Abstract
Human intestinal macrophages contribute to tissue homeostasis in noninflamed mucosa through profound down-regulation of pro-inflammatory cytokine release. Here, we show that this down-regulation extends to Toll-like receptor (TLR)-induced cytokine release, as intestinal macrophages expressed TLR3–TLR9 but did not release cytokines in response to TLR-specific ligands. Likely contributing to this unique functional profile, intestinal macrophages expressed markedly down-regulated adapter proteins MyD88 and Toll interleukin receptor 1 domain-containing adapter-inducing interferon β, which together mediate all TLR MyD88-dependent and -independent NF-κB signaling, did not phosphorylate NF-κB p65 or Smad-induced IκBα, and did not translocate NF-κB into the nucleus. Importantly, transforming growth factor-β released from intestinal extracellular matrix (stroma) induced identical down-regulation in the NF-κB signaling and function of blood monocytes, the exclusive source of intestinal macrophages. Our findings implicate stromal transforming growth factor-β-induced dysregulation of NF-κB proteins and Smad signaling in the differentiation of pro-inflammatory blood monocytes into noninflammatory intestinal macrophages.
Keywords: Cytokines/TGFbeta, Development Differentiation/Tissue, Immunology/Innate Immunity, Immunology/LPS, Immunology/Toll Receptors, Signal Transduction, Intestine, Mucosal
Introduction
The mucosa of the normal human small intestine is remarkable for the absence of inflammation, despite constant exposure to immunostimulatory bacteria and bacterial products. To elucidate the homeostatic mechanisms underlying the absence of intestinal inflammation, we have shown that locally produced IL-83 and TGF-β recruit pro-inflammatory blood monocytes to the lamina propria (1), where extracellular matrix (stromal) TGF-β, and possibly other mucosal factors, induce rapid differentiation of the newly arrived monocytes into intestinal macrophages incapable of inducible cytokine release (1–3). Consistent with their noninflammatory profile (4), intestinal macrophages also do not express many innate response receptors (2, 3, 5) and are down-regulated for chemokine receptors (6–8), triggering receptor expressed on monocytes-1 (TREM-1) (9, 10) and co-stimulatory molecules (3, 11). However, intestinal macrophages retain avid phagocytic and bacteriocidal activity (2, 3, 5), but even phagocytosis of Gram-negative bacteria does not induce pro-inflammatory cytokine release (3). Thus, lamina propria macrophages in the human small intestine are unique among mononuclear phagocytes for the presence of inflammation anergy, a feature that likely evolved to attenuate bacterially induced inflammation in the intestinal mucosa.
The mechanisms underlying the inflammation anergy of intestinal macrophages in normal mucosa are not known. Here, we show that a high proportion of intestinal macrophages displayed TLR3, TLR5, and TLR7–9, and a small proportion expressed TLR4 and TLR6 but did not release pro-inflammatory cytokines in response to the corresponding TLR ligand. Moreover, intestinal macrophages did not express detectable MD-2 and could not respond to rough LPS, even in the presence of MD-2 expression via gene transfection, predicting a downstream block in the signal transduction pathways that regulate cytokine gene transcription. Confirming this prediction, intestinal macrophages expressed barely detectable levels of key proteins in the MyD88-dependent and -independent NF-κB signal pathway and expressed Smad-induced IκBα, the negative regulator of NF-κB, but they did not phosphorylate IκBα or NF-κB and did not translocate NF-κB into the nucleus. Furthermore, intestinal stroma-associated TGF-β profoundly inhibited NF-κB signaling and TLR-mediated cytokine release in blood monocytes, the exclusive source of intestinal macrophages. These findings indicate a mechanism in which stromal TGF-β-induced Smad signaling and dysregulation of NF-κB signal proteins mediate the differentiation of pro-inflammatory blood monocytes into inflammation anergic intestinal macrophages.
EXPERIMENTAL PROCEDURES
Intestinal Macrophages and Blood Monocytes
Macrophages were isolated from intestinal mucosa of subjects undergoing gastrojejunostomy for obesity or healthy organ transplantation donors by enzyme digestion and purified by counterflow centrifugal elutriation, as described previously (5, 12, 13). Blood monocytes from the same donors were purified by elutriation and subjected to the same isolation protocol so that the monocytes and intestinal macrophages were treated similarly. Macrophages and monocytes were routinely >98% pure, 98% viable by propidium iodide staining, and avidly phagocytic, similar to our previously characterized intestinal macrophages (1–3, 5). Human umbilical vein endothelial cells were obtained from American Type Culture Collection. All studies were undertaken with Internal Review Board approval.
Stroma-conditioned Media (S-CM)
Cell-depleted lamina propria extracellular matrix (stroma) was prepared as described previously (1, 3, 14).
Transfection
Using the human macrophage nucleofector kit (Amaxa), intestinal macrophages (7 × 105/100 μl of nucleofector solution) were transfected with 2 μg of pDUO-hMD-2/TLR4 (InvivoGen) according to the manufacturer's instructions. After 24 h of culture, the expression of MD-2 and TLR4 was analyzed by flow cytometry.
Flow Cytometry
Cells (2 × 106) were stained with phycoerythrin-, allophycocyanin-, phycoerythrin-cyanin 5 (PE-Cy5)-, or FITC-conjugated monoclonal antibodies to CD13, CD14, HLA-DR (BD Biosciences), TLR1–9 (eBiosciences), and MD-2 (Santa Cruz Biotechnology) or control monoclonal antibodies of the same isotype and fluorochrome and analyzed by flow cytometry. For intracellular staining, cells were permeabilized prior to staining with the relevant antibodies and matching control monoclonal antibodies. Data were evaluated using CellQuest software (BD Biosciences).
Cytokine Production
Supernatants of cells (2 × 106/ml) cultured for 24 h in the absence or presence of rough LPS (Salmonella minnesota R595; Alexis), smooth LPS (Salmonella abortus equi; Alexis), soluble CD14 (Alexis), or TLR1–9 ligands (Invitrogen) in the absence or presence of IFN-γ, anti-IL-10, or anti-IL-4 antibodies (R & D Systems) were analyzed for TNF-α, IL-1β, IL-6, IL-8, and IL-10 and RANTES by ELISA (R & D Systems). TLR ligands included the following: TLR1, Pam3CSK4 (0.2 μg/ml); TLR2, heat-killed Listeria monocytogenes (5 × 108 cells/ml); TLR3, poly(1:C) (25 μg/ml); TLR4, LPS (1 μg/ml); TLR5, Salmonella typhimurium flagellin (100 ng/ml); TLR6, FLS1 (Pam2CaDPKH PKSF) (10 ng/ml); TLR7, imiquimod (0.5 μg/ml); TLR8, SSRNA40 (0.5 μg/ml); and TLR9, ODN2006 (5 μm).
Real Time PCR
Intestinal macrophages and autologous blood monocytes (1 × 106 cells/ml) stimulated for 2 h (for IL-1, IL-6, IL-8, and TNF-α mRNA) or 12 h (for IL-10 and TGF-β mRNA) with smooth LPS (1 μg/ml) were harvested, and RNA was isolated (QIA RNeasy kit, Qiagen), and cDNA was generated from total RNA (transcriptor first strand cDNA synthesis kit, Roche Applied Science). Target genes were amplified in 25-μl mixtures containing TaqMan Universal PCR master mix and the following 6-carboxyfluorescein/dihydrocyclopyrroloindole tripeptide minor groove binder (FAM/MGB)-labeled primer-probe sets: IL-1β (assay ID Hs00174097_m1, Ref. Seq. NM_000576.2), IL-6 (Hs00985639_m1, Ref. Seq. NM_000600.2), IL-8 (Hs00174103_m1, Ref. Seq. NM_000584.2), and TNF-α (Hs00174128_m1, Ref. Seq. NM_000594.2), all from Applied Biosystems. All primers were chosen to span exon junctions and to exclude nonspecific amplification of genomic DNA. Expression of endogenous control gene 18 S rRNA (Ref. Seq. X03205.1) or glyceraldehyde-3-phosphate dehydrogenase (Ref. Seq. NM_002046.3) was determined simultaneously with VICTM/6-carboxytetramethylrhodamine (VIC/TAMRA)-labeled primer probe sets in a multiplex PCR setup. Real time PCR was run for 40 cycles (15 s at 95 °C, 60 s at 60 °C) on a Chromo4 PCR system (Bio-Rad) and analyzed with Opticon MonitorTM software, version 3.1. Relative expression rates of target genes in unstimulated versus stimulated cells were calculated using the method of Pfaffl (15). All PCRs were performed twice, once with each reference gene, and data are presented as the geometric mean of both reactions.
Microarray Analysis
Total cellular RNA was extracted (RNeasy kit, Qiagen) (16) from intestinal macrophages, and autologous blood monocytes from two donors and cDNA were synthesized (Superscript Choice System, Invitrogen) utilizing an oligo(dT)24 primer. Biotinylated cRNA was synthesized using a BioArray HighYield RNA transcription labeling kit (ENZO Diagnostics) and purified through RNeasy nucleic acid columns. cRNA quality was evaluated by hybridization to Test3 GeneChips (Affymetrix), and only samples whose 3′:5′ ratios were less than three were utilized for subsequent hybridization to HuGene U95-AV2 GeneChips (Affymetrix). After scanning, fluorescence data were processed by the GeneChip operating system (version 1.1, Affymetrix). Background correction, normalization, generation of expression values, and analysis of differential gene expression were performed using dChip analysis software (DNA-Chip analyzer (dChip), version 1.3, Harvard University) in compliance with minimal information about microarray experiment (MIAME) guidelines (www.ncbi.nlm.nih.gov). Fold differences were calculated by comparing the fluorescence intensities of each probe set per gene on the array for intestinal macrophages to blood monocytes. A fold difference ≥2.0 plus p ≤ 0.05 was considered significant. The Affymetrix GeneChip Operating System data files (*.cel. and *.chp) have been deposited in the Gene Expression Omnibus data base (www.ncbi.nlm.nih.gov).
NF-κB Activation
Phosphorylation of NF-κB p65 and IκBα
Cells (1 × 106) were incubated (37 °C) with smooth LPS (1 μg/ml), and at the indicated time cold PBS was added, and the cells were washed, fixed in 1% paraformaldehyde, 0.2% saponin (Cytofix/Cytoperm, BD Biosciences), stained with anti-p-NF-κB p65-FITC or anti-p-IκBα-FITC (Santa Cruz Biotechnology) or control antibodies, and analyzed by flow cytometry. Data were evaluated with CellQuest.
NF-κB p50 ELISA
Nuclear extracts were prepared from 10 × 106 cells treated with smooth LPS (1 μg/ml) at 37 °C using the NE-PER kit (Nuclear and Cytoplasmic Extraction Reagents, Pierce). NF-κB DNA binding was detected using the NF-κB transcription factor ELISA (Panomics, Inc.), based on the ability of activated NF-κB p50 to bind an NF-κB consensus binding site on a biotinylated oligonucleotide immobilized on streptavidin-coated wells in a 96-well plate. Bound NF-κB was detected by anti-NF-κB antibody (R & D Systems), and the signal was quantified by horseradish peroxidase-tetramethylbenzidine (HRP-TMB) binding at 450 nm using an EL800 ELISA reader (Biotek Instruments, Inc.).
Immunocytochemistry for NF-κB p65
Cells were incubated in the presence or absence of S-CM (500 μg of protein/ml) for 1 h at 37 °C, exposed to smooth LPS (1 μg/ml) for 1 h, fixed and permeabilized (20 min with Cytofix/Cytoperm, BD Biosciences), and washed (Cytoperm Buffer, BD Biosciences). After rinsing, cells were blocked with casein protein (DAKO) for 1 h and incubated with rabbit anti-NF-κB p65 antibodies (0.05 mg/ml) for 90 min (Santa Cruz Biotechnology) or irrelevant antibody. Cells were washed, incubated with donkey anti-rabbit IgG-FITC (1:500; Jackson ImmunoResearch) for 30 min, washed with PBS, and counterstained with 4′,6-diamidino-2-phenylindole (5 min) for the identification of nuclei. Cells were visualized by confocal microscopy, and the cytoplasmic and nuclear fluorescence intensity of NF-κB was converted to histograms using IPLab software version 3.6 (BD Biosciences Bioimaging).
Western Blot
Cells (10 × 106/ml) were pelleted, resuspended in cold RIPA buffer (Santa Cruz Biotechnology), homogenized, incubated for 30 min, and supernatants collected by centrifugation, and protein levels were determined. Extracts were diluted in SDS sample buffer (Bio-Rad) and boiled for 3 min, and proteins were separated by acrylamide (12.5%) gel electrophoresis and transferred to nitrocellulose membranes (Bio-Rad). Blots were blocked with PBS, 5% bovine serum albumin, 0.05% Tween 20 (1 h at room temperature) and incubated with antibodies to MyD88, TRIF/TICAM, TRAF6, IκBα, p-IκBα, Smad2, p-Smad2, Smad4, or Smad7 (Santa Cruz Biotechnology) 1:2000 to 1:10,000 in PBS, 0.05% Tween 20 overnight at 4 °C. After washing with PBS/Tween 20, blots were probed with horseradish peroxidase-conjugated secondary antibodies (1:50,000 in PBS, 0.05% Tween 20; 1 h at room temperature) and bands detected using the chemiluminescent method (SuperSignal West Dura, Thermo Scientific). Blots were stripped and reprobed with anti-actin or anti-p53 antibodies (Calbiochem and kind gift of Dr. E. Benveniste, University of Alabama at Birmingham) to ensure equal loading on all gels.
Statistical Analysis
Data were analyzed by t test/Mann-Whitney U test, where appropriate. Data are expressed as mean ± S.E. unless stated otherwise.
RESULTS
Intestinal Macrophages Do Not Respond to Smooth or Rough LPS
Mononuclear phagocytes interact with LPS through CD14 and TLR4. CD14, which is present in membrane-bound and soluble forms, does not transduce the LPS signal, whereas TLR4 in association with the secreted protein MD-2 (TLR4-MD-2) does (17). Smooth LPS, which contains the O-antigen side chain composed of oligosaccharide subunits, binds TLR4-MD-2 only in the presence of CD14 (CD14-dependent), leading to NF-κB activation and pro-inflammatory cytokine production (18). In contrast, rough LPS, which lacks the O-antigen, can bind directly to TLR4-MD-2 and induces NF-κB activation through both CD14-independent and -dependent binding.
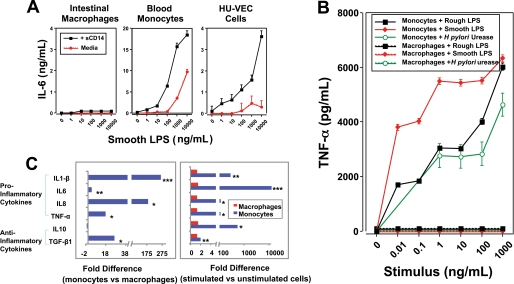
We first investigated the response of intestinal macrophages to smooth LPS. Intestinal macrophages do not express CD14 (2, 3, 5), and the addition of sCD14 to cultures of intestinal macrophages did not induce the cells to produce IL-6 in the presence of smooth LPS (Fig. 1A). However, sCD14 enhanced inducible cytokine release by monocytes and was required for cytokine release by human umbilical vein endothelial cells (Fig. 1A). Intestinal macrophages, unlike autologous monocytes, exposed to increasing concentrations of rough LPS also produced no detectable TNF-α (Fig. 1B). Helicobacter pylori urease, which we have shown induces cytokine production by monocytes through an LPS-independent mechanism (19), also induced no detectable TNF-α from intestinal macrophages but induced abundant TNF-α from blood monocytes (Fig. 1B). As an additional control, we also determined the ability of monocyte-derived macrophages cultured for 1, 2, and 5 days to release TNF-α after stimulation with rough LPS, smooth LPS, or flagellin (the ligand for TLR5). Monocyte-derived macrophages released progressively lower levels of TNF-α over this time course, but even on day 5 the cells released large amounts of inducible TNF-α compared with unstimulated cells (p < 0.01) (supplemental Fig. 1). Thus, monocyte-derived macrophages do not acquire the inflammation anergy characteristic of intestinal macrophages, even after 5 days of culture, unlike monocytes cultured in the presence of normal S-CM over the same period (3). Consistent with these findings, untreated intestinal macrophages spontaneously expressed manyfold lower levels of the pro-inflammatory cytokines IL-1β (265-fold; p < 0.01), IL-6 (3.7-fold; p < 0.09), IL-8 (182-fold; p < 0.03), TNF-α (17-fold; p < 0.24), and the anti-inflammatory cytokine TGF-β (26-fold; p < 0.03), but expressed equivalent levels of IL-10 (p < 0.05), compared with untreated monocytes, from the same donors (Fig. 1C, left panel). In addition, LPS-treated intestinal macrophages displayed a profound inability to up-regulate cytokine gene expression for IL-1β (p < 0.02), IL-6 (p < 0.001), IL-8 (p < 0.05), TNF-α (p < 0.02), IL-10 (p < 0.03), and TGF-β (p < 0.003) compared with LPS-stimulated monocytes (Fig. 1C, right panel). Thus, intestinal macrophages do not respond to either smooth LPS, even in the presence of sCD14, or rough LPS, indicating that intestinal macrophages either do not express TLR4 and/or MD-2 or have aberrant downstream signaling in the NF-κB activation pathway.
FIGURE 1.
Intestinal macrophages are down-regulated for pro-inflammatory cytokine release. A, intestinal macrophage LPS responsiveness is not restored in the presence of sCD14. Intestinal macrophages (2 × 106/ml) incubated with increasing concentrations of smooth LPS in the presence or absence of sCD14 did not release detectable IL-6, but sCD14 potently enhanced IL-6 release from autologous LPS-stimulated blood monocytes and caused inducible IL-6 release from human umbilical vein endothelial cells (HU-VEC). B, intestinal macrophages do not release TNF-α in response to stimulation with LPS or H. pylori urease. Intestinal macrophages (2 × 106/ml) cultured with increasing concentrations of rough LPS, smooth LPS, or H. pylori urease did not release detectable TNF-α, in sharp contrast to autologous blood monocytes, which released high levels of TNF-α in response to each stimulus. C, intestinal macrophage cytokine mRNA levels are sharply reduced compared with the levels in blood monocytes. Cytokine mRNA levels, determined by Affymetrix gene array analysis, were significantly lower in resting intestinal macrophages compared with autologous, unstimulated monocytes (C, left panel); mRNA levels did not increase in intestinal macrophages following LPS treatment but increased sharply in LPS-stimulated blood monocytes (C, right panel). Data are from representative experiments (n = 3) (***, p < 0.001; **, p < 0.01; *, p < 0.05).
Intestinal Macrophages Express TLR4 but Not MD-2, and MD-2 Transfection Does Not Restore LPS Responsiveness
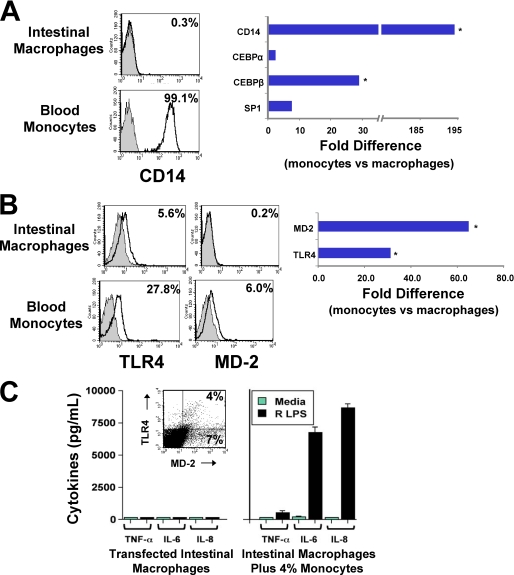
Consistent with our previous finding that intestinal macrophages do not express CD14 protein or mRNA (2, 3), intestinal macrophages expressed reduced mRNA levels of transcription factors involved in CD14 promoter activation, including CEPBα, CEPBβ (p < 0.03), and Sp1, compared with autologous blood monocytes (Fig. 2A). These findings implicate down-regulation of CD14 expression in intestinal macrophages at the level of the CD14 promoter. In addition, intestinal macrophages expressed low levels of TLR4 protein (Fig. 2B), in keeping with our earlier report that they express TLR4 transcripts (2), but did not express detectable MD-2 protein or mRNA (p < 0.01) (Fig. 2B), even after stimulation (p < 0.04) (data not shown). Importantly, monocyte-derived macrophages cultured for up to 5 days in vitro did not acquire the phenotype of intestinal macrophages (supplemental Fig. 1B). Moreover, TLR4-MD-2-transfected intestinal macrophages, 4% of which expressed both TLR4 and MD-2 (Fig. 2C, inset, and supplemental Fig. 2), did not release detectable levels of inducible TNF-α, IL-6, or IL-8 in response to rough LPS (Fig. 2C, left panel). In contrast, control intestinal macrophages spiked with 4% monocytes (as well as HEK293 cells transfected to express TLR4 and MD-2, data not shown) produced substantial levels of cytokine following LPS exposure (Fig. 2C, right panel).
FIGURE 2.
MD-2 gene transfection does not restore LPS-responsiveness to intestinal macrophages. Intestinal macrophages, which are exclusively CD14−, expressed markedly reduced levels of the transcription factors involved in activation of the CD14 promoter (CEBPα, CEBPβ, and Sp1) (A) and low levels of TLR4 protein and no detectable MD-2 protein or MD-2 mRNA (B). MD-2-transfected intestinal macrophages (C, inset) did not release TNF-α, IL-6, or IL-8 after culture for 24 h with rough (R) LPS (1 μg/ml) (C, left panel), whereas a mixture of intestinal macrophages plus 4% blood monocytes released high levels of inducible TNF-α, IL-6, and IL-8 (C, right panel) (values represent mean ± S.E. of triplicate wells). Data are from representative experiments (n = 3) (*, p < 0.005).
Intestinal Macrophages Express TLRs but Do Not Release Inflammatory Cytokines after Exposure to TLR Ligands
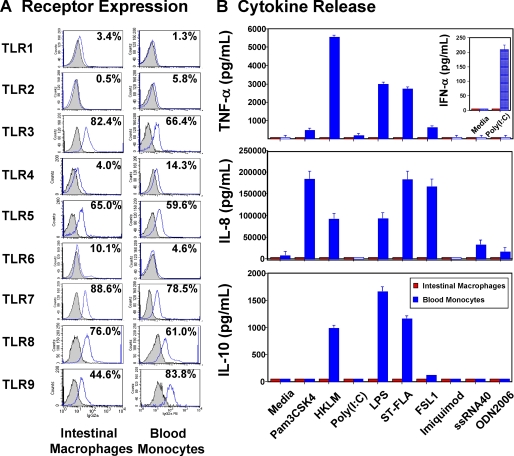
To establish whether the inflammation anergy of intestinal macrophages extends to other TLR-specific ligands, we characterized primary intestinal macrophages for TLR expression and TLR-mediated cytokine responses. Intestinal macrophages expressed TLR3 and TLR5–9 comparable with (or greater than) the expression levels for autologous blood monocytes (Fig. 3A and Table 1), yet did not release detectable TNF-α, IFN-α, IL-8, and IL-10 (Fig. 3B) or IL-1, IL-6, or IFN-β in response to TLR-specific ligands in the presence or absence of IFN-γ, anti-IL-10, or anti-IL-4 antibodies (data not shown). In sharp contrast, autologous blood monocytes stimulated with the same ligands released large amounts of TNF-α, IFN-α via poly(I-C) stimulation of TLR3 (Fig. 3B, upper panel inset), IL-8, and IL-10 (Fig. 3B), as well as IL-1 and IL-6 (data not shown). Thus, compared with blood monocytes, intestinal macrophages are profoundly and globally down-regulated for the production of an array of pro-inflammatory and regulatory cytokines. Because TLR1–9 (excluding TLR3) share the MyD88-dependent signaling pathway, whereas TLR3 and TLR4 signal through a MyD88-independent pathway involving the adapter protein TRIF, we next determined whether the inflammation anergy of intestinal macrophages is due to dysregulated NF-κB activation pathways.
FIGURE 3.
Intestinal macrophages express TLRs but do not respond to TLR ligation. A, intestinal macrophages and blood monocytes analyzed by fluorescence-activated cell sorter expressed TLR1 and TLR3–9 but not TLR2. Data shown are from one representative experiment (n = 6). B, blood monocytes but not intestinal macrophages (2 × 106/ml) incubated 18 h with TLR ligands produced TNF-α, IFN-α, IL-8, and IL-10. Values represent mean ± S.E. of triplicate wells (n = 4).
TABLE 1.
TLR expression by human intestinal macrophages and autologous blood monocytes
| TLR | Intestinal macrophages |
Blood monocytes |
p | ||
|---|---|---|---|---|---|
| Meana | S.E. | Mean | S.E. | ||
| TLR1 | 1.42 | 0.48 | 1.15 | 0.03 | 0.29 |
| TLR2 | 1.08 | 0.048 | 6.35 | 1.72 | 0.001 |
| TLR3 | 79.92 | 9 | 67.1 | 11.28 | 0.165 |
| TLR4 | 5.8 | 1.76 | 21.3 | 4.32 | 0.0001 |
| TLR5 | 45.2 | 12.28 | 48.1 | 10.68 | 0.422 |
| TLR6 | 17.2 | 4.12 | 6.3 | 2.52 | 0.022 |
| TLR7 | 39.1 | 15.8 | 93.1 | 4.96 | 0.015 |
| TLR8 | 63.9 | 18.9 | 58.1 | 13.5 | 0.405 |
| TLR9 | 59.3 | 13.28 | 71.88 | 15.32 | 0.242 |
a Mean percentage of positive cells (n = 6) is shown.
Intestinal Macrophages Exhibit Dysregulated NF-κB and Smad Signaling
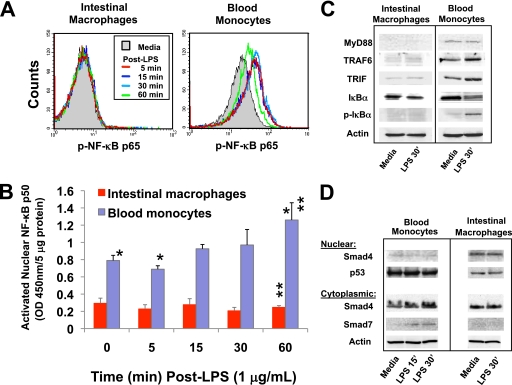
To determine whether the inability of intestinal macrophages to release inducible cytokines was due to ineffective translocation of NF-κB into the nucleus where it binds to target promoters, we first examined intestinal macrophages and autologous blood monocytes for phosphorylated NF-κB (p-NF-κB p65). Intestinal macrophages did not express constitutive or LPS-inducible p-NF-κB p65 (Fig. 4A). In contrast, NF-κB was rapidly phosphorylated in LPS-stimulated blood monocytes, achieving within 5 min maximal expression that was maintained for up to 30 min (Fig. 4A). We next examined intestinal macrophages and autologous blood monocytes for the ability to translocate cytoplasmic NF-κB into the nucleus. Cells were treated with LPS, and cytoplasmic and nuclear preparations were monitored for p50. Blood monocytes displayed a progressive increase in nuclear NF-κB p50, whereas intestinal macrophage nuclear levels of NF-κB p50 did not change after exposure to LPS (Fig. 4B). Moreover, nuclear NF-κB p50 levels were significantly lower in intestinal macrophages compared with blood monocytes at each time point (p < 0.006). These findings indicate that intestinal macrophages neither activate NF-κB nor translocate NF-κB into the nucleus in response to bacterial LPS.
FIGURE 4.
Intestinal macrophages do not phosphorylate NF-κB p65 or translocate NF-κB p50 into the nucleus. A, intestinal macrophages and autologous blood monocytes were stimulated with smooth LPS (1 μg/ml) for 5, 15, 30, or 60 min, and NF-κB p65 phosphorylation was assessed by flow cytometry. B, nuclear transport of NF-κB p50 was determined by comparison of NF-κB p50 levels in nuclear cell extracts by ELISA after exposure to LPS (1 μg/ml). Levels of nuclear NF-κB p50 were significantly lower in intestinal macrophages than blood monocytes at each time point, p < 0.006. C, intestinal macrophages and autologous blood monocytes (10 × 106/ml) were incubated for 0 or 30 min with smooth LPS (1 μg/ml), and extracts were analyzed for MyD88-dependent and -independent NF-κB from signal proteins by Western blot (A). D, macrophages were also analyzed for Smad signal proteins by Western blot. Data are from a representative experiment (n = 3) (**, p < 0.01; *, p < 0.05).
We next investigated whether the inability of intestinal macrophages to activate NF-κB was due to dysfunctional elements in the NF-κB activation pathway. Intestinal macrophages did not express or expressed barely detectable levels of the adapter proteins MyD88 and TRIF and the MyD88 downstream target TRAF6 in the absence or presence of LPS, in sharp contrast to the strong constitutive expression of these proteins in autologous monocytes (Fig. 4C). Importantly, intestinal macrophages expressed but did not phosphorylate IκBα (Fig. 4C), the negative regulator of NF-κB, whereas monocytes constitutively expressed and rapidly phosphorylated IκBα, reflected in strong p-IκBα expression within 30 min of LPS exposure (Fig. 4C). Thus, intestinal macrophages are incapable of transducing a signal through the NF-κB pathway.
Having previously shown that intestinal macrophages express TGF-β receptors I and II (1) and that stromal TGF-β differentiates blood monocytes into cells with the phenotype and function profile of noninflammatory intestinal macrophages (3), we examined intestinal macrophages for Smad proteins, the TGF-β receptor substrates that propagate the TGF-β signal. Smad4, a critical component of the TGF-β signal cascade that associates with the phosphorylated heterodimeric Smad2/3 complex and then translocates into the nucleus to initiate gene transcription, was present in the cytoplasm of both intestinal macrophages and blood monocytes and in the nuclei of freshly isolated unstimulated intestinal macrophages (Fig. 4D), consistent with active TGF-β signaling in intestinal macrophages. In addition, intestinal macrophages did not express cytoplasmic Smad7 (Fig. 4D), the inhibitor of Smad2/3 phosphorylation (20, 21), thereby preventing the nuclear translocation of the Smad2/3-Smad4 complex. The nuclear control p53 was also present at reduced levels in nuclear preparations from intestinal macrophages compared with blood monocytes despite equal protein loading. In this context, p53 mRNA expression was reduced in intestinal macrophages 2.5-fold compared with blood monocytes (data not shown). In contrast, unstimulated blood monocytes expressed barely detectable Smad4 and low levels of Smad7 after stimulation with LPS (Fig. 4D). The expression of nuclear Smad4, but not cytoplasmic Smad7, indicated constitutive TGF-β signaling in human intestinal macrophages.
Genes for MyD88 Signal Pathway Proteins Are Potently Down-regulated in Intestinal Macrophages
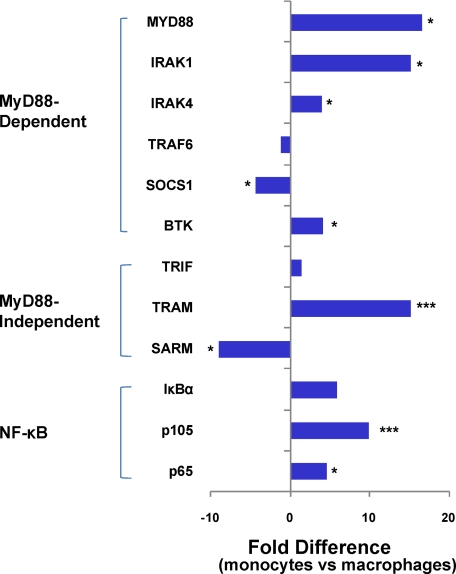
Intestinal macrophages displayed profoundly reduced mRNA levels for other MyD88-dependent signal proteins essential for TLR-mediated NF-κB activation, including MyD88 (p < 0.03), IRAK1 (p < 0.02), IRAK4 (p < 0.05), NF-κB p105 (p < 0.02), and NF-κB p65 (p < 0.001) (Fig. 5). The reduced mRNA expression of NF-κB p105 (and RelB) concurs with the significantly reduced level of NF-κB p50 protein reported here (Fig. 4B). In addition, intestinal macrophages expressed reduced levels of mRNA for IRF2 (p < 0.03), but mRNA levels for IRF1 and IRF3–7 were unchanged compared with autologous monocytes (supplemental Table 1). In addition, mRNA expression of the inhibitor suppressor of cytokine signaling 1 (SOCS1) (22) is up-regulated (p < 0.03) and that of Bruton agammaglobulinemia tyrosine kinase, which promotes TLR2 and TLR4 signaling (23) and NF-κB p65 phosphorylation (24), is reduced (p < 0.05) in intestinal macrophages compared with that of blood monocytes (25). These findings suggest that the inability of intestinal macrophages to activate NF-κB is dysregulated transcriptionally at multiple steps in the MyD88-dependent NF-κB signal pathway. Also, mRNA expression for TRIF was not lower in intestinal macrophages compared with blood monocytes, but the level of TRIF-related adapter molecule mRNA was sharply reduced (p < 0.001) and that of sterile and Armadillo motif-containing protein (SARM), the inhibitor of TRIF/TRIF-related adapter molecule MyD88-independent signaling (26), was markedly increased (p < 0.04) in the macrophages (Fig. 5). These findings suggest that the inability of intestinal macrophages to release IFN-α and TNF-α through TLR3- and TLR4-mediated MyD88-independent TRIF signaling may be dysregulated transcriptionally and through the expression of the SARM inhibitor. Importantly, mRNA levels for IκBα, although lower than in blood monocytes, were not significantly reduced (p < 0.29) (Fig. 5), which is consistent with our finding that intestinal macrophages constitutively express IκBα (Fig. 4).
FIGURE 5.
Gene expression for MyD88 signal pathway proteins in intestinal macrophages. Affymetrix gene array analysis showed undetectable or markedly reduced levels of mRNA for MyD88, the NF-κB components (p105 and p65), and Bruton agammaglobulinemia tyrosine kinase, IRAK1, and IRAK4 genes in intestinal macrophages compared with autologous blood monocytes but increased mRNA levels for SARM, SOCS1, and TRAF6 in the macrophages (***, p < 0.001; *, p < 0.05).
Stroma-derived TGF-β Dysregulates NF-κB Nuclear Transport and IκBα Phosphorylation in Blood Monocytes
Stromal factors, including TGF-β, down-regulate monocyte cytokine production, inducing the inflammation anergy characteristic of intestinal macrophages (3). We now show that stromal products also down-regulate monocyte cytokine (TNF-α and RANTES) release in response to TLR-specific ligands (Fig. 6A). Because NF-κB inactivation is the likely cause of the inflammation anergy in intestinal macrophages, and blood monocytes are the exclusive source of intestinal macrophages (1), we investigated whether stromal TGF-β inactivates NF-κB in blood monocytes. Confocal microscopy showed that NF-κB p65 was located predominantly in the cytoplasm in unstimulated monocytes, in the nuclei of LPS-stimulated monocytes (Fig. 6B, upper panels), and in the presence of S-CM alone (supplemental Fig. 3A); moreover, when the cells were preincubated with S-CM, LPS-stimulated NF-κB translocation into the nuclei was almost completely blocked (Fig. 6B, upper panels). Fluorescence intensity analysis confirmed that NF-κB distributed primarily to the cytoplasm in unstimulated cells and the nucleus in LPS-stimulated cells and remained predominantly in the cytoplasm in stimulated cells pretreated with S-CM (Fig. 6B, lower panels). Consistent with this interpretation, NF-κB remained exclusively in the cytoplasm of intestinal macrophages in each condition (Fig. 6B, insets). S-CM blockade of the nuclear translocation of NF-κB was independent of the stimulus (macrophage-colony-stimulating factor (M-CSF), LPS, H. pylori urease, and IFN-γ), and importantly, S-CM-mediated inhibition of nuclear translocation was reversed when the S-CM was preincubated with anti-TGF-β antibodies (Fig. 6C) but not inhibited when S-CM was preincubated with irrelevant antibody (supplemental Fig. 3B). These findings indicate that among the stromal products, TGF-β is critical for the down-regulation of NF-κB activation. In addition, fluorescence-activated cell sorter analysis of isolated cells showed that intestinal macrophages did not phosphorylate IκBα and that preincubation of monocytes with S-CM inhibited the ability of the cells to phosphorylate IκBα (Fig. 6D). A similar inhibition was observed by Western blot analysis of cell extracts, and this inhibition was reversed when the S-CM was preincubated with anti-TGF-β antibodies, further implicating stroma-derived TGF-β in the dysregulated nuclear translocation of NF-κB in intestinal macrophages. S-CM also induced an increase in inducible IκBα expression in monocytes that was partially reversed by TGF-β antibodies (Fig. 6E). Thus, stromal TGF-β inhibition of IκBα phosphorylation, leading to increased levels of the cytoplasmic NF-κB inhibitor IκBα, indicates an additional mechanism for the stromal inactivation of NF-κB in monocytes newly recruited to the mucosa. Finally, both intestinal macrophages and blood monocytes expressed, but did not phosphorylate, Smad2. However, in the presence of S-CM alone or S-CM plus LPS, monocytes expressed phosphorylated Smad2. Furthermore, in the presence of S-CM preincubated with anti-TGF-β antibodies, this phosphorylation was abrogated, implicating a role for stroma-derived TGF-β in the down-regulation of monocyte pro-inflammatory function (Fig. 6E). These findings were corroborated by additional experiments with control monocytes and monocyte-derived macrophages treated with recombinant human TGF-β.
FIGURE 6.
Stromal factors down-regulate NF-κB translocation and IκBα phosphorylation in blood monocytes. A, intestinal macrophages and blood monocytes (2 × 106/ml), the latter cultured in the presence or absence of S-CM (500 μg protein/ml), were incubated with or without specific TLR ligands for 18 h, and culture supernatants were assayed for TNF-α and RANTES (inset). Values are the means ± S.E. (n = 4). B, blood monocytes and intestinal macrophages incubated in media alone, in media plus smooth LPS (1 μg/ml, 1 h), or treated with S-CM (500 μg/ml, 1 h) and then LPS were analyzed for NF-κB nuclear translocation by immunofluorescence and confocal microscopy (upper panels) and fluorescence intensity (lower panels: green line, NF-κB p65; blue line, nucleus). NF-κB localized predominantly to the cytoplasm of untreated monocytes (left panel) and translocated into the nucleus after LPS treatment (middle panel) but did not translocate when the cells were treated with S-CM prior to the LPS (right panel). NF-κB remained exclusively in the cytoplasm of intestinal macrophages (insets) in each condition. Histograms show distribution of NF-κB (green line) in relation to the nucleus (blue line). C, anti-TGF-β antibodies block S-CM down-regulation of stimulus-driven nuclear translocation of NF-κB in blood monocytes. Blood monocytes preincubated with M-CSF (10 ng/ml) (or LPS, H. pylori urease, or IFN-γ, data not shown) display nuclear NF-κB (left panel), whereas preincubation of M-CSF-pulsed monocytes with S-CM (100 μg/ml) inhibited nuclear translocation of NF-κB (middle panel). However, preincubation of M-CSF-pulsed monocytes with S-CM plus anti-TGF-β antibodies (25 μg/ml) reversed S-CM down-regulation of the stimulus-driven nuclear translocation of NF-κB (right panel). Data are representative of a single experiment for each stimulus (n = 4). Histograms depict NF-κB distribution, as described in B. D, expression and phosphorylation of IκBα in intestinal macrophages and monocytes by flow cytometry. Intestinal macrophages, unlike blood monocytes, did not phosphorylate IκBα after stimulation with LPS, and monocyte phosphorylation of IκBα was inhibited by pretreatment of the cells with S-CM (250 μg/ml). E, expression and phosphorylation of IκBα and Smad2 in intestinal macrophages and blood monocytes. Intestinal macrophages expressed constitutive IκBα but did not phosphorylate IκBα. Blood monocytes also expressed constitutive IκBα but did phosphorylate IκBα after LPS stimulation, and phosphorylation was inhibited by pretreatment of the cells with S-CM. However, S-CM inhibition of inducible IκBα phosphorylation in blood monocytes was reversed when the S-CM (250 μg/ml) was preincubated (1 h) with anti-TGF-β antibodies (25 μg/ml). Coincident with blockade of inducible IκBα phosphorylation in monocytes, exposure of the monocytes to S-CM prior to stimulation caused a sharp increase in IκBα, which also was reversed when the S-CM was preincubated with anti-TGF-β antibodies. Intestinal macrophages expressed, but did not phosphorylate, Smad2. Blood monocytes expressed Smad2 but did not express phosphorylated Smad2 in medium alone, or after LPS stimulation, but in the presence of TGF-β alone (100 ng/500 μl), S-CM alone (250 μg/500 μl), or S-CM + LPS, monocytes expressed phosphorylated Smad2. The S-CM induction of phosphorylated Smad2 was blocked by pretreatment of S-CM with anti-TGF-β antibodies. Data are representative of three independent experiments (n = 3). rh, recombinant human.
DISCUSSION
In contrast to macrophages in other tissues, resident macrophages in normal human intestinal mucosa do not express many innate response receptors, including CD14 and FcαR, and are down-regulated for the production and release of pro-inflammatory cytokines in response to LPS and IgA (2, 3). This phenotype has been shown to persist for at least 2 months (3). Here, we show that intestinal macrophages express TLRs but do not release TLR-inducible cytokines (even in the presence of exogenous CD14, after transfection with MD-2, or following pretreatment with IFN-γ), implicating ineffective downstream signaling as the mechanism of the inflammation anergy characteristic of intestinal macrophages. Consistent with the latter conclusion, intestinal macrophages do not phosphorylate NF-κB p65 or translocate cytoplasmic NF-κB p50 into the nucleus.
Two lines of evidence provide an explanation for the inability of intestinal macrophages to activate NF-κB. First, intestinal macrophages expressed undetectable MyD88 and markedly reduced TRIF adapter proteins, as well as reduced signal protein TRAF6. However, the cells expressed increased levels of mRNA for SOCS1, which promotes the degradation of MAL (MyD88 adaptor-like protein) (22), and SARM, which inhibits TRIF signaling (26). MyD88 is a critical element in the NF-κB activation pathway of all TLRs except TLR3, and TRIF mediates TLR3-induced RANTES and IFN-β production, as well as TLR4-mediated MyD88-independent signaling (27). Thus, the blockade of MyD88-dependent and -independent NF-κB signaling causes ineffective TLR-mediated responses in intestinal macrophages. Second, intestinal macrophages displayed constitutive Smad signaling, reflected in the presence of nuclear Smad4 and the absence of cytoplasmic Smad7 inhibitor in the cells, albeit intestinal macrophages did constitutively express p-Smad2. In support of active Smad signaling, intestinal macrophages expressed constitutive IκBα and did not phosphorylate IκBα for proteosomal degradation, allowing continuous cytoplasmic sequestration of NF-κB, thereby blocking pro-inflammatory cytokine gene transcription. Intestinal macrophages were similarly unable to activate NF-κB through mitogen-activated protein kinase (MAPK) pathways involving p-p38, p-ERK, or p-JNK,4 pathways dependent on TRAF6 (28). Furthermore, inhibition of NF-κB activation in intestinal macrophages would also explain the inability of the cells to respond to phorbol myristate acetate (3) and to express TREM-1 (9, 10), which are transcriptionally regulated by NF-κB and PUI (29). Thus, ineffective NF-κB signaling in intestinal macrophages and TGF-β-mediated Smad-induced IκBα up-regulation together provide a mechanism for the inflammation anergy of intestinal macrophages.
Unlike murine intestinal macrophages, which are CD11b+CD11c− and capable of constitutive and TLR-inducible IL-10 release (30), human intestinal macrophages are CD11b−CD11c− (3) and release neither constitutive nor inducible IL-10. Intestinal macrophages also did not release inducible pro-inflammatory cytokines when cultured with antibodies to IL-10 (or IL-4). Notwithstanding the important role of IL-10 as a key immunoregulatory cytokine in the mucosa (31, 32), our findings indicate that humans and mice may regulate intestinal macrophage responsiveness by different mechanisms.
Pro-inflammatory blood monocytes, which constitutively express TGF-βRI, -II, and the IL-8 receptors CXCR1,2, are potently recruited by stromal TGF-β and IL-8 (1). Once the cells have entered the lamina propria, our in vitro studies (3) suggest that stroma-associated products, in particular TGF-β, induce the monocytes to undergo rapid NF-κB inactivation and inhibition of TLR-mediated signal transduction, leading to down-regulated pro-inflammatory, but not host defense, function. Therefore, we investigated whether products derived from intestinal stroma induce in blood monocytes, the cells from which intestinal macrophages are derived, dysregulated NF-κB signaling similar to that of intestinal macrophages. As we show here, stromal TGF-β inhibited the phosphorylation of IκBα and the nuclear transport of NF-κB p65 in monocytes while inducing the phosphorylation of Smad2.
Dysregulated Smad signaling appears to play an important role in inflammatory bowel disease, where overexpression of Smad7 (33) promotes defective Smad2/3 phosphorylation, diminished Smad signaling (34), and reduced IκBα expression, leading to constitutive NF-κB-dependent gene transcription with sustained NF-κB activation and increased synthesis of pro-inflammatory cytokines (35). Our findings provide a new framework for understanding the discordant NF-κB signaling in inflammatory bowel disease. In contrast to the overexpression of Smad7, which promotes NF-κB signaling in the inflamed mucosa, Smad7 in normal intestinal macrophages is underexpressed and not inducible, leading to persistent IκBα expression and unavailable NF-κB p50/p65 for cytokine gene transcription.
As discussed by Hayden and Ghosh (36), the inducible regulation of gene expression through NF-κB activation is a fundamental element of normal physiology, allowing the host to avert an array of challenges, including microbial perturbation. The findings presented here extend this concept by showing that macrophages in normal human intestinal mucosa are profoundly down-regulated for NF-κB activation, a feature that likely evolved as a selective advantage to limit bacterially induced intestinal inflammation (37). However, intestinal macrophages maintain powerful host defense function, including avid phagocytic and bacteriocidal activities (3). Consistent with these findings, S-CM inhibited blood monocyte NF-κB activation but not phagocytic or bacteriocidal activity for Escherichia coli or S. typhimurium (3). Thus, the notion that “mis-regulation” of NF-κB is associated exclusively with certain inflammatory, immunodeficiency, and cancer-related diseases (38, 39) is modified by our finding that down-regulation of macrophage NF-κB is associated with mucosal homeostasis in uninflamed healthy mucosa. Supporting this concept, the gene array analysis presented here showed profound down-regulation of genes for the CD14 transcription factors CEBPα, CEBPβ, and Sp1; the LPS receptor complex components CD14, TLR4, and MD-2, and the NF-κB family members p105 and p65 in intestinal macrophages but not blood monocytes. Gene expression for the signal proteins IRAK1 and -4 also was down-regulated, whereas gene expression for the inhibitors SARM and SOCS1 was increased. Thus, the inactivation of NF-κB in intestinal macrophages appears to be the consequence of dysregulated gene transcription for multiple proteins in the MyD88-dependent signaling cascade. Consistent with these findings, MyD88 mRNA levels were also reduced in intestinal macrophages, and TRAF6 mRNA expression was unchanged in these cells, but the corresponding proteins were not detectable.
The inability of intestinal macrophages to express CD14 protein or mRNA challenges the concept that CD14 expression is a function of monocyte differentiation into tissue macrophages. In this connection, the gene array studies presented here suggest that the absence of CD14 on intestinal macrophages is regulated at the level of gene transcription and is not the consequence of enzymatic digestion of surface glycoproteins. Moreover, the parallel down-regulation of message for CEBPα and CEBPβ, transcription factors that activate the promoter for CD14 (40), and Sp1, a transcription factor that regulates tissue-specific expression of the CD14 promoter (41), suggests the regulatory elements that control CD14 promoter activity are involved in the down-regulation of CD14 expression in intestinal macrophages. However, the inability of intestinal macrophages to respond to LPS in the presence of exogenous CD14 excludes dysregulated CD14 as the exclusive cause of the LPS unresponsiveness. Furthermore, the expression of TLR3 and TLR5–9 in intestinal macrophages indicates that impaired TLR expression does not account for the inflammation anergy in intestinal macrophages. Rather, the inability of intestinal macrophages to activate NF-κB, mediated through dysregulated NF-κB signal proteins and Smad-mediated IκBα expression, endow intestinal macrophages with inflammation anergy.
In summary, we show here that the profound inflammation anergy of intestinal macrophages in normal intestinal mucosa is mediated through a network of down-regulatory mechanisms. These mechanisms effect the prompt and efficient inactivation of NF-κB, thereby promoting the absence of inflammation in intestinal mucosa despite the close proximity of lamina propria macrophages to lumenal bacteria and food antigens. Thus, a breach in mucosal (epithelial) integrity would be met by potent host defense (phagocytic and bacteriocidal) activity but not an inflammatory response in the normal intestinal mucosa. These findings provide new insight into tissue homeostasis in normal intestinal mucosa and suggest a new perspective from which to experimentally address the regulation of inflammation in mucosal diseases of the human small intestine.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK-74033, DK-47322, DK-54495, AI-83027, DK-61297, DK-43183, AI-83539, and RR-20136. This work was also supported by University of Alabama at Birmingham Digestive Disease Research Developmental Center Grant DK-64400, the Crohn's and Colitis Foundation of America, the Investigator-sponsored Study Program of AstraZeneca, The Bradford Family Fund, and the Research Service of Veterans Affairs.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
L. E. Smythies, manuscript in preparation.
- IL
- interleukin
- TLR
- Toll-like receptor
- IFN
- interferon
- TNF
- tumor necrosis factor
- TGF
- transforming growth factor
- ELISA
- enzyme-linked immunosorbent assay
- LPS
- lipopolysaccharide
- S-CM
- stroma-conditioned media
- RANTES
- regulated on activation normal T cell expressed and secreted
- sCD14
- soluble CD14
- FITC
- fluorescein isothiocyanate
- PBS
- phosphate-buffered saline
- SARM
- sterile and Armadillo motif-containing protein
- M-CSF
- macrophage-colony-stimulating factor
- TRIF
- Toll interleukin receptor 1 domain-containing adapter-inducing interferon β
- Ref. Seq.
- Reference sequence
- CEBP
- CCAAT/enhancer-binding protein.
REFERENCES
- 1.Smythies L. E., Maheshwari A., Clements R., Eckhoff D., Novak L., Vu H. L., Mosteller-Barnum L. M., Sellers M., Smith P. D. (2006) J. Leukocyte Biol. 80, 492–499 [DOI] [PubMed] [Google Scholar]
- 2.Smith P. D., Smythies L. E., Mosteller-Barnum M., Sibley D. A., Russell M. W., Merger M., Sellers M. T., Orenstein J. M., Shimada T., Graham M. F., Kubagawa H. (2001) J. Immunol. 167, 2651–2656 [DOI] [PubMed] [Google Scholar]
- 3.Smythies L. E., Sellers M., Clements R. H., Mosteller-Barnum M., Meng G., Benjamin W. H., Orenstein J. M., Smith P. D. (2005) J. Clin. Invest. 115, 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith P. D., Ochsenbauer-Jambor C., Smythies L. E. (2005) Immunol. Rev. 206, 149–159 [DOI] [PubMed] [Google Scholar]
- 5.Smith P. D., Janoff E. N., Mosteller-Barnum M., Merger M., Orenstein J. M., Kearney J. F., Graham M. F. (1997) J. Immunol. Methods 202, 1–11 [DOI] [PubMed] [Google Scholar]
- 6.Li L., Meng G., Graham M. F., Shaw G. M., Smith P. D. (1999) Gastroenterology 116, 1043–1053 [DOI] [PubMed] [Google Scholar]
- 7.Meng G., Sellers M. T., Mosteller-Barnum M., Rogers T. S., Shaw G. M., Smith P. D. (2000) J. Infect. Dis. 182, 785–791 [DOI] [PubMed] [Google Scholar]
- 8.Shen R., Richter H. E., Clements R. H., Novak L., Huff K., Bimczok D., Sankaran-Walters S., Dandekar S., Clapham P. R., Smythies L. E., Smith P. D. (2009) J. Virol. 83, 3258–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schenk M., Bouchon A., Birrer S., Colonna M., Mueller C. (2005) J. Immunol. 174, 517–524 [DOI] [PubMed] [Google Scholar]
- 10.Schenk M., Bouchon A., Seibold F., Mueller C. (2007) J. Clin. Invest. 117, 3097–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rugtveit J., Bakka A., Brandtzaeg P. (1997) Clin. Exp. Immunol. 110, 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smythies L. E., Wahl L. M., Smith P. D. (2006) Curr. Protocol. Immunol. 7.6B, 1–9 [Google Scholar]
- 13.Wahl L. M., Wahl S. M., Smythies L. E., Smith P. D. (2006) Curr. Protocol. Immunol. 7.6A, 1–10 [Google Scholar]
- 14.Maheshwari A., Smythies L. E., Wu X., Novak L., Clements R., Eckhoff D., Lazenby A. J., Britt W. J., Smith P. D. (2006) J. Leukocyte Biol. 80, 1111–1117 [DOI] [PubMed] [Google Scholar]
- 15.Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankaran S., George M. D., Reay E., Guadalupe M., Flamm J., Prindiville T., Dandekar S. (2008) J. Virol. 82, 538–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagai Y., Akashi S., Nagafuku M., Ogata M., Iwakura Y., Akira S., Kitamura T., Kosugi A., Kimoto M., Miyake K. (2002) Nat. Immunol. 3, 667–672 [DOI] [PubMed] [Google Scholar]
- 18.Jiang Z., Georgel P., Du X., Shamel L., Sovath S., Mudd S., Huber M., Kalis C., Keck S., Galanos C., Freudenberg M., Beutler B. (2005) Nat. Immunol. 6, 565–570 [DOI] [PubMed] [Google Scholar]
- 19.Mai U. E., Perez-Perez G. I., Wahl L. M., Wahl S. M., Blaser M. J., Smith P. D. (1991) J. Clin. Invest. 87, 894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naiki Y., Michelsen K. S., Zhang W., Chen S., Doherty T. M., Arditi M. (2005) J. Biol. Chem. 280, 5491–5495 [DOI] [PubMed] [Google Scholar]
- 21.Imai K., Takeshita A., Hanazawa S. (2000) Infect. Immun. 68, 2418–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansell A., Smith R., Doyle S. L., Gray P., Fenner J. E., Crack P. J., Nicholson S. E., Hilton D. J., O'Neill L. A., Hertzog P. J. (2006) Nat. Immunol. 7, 148–155 [DOI] [PubMed] [Google Scholar]
- 23.Gray P., Dunne A., Brikos C., Jefferies C. A., Doyle S. L., O'Neill L. A. (2006) J. Biol. Chem. 281, 10489–10495 [DOI] [PubMed] [Google Scholar]
- 24.Doyle S. L., Jefferies C. A., O'Neill L. A. (2005) J. Biol. Chem. 280, 23496–23501 [DOI] [PubMed] [Google Scholar]
- 25.O'Neill L. A., Bowie A. G. (2007) Nat. Rev. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 26.Carty M., Goodbody R., Schröder M., Stack J., Moynagh P. N., Bowie A. G. (2006) Nat. Immunol. 7, 1074–1081 [DOI] [PubMed] [Google Scholar]
- 27.Takeda K., Akira S. (2004) Semin. Immunol. 16, 3–9 [DOI] [PubMed] [Google Scholar]
- 28.Gohda J., Matsumura T., Inoue J. (2004) J. Immunol. 173, 2913–2917 [DOI] [PubMed] [Google Scholar]
- 29.Zeng H., Ornatowska M., Joo M. S., Sadikot R. T. (2007) Eur. J. Immunol. 37, 2300–2308 [DOI] [PubMed] [Google Scholar]
- 30.Denning T. L., Wang Y. C., Patel S. R., Williams I. R., Pulendran B. (2007) Nat. Immunol. 8, 1086–1094 [DOI] [PubMed] [Google Scholar]
- 31.Monteleone I., Platt A. M., Jaensson E., Agace W. W., Mowat A. M. (2008) Eur. J. Immunol. 38, 1533–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asseman C., Mauze S., Leach M. W., Coffman R. L., Powrie F. (1999) J. Exp. Med. 190, 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monteleone G., Del Vecchio Blanco G., Monteleone I., Fina D., Caruso R., Gioia V., Ballerini S., Federici G., Bernardini S., Pallone F., MacDonald T. T. (2005) Gastroenterology 129, 1420–1429 [DOI] [PubMed] [Google Scholar]
- 34.Monteleone G., Kumberova A., Croft N. M., McKenzie C., Steer H. W., MacDonald T. T. (2001) J. Clin. Invest. 108, 601–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monteleone G., Mann J., Monteleone I., Vavassori P., Bremner R., Fantini M., Del Vecchio Blanco G., Tersigni R., Alessandroni L., Mann D., Pallone F., MacDonald T. T. (2004) J. Biol. Chem. 279, 3925–3932 [DOI] [PubMed] [Google Scholar]
- 36.Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 37.Smith P., Shen R., Smythies L. (2009) Mucosal Immunol. 2, 378–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Courtois G., Gilmore T. D. (2006) Oncogene 25, 6831–6843 [DOI] [PubMed] [Google Scholar]
- 39.Karin M. (2006) Nature 441, 431–436 [DOI] [PubMed] [Google Scholar]
- 40.Pan Z., Hetherington C. J., Zhang D. E. (1999) J. Biol. Chem. 274, 23242–23248 [DOI] [PubMed] [Google Scholar]
- 41.Zhang D. E., Hetherington C. J., Tan S., Dziennis S. E., Gonzalez D. A., Chen H. M., Tenen D. G. (1994) J. Biol. Chem. 269, 11425–11434 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.