peptide (Kd = 90.5 μm) (Fig. 3C). Thus, the deleterious effects of retro-inverso isomerization on p53(15–29) binding to MDM2 and to MDMX are nearly identical, equivalent to a loss of binding free energy of 3.2 kcal/mol (a 246-fold increase in Kd) to 3.3 kcal/mol (an increase in Kd by 280-fold by SPR or 306-fold by ITC).
Retro-inverso Isomerization of PMI Diminishes Its Binding to MDM2 by 4.9 kcal/mol
The binding affinity of PMI for MDM2 is nearly 2 orders of magnitude higher than that of p53(15–29) (15). We compared the ability of PMI and RI-PMI to bind synMDM2 using the SPR-based competition assay. As shown in Fig. 3D, the C-terminally amidated PMI bound to synMDM2 with a Kd value of 5.2 nm, whereas RI-PMI was 4,250-fold weaker (Kd = 22.1 μm). The Kd difference between PMI and RI-PMI amounts to a loss of binding free energy of 4.9 kcal/mol.
Retro-inverso Isomerization Diminishes CAI Binding to the C-terminal Domain of HIV-1 Capsid Protein by 3.0 kcal/mol
HIV capsid protein (CA) assembles into a cone-shaped core structure encasing the viral RNA (42). The C-terminal one-third of CA (CCA) adopts a four-helix bundle conformation and dimerizes in solution through hydrophobic packing of its second α-helix (29, 43). CCA mediates viral assembly and maturation (44) and is a significant yet largely unexploited antiviral target. Sticht et al. (45) identified via phage display a duodecimal peptide inhibitor, termed CAI (ITFEDLLDYYGP), that inhibited assembly of immature- and mature-like particles in vitro. CAI bound at micromolar affinity as an amphipathic α-helix to a conserved hydrophobic groove of CCA, forming a compact five-helix bundle with altered dimeric interactions (46).
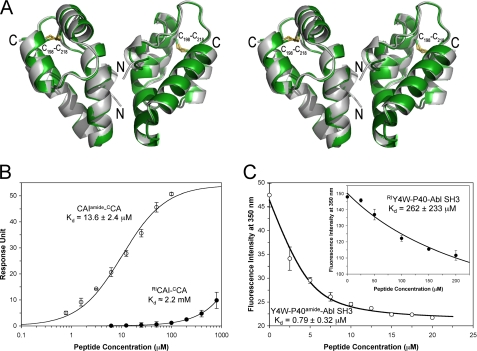
We chemically synthesized CCA via native chemical ligation and determined its crystal structure at 2.05 Å resolution. As expected, dimerization of synthetic CCA is mediated primarily by hydrophobic interactions between the second α-helices (Fig. 4A). Comparative structural analysis indicates that the synthetic CCA dimer is nearly identical to the recombinant CA(146–231) dimer (root mean square deviation(Cα) = 0.47 Å) (29); the latter fits well as a dimeric unit into the cryoelectron microscopy map of the full-length HIV CA capsid lattice (47). To quantify interactions of CAI and RI-CAI with CCA, we immobilized the synthetic protein on a CM5 biosensor chip and obtained steady-state binding kinetics of the two peptides at different concentrations. A non-linear regression analysis yielded Kd values of 13.6 μm and 2.2 mm for CAI and RI-CAI, respectively (Fig. 4B). Thus, retro-inverso isomerization of CAI decreased its binding affinity for CCA by 162-fold or 3.0 kcal/mol.
FIGURE 4.
Effects of retro-inverso isomerization on peptide binding of HIV CCA and Abl SH3 domain. A, stereo view of superimposed crystal structures of synthetic HIV CCA (green) and recombinant HIV CA(146–231) (gray; Protein Data Bank code 1A43) (29). Disulfide bonds are displayed as ball-and-stick representations. The overall structures of dimers are very similar, with the root mean square deviation between equivalent Cα atoms of 0.47 Å. Dimerization is primarily mediated by hydrophobic interactions between the α2 helices in an anti-parallel fashion, which buries 1,500–1,800 Å2 of the surface area. B, SPR quantification of direct binding of CAI and RI-CAI to immobilized synthetic HIV CCA (500 RUs) based on steady-state kinetic assays. C, fluorescence titration of the synthetic Abl SH3 domain by Y4W-P40 and RI-Y4W-P40 in 5 mm phosphate buffer, pH 7.0. Progressive subtractions of the background signal contributed by the Trp-containing peptides were carried out as described (16).
Retro-inverso Isomerization Diminishes Y4W-P40 Binding to Abl SH3 Domain by 3.4 kcal/mol
SH3 domains are small eukaryotic protein modules of ∼60 amino acid residues that intramolecularly regulate the activity of Src family tyrosine kinases and, more generally, target their parent protein molecules to cellular sites of their recognition partners (48, 49). Selectively interfering with SH3-dependent signaling events with peptide ligands is of great interest in biology (50, 51). Most SH3 domains recognize proline-rich peptides that adopt an extended, left-handed polyproline II helical conformation in the complexes (52, 53). The Serrano laboratory previously reported a micromolar affinity decapeptide ligand, termed P40 (APTYSPPPPP), of the SH3 domain of Abl tyrosine kinase (54). We subsequently improved the binding affinity of P40 for Abl SH3 domain through a Y4W mutation, which was identified by affinity panning and mass spectrometric decoding of targeted synthetic peptide libraries (16).
For this study, we measured the binding affinity of Y4W-P40 and its retro-inverso isomer RI-Y4W-P40 for Abl SH3 domain using a previously detailed Trp fluorescence titration method (16). As shown in Fig. 4C, Y4W-P40 bound to Abl SH3 domain at an affinity of 0.79 μm, contrasting with a Kd value of 262 μm of RI-Y4W-P40 for the same protein under identical conditions. Therefore, retro-inverso isomerization weakened Y4W-P40 binding to Abl SH3 domain by 332-fold or 3.4 kcal/mol.
Effects of Reverse Retro-inverso Isomerization of a d-Peptide Ligand of MDM2
Mirror image phage display, a powerful combinatorial technique developed by Kim and co-workers (55), is an elegant tool that enables quick identification of d-peptide ligands of a native protein (56–58). Mirror image phage display screens phage-expressed peptide libraries against the d-enantiomer of a native l-protein of interest, yielding an l-peptide ligand that binds specifically to the d-protein. After enantiomeric inversion, the resultant d-peptide ligand, for reasons of symmetry, binds to the native l-protein at the same affinity. Using mirror image phage display, we recently identified a duodecimal d-peptide inhibitor of MDM2, termed DPMI-α (DTDNDWDYDADNDLDEDKDLDLDR-NH2) and its mutant DPMI-β (DTDADWDYDADNDFDEDKDLDLDR-NH2) (59). DPMI-α and DPMI-β, differing by 2 amino acid residues, bound to synMDM2 at affinities of 219 and 34.5 nm, respectively (59). We used a reverse retro-inverso strategy, converting DPMI-β to its retro-all-l-isomer (RLLKEFNAYWAT-NH2). Quantification of the binding of RI-DPMI-β to synMDM2 by competition SPR resulted in a Kd value of greater than 1 mm, at least a 25000-fold decrease in binding affinity (supplemental Fig. S2).
DISCUSSION
The tumor suppressor protein p53 transcriptionally regulates growth inhibitory and apoptotic responses to prevent stressed cells from proliferating and passing mutations on to the next generation (41, 60, 61). Dubbed the “guardian of the genome,” p53 is critical for maintaining genetic stability and preventing tumor development (62). Not surprisingly, in 50% of human cancers, p53 is either deleted or carries missense mutations primarily in its DNA-binding domain. In many other tumors harboring wild type p53, the oncoprotein MDM2 and its homolog MDMX negatively regulate the activity and stability of the tumor suppressor protein (41, 60), resulting directly in p53 inactivation and malignant progression (41, 60). It has been validated both in vitro and in vivo that MDM2/MDMX antagonists disrupt the p53-MDM2/MDMX interactions and selectively kill tumor cells by reactivating the p53 pathway (63, 64). Due to their high potency and specificity (thus low toxicity), peptides and/or peptidomimetics capable of antagonizing MDM2 to activate p53 are of potential therapeutic value (65). d-Peptide antagonists are particularly attractive because they are fully resistant to proteolytic degradation in vivo, thereby ensuing maximal bioavailability and optimal therapeutic efficacy.
Retro-inverso peptides as antigenic mimicry of their parent l-peptides succeed in some cases yet fail in others (5, 6). By contrast, the retro-inverso strategy has garnered a much less impressive track record in mimicking small, biologically active peptides that become helical upon target binding (9, 10, 66). For these reasons, the report by Sakurai et al. (11) that retro-inverso p53(15–29) retains the same biological activity as its parent l-form is highly unusual and somewhat provocative. Obviously, if the retro-inverso strategy works for the p53-MDM2 interacting system, protease-resistant d-peptide activators of p53 can be readily designed through enantiomeric conversion of high affinity l-peptide ligands of MDM2 selected from phage-displayed peptide libraries (15, 38). More importantly, this simple but powerful approach, if borne out, will have a far reaching impact on the design of potent and stable d-peptide inhibitors for a great variety of therapeutic applications.
Unfortunately, we completely failed on multiple accounts to replicate the finding by Sakurai et al. (11) after subjecting four peptide-protein interacting systems to painstaking scrutiny using a battery of biochemical, biophysical, and structural tools. Compelling evidence from the multifaceted experiments allows us to conclude that retro-inverso isomers are not isofunctional to their parent l-peptides with respect to target protein binding. As has been shown repeatedly in this work, the energetic penalty of peptide retro-inverso isomerization amounts to 3.0–4.9 kcal/mol in binding free energy. The lack of general success with the retro-inverso strategy in molecular mimicry is attributed to the fact that a retro-inverso peptide, with inverted amide peptide bonds, cannot possibly be equivalent to its parent sequence at the secondary and tertiary structure levels, despite similar side chain topologies at the primary structure level (66).
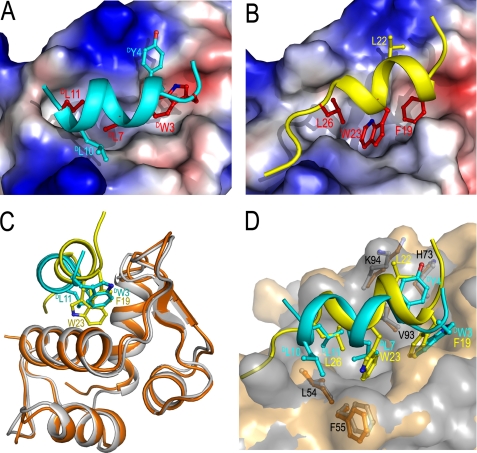
The co-crystal structure of DPMI-α-MDM2(25–109) has recently been determined (59), providing a much needed explanation for the deleterious functional effect of peptide retro-inverso isomerization. Comparative structural analysis indicates that the binding mode of DPMI-α for MDM2 differs significantly from that of p53 or PMI. As expected and, again, confirmed by the CD and NMR spectroscopic studies, the d-peptide ligand adopts an amphipathic left-handed helical conformation in the complex (Fig. 5A), contrasting with the right-handed α-helical p53(15–29) (Fig. 5B). This finding comes as no surprise because d-peptides, contrary to the suggestion by Sakurai et al. (11), are never known to exist in right-handed helical conformations due to highly unfavorable energy (67, 68). Superposition of DPMI-α-MDM2(25–109) and p53(15–29)-MDM2(17–125) unveils a substantial positional difference at the binding interface between the two helical peptide ligands of opposite handedness, despite similar overall structures of MDM2 from the two complexes (root mean square deviation(Cα) = 0.7 Å). As shown in Fig. 5C, the d-peptide shifts, in relation to p53(15–29), 3.8 Å toward the α2 helix of MDM2, accompanying a “close-in” movement of the opposite edge of the binding pocket. It is therefore inconceivable that RI-p53(15–29) would be able to structurally mimic the binding of p53(15–29) to MDM2 in the same handedness and with a similar conformation.
FIGURE 5.
The p53 binding domain of MDM2 in complex with peptide inhibitors. A, close-up view of the binding interface of DPMI-α and synMDM2 (Protein Data Bank code 3LNJ) (59). The electrostatic potential is displayed on the molecular surface of MDM2 and colored red for acidic, blue for basic, and white for apolar residues. The energetically most important residues, DTrp3, DLeu7, and DLeu11, are shown as red sticks. B, close-up view of the binding interface of p53(15–29) and MDM2(17–125) (Protein Data Bank code 1YCR) (14). C, superimposition of the overall structures of the synMDM2-DPMI-α and MDM2(17–125)-p53(15–29) complexes. synMDM2-DPMI-α is colored gray/cyan, whereas MDM2(17–125)-p53(15–29) is orange/yellow. The MDM2 molecules are depicted by ribbons, and the side chains of DTrp3 (Phe19), DLeu7 (Trp23), and DLeu11 (Leu26) are shown as ball-and-stick representations. D, close-up view of superimposed DPMI-α and p53(15–29) binding interfaces on a molecular surface of MDM2. DTyr4 and DLeu10 side chains are displayed together with the residues forming DTyr4 and DLeu10 binding pockets.
The change of helical peptide handedness alters protein-binding energetics as well. Occupying the same recognition pockets of MDM2, DTrp3, DLeu7, and DLeu11 of DPMI-α are topologically equivalent to Phe19, Trp23, and Leu26 of p53 (Fig. 5D). In p53 and p53-like peptides, Trp23, as a functional hot spot residue, contributes the greatest binding free energy to MDM2 association (69–71). In contrast, DPhe7 is most preferred at the equivalent position of DPMI-α, roughly 6-fold better than DTrp7 (59). Further, although the hydrophobic triad Phe19/Trp23/Leu26 of p53 dominates MDM2 recognition, energetic contributions to MDM2 binding are more evenly distributed among DTrp3, DTyr4, DLeu7, DGlu8, and DLeu11 of DPMI-α (Fig. 5D) (59). Importantly, despite the fact that DPMI-α and p53(15–29) or p53(17–28) had similar binding affinities for MDM2 (15, 59), the retro-inverso isomer of p53(17–28) (DEDPDLDLDKDWDLDDDSDFDTDE) or RI-p53(15–29) shares no sequence identity and only limited sequence similarity to DPMI-α (DTDNDWDYDADNDLDEDKDLDLDR). Assuming that RI-p53(17–28) or RI-p53(15–29) binds to MDM2 similarly to DPMI-α, the d-peptide isomer would be obligated to present contact residues significantly different from DTrp3, DTyr4, DLeu7, DGlu8, and DLeu11 of DPMI-α. In the two most probable modes of interaction with MDM2, these corresponding residues would be 1) DLeu, DLeu, DLeu, DAsp, and DThr or 2) DPro, DLeu, DTrp, DLeu, and DPhe. In either case, the binding of the retro-inverso peptide to MDM2 would be decimated due to less favorable subsite interactions. As was the case with RI-p53(15–29) and p53(15–29) structurally, it is equally implausible that these two peptides would be functionally equivalent.
It remains unclear why a stark discrepancy exists between our findings and the results reported by Sakurai et al. (11), who based their conclusion solely on an inhibition ELISA. The IC50 values of p53(15–29) and RI-p53(15–29) determined by Sakurai et al. (11) are 17.4 and 15.4 μm, respectively. Their IC50 value of p53(15–29) is significantly higher than ours (∼2 μm). Notably, the Kd values of wild type p53 peptides interacting with MDM2 have been determined by a number of laboratories using various (more accurate) biochemical and biophysical techniques. These values, including ours, generally fall between 60 to 700 nm (72, 73), depending on the method as well as the length of peptide/protein constructs. For example, Schon et al. (32) reported a Kd value of 220 nm for p53(15–29) and recombinant MDM2(2–125) using stopped-flow fluorescence spectroscopy and of 575 nm using isothermal titration calorimetry. Additional studies are warranted to clarify this important discrepancy for both conceptual and practical reasons.
In conclusion, although successful use of retro-inverso isomers as antigenic mimicry of their parent l-peptides is well documented, failed applications amply exist in the literature, reflecting known structural plasticity of antibody-antigen recognition. As molecular mimicry of biologically active peptide ligands of a helical nature, however, retro-inverso isomers are significantly inferior to their parent l-peptides. A clear limitation exists to the use of the retro-inverso strategy for peptide-based molecular design. Our findings, while contradicting an earlier report, are entirely consistent with the current understanding of how d-peptide ligands recognize native proteins at the molecular level.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grants AI072732 and AI061482 (to W. L.) and the NIH Intramural Research Program (to S. G. T.). This work was also supported by American Cancer Society Research Scholar Grant CDD112858 (to W. L.), the China Scholarship Council, and National Basic Research Program of China Grant 2007CB935800 (to C. L.).
The atomic coordinates and structure factors (code 3LRY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- HIV
- human immunodeficiency virus
- CA
- capsid protein
- CCA
- C-terminal domain of CA
- SH3
- Src homology 3
- HPLC
- high pressure liquid chromatography
- PBS
- phosphate-buffered saline
- TCEP
- tris(2-carboxyethyl)phosphine
- SPR
- surface plasmon resonance
- RU
- resonance unit
- ITC
- isothermal titration calorimetry
- ELISA
- enzyme-linked immunosorbent assay
- TFE
- trifluoroethanol
- RI
- retro-inverso isomer.
REFERENCES
- 1.Arkin M. R., Wells J. A. (2004) Nat. Rev. Drug Discov. 3, 301–317 [DOI] [PubMed] [Google Scholar]
- 2.Wells J. A., McClendon C. L. (2007) Nature 450, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 3.Vagner J., Qu H., Hruby V. J. (2008) Curr. Opin. Chem. Biol. 12, 292–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chorev M., Goodman M. (1979) Acc. Chem. Res. 12, 1–7 [Google Scholar]
- 5.Van Regenmortel M. H., Muller S. (1998) Curr. Opin. Biotechnol. 9, 377–382 [DOI] [PubMed] [Google Scholar]
- 6.Nair D. T., Kaur K. J., Singh K., Mukherjee P., Rajagopal D., George A., Bal V., Rath S., Rao K. V., Salunke D. M. (2003) J. Immunol. 170, 1362–1373 [DOI] [PubMed] [Google Scholar]
- 7.Guichard G., Benkirane N., Zeder-Lutz G., van Regenmortel M. H., Briand J. P., Muller S. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9765–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundberg E. J., Mariuzza R. A. (2002) Adv. Protein Chem. 61, 119–160 [DOI] [PubMed] [Google Scholar]
- 9.Pal-Bhowmick I., Pandey R. P., Jarori G. K., Kar S., Sahal D. (2007) Biochem. Biophys. Res. Commun. 364, 608–613 [DOI] [PubMed] [Google Scholar]
- 10.Rai J. (2007) Chem. Biol. Drug Des. 70, 552–556 [DOI] [PubMed] [Google Scholar]
- 11.Sakurai K., Chung H. S., Kahne D. (2004) J. Am. Chem. Soc. 126, 16288–16289 [DOI] [PubMed] [Google Scholar]
- 12.Richards F. M. (1963) Annu. Rev. Biochem. 32, 269–300 [DOI] [PubMed] [Google Scholar]
- 13.Brown C. J., Lain S., Verma C. S., Fersht A. R., Lane D. P. (2009) Nat. Rev. Cancer 9, 862–873 [DOI] [PubMed] [Google Scholar]
- 14.Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. (1996) Science 274, 948–953 [DOI] [PubMed] [Google Scholar]
- 15.Pazgier M., Liu M., Zou G., Yuan W., Li C., Li C., Li J., Monbo J., Zella D., Tarasov S. G., Lu W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4665–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santamaria F., Wu Z., Boulègue C., Pál G., Lu W. (2003) J. Mol. Recognit. 16, 131–138 [DOI] [PubMed] [Google Scholar]
- 17.Dawson P. E., Kent S. B. (2000) Annu. Rev. Biochem. 69, 923–960 [DOI] [PubMed] [Google Scholar]
- 18.Dawson P. E., Muir T. W., Clark-Lewis I., Kent S. B. (1994) Science 266, 776–779 [DOI] [PubMed] [Google Scholar]
- 19.Schnölzer M., Alewood P., Jones A., Alewood D., Kent S. B. (1992) Int. J. Pept. Protein Res. 40, 180–193 [DOI] [PubMed] [Google Scholar]
- 20.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piotto M., Saudek V., Sklenár V. (1992) J. Biomol. NMR 2, 661–665 [DOI] [PubMed] [Google Scholar]
- 22.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 23.Wuthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, Inc., New York [Google Scholar]
- 24.Keller R. (2004) The Computer Aided Resonance Assignment Tutorial, 1st Ed., CANTINA Verlag, Goldau, Switzerland [Google Scholar]
- 25.Herrmann T., Güntert P., Wüthrich K. (2002) J. Biomol. NMR 24, 171–189 [DOI] [PubMed] [Google Scholar]
- 26.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 29.Worthylake D. K., Wang H., Yoo S., Sundquist W. I., Hill C. P. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 85–92 [DOI] [PubMed] [Google Scholar]
- 30.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 31.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32.Schon O., Friedler A., Bycroft M., Freund S. M., Fersht A. R. (2002) J. Mol. Biol. 323, 491–501 [DOI] [PubMed] [Google Scholar]
- 33.Lai Z., Auger K. R., Manubay C. M., Copeland R. A. (2000) Arch. Biochem. Biophys. 381, 278–284 [DOI] [PubMed] [Google Scholar]
- 34.Wells M., Tidow H., Rutherford T. J., Markwick P., Jensen M. R., Mylonas E., Svergun D. I., Blackledge M., Fersht A. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C., Liu M., Monbo J., Zou G., Li C., Yuan W., Zella D., Lu W. Y., Lu W. (2008) J. Am. Chem. Soc. 130, 13546–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Z., Prahl A., Powell R., Ericksen B., Lubkowski J., Lu W. (2003) J. Pept. Res. 62, 53–62 [DOI] [PubMed] [Google Scholar]
- 37.Li C., Pazgier M., Liu M., Lu W. Y., Lu W. (2009) Angew Chem. Int. Ed. Engl. 48, 8712–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu B., Gilkes D. M., Chen J. (2007) Cancer Res. 67, 8810–8817 [DOI] [PubMed] [Google Scholar]
- 39.Hu B., Gilkes D. M., Farooqi B., Sebti S. M., Chen J. (2006) J. Biol. Chem. 281, 33030–33035 [DOI] [PubMed] [Google Scholar]
- 40.Phan J., Li Z., Kasprzak A., Li B., Sebti S., Guida W., Schönbrunn E., Chen J. (2010) J. Biol. Chem. 285, 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marine J. C., Dyer M. A., Jochemsen A. G. (2007) J. Cell Sci. 120, 371–378 [DOI] [PubMed] [Google Scholar]
- 42.Freed E. O. (1998) Virology 251, 1–15 [DOI] [PubMed] [Google Scholar]
- 43.Gamble T. R., Yoo S., Vajdos F. F., von Schwedler U. K., Worthylake D. K., Wang H., McCutcheon J. P., Sundquist W. I., Hill C. P. (1997) Science 278, 849–853 [DOI] [PubMed] [Google Scholar]
- 44.Dorfman T., Bukovsky A., Ohagen A., Höglund S., Göttlinger H. G. (1994) J. Virol. 68, 8180–8187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sticht J., Humbert M., Findlow S., Bodem J., Müller B., Dietrich U., Werner J., Kräusslich H. G. (2005) Nat. Struct. Mol. Biol. 12, 671–677 [DOI] [PubMed] [Google Scholar]
- 46.Ternois F., Sticht J., Duquerroy S., Kräusslich H. G., Rey F. A. (2005) Nat. Struct. Mol. Biol. 12, 678–682 [DOI] [PubMed] [Google Scholar]
- 47.Ganser-Pornillos B. K., Cheng A., Yeager M. (2007) Cell 131, 70–79 [DOI] [PubMed] [Google Scholar]
- 48.Cohen G. B., Ren R., Baltimore D. (1995) Cell 80, 237–248 [DOI] [PubMed] [Google Scholar]
- 49.Pawson T. (1995) Nature 373, 573–580 [DOI] [PubMed] [Google Scholar]
- 50.Dalgarno D. C., Botfield M. C., Rickles R. J. (1997) Biopolymers 43, 383–400 [DOI] [PubMed] [Google Scholar]
- 51.Mayer B. J. (2001) J. Cell Sci. 114, 1253–1263 [DOI] [PubMed] [Google Scholar]
- 52.Lim W. A., Richards F. M., Fox R. O. (1994) Nature 372, 375–379 [DOI] [PubMed] [Google Scholar]
- 53.Yu H., Chen J. K., Feng S., Dalgarno D. C., Brauer A. W., Schreiber S. L. (1994) Cell 76, 933–945 [DOI] [PubMed] [Google Scholar]
- 54.Pisabarro M. T., Serrano L. (1996) Biochemistry 35, 10634–10640 [DOI] [PubMed] [Google Scholar]
- 55.Schumacher T. N., Mayr L. M., Minor D. L., Jr., Milhollen M. A., Burgess M. W., Kim P. S. (1996) Science 271, 1854–1857 [DOI] [PubMed] [Google Scholar]
- 56.Eckert D. M., Malashkevich V. N., Hong L. H., Carr P. A., Kim P. S. (1999) Cell 99, 103–115 [DOI] [PubMed] [Google Scholar]
- 57.Wiesehan K., Buder K., Linke R. P., Patt S., Stoldt M., Unger E., Schmitt B., Bucci E., Willbold D. (2003) Chembiochem 4, 748–753 [DOI] [PubMed] [Google Scholar]
- 58.Welch B. D., VanDemark A. P., Heroux A., Hill C. P., Kay M. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16828–16833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li C., Pazgier M., Liu M., Yuan W., Li C., Lu W. (2010) Angew Chem. Int. Ed. Engl., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toledo F., Wahl G. M. (2006) Nat. Rev. Cancer 6, 909–923 [DOI] [PubMed] [Google Scholar]
- 61.Vousden K. H., Lane D. P. (2007) Nat. Rev. Mol. Cell Biol. 8, 275–283 [DOI] [PubMed] [Google Scholar]
- 62.Lane D. P. (1992) Nature 358, 15–16 [DOI] [PubMed] [Google Scholar]
- 63.Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 64.Shangary S., Qin D., McEachern D., Liu M., Miller R. S., Qiu S., Nikolovska-Coleska Z., Ding K., Wang G., Chen J., Bernard D., Zhang J., Lu Y., Gu Q., Shah R. B., Pienta K. J., Ling X., Kang S., Guo M., Sun Y., Yang D., Wang S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3933–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murray J. K., Gellman S. H. (2007) Biopolymers 88, 657–686 [DOI] [PubMed] [Google Scholar]
- 66.Fischer P. M. (2003) Curr. Protein Pept. Sci. 4, 339–356 [DOI] [PubMed] [Google Scholar]
- 67.Nanda V., DeGrado W. F. (2006) J. Am. Chem. Soc. 128, 809–816 [DOI] [PubMed] [Google Scholar]
- 68.Sia S. K., Kim P. S. (2001) Biochemistry 40, 8981–8989 [DOI] [PubMed] [Google Scholar]
- 69.Böttger A., Böttger V., Garcia-Echeverria C., Chène P., Hochkeppel H. K., Sampson W., Ang K., Howard S. F., Picksley S. M., Lane D. P. (1997) J. Mol. Biol. 269, 744–756 [DOI] [PubMed] [Google Scholar]
- 70.Kortemme T., Baker D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14116–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Massova I., Kollman P. A. (1999) J. Am. Chem. Soc. 121, 8133–8143 [Google Scholar]
- 72.Chène P. (2003) Nat. Rev. Cancer 3, 102–109 [DOI] [PubMed] [Google Scholar]
- 73.Li C., Pazgier M., Li C., Yuan W., Liu M., Wei G., Lu W.-Y., Lu W. (2010) J. Mol. Biol. 398, 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.