Abstract
Five highly conserved polar residues connected by a number of structural water molecules together with two rotamer micro-switches, TrpVI:13 and TyrVII:20, constitute an extended hydrogen bond network between the intracellular segments of TM-I, -II, -VI, and -VII of 7TM receptors. Molecular dynamics simulations showed that, although the fewer water molecules in rhodopsin were relatively movable, the hydrogen bond network of the β2-adrenergic receptor was fully loaded with water molecules that were surprisingly immobilized between the two rotamer switches, both apparently being in their closed conformation. Manipulations of the rotamer state of TyrVII:20 and TrpVI:13 demonstrated that these residues served as gates for the water molecules at the intracellular and extracellular ends of the hydrogen bond network, respectively. TrpVI:13 at the bottom of the main ligand-binding pocket was shown to apparently function as a catching trap for water molecules. Mutational analysis of the β2-adrenergic receptor demonstrated that the highly conserved polar residues of the hydrogen bond network were all important for receptor signaling but served different functions, some dampening constitutive activity (AsnI:18, AspII:10, and AsnVII:13), whereas others (AsnVII:12 and AsnVII:16) located one helical turn apart and sharing a water molecule were shown to be essential for agonist-induced signaling. It is concluded that the conserved water hydrogen bond network of 7TM receptors constitutes an extended allosteric interface between the transmembrane segments being of crucial importance for receptor signaling and that part of the function of the rotamer micro-switches, TyrVII:20 and TrpVI:13, is to gate or trap the water molecules.
Keywords: Adrenergic Receptor, Computer Modeling, G-protein-coupled Receptors (GPCR), Molecular Dynamics, Mutagenesis Mechanisms, Protein Conformation, Receptors
Introduction
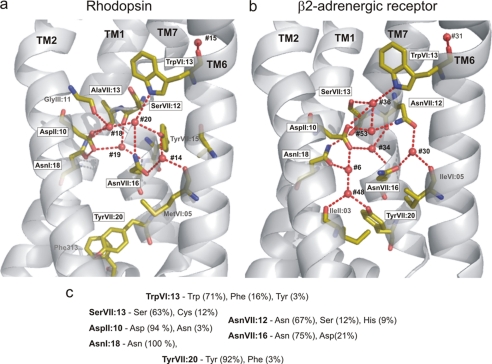
In recent years several structures of 7TM2 receptors have been published, making it possible to compare conserved regions in different receptors and study their putative functional importance (1–7). The most extended of these highly conserved regions is the water hydrogen bond network located between the intracellular segments of TM-I, TM-II, TM-VI, and TM-VII reaching from TrpVI:13 of the CWXP motif at the bottom of the main ligand-binding pocket to TyrVII:20 of the NPXXY motif on the intracellular side of the receptor (Fig. 1). A number of conserved polar residues in these transmembrane segments have for many years been suspected to form a hydrogen bond network of likely functional importance. The recent x-ray structures have confirmed this notion but have also revealed that very few hydrogen bonds are in fact found directly between the side chains of these polar residues. Instead, a number of structural water molecules function as integral connecting parts of the hydrogen bond network (Fig. 1). Thus, in the B2AR structure (2, 3) six internal water molecules form hydrogen bonds with the highly conserved polar amino acid residues located between TrpVI:13 and TyrVII:20, whereas in the rhodopsin structures (8–10) only four water molecules have been identified in this network (Fig. 1).3
FIGURE 1.
The hydrogen bond network between TM-I, -II, -VI, and -VII in rhodopsin compared with B2AR. a, the hydrogen bond network in rhodopsin (1GZM) as viewed from between TM-III and -V (TM-III to TM-V are not shown). Only key residues involved in the hydrogen bond network are indicated as sticks. Water molecules are indicated by red spheres. Note that TyrVII:20 in rhodopsin is turned “down” toward helix VIII and that no waters are found in the rhodopsin structure between TyrVII:20 and AspII:10/AsnI:18. b, hydrogen bond network in B2AR (2RH1) from the same angle as in panel a (rhodopsin). Note that TyrVII:20 in B2AR is rotated “upward” and that two water molecules (#6 and #48) are found between TyrVII:20 and AspII:10/AsnI:18. The water molecules are numbered as in the two original x-ray structures (see supplemental Table S1 for comparison of water numbers).
Several studies have suggested that TrpVI:13 functions as a rotamer switch where the inactive conformation is the “vertical” conformation (g+ conformation) observed in all published crystal structures, and where the putative active conformation is the trans conformation with the indole ring rotated toward TM-V to form an aromatic interaction with PheV:13, an equally highly conserved residue (11–13). In the inactive conformation the NH of the indole ring of TrpVI:13 forms a hydrogen bond to the most extracellular located water molecule of the hydrogen bond network. TyrVII:20 is found at the intracellular side, and contrary to TrpVI:13 it is found in various conformations in various x-ray structures. Thus, in all the bovine rhodopsin structures TyrVII:20 forms a face-to-face aromatic stacking with PheVIII:04 of helix VIII, whereas in the adrenergic structures as well as the adenosine A2a and squid rhodopsin structures TyrVII:20 is rotated into the receptor structure to form a hydrogen bond with the most intracellularly located water molecule of the hydrogen bond network (5, 14). In the opsin structures, one of which is a presumed active conformation in complex with a C-terminal peptide fragment of the G-protein transducin, TyrVII:20 holds a third conformation, where it is further rotated toward TM-VI to engage in a hydrophobic cluster between TM-VI and TM-VII (6, 7). These observed differences suggest that TyrVII:20 changes conformation during activation and that this residue, together with TrpVI:13, appears to function as micro-switches both being involved in the water hydrogen bond network.
It is generally assumed that the water hydrogen network is important in receptor activation, although the precise mechanism is rather unclear (15, 16). For example, solid state NMR spectroscopy and micro-second molecular dynamics (MD) studies of rhodopsin have suggested that the degree of hydration of the receptor changes during activation (17). These studies suggest that structural water molecules have an important role in activation of rhodopsin and possibly also for other 7TM receptors. Recently Angel and coworkers used radiolysis to study the residues in close contact to water molecules in rhodopsin and showed activation-induced changes in labeling mediated by water molecules, suggesting that the structural water molecules seen in the crystal structure of rhodopsin have different dynamic and structural properties dependent on the activation state of the receptor (18, 19).
In the present investigation we use MD simulation of rhodopsin and the B2AR combined with mutational analysis of the B2AR to study the hydrogen bond network. MD studies have previously been performed on rhodopsin but not with a focus on the structural water molecules and the hydrogen bond network (17, 20–27). Recently, MD studies of the B2AR have been published focusing on receptor mobility, ligand binding, and on the so-called “ionic lock” between TM-III and TM-VI (28–30). The large scale, rigid body movements of helices, which are believed to be part of the activation mechanism for 7TM receptors (1, 31), will likely occur on a time scale in the range of microseconds to milliseconds and are consequently not expected to be observed during normal MD simulations (31, 32). Thus, in the present study we focus on the MD of the water hydrogen bond network. MD simulations are particularly suited to study movements of side chains and, for example, water molecules, which occur at a time scale of relatively few nanoseconds. By comparing MD simulations performed with rhodopsin and B2AR placed in a lipid membrane surrounded by a water-box, we observed that the hydrogen bond network is surprisingly stable in B2AR as compared with rhodopsin and show that this is likely due to the closed, gating function of the two rotamer switches, TrpVI:13 and TyrVII:20. Interestingly, Ala substitution of the five conserved polar residues located between these rotamer switches had major effects on both the constitutive and the agonist-induced signaling of the B2AR.
EXPERIMENTAL PROCEDURES
B2AR Simulation Setup
The recently published high resolution x-ray structure of B2AR (2RH1) (2, 3) was used as a starting structure in the B2AR simulations. The structure was solved to a resolution of 2.4 Å. In the simulations the T4-lysozyme part of the structure was truncated and the intracellular parts of TM5 and TM6 were covalently connected. Because the structure contained coordinates for residue 29–230 and 263–342 only these residues were considered in the simulation. As it is seen in the x-ray structure, Cys-341 was also palmitoylated in the simulations. Hydrogens were added using the HBUILD facility in CHARMM (33), and protonation states according to pH 7 were used for all aspartic acids. Glu-122 was kept protonated, because this residue contacts the hydrophobic part of the membrane. Fourier transformed infrared spectroscopy studies have suggested that the conserved residue AspII:10 is protonated in rhodopsin, and this residue (Asp-79) was therefore also kept protonated in the B2AR simulation (34).
Rhodopsin and Opsin Simulation Setup
The high resolution x-ray structure of rhodopsin (1GZM) (8) solved to a resolution of 2.65 Å was used as a starting structure in the simulations of bovine rhodopsin and opsin. The opsin model was constructed from bovine rhodopsin by removing 11-cis-retinal. Residues 1–324 were included in both systems, and the C-terminal tail of the rhodopsin receptor was excluded, because no well defined three-dimensional structure has been shown for this part of the structure. Glycosylations of the extracellular part of the receptor were not considered, but both Cys-322 and Cys-323 were palmitoylated. Protonation states according to pH 7 were used for all aspartic acids, glutamic acids, and lysine residues except Asp-83 (AspII:10) and Glu-122, which were both protonated. Nϵ protonation was chosen for all histidine residues for both receptors.
General Setup
In addition to the receptors the systems contained membrane and water molecules. The Membrane Builder in VMD (35) was used to build a 1-palmitoyl,2-oleoyl-sn-glycero-3-phosphocholine membrane slice, which was subsequently equilibrated. The equilibration was done by first running a 0.25 ns simulation with the headgroups fixed and after that running a 0.25 ns simulation with all atoms free to move. This resulted in a membrane slice with a thickness of 38 Å measured from the phosphor atoms of the headgroups (36).
The receptor models were manually inserted into the membrane, and the lipid molecules overlapping with the receptors were deleted. The membrane plane was parallel to the x,y plane, and the plug-in SOLVATE in VMD (35) was used to add water molecules in the z-direction around the membrane and the solvent-exposed parts of the receptor. Water molecules added in the hydrophobic part of the membrane were subsequently deleted. In all simulations the water molecules seen in the x-ray crystal structures were also present in the simulation. To compensate for the net charge of the protein a number of water molecules were replaced with chloride ions to ensure a zero net charge of the system. A hexagonal simulation cell was used for all the simulations. The minimum distance between the protein and the edge of the hexagonal cell was 16 Å. Parameters for retinal and palmitoyl were provided to the nanoscale molecular dynamics homepage by Jan Saam, and these parameters were used in the simulations.
Simulation
All calculations were carried out with the nanoscale molecular dynamics simulation package (37) using the CHARMM27 force field (33). The temperature and the pressure were kept constant throughout the simulation. The system was simulated using Langevin dynamics with an MD step of 1 fs, and coordinates were saved every 1 ps.
The particle mesh Ewald method was used to calculate electrostatic interaction using a cut-off of 12 Å and a switching function of 10 Å. Periodic boundary conditions were used during all the simulations.
To equilibrate the system it was minimized while keeping the protein backbone fixed for 1000 steps. Subsequently the system was minimized for 2000 steps with all atoms free to move. Then 250 ps of MD simulation was run with the protein backbone fixed, followed by 2000 steps minimization with all atoms free to move.
Hydrogen Bonds
The hydrogen bond analysis was performed using the HBOND routine (35) in VMD using a distance cut-off of 3.5 and an angle cut-off of 30.
Ligand
Pindolol was purchased from Sigma.
Molecular Biology
The B2AR cDNA was cloned into the eukaryotic expression vector pCMV-Tag(2B) (Stratagene, La Jolla, CA). Mutations were constructed by PCR using the overlap extension method as previously described (38). The PCR products were digested with appropriate restriction endonucleases (BamHI and EcoRI), purified and cloned into the pCMV-Tag(2B) vector. All PCR experiments were performed using Pfu polymerase (Stratagene) according to the instructions of the manufacturer. All mutations were verified by restriction endonuclease mapping and subsequent DNA sequence analysis using an ABI 310 automated sequencer.
Cell Biology
COS-7 cells were grown in Dulbecco's modified Eagle's medium 1885 supplemented with 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected using the calcium phosphate precipitation method with chloroquine addition as previously described (39). The amount of cDNA (20 μg/75 cm2) resulting in maximal basal signaling was used for the dose-response curves.
cAMP Assay
One day after transfection, COS-7 cells were plated into white 96-well plates (20,000 cells/well). The day after cAMP assay was performed using DiscoverRX HitHunterTM cAMPxs + kit (Freemont, CA) according to the manufacturer's protocol.
Data Analysis
The cAMP curves were generated, and EC50 values were determined by nonlinear regression using Prism (GraphPad, San Diego, CA).
RESULTS
Overall Characteristics of the MD Simulations
MD simulations were performed and compared for rhodopsin (1GZM) (8) and for B2AR generated by removal of the T4L part of the B2AR-T4L fusion protein (2RH1) (2, 3) as described under “Experimental Procedures.” Several of the MD simulations were performed on structures where the ligands had been removed, that is, all the B2AR simulations where performed without carazolol. In all cases the MD simulations were performed with the receptors placed in a 1-palmitoyl,2-oleoyl-sn-glycero-3-phosphocholine membrane and surrounded by water on both sides in a hexagonal simulation box.
The root-mean square deviations (r.m.s.d. values) for the Cα atoms were calculated both for the whole protein and for the transmembrane helical segments of all three receptors (Fig. 2c). The r.m.s.d. values found in this study for rhodopsin are comparable to those observed in previous molecular dynamics studies performed on this molecule (20, 23). For rhodopsin with 11-cis-retinal removed the overall dynamics of the system was similar to the r.m.s.d. of rhodopsin, which stabilized at ∼2 Å for the whole structure and 1.4 Å for the TM regions, indicating no major difference in the overall dynamics of this system with and without the 11-cis-retinal ligand present (Fig. 2, a and b). The B2AR system was also surprisingly stable during the MD simulation with an r.m.s.d. of ∼1.6 Å for the whole molecule and ∼1 Å for the TM regions, indicating that the structure of the B2AR without the T4L is even more stable than rhodopsin and rhodopsin without 11-cis-retinal.
FIGURE 2.
r.m.s.d. and r.m.s.f. values for rhodopsin, rhodopsin without 11-cis-retinal, and B2AR during MD simulations. a–c, r.m.s.d. of Cα for rhodopsin (a), rhodopsin without 11-cis-retinal (b), and B2AR (c), as a function of time during MD simulations for 8, 20, and 8 ns, respectively. For each structure, the r.m.s.d. for the Cα of the whole structure (in red) and for the Cα of the transmembrane helices only (in black), are shown. d–f, r.m.s.f. calculated after 8 ns of MD simulation for each residue in: rhodopsin (d), rhodopsin without 11-cis-retinal (e), and B2AR (f). The transmembrane helices are indicated with red cylinders. The r.m.s.f. was calculated after the start structure and the structure after 8 ns were aligned; this alignment was only based on the structure of the transmembrane helices with the lengths as indicated in the figure.
The root mean square fluctuation (r.m.s.f.) values for each residue in the three structures are shown in Fig. 1 (d–f). As expected the loops, the extracellular and the intercellular extensions, were much more dynamic than the helical segments during the MD simulation. In B2AR especially residues located in the intracellular loop 3 connecting TM5 and TM6 displayed high r.m.s.f. values (Fig. 2f). This was expected, because this loop was modified compared with the x-ray structure through removal of the T4L and connecting the free ends.
Dynamics of the Hydrogen Bond Network between TM-I, -II, -VI, and -VII
This hydrogen bond network consists of five conserved polar side chains one of which, SerVII:13, is missing in rhodopsin (Fig. 1) as well as a number of exposed backbone carbonyls and NH groups corresponding to irregularities in the helical structure plus a number of structural water molecules. In the B2AR (2RH1) a total of six structural water molecules are found in the hydrogen bond network between TM-I, -II, -VI, and -VII (Fig. 1b), whereas only four of these, i.e. the most extracellular located ones, are present in rhodopsin (Fig. 1a). The structural water molecules found in the crystal structures were all included in the MD simulations. In all the experiments the water molecules moved around and changed hydrogen bonding partners throughout the simulations; however, the degree and pattern of movement was very different in rhodopsin versus B2AR.
The Hydrogen Bond Network of Rhodopsin
In rhodopsin where 11-cis-retinal had been removed but, somewhat surprisingly, also in the rhodopsin structure where the ligand was still present the structural water molecules moved relatively freely during the MD simulations (Fig. 3). For example, in the intact rhodopsin simulation water #18 rapidly moved out of the hydrogen bond network toward the binding pocket, i.e. passing TrpVI:13, which cannot rotate away when retinal is present (13). Also waters #19 and #14 moved several Å down and up, respectively, but did not leave the hydrogen bond network (Fig. 3A). These movements are also shown in supplemental Fig. 1, where the z-coordinate of the water molecules is depicted.4 In the rhodopsin structure without 11-cis-retinal, water #20, which is the one making the hydrogen bond to the indole side chain of TrpV:13, rapidly moved away up into the binding pocket, whereas water #14 moved around until it, in this simulation, was trapped at another position in the hydrogen bond network as indicated by the stable r.m.s.d. of ∼8 Å (Fig. 3B). The simulations of rhodopsin and rhodopsin without 11-cis-retinal were run twice using different random seeds, and in both cases the water molecules were much more movable as compared with the waters of the B2AR (see below).
FIGURE 3.
r.m.s.d. values for water molecules of the hydrogen bond network between TM-I, -II, -VI, and -VII in rhodopsin (a) and rhodopsin without 11-cis-retinal (b) during MD simulation. Water #15 in rhodopsin and rhodopsin without 11-cis-retinal is the “TM-VI kink water” making hydrogen bond to the backbone of CysVI:12 and TyrVI:16 and the backbone carbonyl of ProVII:05 (see Fig. 1). The rest of the waters are those located within the hydrogen bond network between TM-I, -II, -VI, and -VII (see Fig. 1).
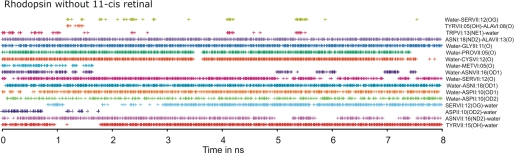
The dynamic structure of the hydrogen-bonding network is further illustrated in Fig. 4, where the occurrence of the individual hydrogen bonds in rhodopsin with 11-cis-retinal removed are indicated as a function of time throughout the MD simulation, i.e. independent of which water molecule is involved in the given hydrogen bond (shown for rhodopsin and B2AR in the supplemental materials). For example, the conceivably very important hydrogen bond between the side chain of the almost obligatory AsnI:18 (Asn-1.50)5 and the backbone carbonyl oxygen of AlaVII:13 (Ala-7.46) is found basically throughout the whole simulation (Fig. 4, dark blue symbols in fifth row from top). In contrast, the hydrogen bond from the indole NH of TrpVI:13 and a water molecule (#20 in the start structure) was observed relatively frequently in the beginning of the simulation but only intermittently later on where it is correlated to a hydrogen bond between the side-chain hydroxyl of SerVII:12 (Ser-7.45) as acceptor and a water molecule (Fig. 4, second and fourth rows from top). Thus, after ∼4 ns of simulation a water molecule becomes trapped for around 200 ps between these two residues, which happens again after 5.5 ns where these two hydrogen bonds are present for ∼400 ps. A similar correlation is also seen, for example, for the backbone carbonyl of SerVII:12 (Ser-7.45) and a water “shared” with the side-chain amino group of AsnVII:16 (Asn-7.49). We conclude that in the inactive, dark state of rhodopsin several of the water molecules of the hydrogen bond network are relatively moveable even in the presence of the 11-cis-retinal ligand.
FIGURE 4.
Dynamics of the hydrogen bonds in the hydrogen bond network between TM-I, -II, -VI, and -VII in rhodopsin without 11-cis-retinal during MD simulations. Hydrogen bonds observed during the first 8 ns of simulation of rhodopsin without 11-cis-retinal are shown (frames were written every 10 ps). Each time a hydrogen bond was present this was marked with a “+.” A hydrogen bond was defined as described under “Experimental Procedures.” In this figure the identity of the water molecule involved in the individual hydrogen bonds, which may vary over time, is not indicated.
The Hydrogen Bond Network of B2AR
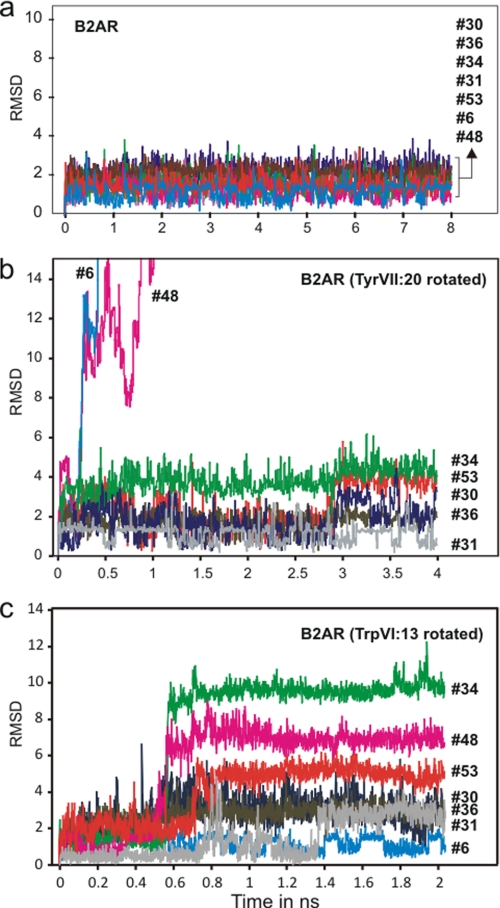
In sharp contrast to the relatively large excursions observed for the structural water molecules in rhodopsin with or without 11-cis-retinal, the six water molecules in the corresponding hydrogen bond network of the B2AR structure all moved surprisingly little throughout the MD simulations (Fig. 5a). Although the water molecules did change hydrogen bond interaction partners, back and forth (supplemental Fig. 2), they only moved very little away from their starting position as indicated in their stable r.m.s.d. values and z-coordinates (Figs. 5a and 6a).
FIGURE 5.
r.m.s.d. values for water molecules of the hydrogen bond network between TM-I, -II, -VI, and -VII in B2AR during MD simulation. a, B2AR with the rotamer switches TrpVI:13 and TyrVII:20 in the position found in the x-ray structure (2RH1). b, B2AR in a starting position with TyrVII:20 manually rotated to a conformation similar to that found in the rhodopsin (1GZM), i.e. toward helix VIII and away from the hydrogen bond network. c, B2AR in a starting position with TrpVI:13 manually rotated to its presumed active, trans conformation (χ1, −180) pointing toward TM-V. To be able to rotate TrpVI:13, PheVI:17 was rotated away from the binding pocket to disrupt the aromatic stacking of PheV:13 and PheVI:17. Water #31 is the TM-VI kink water making hydrogen bond to the backbone of CysVI:12 and PheVI:16 besides the backbone carbonyl of IleVII:05 (see Fig. 1). The rest of the waters are those located directly within the hydrogen bond network between TM-I, -II, -VI, and -VII (see Fig. 1). Note that especially waters #6 and #48, i.e. those that are not observed in rhodopsin, move away when TyrVII:20 is rotated, and that several of the waters become mobile when TrpVI:13 is rotated.
FIGURE 6.
z-Coordinates and surfaces of water molecules in the hydrogen bond network between TM-I, -II, -VI, and -VII in B2AR during the MD simulations shown in Fig. 5. a and b, B2AR with TrpVI:13 and TyrVII:20 in the conformations found in the crystal structure (2RH1). c and d, B2AR with TyrVII:20 manually rotated to a conformation similar to that found in the rhodopsin (1GZM), i.e. toward helix VIII and away from the hydrogen bond network. e and f, B2AR with TrpVI:13 manually rotated to its presumed active, trans conformation (χ1, −180) pointing toward TM-V. In the left column of a, c. and e are shown the z-coordinates (i.e. perpendicular to the membrane) of the water molecules. In the right column of b, d, and f are shown the surfaces of the water molecules from all frames as one combined surface with the different colors representing the different water molecule using the same color code as shown in the panels to the left.
Water Dynamics in B2AR after Rotation of TyrVII:20
One of the major differences between rhodopsin and B2AR in this area of the seven helical bundle is the rotamer conformation of the highly conserved TyrVII:20 of the NPXXY motif in TM-VII. In rhodopsin, TyrVII:20 is rotated “down” toward helix VIII to make a face-to-face aromatic interaction with PheVIII:04 (Phe-313) (Fig. 1a). In contrast, in B2AR the hydroxyl group of TyrVII:20 points “up” toward the ligand-binding pocket and forms a hydrogen bond to water molecule #48, i.e. the one closest to the cytosol in the water hydrogen bond network (Fig. 1b). Thus, it appears that TyrVII:20, in analogy with TrpVI:13, constitutes a rotamer micro-switch, which has been trapped in two different states in rhodopsin and B2AR, respectively. However, in none of the MD simulations did we observe any major changes in the rotamer state of TyrVII:20 independent upon the starting position (data not shown). In the conformation that TyrVII:20 adopts in B2AR it could very well be responsible for “trapping” the water molecules within the hydrogen bond network and preventing them from moving toward the cytosolic side.
To test this hypothesis we repeated the MD simulation of the B2AR with TyrVII:20 manually rotated into the conformation observed in rhodopsin, i.e. facing toward helix VIII and here making an aromatic-aromatic interaction with PheVIII:04. As expected, after only ∼200 ps the two water molecules closest to the cytosol, #48 and #6, both moved toward the intracellular part of the receptor and out of the hydrogen bond network (Figs. 5b, 6c, and 6d). However, as in rhodopsin TyrVII:20 did not change rotamer state during these MD simulations either conceivably due to its favorable aromatic interaction.
These experiments indicate that the two different rotamer states observed for TyrVII:20 in rhodopsin and in B2AR, respectively, are both rather stable and that TyrVII:20 in the state observed in the B2AR is partly responsible for keeping the “water filled,” highly stable version of the hydrogen bond network observed in this structure.
Water Dynamics in B2AR after Rotation of TrpVI:13
As previously described the rotamer state of the TrpVI:13 micro-switch is very stable in the B2AR structure conceivably due to its interaction with water #36 of the hydrogen-bonding network (13) (Fig. 1b). To study whether TrpVI:13 in a similar manner as TyrVII:20 has a gating function at the “top” of the water hydrogen bond network, we manually rotated TrpVI:13 to its proposed active conformation, i.e. the trans conformation. However, it is not possible to rotate TrpVI:13 to the trans conformation without first breaking the blocking aromatic stacking between PheV:13 and PheVI:17 (13). We therefore also rotated PheVI:17 to a favorable conformation pointing away from the binding pocket before starting the MD experiment.
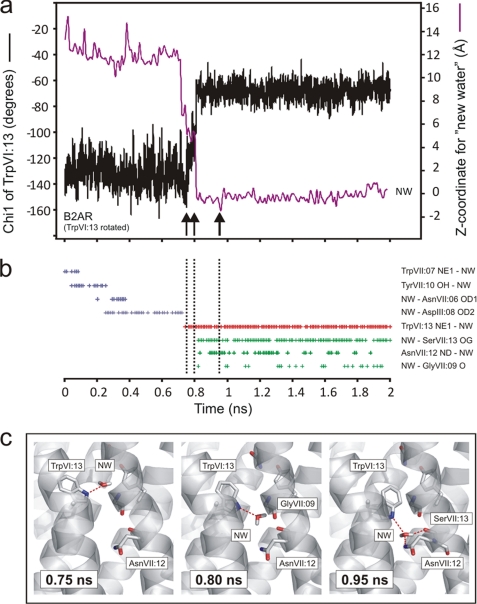
In the B2AR structure, where no change in the overall helical bundle structure had been made, TrpVI:13 was not stable in its proposed active rotamer state, but it did remain in the trans conformation for 0.8 ns of the simulation (Fig. 7a). However, after ∼0.5 ns water #34 left the hydrogen bond network and moved up into the binding pocket where, for the rest of the simulation (shown in Fig. 6E), it formed a stable hydrogen bond with AspIII:08 as indicated by its stable z-coordinate (Fig. 6E). Importantly, after ∼0.75 ns one of the added bulk water molecules, which had moved around in the main ligand-binding pocket making hydrogen bonds to, for example AsnVII:06 and AspIII:08 engaged in a hydrogen bond interaction with the NH group of the side chain of TrpVI:13 (Fig. 7, b and c). When TrpVI:13 subsequently rotated back to the g+ conformation seen in the crystal structure, it dragged the hydrogen bond-attached water molecule along into the hydrogen bond network (Fig. 7c). Through this rotation of TrpVI:13 the “new water molecule” became trapped in the network making hydrogen bonds with the side chains of SerVII:13 and AsnVII:12, and the backbone carbonyl of GlyVII:09 (Fig. 7, b and c). No water molecule from the water-filled main ligand-binding pocket had, during the 0.8 ns previous to the back rotation of TrpVI:13, visited the hydrogen bond network on its own.
FIGURE 7.
Movements and interactions of TrpVI:13 and a bulk water molecule (“new water”) being transported into the water hydrogen bonding network during MD simulation. a, χ1 of TrpVI:13 (in black) and z-coordinate of new water (NW) molecule (in purple) over the course of the simulation with TrpVI:13 in a starting position manually rotated into its presumed active conformation. b, hydrogen bonds observed between new water and polar residues in the receptor during the simulation. Each time a hydrogen bond was present this was marked with a “+.” A hydrogen bond was defined as described under “Experimental Procedures.” The hydrogen bonds colored blue represent hydrogen bonds to residues in the binding pocket, the purple hydrogen bonds are hydrogen bond to TrpVI:13, and the green hydrogen bonds are to polar residues in the hydrogen bonding network. c, snapshots of the hydrogen bonds between new water and selected residues in the receptor at key points in time as indicated by vertical arrows and dotted lines in a and b: 0.75 ns, hydrogen bond to TrpVI:13 still in the trans conformation; 0.80 ns, new water following TrpVI:13 as it rotates back to its inactive conformation; 0.95 ns, TrpVI:13 closing off the water hydrogen bond network and new water making hydrogen bonds to polar residues of the network.
These experiments indicate that in the B2AR the rotamer state of TrpVI:13 determines whether water molecules can move in and out of the water hydrogen bond network at the extracellular side, and that TrpVI:13 apparently is responsible for “catching” water molecules and in its inactive rotamer conformation trapping them within the hydrogen bond network, i.e. by preventing them from moving “back up” into the binding pocket .
Mutational Analysis of the Hydrogen Bond Network in B2AR
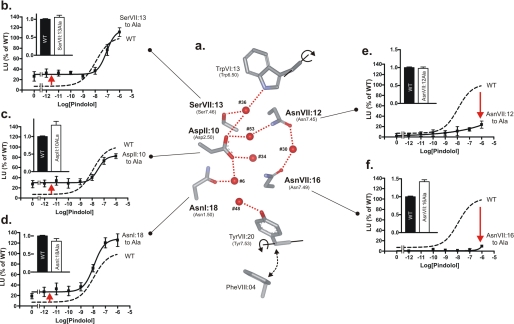
To determine the functional importance of each of the conserved, polar residues of the hydrogen bond network in the B2AR we substituted these individually with alanine and studied the effect on basal and agonist-induced signaling as reflected by cAMP production in transiently transfected COS-7 cells. As shown in Fig. 8, a rather similar effect was observed upon Ala substitution of the three polar residues contributing to the hydrogen bond network between TM-I, TM-II, and TM-VII, i.e. AsnI:18, AspII:10, and SerVII:13. That is, in all of these cases, removal of the polar side chain led to a gain-of-function in respect to an increase in the ligand-independent, constitutive signaling activity of the B2AR to ∼30% of the Emax (Fig. 8, b–d). The potency and maximal efficacy of the agonist-induced signaling was similar to that observed in the wild-type receptor. However, a very different, loss-of-function pattern was observed upon Ala substitution of the two remaining, polar residues, AsnVII:12 and AsnVII:16, which are located one helical turn apart in TM-VII and are connected though hydrogen bond formation to a common water molecule, #30 situated between TM-VI and -VII (Figs. 1b and 8a). In both of these mutants, i.e. AsnVII:12 to Ala and AsnVII:16 to Ala, the agonist-induced signaling was almost eliminated (Fig. 7, e and f). All of the mutated receptors were expressed to a similar degree as the wild-type receptor determined by cell surface enzyme-linked immunosorbent assay (insets in Fig. 8, b–f).
FIGURE 8.
Functional consequence of Ala substitutions of the conserved polar residues of the water hydrogen bond network between transmembrane segments I, II, VI, and VII in the B2AR. a, structure of the hydrogen bond network of B2AR (2RH1) with only key side chains shown as stick models and water molecules as red spheres. Only key hydrogen bonds are indicated for simplicity. Panels b–f, basal and agonist (pindolol)-induced cAMP production in COS-7 cells transiently transfected with either wild-type (dotted lines) B2AR or mutant forms of the following: SerVI:13 to Ala (b), AspII:10 to Ala (c), AsnI:18 to Ala (d), AsnVII:12 to Ala (e), and AsnVII:16 to Ala (f). Cell surface receptor expression, measured by enzyme-linked immunosorbent assay, is shown in the inserted column diagrams in each panel. Note that the constitutive activity is increased by substitutions of polar residues interacting with the “movable waters” (see Figs. 5 and 6), whereas the agonist-induced signaling is impaired by substitution of the two residues, AsnVII:12 and AsnVII:16, located one helical turn apart and interacting with the TM-VII kink water.
We conclude that the polar residues of the hydrogen bond network are highly important for receptor function, but that different residues and different parts of the network apparently play different roles, some being permissive for agonist signaling and some dampening for the constitutive activity.
DISCUSSION
Although the residues involved in the water hydrogen bond network between TM-I, -II, -VI, and -VII are generally conserved among 7TM receptors, it is only AsnI:18, that is truly obligatory (Fig. 1c), i.e. conserved close to 100%, probably because AsnI:18 makes the important hydrogen bond to the exposed backbone carbonyl at the kink in TM-VII. However, the B2AR offers an ideal model system to study this hydrogen bond network, because in contrast to for example rhodopsin the B2AR displays a network with each position being the most conserved among 7TM receptors (Fig. 1, b and c). Moreover, in the high resolution x-ray structures of B2AR the hydrogen bond network appears to be “fully loaded” with water molecules (2, 3). These water molecules have B-factors that are comparable to the B-factors of the side chains of the polar residues of the hydrogen bond network (2, 3) probably due to the fact that they are relatively immobilized in the B2AR structure as demonstrated by the MD simulations of the present study (Fig. 5a). Thus, a “vertical” string of five water molecules is located between TM-I, -II, and -VII connecting the two rotamer switches, TrpVI:13 and TyrVII:20. The x-ray structure and our MD simulations show that the side chains of all five polar residues of the network are interacting with these five water molecules. A sixth water molecule, #30, is located between TM-VI and -VII and is “shared” by two of the polar residues, AsnVII:12 and AsnVII:16, in the NPXXY motif in TM-VII (Fig. 1b). During the MD simulations this water molecule, which is further stabilized by a third hydrogen bond to the backbone carbonyl at position VI:05, never leaves its original position. Because the conserved AsnVII:12 and AsnVII:16 are located one helical turn apart and, importantly, on each side of the kink in TM-VII (Fig. 1b), it appears that water #30 plays an essential role in stabilizing the kink in TM-VII. This stable, “triangulated” water molecule at the TM-VII kink is clearly identified even in for example the A2a receptor structure, where otherwise rather few water molecules are identified in this network (5). Another water molecule being important for helical kink formation is #31 (in the B2AR), which is not directly connected to the hydrogen bond network discussed here but instead is hydrogen bonding to the free, exposed carbonyl of CysVI:12 of the CWXP motif plus other backbone elements of both TM-VI and -VII (Fig. 1b, top right corner). During the MD simulations of the present study water #31 was also highly immobilized, although it is directly exposed to the main ligand-binding pocket (Figs. 5 and 6).
Thus a picture is emerging where certain structural water molecules are more strongly bound to the receptor protein, conceivably being involved in helical kink formation, i.e. #30 and #31 (of the B2AR), whereas other water molecules clearly are more mobile and together with the polar side chains are forming an extended flexible, allosteric interface between the transmembrane helices.
Functional Analysis of the Polar Residues
The mutational analysis of the polar residues in B2AR clearly distinguished between residues involved in hydrogen bond formation with the “flexible” part of the hydrogen bond network and those interacting with the “stable” parts. Thus Ala substitution of AsnI:18, AspII:10, and SerVII:13, which are connected through the vertical string of “movable” water molecules, in all cases resulted in a similar increase in constitutive activity combined with a rather normal agonist-induced signaling (Fig. 8). It could be reasoned that the increase in constitutive activity is a result of a weakening of the flexible hydrogen bond network, which through the most extracellularly located water molecule normally would stabilize the inactive conformation of the TrpVI:13 rotamer switch. It was, however, surprising that Ala substitution of one of the most highly conserved residues among 7TM receptors, AsnI:18, only had such a relatively small effect, especially because the major role of AsnI:18 would appear to be to stabilize the kink in TM-II (see above). Nevertheless, the conclusion will be that a major role of the flexible part of the hydrogen bond network is to stabilize the inactive state of the receptor, at least in the B2AR.
In contrast, removal of the polar side chain of either AsnVII:12 or AsnVII:16 by Ala substitution in both cases basically eliminated agonist-induced signaling (Fig. 8). Because these two residues are stabilizing the kink in TM-VII through the hydrogen bond “bridge” over water #30, these results indicate that the special kink in TM-VII is crucial for receptor activation.
AsnVII:16 (Asn-7.49) of the NPXXY was an early motif among popular targets for mutations, and it has therefore previously been reported that elimination of this polar side chain by Ala substitution eliminates agonist-induced signaling, for example in the 5HT2a, the NK2, and the thyroid stimulation hormone receptors (40–42). This has also been observed in the present study also for the B2AR. AsnVI:16 and AspII:10 have in several cases been successfully swapped (40, 41, 43). In this connection it was shown that introduction of an acidic Asp residue in position VII:16 resulted in increased constitutive and agonist-induced signaling, for example in the thyroid stimulation hormone receptor (44).
Function of the Rotameric Switches
TyrVII:20
The MD simulations of the present study demonstrated that TyrVII:20 functions as a gate for the water molecules of the hydrogen bond network as the most cytosolic of these wander or diffuse out of the network when the side chain of TyrVII:20 in the B2AR was manually rotated out into the position observed in bovine rhodopsin: interacting with helix VIII (Figs. 5 and 6).
From a structural point of view, TyrVII:20 constitutes the aromatic, rotamer switch or gate in a ring of four highly conserved, interdigitating, hydrophobic residues that besides TyrVII:20 (92%) comprises: LeuII:06 (91%), LeuIII:19 (74%), and IleVI:05 (Val 42% and Ile 28%). In its closed form, this hydrophobic ring shields the water hydrogen bond network toward the cytosol. In the x-ray structures of the B2AR, B1AR, the A2a receptor, and squid rhodopsin the hydrophobic ring is in all cases closed with TyrVII:20 rotated into a position where its phenolic OH group interacts with the water hydrogen bond network (Fig. 1b). In the supposedly active form of opsin in complex with the Gα peptide fragment, the hydrophobic ring is also closed as TyrVII:20 interacts even more tightly with the other hydrophobic residues albeit in another conformation and from an altered backbone structure in TM-VII (1, 6, 7). In contrast, in all the structures of bovine rhodopsin the gate to the hydrogen bond network is open as the TyrVII:20 rotamer switch is rotated into a close aromatic interaction with PheVIII:04 in helix VIII, and the most cytosolic water molecules are absent or not identified in these structures (8–10).
TrpVI:13
Much focus has for many years been on the potential important functional role of TrpVI:13 (Trp-6.50) based on various forms of biophysical evidence from especially rhodopsin indicating that TrpVI:13 would change conformation upon receptor activation (45, 46). This was recently further substantiated by NMR spectroscopic analysis (47). In the B2AR TrpVI:13 has, based on MD simulations in vacuo of the isolated TM-VI, been described as a rotamer switch that was suggested to be synchronized with similar rotations of CysVI:12 and PheVI:9, which combined was described as a “toggle switch” (11). Our previous work supports the notion that TrpVI:13 is crucially involved in receptor activation in a number of 7TM receptors and that it probably is stabilized in its active conformation by an aromatic interactions with the equally highly conserved PheV:13 (13).
The present MD simulation adds another property to TrpVI:13, because it appears to function as a “water-catching gate” for the hydrogen bond network between TM-I, -II, -VI, and -VII. Thus, the data presented in Fig. 7 suggest that TrpVI:13 in its supposedly active rotamer conformation is able to bind and drag a bulk water molecule into the hydrogen bond network when it rotates back into its inactive conformation. The new water molecule is thereby trapped and becomes the most extracellularly located water molecule in the network. Here it has a number of choices in respect of new hydrogen bond partners, i.e. especially SerVII:13 but also AsnVII:12 and AspII:10, as well as the backbone carbonyl at position VII:09 and the other water molecules in the network (Figs. 1b, 7b, and 7c). This water molecule, #36 (in B2AR), plus its secondary hydrogen bond partners, apparently locks TrpVI:13 in its inactive rotamer state (1). It is interesting that in all the x-ray structures of 7TM receptors yet available, including the supposedly active opsin structure in complex with the G protein fragment, the TrpVI:13 rotamer switch is found in the inactive closed conformation (1). This is in contrast to other micro-switches such as ArgIII:26 (Arg-3.50) and TyrVII:20 (Tyr-7.53), which are found in different states in different x-ray structures. It should be noted that we are still lacking an x-ray structure of a 7TM with an agonist bound and with the intracellular part of the receptor in its fully active overall conformation.
MD Simulations and Receptor Function
Regarding the MD simulations performed in the present study it was not expected that we would be able to observe major conformational changes occurring between, for example the helical segments as described in the “Global Toggle Switch Model,” because this type of movements occurs on a much longer time scale (31, 32). Previously we have instead used Monte Carlo type of MD simulations to probe for active conformations that would satisfy experimentally determined distance constraints between helices as based especially on activating metal-ion sites (48, 49). Recently special customized computers and algorithms have been developed that allow for classic MD simulations to be performed on the millisecond time scale which should in principle make it possible to study for example helical movements in 7TM receptors during agonist-induced receptor activation (50, 51). Although larger conformational changes are expected to occur on millisecond time scales running a simulation for >1 ms does not guarantee observation of large conformational changes. In studies where purified B2ARs have been labeled with fluorescence probes and different fluorescent profiles were observed when adding different agonists, the purified receptor is incubated for 30 min before observing the full effect of the agonist (52, 53). In other words, receptors are found in equilibrium of conformations and addition of an agonist does not change receptor conformation of all receptors in a sample on a millisecond time scale.
Allosteric Interface versus “Molecular Domino Bricks”?
As shown in Fig. 8a, the elements of the hydrogen bond network appear to connect the bottom of the main ligand-binding pocket, TrpVI:13, with the intracellular face of the receptor, which interacts with G-proteins, i.e. TyrVII:20 and helix VIII. However, especially the fact that most of these elements, including the rotamer switches, are dispensable, i.e. not truly conserved, argues against the notion that they function as an assembly of “molecular domino bricks,” which in a sequential way mediates the intramolecular signal transduction process (1, 13). Instead a “concerted action” allosteric model of the Monod-Wyman-Changeux type is favored where the rotamer switches together with the polar residues and water molecules of the hydrogen bond network form an extended allosteric interface between the transmembrane helices, which perform the global toggle switch movements (1, 13, 54). In such a model any of the micro-switches are in principle dispensable, provided that other parts of the allosteric interface are able to stabilize a global, active conformation. Thus, during the gradual evolutionary process different parts of the allosteric interface could in certain receptors have been strengthened and thereby allow other parts of the interface to be weakened (13). However, “acute” substitutions of the various elements could have a dramatic effect on either spontaneous and/or agonist-induced activation as shown for the polar residues of the hydrogen bond network in the present study (Fig. 8) (40–42, 44).
Variations in the setting of the different micro-switches and other elements of the hydrogen bond network constituting this allosteric interface may be highly important for fine tuning the signal or be involved in controlling, for example, signal transduction pathway-biased signaling (1).
Supplementary Material
The work was supported by grants from The Danish Medical Research Council, The Novo Nordisk Foundation, The Lundbeck Foundation, and European Union's Seventh Framework Programme (Grant Agreement 223057-GIPIO).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table S1.
The number of structural water molecules identified in these structures is dependent upon the resolution and how well defined the water molecules are.
Because the z-axis is perpendicular to the membrane slice where the receptors are inserted into in the simulation, the z-coordinate for the water molecules describes their movement towards the intracellular or the extracellular part of the membrane.
- 7TM
- 7 transmembrane segment 7
- MD
- molecular dynamics
- CMV
- cytomegalovirus
- r.m.s.d.
- room mean square deviation
- r.m.s.f.
- room mean square fluctuation
- B2AR
- β-2 adrenergic receptor.
REFERENCES
- 1.Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Trends Pharmacol. Sci. 30, 249–259 [DOI] [PubMed] [Google Scholar]
- 2.Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Kobilka B. K. (2007) Science 318, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 4.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 7.Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 8.Li J., Edwards P. C., Burghammer M., Villa C., Schertler G. F. (2004) J. Mol. Biol. 343, 1409–1438 [DOI] [PubMed] [Google Scholar]
- 9.Okada T., Sugihara M., Bondar A. N., Elstner M., Entel P., Buss V. (2004) J. Mol. Biol. 342, 571–583 [DOI] [PubMed] [Google Scholar]
- 10.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 11.Shi L., Liapakis G., Xu R., Guarnieri F., Ballesteros J. A., Javitch J. A. (2002) J. Biol. Chem. 277, 40989–40996 [DOI] [PubMed] [Google Scholar]
- 12.Mirzadegan T., Benkö G., Filipek S., Palczewski K. (2003) Biochemistry 42, 2759–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holst B., Nygaard R., Valentin-Hansen L., Bach A., Engelstoft M. S., Petersen P. S., Frimurer T. M., Schwartz T. W. (2009) J. Biol. Chem. 285, 3973–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami M., Kouyama T. (2008) Nature 453, 363–367 [DOI] [PubMed] [Google Scholar]
- 15.Pardo L., Deupi X., Dölker N., Lüpez-Rodríguez M. L., Campillo M. (2007) Chembiochem. 8, 19–24 [DOI] [PubMed] [Google Scholar]
- 16.Schertler G. F. (2008) Nature 453, 292–293 [DOI] [PubMed] [Google Scholar]
- 17.Grossfield A., Pitman M. C., Feller S. E., Soubias O., Gawrisch K. (2008) J. Mol. Biol. 381, 478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angel T. E., Chance M. R., Palczewski K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8555–8560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angel T. E., Gupta S., Jastrzebska B., Palczewski K., Chance M. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14367–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saam J., Tajkhorshid E., Hayashi S., Schulten K. (2002) Biophys. J. 83, 3097–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kholmurodov KhT., Fel'dman T. B., Ostrovskii M. A. (2007) Neurosci. Behav. Physiol. 37, 161–174 [DOI] [PubMed] [Google Scholar]
- 22.Crozier P. S., Stevens M. J., Forrest L. R., Woolf T. B. (2003) J. Mol. Biol. 333, 493–514 [DOI] [PubMed] [Google Scholar]
- 23.Schlegel B., Sippl W., Höltje H. D. (2005) J. Mol. Model 12, 49–64 [DOI] [PubMed] [Google Scholar]
- 24.Huber T., Botelho A. V., Beyer K., Brown M. F. (2004) Biophys. J. 86, 2078–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crozier P. S., Stevens M. J., Woolf T. B. (2007) Proteins 66, 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau P. W., Grossfield A., Feller S. E., Pitman M. C., Brown M. F. (2007) J. Mol. Biol. 372, 906–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khelashvili G., Grossfield A., Feller S. E., Pitman M. C., Weinstein H. (2009) Proteins 76, 403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber T., Menon S., Sakmar T. P. (2008) Biochemistry 47, 11013–11023 [DOI] [PubMed] [Google Scholar]
- 29.Vanni S., Neri M., Tavernelli I., Rothlisberger U. (2009) Biochemistry 48, 4789–4797 [DOI] [PubMed] [Google Scholar]
- 30.Dror R. O., Arlow D. H., Borhani D. W., Jensen M. Ø., Piana S., Shaw D. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4689–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbell W. L., Altenbach C., Hubbell C. M., Khorana H. G. (2003) Adv. Protein Chem. 63, 243–290 [DOI] [PubMed] [Google Scholar]
- 32.Knierim B., Hofmann K. P., Ernst O. P., Hubbell W. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20290–20295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacKerell A. D., Jr., Banavali N., Foloppe N. (2000) Biopolymers 56, 257–265 [DOI] [PubMed] [Google Scholar]
- 34.Fahmy K., Jäger F., Beck M., Zvyaga T. A., Sakmar T. P., Siebert F. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10206–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–8, 27–8 [DOI] [PubMed] [Google Scholar]
- 36.Nygaard T. P., Rovira C., Peters G. H., Jensen M. Ø. (2006) Biophys. J. 91, 4401–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kale L., Schulten K. (2005) J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holst B., Hastrup H., Raffetseder U., Martini L., Schwartz T. W. (2001) J. Biol. Chem. 276, 19793–19799 [DOI] [PubMed] [Google Scholar]
- 39.Holst B., Zoffmann S., Elling C. E., Hjorth S. A., Schwartz T. W. (1998) Mol. Pharmacol. 53, 166–175 [DOI] [PubMed] [Google Scholar]
- 40.Sealfon S. C., Chi L., Ebersole B. J., Rodic V., Zhang D., Ballesteros J. A., Weinstein H. (1995) J. Biol. Chem. 270, 16683–16688 [DOI] [PubMed] [Google Scholar]
- 41.Govaerts C., Lefort A., Costagliola S., Wodak S. J., Ballesteros J. A., Van Sande J., Pardo L., Vassart G. (2001) J. Biol. Chem. 276, 22991–22999 [DOI] [PubMed] [Google Scholar]
- 42.Donnelly D., Maudsley S., Gent J. P., Moser R. N., Hurrell C. R., Findlay J. B. (1999) Biochem. J. 339, 55–61 [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou W., Flanagan C., Ballesteros J. A., Konvicka K., Davidson J. S., Weinstein H., Millar R. P., Sealfon S. C. (1994) Mol. Pharmacol. 45, 165–170 [PubMed] [Google Scholar]
- 44.Urizar E., Claeysen S., Deupí X., Govaerts C., Costagliola S., Vassart G., Pardo L. (2005) J. Biol. Chem. 280, 17135–17141 [DOI] [PubMed] [Google Scholar]
- 45.Spooner P. J., Sharples J. M., Goodall S. C., Bovee-Geurts P. H., Verhoeven M. A., Lugtenburg J., Pistorius A. M., Degrip W. J., Watts A. (2004) J. Mol. Biol. 343, 719–730 [DOI] [PubMed] [Google Scholar]
- 46.Borhan B., Souto M. L., Imai H., Shichida Y., Nakanishi K. (2000) Science 288, 2209–2212 [DOI] [PubMed] [Google Scholar]
- 47.Crocker E., Eilers M., Ahuja S., Hornak V., Hirshfeld A., Sheves M., Smith S. O. (2006) J. Mol. Biol. 357, 163–172 [DOI] [PubMed] [Google Scholar]
- 48.Elling C. E., Frimurer T. M., Gerlach L. O., Jorgensen R., Holst B., Schwartz T. W. (2006) J. Biol. Chem. 281, 17337–17346 [DOI] [PubMed] [Google Scholar]
- 49.Rosenkilde M. M., Andersen M. B., Nygaard R., Frimurer T. M., Schwartz T. W. (2007) Mol. Pharmacol. 71, 930–941 [DOI] [PubMed] [Google Scholar]
- 50.Maragakis P., Lindorff-Larsen K., Eastwood M. P., Dror R. O., Klepeis J. L., Arkin I. T., Jensen M. Ø., Xu H., Trbovic N., Friesner R. A., Iii A. G., Shaw D. E. (2008) J. Phys. Chem. B 112, 6155–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freddolino P. L., Arkhipov A. S., Larson S. B., McPherson A., Schulten K. (2006) Structure 14, 437–449 [DOI] [PubMed] [Google Scholar]
- 52.Yao X., Parnot C., Deupi X., Ratnala V. R., Swaminath G., Farrens D., Kobilka B. (2006) Nat. Chem. Biol. 2, 417–422 [DOI] [PubMed] [Google Scholar]
- 53.Yao X. J., Vélez Ruiz G., Whorton M. R., Rasmussen S. G., DeVree B. T., Deupi X., Sunahara R. K., Kobilka B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9501–9506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz T. W., Frimurer T. M., Holst B., Rosenkilde M. M., Elling C. E. (2006) Annu. Rev. Pharmacol. Toxicol. 46, 481–519 [DOI] [PubMed] [Google Scholar]
- 55.Baldwin J. M. (1993) EMBO J. 12, 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ballesteros J. A., Weinstein H. (1995) in Methods in Neurosciences (Stuart C. S. ed) pp. 366–428, Academic Press, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.