Abstract
Naturally occurring IgG antibodies are bivalent and monospecific. Bispecific antibodies having binding specificities for two different antigens can be produced using recombinant technologies and are projected to have broad clinical applications. However, co-expression of multiple light and heavy chains often leads to contaminants and pose purification challenges. In this work, we have modified the CH3 domain interface of the antibody Fc region with selected mutations so that the engineered Fc proteins preferentially form heterodimers. These novel mutations create altered charge polarity across the Fc dimer interface such that coexpression of electrostatically matched Fc chains support favorable attractive interactions thereby promoting desired Fc heterodimer formation, whereas unfavorable repulsive charge interactions suppress unwanted Fc homodimer formation. This new Fc heterodimer format was used to produce bispecific single chain antibody fusions and monovalent IgGs with minimal homodimer contaminants. The strategy proposed here demonstrates the feasibility of robust production of novel Fc-based heterodimeric molecules and hence broadens the scope of bispecific molecules for therapeutic applications.
Keywords: Antibodies, Cancer Therapy, Immunology, Protein Structure, Protein-Protein Interactions, Antibody Engineering, Bispecific Single Chain Fv, Fc Heterodimer, Monovalent IgG, Protein-Protein Interface Engineering
Introduction
Monoclonal antibodies have become an increasingly important class of therapeutic molecules for numerous indications including cancer, inflammatory diseases, and viral infections (1). By specifically binding to their targets such as cytokines in circulation or receptors on the cell surface, antibodies can either block or activate certain biochemical pathways. In addition, antibodies can also recruit other effector cells from the immune system for the selective destruction of cells expressing their target antigens. Modifications of the antibody IgG format differing in valence level or the degree of specificity or both, through protein engineering, offer the promise of extending the useful therapeutic properties of antibodies even further (2).
In IgG format antibodies, the two antigen binding fragment (Fab) arms are connected by a flexible hinge region to the homodimeric Fc fragment. The Fc region of IgG mediates antibody effector functions by its interactions with Fc receptors on effector cells and confers a long serum half-life in vivo through its interactions with neonatal Fc receptors (3). In addition, Fc can be easily produced in large quantities from mammalian cells and other hosts. For these reasons, the Fc fragment is commonly used as a partner to make fusions with other therapeutic proteins for in vivo applications (4). The Fc is often capable of bringing antibody-like qualities to the fusion protein. Due to the inherent dimeric nature of the Fc fragment, Fc fusions are produced as bivalent homodimeric proteins. However, for certain applications such as the production of bispecific antibodies where heterodimeric assembly between two different molecules is required, it is desirable to have Fc fragments that can efficiently heterodimerize. Although simple coexpression of two different Fc heavy chains can lead to the formation of some heterodimer, the resulting products also contain significant amounts of homodimers that need to be purified away (5). Methods that can reliably promote Fc heterodimer formation and efficiently suppress the homodimer forms are thus required to facilitate scalable and efficient productions to support clinical trails and commercial marketing of bispecific protein-based therapies.
A strategy was proposed earlier by Carter and co-workers (5–8) to produce a Fc heterodimer using a set of “knob-into-hole” mutations in the CH3 domain of Fc. These mutations lead to the alteration of residue packing complementarity between the CH3 domain interface within the structurally conserved hydrophobic core so that formation of the heterodimer is favored compared with homodimers. Although the strategy led to higher heterodimer yield, the homodimers were not completely suppressed (7). In this work, we explored the feasibility of retaining the hydrophobic core integrity whereas driving the formation of Fc heterodimer by changing the charge complementarity at the CH3 domain interface. Taking advantage of the electrostatic steering mechanism, we were able to efficiently promote Fc heterodimer formation with minimum contamination of homodimers through mutation of two pairs of peripherally located charged residues. In contrast to the knob-into-hole design, the homodimers were evenly suppressed due to the nature of the electrostatic repulsive mechanism. This new form of Fc heterodimer greatly broadens our options for making asymmetrical Fc fusion proteins, where each Fc chain is fused with a different function group such as antibody fragment, peptide, soluble receptors, etc.
EXPERIMENTAL PROCEDURES
Computational Analyses
The computational scheme used to analyze the CH3 domain interface and to identify the charged residue pair that is likely to significantly impact dimer formation is shown in supplemental Fig. S1. A total of 27 (46 chains; some were deposited as dimer, whereas others as monomer) antibody crystal structures that had coordinates corresponding to the Fc region were identified from the Protein Data Bank (PDB)4 (9) using a structure-based search algorithm (10). Examination of the identified Fc crystal structures revealed that the structure determined at the highest resolution corresponds to the Fc fragment of Rituximab bound to a minimized version of the B-domain from protein A called Z34C (PDB code 1L6X) (11). The biological Fc homodimer structure for 1L6X was generated using the deposited Fc monomer coordinates and crystal symmetry. Two methods were used to identify the residues involved in the CH3-CH3 domain interaction: (i) contact as determined by distance limit criterion and (ii) solvent accessible surface area analysis. According to the contact-based method, interface residues are defined as residues whose side chain heavy atoms are positioned closer than a specified limit (4.5 Å) from the heavy atoms of any residues in the second chain. The second method involves calculating solvent accessible surface area (ASA) of the CH3 domain residues in the presence and absence of the second chain (12). The residues that show a difference (>1 Å2) in ASA between the two calculations are identified as interface residues. Both methods identified a similar set of interface residues. Furthermore, they were consistent with the published work (13). Supplemental Table S1 lists the 24 interface residues identified based on the contact criterion method, using the distance limit of 4.5 Å. These residues were further examined for structural conservation. For this purpose, 46 Fc crystal structures identified from the PDB were superimposed and analyzed by calculating root mean square deviation for the side chain heavy atoms. The CH3 domain interface buried (% ASA ≤ 10; % ASA refers to ratio of observed ASA to the standard ASA of amino acids (12)) and exposed (% ASA > 10) residue positions were identified using the solvent accessible surface area calculation.
Generation of Heterodimer Constructs Using Fc Charge-pair Mutation
A rat anti-mouse NKG2D antibody, designated M315, was generated through conventional hybridoma fusions and the DNA sequences encoding the variable heavy chain (VH) and variable light chain (VL) were used to construct M315 scFv-Fc using a previously described method (14). The M315 scFv-Fc and huIgG1Fc were cloned into the pTT5 mammalian expression vector and the two constructs were used to co-transfect 293 cells to assess formation of the Fc/scFv-Fc heterodimer relative to the Fc homodimer and scFv-Fc homodimer. The charge residue pairs in the CH3 region identified through computational analysis were changed to amino acids of opposite the charge polarity on either human IgG1 Fc (dummy) or M315 scFv-Fc constructs. The mutations were generated using the QuikChange® mutagenesis kit from Stratagene and verified by DNA sequencing. The mutations are denoted by wild type residues followed by the position using the Eu numbering system and then the replacement residue in single letter code. The Fc sequence used in these two constructs was derived from the human IgG1 non-(a) allotype, which has a Glu at position 356 and a Met at position 358. The CH3 sequences from the crystal structure are from a different IgG1 allotype, which has an Asp at position 356 and a Leu at position 368.
14D2 hamster anti-mouse p55TNFR antibody was obtained from Hiko Kohno. The VH and VL sequences were cloned from total RNA obtained from the hybridoma cells using degenerate primers to the 5′ sequence determined from Edman N-terminal sequence of purified 14D2 antibody and 3′ primers complimentary to sequence in the heavy chain constant region of hamster IgG1. The 14D2 VL-hu light chain κ fusion was made by amplifying the 14D2 VL with the following primers: 5′-CTGGTGCTAGCGATATAGTGATGTCGCAG and 5′-CAGCCACCGTACGTTTGATTTCCAGCTTGG. The amplicon was subcloned into a vector containing the sequence of the hu κ light chain constant sequence. The 14D2VL-hu κLC-huIgG1Fc fusion was spliced together by overlap extension using the following primers: 5′-CGTTTAAACGTCGACGTTTAAACGCCGCCAG, 5′-GGCATGTGTGAGTTTTGTCACACTCTCCCCTGTTG, 5′-CAACAGGGGAGAGTGTGACAAAACTCACACATGCC, 5′-GTTTAAACAGATCCGCGGCCGCTCTAGCCCC. The 14D2VH-huIgG1-His6 fusion was constructed by amplification of the 14D2VH with the following primers to add AscI and NheI restriction sites 5′ and 3′, respectively, to ligate the 14D2VH directly upstream to the huIgG1-His6 constant sequence in a mammalian expression vector.
To reduce the formation of light chain-Fc homodimer, charge mutations K409E:K392D were introduced on the Fc attached to the light chain and D399R:E356K were introduced on the Fc of the heavy chain. The heavy chain and light chain constructs were cotransfected into Chinese hamster ovary cells to generate a stable pool. A similar set of constructs were also generated for 14D2 using the mouse Fc backbone in which N399K:E356K (equivalent to D399K:E356K in human Fc) mutations were introduced on the heavy chain mouse IgG1 Fc and K409D:K392D (equivalent to K409D:K392D in human Fc) mutations were introduced on the light chain mouse IgG1 Fc fusion.
Bispecific scFv-Fc constructs were also generated using the Fc charge mutations. scFv constructs were first generated using the variable heavy and light chain sequences of OKT3 and anti-tumor-associated receptor tyrosine kinase (TARTK) IgG1. The scFv fragments were cloned into a mammalian expression vector to generate scFv-Fc expression constructs (14). D399K:E356K mutations were introduced on the Fc backbone of OKT3 scFv-Fc vector. K409D:K392D mutations were introduced on the TARTK scFv-Fc construct. The two vectors were cotransfected to 293 cells and the scFv-Fc was purified from culture medium using a monoclonal antibody Select Sure column and SEC then subjected to mass spectrometric analysis.
Protein Production and Analysis
DNA was transfected into human embryonic kidney cell line 293 using LipofectamineTM 2000 reagent (Invitrogen) or polyethylenimine depending on the volume used. The cell culture supernatant was harvested 5–6 days after transfection and run on SDS-PAGE gels under non-reduced conditions. The gel was then transferred to a nitrocellulose membrane and subject to Western analysis using peroxidase-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories).
Mass Spectrometric Analysis of Bispecific scFv-Fc and Monovalent IgG
The bispecific scFv-Fc and monovalent IgG samples were deglycosylated and followed by LC/MS analysis. About 20 μg of bispecific scFv-Fc was deglycosylated by incubation with 1 μl of peptide:N-glycosidase F (New England) overnight at 37 °C in its native solution. After deglycosylation, the bispecific scFv-Fc was analyzed by high pressure liquid chromatography ESI-time-of-flight analysis (Agilent 6210 time-of-flight mass spectrometer in combination with an Agilent 1200 LC system, Santa Clara, CA). A Varian Pursuit 5μ Diphenyl 150 × 2.1-mm column was connected to the LC system and operated at 400 μl/min. The column temperature was 75 °C, solvent A was 0.1% trifluoroacetic acid in water, and solvent B was 0.1% trifluoroacetic acid in acetonitrile. The gradient started at 25% B and increased to 80% B over 55 min. The time-of-flight mass spectrometer was tuned and calibrated in the range of 100 to 4500 m/z. It has better than 3 ppm mass accuracy through continuous and automated reference mass introduction.
T-cell-dependent Cytotoxicity Assay
U87 or U87-TARTK cells (target cells) were seeded in full growth medium at 10,000 cells/well (100 μl) in clear-bottom blackwell 96-well assay plates (Costar 9927) leaving one row empty for effector cell control. The cells were incubated at 37 °C in 5% CO2 overnight. Peripheral blood mononuclear cells (effector cells) were quickly thawed and washed 3 times at 200 × g for 10 min in pre-warmed immune cell medium (RPMI 1640, 10% fetal bovine serum, 55 μm 2-mercaptoethanol, 1 mm sodium pyruvate, 2 mm l-glutamine (10% ICM)) and resuspended at 2 × 10−6/ml. Bispecific antibodies were diluted in ICM at 4 times their final concentration and then titrated 1:10 across a 96-well microplate. 50 μl of the bispecific antibody dilution or control medium were added to parental and TARTK expressing cells in triplicate spots. Duplicate plates were set up to allow the determination of the experimental release of total lysis. 50 μl of effector cells were added to each well except the target cell alone control wells. Each well contained a total of 200 μl. The plates were incubated at 37 °C in 5% CO2 for 48 h. Cytotoxicity was assessed using bioluminescent ToxiLight® Bioassay Kit (Lonza, Rockland, ME). Briefly, 10 μl of 10% Triton X-100 was added to each well of the total lysis plates and 10 μl of phosphate-buffered saline was added to each well of the assay plates. Plates were incubated at 37 °C in 5% CO2 for 10 min. 100 μl was carefully removed from each well of each plate. 50 μl of ToxiLight® substrate was added to each well and incubated for 5 min at room temperature. Luminescence was detected using a Tecan GENios plate reader (1 s integrated reading). Percent cytotoxicity was calculated from the relative light units according to the formula: % cytotoxicity = ((experimental release) − (spontaneous release))/((total lysis) − (total spontaneous release)). Experimental release was the data obtained from each experimental condition. Spontaneous release was the average reading obtained from the effector + target cell wells without bispecific antibody.
Cell Culture and Peripheral Blood Mononuclear Cells
U87 and U87-TARTK cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat inactivated fetal bovine serum, 2 mm l-glutamine, 1 mm sodium pyruvate, and 0.1 mm minimal essential medium non-essential amino acids. Cultures were split every 3 to 4 days at 1:5 and 1:10 ratios. Peripheral blood mononuclear cell-enriched Leukopacks were obtained from Biological Specialty Corporation (Colmar, PA) and frozen at 1 × 108/ml in 90% fetal bovine serum, 10% Me2SO prior to use.
TNF-mediated L929 Cytotoxicity Assay
Murine fibroblast L929 cells (ATCC CCL-1) were maintained in Eagle's minimal essential medium supplemented with 10% horse serum and 1% penicillin-streptomycin-glutamine (100x, Life Technology), 1% non-essential amino acid (Life Technology), 1% sodium pyruvate, and 1% HEPES, and maintained at 37 °C with 5% CO2 in a humidified incubator. For the cytotoxicity assay, cells were plated in 96-well plates at 70,000 cells/well in 100 μl of medium supplemented with actinomycin D (2 μg/ml, Sigma catalog number A9415) and allowed to incubate for 2 h. The indicated amounts of antibodies (0.4–50 μg/ml) were added to the wells and plates were incubated 18 h. Following a wash with phosphate-buffered saline, Alamar Blue (Invitrogen DAL1025) was added at 1:10 (v/v) dilution in complete Eagle's minimal essential medium and cells were incubated an additional 4 h. Plates were read on a fluorescence plate reader (excitation 544 nm and emission 595 nm). To assess antagonism of TNF signaling, antibodies were preincubated with the cells at the indicated concentrations (1 μm to 50 pm) for 30 min, after which the cells were stimulated with 5 pm muTNF (R&D Systems catalog number 410-MT) for 18 h. Cellular viability was assessed with Alamar Blue as described for the agonist assay above. Percent inhibition of cell death was calculated using medium-stimulated and TNF-stimulated control wells.
Pharmacokinetics Study
Experimental cohorts of C57BL/6 mice were intraperitoneally administered a single 100-μg dose of various anti-murine p55 constructs. At multiple time points following injection, animals were euthanized and blood was collected for analysis.
Serum Antibody Concentrations and Pharmacokinetic Analysis
Serum concentrations of various 14D2 constructs were determined using a sandwich enzyme-linked immunosorbent assay method. Recombinant murine sTNFRI (murine p55; R&D Systems) and horseradish peroxidase-conjugated goat anti-murine IgG Fc fragment-specific polyclonal sera (Pierce) were used as the capture and the detection antibodies, respectively. Plate wells were developed using tetramethylbenzidine and peroxide substrate solution (Kirkegaard & Perry). The resulting colorimetric reaction was quenched with 10% phosphoric acid (VWR) and optical densities were determined at a wavelength of 450–650 nm. The conversion of OD values into concentrations for the quality control samples and serum specimens was achieved through Watson LIMSTM software mediated comparison to a concurrently analyzed standard curve, which was regressed according to a four-parameter logistic model. Non-compartmental pharmacokinetic analyses, including half-life determinations, were performed on mean concentration-time data using Watson software. The area under the curve values were calculated using the log-linear trapezoidal method.
Mouse Xenograft Studies
Seven-week-old NOD.SCID female mice were obtained from Harlan Laboratories and maintained under sterile conditions. Animals were provided food (Teklab 2920) and acidified water ad libitum. All animal care complied with the Canadian Council on Animal Care guidelines. U87-TARTK tumor cells were mixed with purified T-cells (Biological Specialty Corporation, 215-01-10) in serum-free RPMI 1640 and mixed 1:1 with Matrigel (BD Biosciences, 356237). The final concentration was 20 × 106 cells/ml of tumor and 25 × 106 cells/ml of T cells. Mice were implanted with 0.1 ml subcutaneously in the right flank on day 0. Treatments with anti-CD3/TARTK bispecific scFv-Fc (14 or 140 μg) were administered intravenously in 200 μl of Dulbecco's phosphate-buffered saline via the tail vein. Initial treatments were given 3 h after tumor implantation and a second treatment was administered 4 days later. Tumors were measured twice weekly and tumor volume was calculated using the formula: (width 2 × length)/2. Mean tumor volumes and standard error for each measurement data were determined and used to generate the graphs.
RESULTS
Enhancing Antibody Fc Heterodimer Formation through Electrostatic Steering Effects
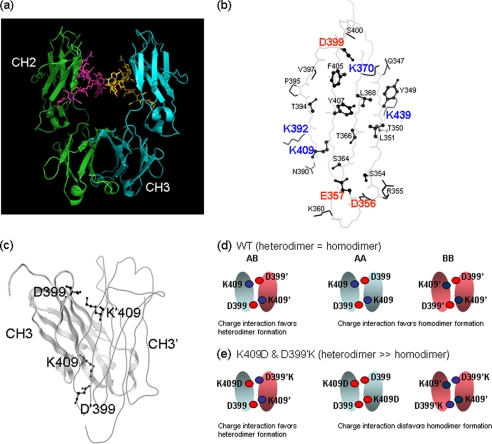
Crystal structures of the Fc region of the antibodies show extensive protein-protein interactions (surface area buried = 2469 Å2) occurring between the CH3 domains (13, 15) and the high affinity (Kd = 10 pm) CH3-CH3 interaction is the primary driver for Fc dimerization (Fig. 1a) (16). A total of 27 (46 chains) antibody crystal structures with coordinates corresponding to the Fc region were identified in the PDB (9) using a structure-based search algorithm (10). The structure determined at the highest resolution (PDB code 1L6X (11), 1.6 Å) was used to identify the CH3-CH3 domain interface residues (24 residues; supplemental Table S1). These residues were further examined for structure and sequence conservation (supplemental Fig. S1). For this purpose, the identified 46 Fc crystal structures were superimposed and analyzed by calculating the root mean square deviation for the side chain heavy atoms. The analysis revealed that the CH3 domain interface includes a central hydrophobic core region surrounded by charged residues that interact to promote dimer formation through favorable electrostatic interactions around the rim of the interface (Fig. 1b and supplemental Fig. S2). High structural conservation for the hydrophobic residues at the core of the interface as well as for a few of the charged residues was noted. Further examination of the residues involved in the CH3-CH3 domain interactions (supplemental Table S1) revealed four pairs of oppositely charged residues along the interface (Asp356–Lys439′, Glu357–Lys370′, Lys392–Asp399′, and Asp399–Lys409′). Comparison of human and mouse IgG subclasses also reveals high conservation for these charged residues (supplemental Fig. S3). We hypothesized that the overall charge polarity could be altered through mutagenesis of specific residues such that attractive electrostatic interactions would favor heterodimer formation, whereas repulsive charged interactions would disfavor homodimer formation. In addition, exploiting charged residues as opposed to hydrophobic residues at the CH3 domain interface may have benefits in terms of retaining the generally favorable biophysical properties of the Fc. It has long been established that the hydrophobic core of protein domains plays an important role in protein folding and stability (17).
FIGURE 1.
a, ribbon representation of an antibody Fc region crystal structure (PDB code 1L6X) (21). Only the CH3 domain is involved in protein-protein interaction. b, the figure shows the backbone trace of the CH3 domain interface structure with residues involved in the domain-domain interaction. The interface residues were identified using a distance cutoff (4.5 Å) method. Structurally conserved and buried (solvent accessible surface area ≤10%) residues are shown in the ball-and-stick model. Solvent-exposed or structurally not conserved residues are shown in stick representation. The analysis is based on the IgG1 crystal structure that determined at high resolution (1.65 Å; 1L6X). c, crystal structure of the CH3 domain homodimer with one domain shown in ribbon representation and the other domain shown in wire model. The Lys409 (K409′ in the second domain) and Asp399 (D399′ in the second) residues are shown in ball-and-stick model to illustrate each pairwise interaction that is represented twice in the structure. This is due to the 2-fold symmetry present in the CH3-CH3 domain interaction. Schematics showing electrostatic interactions in the wild type (d) and charge-pair mutant (e) were designed as an example to enhance heterodimer formation and hinder homodimer formation. In the case of wild type, the charge-charge interaction favors both heterodimer and homodimer formation giving them equal probability. In the case of charge-pair mutant, the charge-charge interaction favors heterodimer and disfavors homodimer formation.
Examination of the four charged residue pairs reveals that in the case of the Lys409–Asp399′ pair both residues are structurally conserved and buried (Table 1). Studies have suggested that structurally conserved and buried residues contribute significantly to protein-protein association (18, 19). Therefore, the Lys409–Asp399′ pair was chosen for a proof-of-concept experimental study. It must also be noted here that due to the 2-fold symmetry present in the CH3-CH3 domain interaction, each unique interaction will be represented twice in the structure (for example, Asp399–Lys409′ and Lys409–Asp399′; Fig. 1c). The strategy to enhance heterodimer formation using the charged residue pair is schematically shown in Fig. 1d. In the wild type Fc, the Lys409–Asp399′ (K409–D399′) interaction favors both heterodimer and homodimer formation. A double mutation switching the charged residue polarity (K409D in first chain and D399′K in the second chain) leads to unfavorable repulsive interactions for the formation of homodimer whereas preserving favorable interaction for the heterodimer formation. The strong repulsive interactions (K409D–D399 and K409′–D399′K) between the charged residues may hinder homodimer formation in the kinetic pathway of dimer assembly.
TABLE 1.
Complementary charged residue pairs at the CH3 domain interface and their structural properties
Solvent accessible surface area (%), calculated using the available highest resolution Fc crystal structure (1L6X), and root mean square deviation (Å2) of the side chain atoms, calculated using the identified 46 Fc crystal structures, are shown in parentheses. ASA ≤10% and root mean square deviation >1.0Å2 are highlighted in bold face.
| Residue in chain A—residue in chain B |
|---|
| ASP A 356 (30.7%, 0.99Å2)—LYS B 439′ (32.9%, 1.99Å2) |
| GLU A 357 (0.0%, 1.28Å2)—LYS B 370′ (13.4%, 0.94Å2) |
| LYS A 392 (39.2%, 1.21Å2)—ASP B 399′ (7.9%, 0.79Å2) |
| LYS A 409 (0.6%, 0.91Å2)—ASP B 399′ (7.9%, 0.79Å2) |
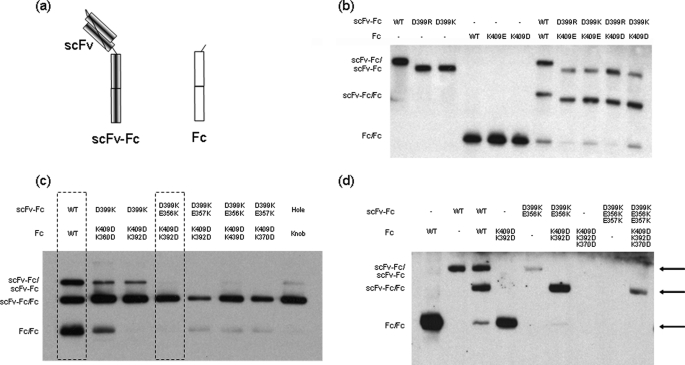
To assess relative yields of heterodimer and homodimer, a model system was used in which two Fc fusions with significant size difference, a scFv-Fc fusion and a Fc fragment (Fig. 2a), were coexpressed in 293 cells. The secretion of the three possible dimeric products (scFv-Fc/scFv-Fc and Fc/Fc homodimers, and scFv-Fc/Fc heterodimer) could be assessed on SDS-PAGE under non-reduced conditions. When the two Fc fusion constructs were coexpressed, less than 50% of the product was scFv-Fc/Fc heterodimer. The introduction of a single charge-pair alteration (K409D on the Fc fragment and D399′K on the scFv-Fc) significantly increased the ratio of the scFv-Fc/Fc heterodimer relative to both scFv-Fc and Fc homodimers (Fig. 2b). Similar enhancement of heterodimer formation was also observed for other similar charge mutant combinations such as K409D/D399′R, K409E/D399′K, and K409E/D399′R (Fig. 2b), underscoring the importance of charge polarity complementation for the formation of Fc heterodimer.
FIGURE 2.
a, schematic drawing of the two constructs used in the co-transfection experiments in 293 cells. The first construct encodes a scFv-Fc fusion. The second construct encodes a dummy Fc, which does not have any domain attached to it. The products of the co-transfection experiments with different Fc variants were analyzed by SDS-PAGE under non-reduced conditions and Western blot as shown in b–d. b, effects of single charge-pair mutations of the Asp399–Lys409′ interaction pair on Fc heterodimerization. Products of either single transfection or cotransfections of Fc variants are shown. All four single charge-pair variants (D399R/K409′E, D399K/K409′E, D399R/K409′D, and D399K/K409′D) led to higher yield for the scFv-Fc/Fc heterodimer (49 to 60%) compared with the homodimers (7 to 23%). The construct for the wild type Fc contained an extra tag of 33 amino acids, so its products appeared larger on SDS-PAGE. c, effects of charge-pair mutation combinations on Fc heterodimerization. The K409D:K392D/D399′K:E356′K mutation combination and the WT/WT are highlighted. The T366W (knob) and T366′S:L368′A:Y407′V (hole) mutation combination was included as a control. d, the effects of charge-pair mutation combinations on Fc homodimer and heterodimer formations. The products of single transfection (homodimer formation) and cotransfection (homodimers and heterodimer) for each Fc variants were shown. The single transfection of the triple mutant constructs (K409D:K392D:K370D and D399′K:E356′K:E357′K) did not yield any product of homodimer. However, cotransfection of the two triple mutant constructs led a heterodimer formation. Note the Fc sequence used in the constructs is derived from human IgG1 non- (a) allotype, which has a Glu at position 356. The crystal structure (1L6X) used for the computational analysis is from a different IgG1 allotype, which has Asp at position 356.
When additional charge-pair mutations were combined with the K409D/D399′K mutation, a further increase of heterodimer formation and decrease of homodimers was observed. The combination of K409D:K392D in the Fc fragment with D399′K:E356′K in the scFv-Fc resulted in heterodimer formation almost exclusively. Other combinations such as K409D:K392D/D399′K:D357′K and K409D:K370D/D399′K:D357′K also offered significant improvement over the K409D:D399′K variant alone (Fig. 2c). Analogous constructs with the knobs-into-holes mutations (T366W/T366′S:L368′A:Y407′V) also formed primarily heterodimer, although a small amount of scFv-Fc homodimers could still be detected under the same experimental conditions (Fig. 2c and supplemental Fig. S4a and Table S2). It may be noted here that the input DNA ratio can influence the relative proportions of the Fc dimer formation as emphasized in earlier work (7).
The effects of charge-pair combinations on the formation of homodimers and heterodimer were further evaluated separately in a different experiment. As shown in Fig. 2d, although both K409D:K392D and D339′K:E356′K double mutants can still form homodimers when expressed alone, they preferentially form heterodimers when expressed together. Interestingly, Fc molecules with three mutations wherein positive-charged residues were changed to negative-charged residues (K409D:K392D:K370D) or negative-charged residues were changed to positive-charged residues (D399K:E356K:E357K) could not be detected as homodimers. However, heterodimers could be produced when the two electrostatically matched triple mutant variants were co-expressed together, although the overall product yield was significantly reduced (Fig. 2d). This observation suggests that charge complementation can critically influence Fc dimer formation. After a survey of many variants, we have determined that overall the combination of two pairs of charge mutations (K409D:K392D/D399′K:E356′K) on two respective Fc chains gave the best results for producing predominantly Fc heterodimers without a significant reduction of product yield.
Production of Bispecific scFv-Fc using Engineered Fc Heterodimer
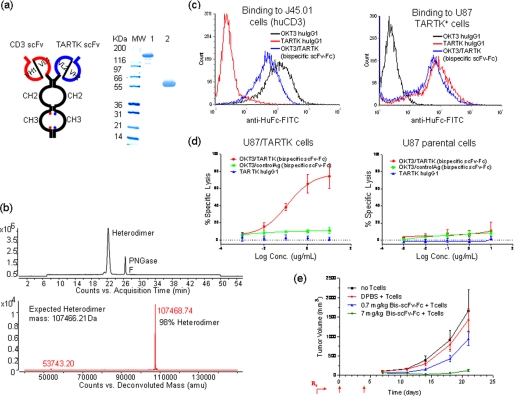
A bispecific molecule based on Fc heterodimer with charge-pair mutations was constructed by fusing two scFv fragments with different binding specificities, one against human CD3 (OKT3)(20) and one against a TARTK,5 to the Fc heterodimer (Fig. 3a). Co-transfection of mammalian cells with expression vectors encoding the two scFv-Fc fusions led to the secretion of single species production with a molecular mass of 120 kDa into the culture medium (Fig. 3a). The product was purified, deglycosylated, and subjected to intact mass analysis by mass spectrometry. As shown in Fig. 3b, the whole mass of the predominant fraction, representing more than 98% of product, matches the expect mass of the bispecific scFv-Fc heterodimer. The bispecific antibody was shown to be able to bind both CD3 on human T cells and TARTK expressed on cancer cells (Fig. 3c). When incubated together with human peripheral blood mononuclear cells and the human glioma cell line U87 that has been transduced to stably express TARTK, the bispecific antibody can specifically induce the killing of U87-TARTK cells but not parental U87 cells at a very low dose (Fig. 3d). The killing activity is not due to the activity of conventional antibody-dependent cellular cytotoxicity as the anti-TARTK human IgG1 failed to kill the U87-TARTK cells. A similar control bispecific antibody that binds to CD3 and an irrelevant target also failed to kill the U87-TARTK cells (Fig. 3d). The CD3xTARTK bispecific antibody also showed a dose-dependent inhibition tumor growth of the U87-TARTK cell in a xenograft model using NOD/SCID mice. Due to the presence of the Fc region, the bispecific scFv-Fc fusion has a much longer in vivo half-life in mouse and can be dosed more infrequently than other types of CD3 bispecific scFv fragments. With two doses of 140 μg of antibody (7 mg/kg) intravenously on days 0 and 4, the bispecific antibody completely suppressed the growth of the TARTK-positive glioma in a xenograft model where human T cells were implanted together with the tumor (Fig. 3e).
FIGURE 3.
a, schematic drawing of bispecific scFv-Fc generated by using the Fc heterodimer with charge-pair mutations (left panel) and SDS-PAGE analysis (right panel) of the purified bispecific scFv-Fc heterodimer in non-reduced (lane 1) and reduced (lane 2) conditions. Anti-CD3 scFv OKT3 (VH1/VL1 in red) was fused to the human Fc backbone with D399′K:E356′K mutations. Anti-TARTK scFv (VH2/VL2 in blue) was fused to human Fc with K409D:K392D mutations. b, the total ion chromatogram of deglycosylated bispecific scFv-Fc heterodimer using peptide:N-glycosidase F (PNGase F). Both heterodimer and peptide:N-glycosidase F peaks were seen in the 55-min gradient using a diphenyl. The deconvoluted mass spectrum result of deglycosylated bispecific scFv-Fc heterodimer is shown in the lower panel. The heterodimer with average mass of 107468.74 is seen, and no homodimers were detected in the mass analysis. The calculated heterodimer average mass is 107466.21. c, the binding of bispecific TARTKxCD3 scFv-Fc heterodimer to U87-TARTK cells and T cells (J45.01) by fluorescence-activated cell sorter. OKT3 huIgG1 and TARTK huIgG1 were used as controls. d, specific cytotoxic activity of bispecific TARTKxCD3 scFv-Fc on TARTK expressing cells. U87-TARTK cells (left panel) or U87 cells (right panel) were incubated with peripheral blood mononuclear cells in the presence of serial dilutions of bispecific TARTKxCD3 scFv-Fc heterodimer (red line), a control bispecific scFv-Fc heterodimer that binds to CD3 and an unrelated antigen (green line), and anti-TARTK huIgG1 (blue line) for 48 h. Percent of cell lysis was calculated and plotted. e, dose-dependent effect of bispecific TARTKxCD3 scFv-Fc heterodimer on the outgrowth of U87-TARTK glioma in NOD/SCID mice. U87-TARTK tumor cells were mixed 1:1 with purified T cells and innoculated subcutaneously. The indicated dose of bispecific TARTKxCD3 scFv-Fc heterodimer or Dulbecco's phosphate-buffered saline (DPBS) control were administrated via tail vein injections on days 0 and 4. Tumor growth curve derived from 10 animals is shown.
Production of Monovalent IgG using Engineered Fc Heterodimer
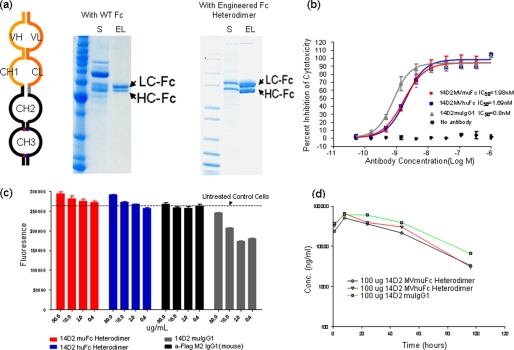
The charge-pair-based Fc heterodimer was also used to generate a novel form of monovalent IgG. As illustrated in Fig. 4a, monovalent IgG is a product of a single Fab fusion to the heterodimer Fc. Compared with the bivalent four-chain structure of full IgG, the monovalent IgG is a two-chain molecule with only a single antigen-binding moiety. The heavy chain (HC) of this molecule is the same as in a standard IgG. The light chain (LC) is further extended by fusion with Fc using a modified hinge sequence. The resulting molecule should bind to antigen monovalently, similar to a Fab molecule, and should retain a pharmacokinetic profile similar to an IgG.
FIGURE 4.
a, schematic drawing of the monovalent IgG (left panel) and SDS-PAGE analysis of monovalent IgG preparations using wild type Fc (middle panel) and heterodimer Fc with charge-pair mutations (right panel). The content of culture supernatant (S) and eluted fractions (EL) after the protein A column of each preparation were examined under reduced conditions. The bands corresponding to LC-Fc and HC-Fc are indicated by arrows, respectively. b, anti-mouse TNFR1 14D2 monovalent IgG, either as human Fc (blue) or mouse Fc heterodimer (red), exhibited good potency in blocking TNF-mediated cytotoxicity in L929 assays. 14D2 mouse IgG1 was used as a control. IC50 values were estimated from the percentage cell lysis curves and listed. c, 14D2 monovalent IgGs were free of agonistic activity. The cellular viability of L929 cells was assessed by fluorescence readout in the presence of 14D2 monovalent IgG, as human Fc (blue) or mouse Fc heterodimer (red), and 14D2 mouse IgG1 and control IgG (anti-FLAG) without TNF. d, 14D2 monovalent IgGs had a similar pharmacokinetics profile as full IgG in mouse serum.
14D2 IgG is a potent antagonistic antibody to mouse TNF receptor 1 (TNFR1). It can efficiently block TNF-mediated cytotoxic activity in L929 cells. However, it also activates TNFR1 at low doses in the absence of TNF. The agonistic activity of 14D2 IgG is due to the cross-linking of TNFR1 by the full IgG molecule, as the Fab fragment of 14D2 is free of agonistic activity (data not shown). To generate a pure antagonistic antibody with long serum half-life for in vivo studies, a monovalent form of the 14D2 IgG molecule was constructed. When the two transcripts were introduced into mammalian cells, only two of three possible products, HC/LC-Fc heterodimer and LC-Fc homodimer, were detected in the medium (supplemental Fig. S5). The HC homodimer cannot be secreted due to the unpaired constant heavy 1 domain (21). When the wild type Fc sequence was used, only 50–60% of the product was the desired monovalent IgG form with the rest consisting of the LC-Fc homodimer. As a result, three times more LC-Fc was seen on the SDS-PAGE gel than HC under reduced conditions (Fig. 4a). However, when charge-pair mutations were used in the Fc constructs, the predominant product in the medium was the HC/LC-Fc heterodimer. As a result, the overall ratio of heavy chain to LC-Fc of the purified product on the reduced SDS-PAGE gel is close to 1:1. This was confirmed by mass spectrometric analysis. More than 90% of the product after protein A purification was the HC/LC-Fc heterodimer (data not shown). The final purified yield of the 14D2 monovalent IgG from a stable pool of Chinese hamster ovary cells was ∼100 mg/liter.
The agonistic and antagonistic activity of the purified monovalent IgG was evaluated in L929 cytotoxicity assays. As shown in Fig. 4b, monovalent IgG, either as human Fc or mouse Fc heterodimer, exhibited good potency in blocking TNF-mediated cytotoxicity with an IC50 value similar to that of the bivalent IgG. However, the two monovalent IgG preparations were free of agonist activity. As shown in Fig. 4c, the monovalent IgG did not lead to any decreased cell viability when incubated with L929 cells. This was in contrast to the 14D2 full IgG, which could induce killing of L929 cells in the absence of TNF. The agonistic activity of 14D2 was not detected for the Fab format, indicating its dependence on the bivalency of IgG. Importantly, both forms of the 14D2 monovalent IgG exhibited similar pharmacokinetic profiles as the full-length IgG when injected in mice (Fig. 4d), demonstrating that incorporation of the charge-pair mutations did not compromise the ability of the Fc to mediate a long serum half-life. Consistent with the in vitro analysis, 14D2 monovalent IgG could also block TNF-mediated chemokine induction in vivo and demonstrated reduced agonist activity in vivo relative to the full-length 14D2 IgG (data not shown). Overall, these observations suggest that monovalent IgG is capable of maintaining its stability and biological activity in vivo and the charge-pair-based Fc fusion proteins can serve as a general platform for making heterodimer-based proteins for in vivo applications.
DISCUSSION
In this work, we have demonstrated that Fc heterodimers can be effectively produced by taking advantage of the electrostatic steering mechanism. Studies have shown that electrostatic steering effects can be exploited to alter the rate of association between proteins (22, 23). For example, protein design and selection techniques have been successfully employed to produce novel heterodimeric coil-coils (24–26). Although the design strategy in these studies included directed pairing of oppositely charged amino acid residues, further work supported the conclusion that formation of the most successful heterodimeric variants depends on interactions beyond electrostatics (27). In other studies, novel heterodimeric zinc finger nucleases have been engineered by introducing oppositely charged residues at the α-helical dimer interface thereby minimizing unwanted off-target DNA cleavage due to homodimers (28, 29). However, near complete suppression of homodimers also required altering hydrophobic interaction (28). These examples involve packing between two helices and the size of the protein-protein interfaces effected are much smaller than that of the ∼2500 Å2 CH3 dimer interface. It is notable that, despite the high affinity interaction between the two CH3 domains and the larger interface, only a few charged residue mutations can promote Fc heterodimer formation while effectively suppressing formation of homodimers. The study presented here suggests the electrostatic steering-based mechanism can be used to engineer large protein-protein interfaces for heterotypic interactions.
In a pioneering study, Carter and co-workers (5–8) engineered the antibody heavy chain for Fc heterodimerization using a knobs-into-holes strategy. The basis for creating knob and hole in the juxtaposed positions is that the knob and hole interaction will favor heterodimer formation, whereas the knob-knob and the hole-hole interactions will hinder homodimer formation due to the steric clash and deletion of favorable interactions, respectively. Although the knobs-into-holes strategy increased the production of heterodimer significantly, monomer and homodimer formations were not suppressed completely (6). The addition of inter-CH3 domain disulfide bond to the knobs-into-holes mutations further enhanced heterodimer formation up to 95% (7, 30), approaching the selectivity achieved here in the charge-pair strategy. The knob-into-hole mutations involve structurally conserved buried residues at the core of the interface. In contrast, the charge residues altered in this study are positioned around the rim of the interface. Unlike the knobs-into-holes method where a large amino acid is mutated to a smaller amino acid in the protein core (or vice versa for knob), the strategy involving charged residues is less likely to affect the packing of atoms at the hydrophobic core of the interface. For both strategies, a concern may arise that changing structurally conserved residues could lead to lower stability and affect other biophysical properties. Our qualitative comparison of differential scanning calorimetric profiles reveal similar melting temperature but higher enthalpy (area under the curve) for the charge-pair design compared with the knobs-into-holes strategy (supplemental Fig. S4). Although for both strategies the CH3 domain melting temperature (Tm) was decreased relative to WT, it still is well within a range that will support viable therapeutic protein productions as suggested by the pharmacokinetic analysis (Fig. 4d). Free energy calculations using a computational method (EGAD (31)) also showed that the charge-pair mutation has similar binding free energy (ΔΔGmut) compared with the knobs-into-holes mutations (supplemental Table S3). A more detailed assessment of free energy comparison is complicated due to the lack of a folding model (e.g. two-state versus three-state) and multidomain nature of the scFv-Fc.
Due to the 2-fold symmetry present in the Fc crystal structure, each unique interaction at the CH3-CH3 domain interface is represented twice in the structure. The electrostatic steering mechanism proposed here exploits the same 2-fold symmetry to effectively hinder the homodimer formation. Fig. 1d shows how a single mutation (K409D in the first chain or D399′K in the second chain) makes use of the symmetry to impart a repulsive electrostatic interaction in the homodimer setting. This repulsive effect could be further enhanced when different charge mutations were combined, particularly when these mutations were spread to different contact region in the CH3 dimer interface (supplemental Fig. S6). In the extreme case where three charged residues were changed to oppositely charged amino acids, no Fc homodimers could be detected when the single construct was expressed alone (Fig. 2d). In addition, the suppression of homodimers was equally effective for both “positively charged” and “negatively charged” homodimers by the nature of electrostatic repulsive mechanism. This is in contrast to the knob into hole design where knob-knob and hole-hole homodimers were not equally suppressed, due to the difference in shape complimentarity.
The ability to efficiently produce Fc heterodimers using the charge-pair mutations potentially allows the generation of many different types of heterodimeric proteins by fusing various functional groups such as peptides, antibody fragments, protein ligands, or receptors to both ends of the Fc polypeptide chains. For example, a biologically active bispecific CD3xTARTK antibody was produced in the form of a bispecific scFv-Fc fusion (Fig. 3). Using this format, in principle any two antibodies can be combined into a single protein by making the appropriate scFv forms of the antibodies and fusing them to the heterodimeric Fc segments. In practice, the stability and expression levels of the individual scFv components have a large influence on the expression levels for the bispecific molecules, in agreement with general observations regarding scFv expression (32). Conceptually, Fc heterodimers could also be used to generate full bispecific IgGs. However, additional engineering work will be required to efficiently control correct pairing of the LC with its HC, respectively. This issue could be circumvented by selecting a promiscuous LC that can pair with both HCs and still allow binding, as suggested previously (7).
We also have demonstrated that homogenous production of monovalent antibodies (Fab-Fc fusions) can be facilitated by using the heterodimeric Fc format (Fig. 4). This format is projected to be especially useful in situations where the goal is to antagonize a membrane-bound receptor (e.g. TNF receptor family molecules) without causing activation of the receptor as conventional bivalent antibodies often do via cross-linking of the receptor. We show here that the monovalent form of anti-TNFR1 IgG based on the charge-pair Fc heterodimer strategy can be produced at high levels and antagonize the receptor without residual agonist activity (Fig. 4). Compared with other forms of monovalent antibodies, such as Fab fragments, this Fab-Fc heterodimer fusion retains a IgG-like long serum half-life in vivo. This format provides a viable option for therapeutic applications where a monovalent antibody form with extended serum half-life is desired. A similar one-armed IgG construct using the knob and hole strategy was also developed for c-MET, a target to which a pure antagonist needs to be in the monovalent form (33).
In summary, we have demonstrated that the charged residues located around the rim of the CH3 domain interface can be engineered to promote Fc heterodimer formation. Using this engineered format, bispecific scFv-Fc as well as a monovalent IgG molecule having the desired in vitro and in vivo activities were effectively produced. Furthermore, pharmacokinetic analysis of the engineered human and mouse Fc heterodimers has shown that the serum half-life, which is an important property of the wild type homodimer Fc, is retained in the designed molecules. The work presented here may have broad implications for engineering relatively large protein-protein interactions where electrostatic steering effects can be harnessed to effectively suppress unwanted homodimer forms whereas promoting the robust production of useful novel heterodimeric molecules.
Supplementary Material
Acknowledgments
We sincerely acknowledge Salman Muzammil and Vladimir Razinkov for carrying out the DSC experiments. We thank Phillip Liu, Neha Patel, Nancy Sun, Sonal Patel, Esther Leng, Randal R. Ketchem, and Bill Fanslow for help with various studies presented in this article. We are grateful to Tom Boone for providing support and encouragement throughout this work.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S6.
I. N. Foltz, unpublished data.
- PDB
- protein data bank
- VH
- variable heavy
- VL
- variable light
- ASA
- accessible surface area
- TNF
- tumor necrosis factor
- HC
- heavy chain
- LC
- light chain
- TARTK
- tumor-associated receptor tyrosine kinase
- hu
- human
- mu
- mouse.
REFERENCES
- 1.Carter P. J. (2006) Nat. Rev. Immunol. 6, 343–357 [DOI] [PubMed] [Google Scholar]
- 2.Holliger P., Hudson P. J. (2005) Nat. Biotechnol. 23, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 3.Presta L. G. (2008) Curr. Opin. Immunol. 20, 460–470 [DOI] [PubMed] [Google Scholar]
- 4.Dumont J. A., Low S. C., Peters R. T., Bitonti A. J. (2006) BioDrugs 20, 151–160 [DOI] [PubMed] [Google Scholar]
- 5.Carter P. (2001) J. Immunol. Methods 248, 7–15 [DOI] [PubMed] [Google Scholar]
- 6.Ridgway J. B., Presta L. G., Carter P. (1996) Protein Eng. 9, 617–621 [DOI] [PubMed] [Google Scholar]
- 7.Merchant A. M., Zhu Z., Yuan J. Q., Goddard A., Adams C. W., Presta L. G., Carter P. (1998) Nat. Biotechnol. 16, 677–681 [DOI] [PubMed] [Google Scholar]
- 8.Atwell S., Ridgway J. B., Wells J. A., Carter P. (1997) J. Mol. Biol. 270, 26–35 [DOI] [PubMed] [Google Scholar]
- 9.Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr., Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. (1977) J. Mol. Biol. 112, 535–542 [DOI] [PubMed] [Google Scholar]
- 10.Ye Y., Godzik A. (2004) Nucleic Acids Res. 32, W582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Idusogie E. E., Presta L. G., Gazzano-Santoro H., Totpal K., Wong P. Y., Ultsch M., Meng Y. G., Mulkerrin M. G. (2000) J. Immunol. 164, 4178–4184 [DOI] [PubMed] [Google Scholar]
- 12.Lee B., Richards F. M. (1971) J. Mol. Biol. 55, 379–400 [DOI] [PubMed] [Google Scholar]
- 13.Miller S. (1990) J. Mol. Biol. 216, 965–973 [DOI] [PubMed] [Google Scholar]
- 14.Fredericks Z. L., Forte C., Capuano I. V., Zhou H., Vanden Bos T., Carter P. (2004) Protein Eng. Des. Sel. 17, 95–106 [DOI] [PubMed] [Google Scholar]
- 15.Deisenhofer J. (1981) Biochemistry 20, 2361–2370 [PubMed] [Google Scholar]
- 16.Ellerson J. R., Yasmeen D., Painter R. H., Dorrington K. J. (1976) J. Immunol. 116, 510–517 [PubMed] [Google Scholar]
- 17.Matthews B. W. (1995) Adv. Protein Chem. 46, 249–278 [DOI] [PubMed] [Google Scholar]
- 18.Bogan A. A., Thorn K. S. (1998) J. Mol. Biol. 280, 1–9 [DOI] [PubMed] [Google Scholar]
- 19.Ma B., Elkayam T., Wolfson H., Nussinov R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5772–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kung P., Goldstein G., Reinherz E. L., Schlossman S. F. (1979) Science 206, 347–349 [DOI] [PubMed] [Google Scholar]
- 21.Feige M. J., Groscurth S., Marcinowski M., Shimizu Y., Kessler H., Hendershot L. M., Buchner J. (2009) Mol. Cell 34, 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marvin J. S., Lowman H. B. (2003) Biochemistry 42, 7077–7083 [DOI] [PubMed] [Google Scholar]
- 23.Sheinerman F. B., Norel R., Honig B. (2000) Curr. Opin. Struct. Biol. 10, 153–159 [DOI] [PubMed] [Google Scholar]
- 24.O'Shea E. K., Lumb K. J., Kim P. S. (1993) Curr. Biol. 3, 658–667 [DOI] [PubMed] [Google Scholar]
- 25.Arndt K. M., Pelletier J. N., Müller K. M., Alber T., Michnick S. W., Plückthun A. (2000) J. Mol. Biol. 295, 627–639 [DOI] [PubMed] [Google Scholar]
- 26.Arndt K. M., Müller K. M., Plückthun A. (2001) J. Mol. Biol. 312, 221–228 [DOI] [PubMed] [Google Scholar]
- 27.Arndt K. M., Pelletier J. N., Müller K. M., Plückthun A., Alber T. (2002) Structure 10, 1235–1248 [DOI] [PubMed] [Google Scholar]
- 28.Miller J. C., Holmes M. C., Wang J., Guschin D. Y., Lee Y. L., Rupniewski I., Beausejour C. M., Waite A. J., Wang N. S., Kim K. A., Gregory P. D., Pabo C. O., Rebar E. J. (2007) Nat. Biotechnol. 25, 778–785 [DOI] [PubMed] [Google Scholar]
- 29.Szczepek M., Brondani V., Büchel J., Serrano L., Segal D. J., Cathomen T. (2007) Nat. Biotechnol. 25, 786–793 [DOI] [PubMed] [Google Scholar]
- 30.Sowdhamini R., Srinivasan N., Shoichet B., Santi D. V., Ramakrishnan C., Balaram P. (1989) Protein Eng. 3, 95–103 [DOI] [PubMed] [Google Scholar]
- 31.Pokala N., Handel T. M. (2005) J. Mol. Biol. 347, 203–227 [DOI] [PubMed] [Google Scholar]
- 32.Wörn A., Plückthun A. (1999) Biochemistry 38, 8739–8750 [DOI] [PubMed] [Google Scholar]
- 33.Jin H., Yang R., Zheng Z., Romero M., Ross J., Bou-Reslan H., Carano R. A., Kasman I., Mai E., Young J., Zha J., Zhang Z., Ross S., Schwall R., Colbern G., Merchant M. (2008) Cancer Res. 68, 4360–4368 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.