Abstract
Objectives
To use clinical and genetic analyses to determine the mutation causing autosomal recessive non-syndromic hearing loss (ARNSHL) segregating in two consanguineous Iranian families.
Study Design
Family study.
Methods
Members of each family received otologic and audiometric examination for the type and extent of hearing loss. Linkage mapping using Affymetrix 50K GeneChips and STRP analysis localized the hearing loss in both families to the DFNB3 locus. Direct sequencing of the MYO15A gene was completed on affected members of both families.
Results
Family L-3165 segregated a novel homozygous missense mutation (c.6371G>A) that results in a p.R2124Q amino acid substitution in the myosin XVa protein. While family L-896 segregated a novel homozygous missense (c.6555C>T) mutation resulting in a p.P2073S amino acid change.
Conclusions
These are the first MYO15A mutations reported to cause DFNB3 sensorineural hearing loss in the Iranian population. Like other mutations located in the myosin tail homology 4 (MyTH4) domain, the p.R2124Q and p.P2073S mutations are predicted to disrupt the function of the myosin XVa protein, which is integral to the mechanosensory activity of hair cells in the inner ear.
Keywords: DFNB3, MYO15A gene, missense mutation
INTRODUCTION
Sensorineural hearing loss (SNHL) is the most prevalent genetic sensory defect in humans. It is estimated that globally 4 out of every 10,000 children born have profound hearing loss1. Non-syndromic SNHL accounts for ~70% of hereditary hearing loss and 80% of SNHL cases have an autosomal recessive mode of inheritance (ARNSHL). To date, 25 genes and 67 loci have been implicated in ARNSHL2.
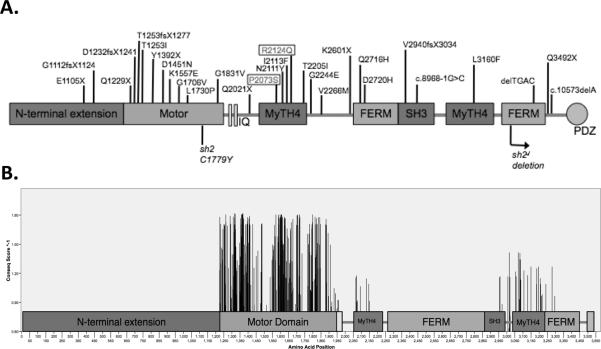
The first report of hearing loss at the DFNB3 locus was from an isolated village in Indonesia where 2% of the population was hearing impaired3. Subsequently, it was shown that the causative mutation in this village and two unrelated families resided in the MYO15A gene4. Since these original reports, 24 additional DFNB3-causing mutations have been identified in MYO15A, which spans 71 kb on chromosome 17p11.2 and is comprised of 66 exons that encode a 3,530 amino acid protein in its longest form. (Table 1 and Fig.1)5–8.
Table 1.
DFNB3-causing MYO15A mutations.
| Exon (domain) | Amino Acid Change | Mutation Type | Origin of Family (#) | Reference |
|---|---|---|---|---|
| Exon 2 (N-terminal) | E1105X | nonsense | Pakistan (1) | 6 |
| Exon 2 (N-terminal) | G1112fsX1124 | deletion frameshift | Pakistan (1) | 6 |
| Exon 3 (motor) | Q1229X | Nonsense | Pakistan (1) | 5 |
| Intron 4 | D1232fsX1241 | splice donor site | Pakistan (1) | 5 |
| Intron 4 | T1253fsX1277 | splice donor site | Pakistan (1) | 6 |
| Exon 5 (motor) | T1253I | missense | India (1) | 6 |
| Exon 10 (motor) | Y1392X | nonsense | Pakistan (1) | 6 |
| Exon 12 (motor) | D1451N | missense | India (1) | 6 |
| Exon 15 (motor) | K1557E | missense | Pakistan (1) | 6 |
| Exon 18 (motor) | G1706V | missense | Pakistan (1) | 6 |
| Exon 19 (motor) | L1730P | missense | Pakistan (1) | 6 |
| Exon 21 (motor) | G1831V | missense | Turkey (1) | 7 |
| Exon 28 | Q2021X | nonsense | Pakistan (1) | 6 |
| Exon 30 (MyTH4 a) | N2111Y | missense | India (1) | 4 |
| Exon 30 (MyTH4 a) | I2113F | missense | Bali (1) | 4 |
| Exon 30 (MyTH4 a) | R2124Q | missense | Iran (1) | This study |
| Exon 30 (MyTH4 a) | P2073S | missense | Iran (1) | This study |
| Exon 31 (MyTH4 a) | T2205I | missense* | North America (1) | 5 |
| Exon 32 | G2244E | missense | Pakistan (1) | 6 |
| Exon 33 | V2266M | missense | Pakistan (1), Turkey (1) | 6 |
| Exon 41 | K2601X | nonsense | India (1) | 4 |
| Exon 44 (FERM a) | Q2716H | missense | Pakistan (1) | 5 |
| Exon 45 (FERM a) | D2720H | missense | Pakistan (4) | 6 |
| Exon 51 (SH3) | V2940fsX3034 | frameshift | Pakistan (1) | 6 |
| Intron 50 | - | splice donor site | Turkey (1) | 7 |
| Exon 57 (MyTH4 b) | L3160F | missense | Pakistan (1) | 6 |
| Exon 62 (FERM b) | - | deletion frameshift** | Brazil (1) | 8 |
| Exon 65 | Q3492X | nonsense | Pakistan (1) | 6 |
| Exon 66 (PDZ ligand) | - | deletion frameshift | Brazil (1) | 8 |
Mutation was found in a patient hemizygous at the DFNB3 locus with Smith-Magenis Syndrome
Mutation was found in a hemizygous individual.
Figure 1.
Domains of the myosin XVa protein. A. Diagram showing the location of human (top) and mouse (bottom) MYO15A mutations. The p.R2124Q and p.P2073S mutations are boxed Figure is not to scale. B. Diagram showing the amino acid positions in each domain of myosin XVa (x-axis) and Conseq conservation scores for each residue (y-axis). Any residue with a Conseq score less than 7 (E-value >0) was excluded. Figure is to scale.
The encoded protein is the unconventional myosin XVa. Myosin XVa is unique among unconventional myosins in that it includes a long N-terminal domain (coded by exon 2) that is alternatively spliced to generate distinct class 1 and class 2 protein isoforms10, 11. The N-terminal domain is required for normal hearing, as premature stop mutations that result in loss of this domain cause DFNB3 hearing loss6. Myosin XVa also contains domains that are conserved within the myosin protein family, including the motor domain, IQ motifs (calmodulin/myosin light chain binding), MyTh4 domains (Myosin-Tail like Homology region 4), FERM motifs (4.1 protein, Ezrin, Radixin, and Moesin), SH3 domain (Src Homology 3), and the PDZ ligand domain (Fig.1A)10, 12.
DFNB3-causing mutations have been identified in all domains, with the exception of the IQ domains in the neck region, in families from Pakistan, India, Turkey, Indonesia, Brazil and North America (Table 1). In this study, we have characterized two Iranian ARNSHL families and identified two novel DFNB3-causing mutations in MYO15A. These novel missense mutations are both located in the first MyTh4 domain and are predicted to disrupt normal myosin XVa function.
MATERIALS AND METHODS
Family Reports
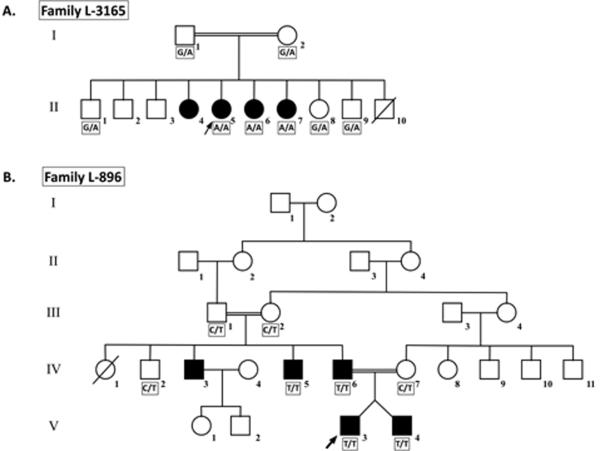
Family L-3165 is a five-generation consanguineous Iranian family segregating apparent ARNSHL (Fig. 2A). Hearing impaired persons appear in the 4th and 5th generations, consistent with autozygosity by descent. Family L-896 (Fig. 2B) is also a five-generation consanguineous Iranian family segregating apparent ARNSHL. In contrast to family L-3165, the inheritance pattern in this family appears to be pseudo-dominant.
Figure 2.
Pedigrees of the Iranian families affected with DFNB3 ARSNHL A. Family L-3165; Genotypes for individuals carrying the c.6371G>A mutation are shown. B. Family L-896; Genotypes for individuals carrying the c.6555C>T mutation are shown. Open symbols = unaffected; filled symbols = affected; diagonal line = deceased.
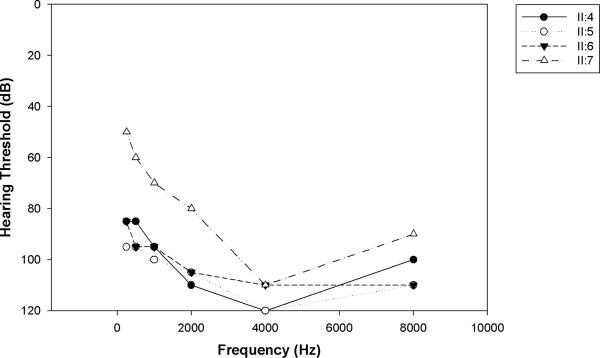
For consenting persons in each family, audiologic testing was completed to document the degree of hearing loss. The test results for family L-3165 are displayed (Fig.3). Examination by an otolaryngologist and clinical geneticist excluded a syndromic presentation. Ten milliliters of whole blood were obtained as a DNA source. Human research institutional review boards at the Welfare Science and Rehabilitation University and the Iran University of Medical Sciences, Tehran, Iran, and the University of Iowa, Iowa City, Iowa, USA approved all procedures.
Figure 3.
Audiograms of affected members of the family L-3165. All affected individuals displayed severe-to-profound bilateral SNHL.
SNP Genotyping and Linkage Analysis of Family L-3165
Genomic DNA from individuals I-1, I-2, II-1, II-4, II-5, II-6, II-7, II-8, II-9 in family L-3165 (Fig. 2) was genotyped for 50,000 SNPs using Affymetrix 50K XBA GeneChips at the Translational Genomics Research Institute (TGEN, Phoenix, Arizona). Genotypes were determined using the BRLMM genotyping algorithm13, 14. Genotyping data were examined with PEDSTATS15 for Mendelian inheritance errors, and MERLIN16 for errors based on inferred double recombination events between tightly linked markers.
An autosomal, genome-wide parametric linkage analysis was performed since males and females appeared equally affected. All linkage analysis was performed with MERLIN. Since the pedigree had 18 bits it was analyzed using exact multipoint linkage analysis16. A subset of 6432 single nucleotide polymorphisms (SNPs) spaced approximately 0.5 cM apart across the genome and with an average heterozygosity of 0.43 was chosen from the 50K XBA set to satisfy the linkage equilibirium requirements of the Lander-Green algorithm for linkage analysis17. The selection and assembly of the data files were performed with an in-house PERL script (linkdatagen.pl, http://bioinf.wehi.edu.au/software). An initial parametric linkage analysis was run assuming a fully penetrant autosomal recessive model with a disease allele frequency of Pr(a)=0.0001 and penetrances of pr(disease|aa)=1=pr(disease|aA), Pr(disease|AA)=0. Haplotypes inferred with Merlin were imported into Haplopainter18.
STRP analysis of Family L-896
Short tandem repeat (STRP) analysis was completed using markers developed by the Cooperative Human Linkage Center and made available through Research Genetics Incorporated (Invitrogen, Carlsbad, CA) according to a published methodology1. Markers with heterozygosity > 50% were selected for this analysis.
PCR and Sequencing
The MYO15A gene was amplified using gene-specific primers (Table 2). Amplification reactions were cycled using a standard protocol on a GeneMate Genius thermocycler (ISC BioExpress, UT, USA). Sequencing was completed with BigDye™ v3.1 Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA), according to the manufacturer's instructions. Sequencing products were read using an ABI 3730s sequencer (Perkin Elmer, Waltham, MA). All sequencing chromatograms were compared to published cDNA sequence; nucleotide changes were detected using Sequencher v4.5 (Gene Code Corporation, Ann Arbor, MI).
Table 2.
Oligonucleotides used for amplification of the MYO15A gene.
| Exon | Forward Primer (5'–3') | Reverse Primer (5'–3') |
|---|---|---|
| 2 | MULTIPLE – AVAILABLE UPON REQUEST | |
| 3 | ATG ACC AAG CCA GGG GTC | CTC TGG CTG GGA GGG TG |
| 4 | GAC CCA TGC CAG AAC CAG | AGA AAT CTG TGC GTC CCA CC |
| 5 | ATC TGT CCG GAT GGA AAC AG | TCT GAC TCA TGG CTC AGG TG |
| 6--7 | GGG AGG TGT GGG AGC TTA G | TCG GGA GTA CAT GAG GTG TG |
| 8 | TCC TGG AGA GAG TGG TGG TC | CTA GGA CAG GCC TTT GGA TG |
| 9--10 | GGG TGT CCC CAG CTA TGC | TAT CTG TAC CTC CCA CCC CG |
| 11 | GTT CTC ATC TGC AGC CCA CT | AAA CTC ACC CTC CCC AAA TC |
| 12 | CAA CTC AGG CCA CCA CAC TA | AAA ACA GGA ACA AGT GAT ATG TGC |
| 13 | GAC TAC TGG CAT GAG CCA CA | TGA CCC AGG GAC AGA GAG AG |
| 14--15 | GCT TTC CGG AGG CAG AG | GAG GGA GGC GAG ACC TTG |
| 16 | AGG GAA GGT AGG GGC AAA | CTG TCT CCA AGG AGG TCC AC |
| 17 | ATT CAA CAT GGG AGG GAG G | TGA GGA CAT GAG GCT GAG AG |
| 18 | ATA GTG AGG TTG CCA CCA GG | TCT CCA ACA GCT AGC AGC AC |
| 19 | TCC CTC CTA GGA TAG ACA GAG AG | AAG GCA GGC TGG GTG TG |
| 20 | TTC CTC CTC ATT TCG GTC TC | CAA GGT CAC ACA GCA TGG G |
| 21–22 | TTC CTC CTC ATT TCG GTC TC | CAA GGT CAC ACA GCA TGG G |
| 23 | TAG CAG ACA CCT CGG GTA GG | GAC TCA GTA GTT GTG GAC CCC |
| 24 | CTT AGT CCA GCC TCC TGG C | TTC AGG CGT GAC CTC TCC |
| 25 | AGG GCC TCT CTA CCT TTT GG | CTA AGT GCC CTT TCC CCT TC |
| 26--28 | GTG CCG GTC GTC ACC TC | CCC AGG GCA AGG ACA ATG |
| 29 | CAC AGA GCA GTG GGT CCA G | CTC ATG GCC CAG TTT CAG G |
| 30 | GGG GAC TGG AAG GAA CAA C | CTT TAA GAC CCT GCC TTG GG |
| 31 | CAG CCC TCA GCC CCA AG | ACT GGG CCC TGC TGA CTC |
| 32 | GCA CAG CCA AAC TGG ACT C | CCT TCT GCC TGG GAG TGG |
| 33 | TCT GTT CAT GTT TAG GGT CTG G | CTC AGC CTG TCC CAG CAG |
| 34--35 | GGA GAA AGC CAC TGA ATA CCA G | GAG AAG CTC TCA GGT CAC CC |
| 36--38 | AGT GTC AGG TGC CTG TTG C | TCC TCT TTA CAG CTT GTG TCT CC |
| 39--40 | TCT GGA GTC CCA GAG AGC AG | GGG CCA TGA TGG ACA CTC |
| 41--42 | ATG TGA TGG GAA AGG GAG AC | CTG TGC CCA CAG ACT TCC TC |
| 43 | ACT CTA GCC TGG GGG ACA AC | CCC AAG TCC TAG ACC CTC CT |
| 44 | CCC AGG AGG ACA GAA AAA GG | GGG AGG GGG AGA TTC AAT AA |
| 45 | AGT ATA GTC CAG CCT GGG TCC | CTG GCT GTG CCT CTG ACT G |
| 46 | TGG CCA TCT CAT CCA TTT CT | CAC AGC TAG GAG CTG CAC AC |
| 47 | GAA CCA GCT GGA CAC ACA GA | AAA TGG GTT TGC TTC AAT GG |
| 48 | GGG CAG GAC AGG ATC AGA AG | AGG GAG ATC CCT GTT GCT G |
| 49--50 | CTA GGC CTC TGG GAG TGG | CAC CAC GAG TGG GTG AAA C |
| 51 | CCC CTT AGT CAC AAG ACA AGA C | TTA TCC CCA CTC GCC TCA C |
| 52 | CTA GGG GTT CGC TTG TCA GT | AGT GGG GCC TCC GAG ACT |
| 53 | TGT GAG GCT CAT TTC AGT GC | AGG GTG CTG AGA ATC AGA GG |
| 54--55 | TGT GTC CCC TTT CTG TTC TG | TGA TAG ATG GGG AAA CTG AAC C |
| 56 | GTG CCC ACC CTG TTC TTA TG | CCT CCT GGA GCA TGG ACA C |
| 57 | TCT CAG CTC AAT CCC AGG AG | TCC ACC CAG TCC CCA AG |
| 58 | ATG GGG GAG TAA ATG CCT TC | GGC TTG TGT CTC CCA TTC AT |
| 59 | CAG GAG ACA AGG GCT GTC C | CTG GAG CCT GGG CTG TC |
| 60 | AGA AGG ACA GAG GTC AAG CC | AAA TCT GGG TGG AGG GC |
| 61 | AAG CTG TGT CCC AGA ACA GG | ACA GGG CCT GAA TCA TGA AC |
| 62 | TGA GAG GGC AGG GTT GC | CAT GCA TGT CCC CAG GTC |
| 63 | ACA GTG AGG ATT GCC TGA GC | TAC CCA TCC TCC ATG ACC AC |
| 64 | AGC CCA GAG AAG CTA TGC AG | AGG CTC AGA GGA GGG AAG AG |
| 65 | TGG TTG AGA CTA TCC TCG CC | GAC CTG ACC TAT CTT GGA GCC |
| 66 | CAA GGT AAG AGC TGG GGA AG | TTG ATC CTG AGA GGT TCA GTG |
Conseq Analysis
Conservation scores for each amino acid in myosin XVa when compared to 50 similar protein sequences was determined using the Conseq program (http://conseq.tau.ac.il/). Any conservation score greater than 1 standard deviation above the mean for the myosin XVa amino acid sequence was regarded as highly conserved.
RESULTS
Linkage Mapping to the DFNB3 Locus
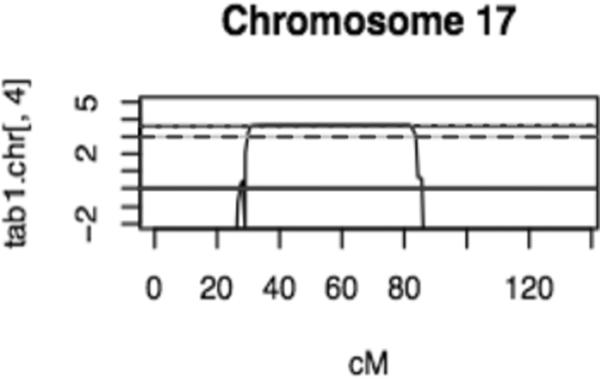
Linkage analysis of family L-3165 detected one hundred and thirty nine Mendelian (0.24% error) and 465 double recombinant (0.8%) errors in the genome-wide analysis. The highest parametric LOD score of 3.7 was achieved for a region on chromosome 17 (Fig. 4). There were no other regions in the genome where the LOD score exceeded 3. The 1-LOD-drop region spanned approximately 56 cM flanked by markers SNP_A-1671362 and SNP_A-1663708 at chromosomal position 17p13.1-q24.3 (9.9–52 Mb). Haplotype analysis reveals that the critical region coincides with the region identified in the linkage analysis (Fig.5). STRP analysis revealed DFNB3 was also the likely disease locus for family L-896 (data not shown).
Figure 4.
Linkage Mapping of family L-3165. Results from chromosome 17 showing significant LOD score of 3.7 (dotted line). LOD scores of 1 (solid line) and 3 (dashed line) are also indicated
Figure 5.
Haplotype analysis of family L-3165. Affymetrix SNP markers on chromosome 17 are shown.
Mutations in MYO15A
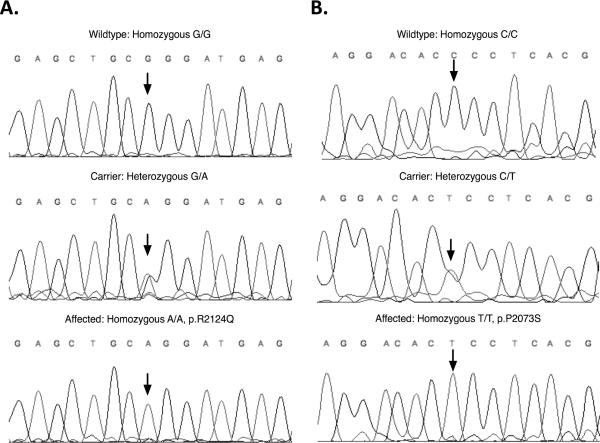
The critical region in both families contained the known ARNSHL locus DFNB3. Direct sequencing of the 65 exons of MYO15A identified novel homozygous missense mutations. In family 3165 a homozygous c.6371G>A mutation was identified that substitutes a glutamine for an arginine (p.R2124Q) in the first MyTH4 domain (Fig.2A and 6A). In family L-896 a novel homozygous missense mutation (c.6555C>T) was also detected in the first MyTH4 domain that results in substitution of a proline for a serine (p.P2073S) (Fig. 2B and 6B).
Figure 6.
Mutations in the MYO15A gene. Chromatograms showing the c.6371G>A mutation found in family L-3165 (A) and c..6555C>T mutation found in family L-896 (B).
The R2124 and P2073 residues are both located in the first MyTH4 domain of the MYO15A protein and are conserved between human and mouse. The substitution of an uncharged, polar glutamine for arginine results in loss of the positive charge on the arginine sidechain. The bulky, non-polar sidechain of proline is replaced with a polar serine sidechain. The c.6371G>A mutation was not observed in 42 (84 chromosomes) Iranian control or 94 (188 chromosomes) CEPH control individuals. The c.6555C>T mutation was not observed in 43 Iranian control (86 chromosomes) or 93 (186 chromosomes) CEPH control individuals.
DISCUSSION
The unconventional myosin XVa protein is required for normal auditory function as its mutation leads to SNHL at the DFNB3 locus. Two other unconventional myosin proteins, VI and VIIa, are similarly required for normal auditory function, and their mutation also leads to hearing impairment (reviewed in Brown 2008). Myosins are molecular motor proteins that drive the movement of actin filaments via ATP hydrolysis to facilitate muscle contraction, organelle trafficking, cell movement, cytokinesis, and signal transduction12.
Hair bundles located at the apex of sensory hair cells in the cochlea are responsible for the mechano-electrical transduction of sound waves in humans. Each hair bundle consists of up to 300 extracellular projections called stereocilia that contain an actin core and are arranged in rows of increasing height, forming a characteristic staircase architecture19. Extracellular lateral projections (side-links) hold the stereocilia of hair bundles together while tip-links at the apex of the stereocilia facilitate signal transduction19, 25. Myosin XVa immunoreactivity has been observed in stereocilia tips at E18.5 in the mouse, coinciding with formation of the hair bundles of the inner ear. Myosin XVa is located at the stereocilia tips following hair bundle maturation and the amount present is directly proportional to the length of the stereocilia11, 20.
Homozygous Shaker-2 (sh2) mice are deficient in myosin XVa due to a premature stop codon in the motor domain of the protein9 (Fig.1). These mice exhibit profound deafness similar to that observed in human DFNB3 patients, in addition to vestibular defects that are not present in human patients. Hair cell stereocilia in sh2 mice are correctly positioned during development, but are much shorter and do not display the characteristic staircase architecture when compared to those of wild-type mice9. Originally, it was hypothesized that the absence of myosin XVa in hair bundles resulted in a defect in mechanosensation21. However, the abnormally short stereocilia of deaf sh2 mice retain mechanosensory activity suggesting that stereocilia length and the architecture of the hair bundle are crucial in maintaining normal hearing function22.
The whirler (whi) mouse, which contains a mutation in the whirlin gene, shows profound hearing loss similar to that of shaker mice[26]. Homozygous whirler mice have a deficiency in the whirlin protein and their stereocilia appear similar to those in shaker mice: abnormally short with no staircase formation4, 9. The tail of myosin XVa, of which the MyTH4 domains form part, interacts with whirlin, suggesting that these two proteins are partners20, 21. The PDZ ligand domain of myosin XVa and two of the three PDZ binding domains of whirlin are required for this interaction. Myosin XVa is the only myosin in mammals known to contain a PDZ domain21.
Homozygous whirler mice (lacking whirlin) have myosin XVa present at their stereocilia tips, while homozygous shaker mice (lacking myosin XVa) have neither whirlin nor myosin XVa present at their stereocilia tips11,21. Myosin XVa has been observed migrating towards the plus ends of actin filaments at stereocilia tips, confirming myosin XVa activity as a motor protein21. These data support the hypothesis that myosin XVa acts as a transport protein and whirlin is its cargo during development of hair bundles in the cochlea.11, 20, 21 This transport most likely occurs during lengthening and formation of stereocilia staircase morphology, given the localization of myosin XVa during this period20,21.
Mutations in all but one protein domain of myosin XVa have been shown to be causative of DFNB3 hearing loss. Three missense mutations in the first MyTH4 domain of the protein have been described previously: two homozygous mutations in Indian and Balinese families, and one hemizygous mutation in a family affected by Smith-Magenis syndrome from North America (Table 1). Here we report two further mutations in the first MyTH4 domain and the first DFNB3 mutations segregating in Iranian families. The arginine-to-glutamine and proline-to-serine substitutions result in the loss of positively charged and bulky, non-polar sidechains and replacement with residues containing polar sidechains. These alterations are predicted to negatively affect the function of the myosin XVa protein, perhaps by disrupting its interaction with whirlin.
Given the large number of DFNB3-causing MYO15A mutations identified in various populations (Fig.1 and Table 1) and the size of the gene, it would be useful to develop a more targeted screening approach for suspected DFNB3 ARNSHL families. In Table 3 and Figure 1 all known MYO15A mutations and the relative conservation of amino acids in each domain of the protein are depicted. A targeted sequencing strategy would focus primarily on the domains with the highest mutation rate and highest percentage of conserved amino acids. The motor domain is the most highly conserved (84.5%) domain and the site of ten mutations. Conversely, the N-terminal domain, which is unique among myosins, contains the least conserved residues and only three mutations. Therefore, sequencing of the motor domain should be a priority.
Table 3.
Characteristics of myosin XVa domains
| Region: | Domain: | Amino Acids | Number of amino acids in domain | Encoded by: | Number ofexons in domain | Number of known mutations in domain | Mutations per exon | %of highly conserved amino acids |
|---|---|---|---|---|---|---|---|---|
| Head | N-terminal extension | 1–1230 | 1231 | Exons 2–3 | 2 | 2 | 1.00 | 1.2 |
| Motor Domain | 1231–1942 | 712 | Exons 3–24 | 22 | 10 | 0.45 | 84.5 | |
| Neck | IQ domains | 1943–1970 | 28 | Exons 24–25 | 2 | 0 | 0.00 | 0.9 |
| Other domain | 1971–2059 | 89 | Exons 25–28 | 4 | 1 | 0.25 | 0.0 | |
| Tail | MyTH4 (a) | 2060–2230 | 171 | Exons 29–31 | 3 | 5 | 1.67 | 3.2 |
| Other domain | 2231–2255 | 25 | Exons 32–42 | 11 | 3 | 0.27 | 0.0 | |
| FERM (a) | 2256–2857 | 602 | Exons 43–48 | 6 | 2 | 0.33 | 0.0 | |
| SH3 | 2858–2989 | 132 | Exons 49–51 | 3 | 2 | 0.67 | 1.2 | |
| Other domain | 2990–3028 | 39 | Exon 52 | 0 | 0 | 0.00 | 1.8 | |
| MyTH4 (b) | 3029–3230 | 202 | Exons 53–59 | 7 | 1 | 0.14 | 6.5 | |
| FERM(b) | 3231–3450 | 220 | Exons 59–64 | 6 | 1 | 0.17 | 0.9 | |
| Other domain | 3451–3497 | 47 | Exon 65 | 1 | 1 | 1.00 | 0.0 | |
| PDZ ligand | 3498–3531 | 34 | Exon 66 | 1 | 1 | 1.00 | 0.0 |
When the mutations identified in this study are included, the first MyTH4 domain contains a relatively high number of known mutations (5). This domain also contains a high percentage of conserved amino acids relative to other domains of the protein. We therefore recommend prioritizing screening of exons encoding the motor and first MyTH4 domains in ARNSHL families linked to the DFNB3 locus.
The structure of the MyTH4 domain has not been characterized. In other myosins, the MyTH4 domain has been implicated in microtubule binding as well as actin binding to the plasma membrane24. Some data suggest that the MyTH4/FERM domains in myosin XVa are required for localization of myosin XVa to stereocilia tips, although their specific function has not been elucidated25. The co-localization of myosin XVa and whirlin proteins appears essential to form the transmembrane actin microfilament assembly complex at the stereocilia tips. We speculate that the p.R2124Q and p.P2073S mutations interfere with the interaction between myosin XVa and whirlin, thereby preventing the formation of this complex which appears to be required for normal hearing.
V. CONCLUSION
We have identified novel mutations in the MYO15A gene in two Iranian DFNB3 families. These mutations are the first to be described in the Iranian population and both affect the first MyTH4 domain of the myosin XVa protein. The first MyTH4 and motor domains are highly conserved at the protein level and constitute common sites for DFNB3-causing mutations. As such they should be preferentially screened in DFNB3 ARNSHL families. Hearing impairment in a mouse model of DFNB3 has been rescued by supplementation of wild-type MYO15A21. Given advances in genetic therapies for hearing loss27, a similar gene replacement strategy could be developed for DFNB3 families like the ones described in this report.
ACKNOWLEDGEMENTS
The authors sincerely thank the family for their participation in this study. RJH Smith is the Sterba Hearing Research Professor, University of Iowa College of Medicine, who supported the project with National Institutes of Health (NIH) grant RO1 DCOO3544. No researchers involved in this study report a conflict of interest.
Funding: National Institutes of Health (NIH) grant RO1 DCOO3544.
Abbreviations
- MYO15A
myosin 15A gene
- MYO15A
myosin 15A protein
- ARNSHL
autosomal recessive non-syndromic hearing loss
- SNHL
sensorineural hearing loss
- MYTH4
myosin tail homology 4 domain
REFERENCES
- 1.Smith RJ, ale J, James F, White KR. Sensorineural hearing loss in children. The Lancet. 2005;365(9462):879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- 2.Van Camp G, Smith RJH. [Accessed June 17, 2008]; Hereditary Hearing Lost HomepageAvailable at: http://webh01.ua.ac.be/hhh/.
- 3.Friedman TB, Liang Y, Weber JL, et al. A gene for congenital, recessive deafness DFNB3 maps to the pericentromeric region of chromosome 17. Nat Genet. 1995;9(1):86–91. doi: 10.1038/ng0195-86. [DOI] [PubMed] [Google Scholar]
- 4.Wang A, Liang Y, Fridell RA, et al. Association of Unconventional Myosin MYO15 Mutations with Human Non-syndromic Deafness DFNB3. Science. 1998;280(5368):1447–1451. doi: 10.1126/science.280.5368.1447. [DOI] [PubMed] [Google Scholar]
- 5.Liburd N, Ghosh M, Riazuddin S, et al. Novel mutations of MYO15A associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with Smith-Magenis syndrome. Hum Genet. 2001;109(5):535–541. doi: 10.1007/s004390100604. [DOI] [PubMed] [Google Scholar]
- 6.Nal N, Ahmed ZM, Erkal E, et al. Mutational spectrum of MYO15A: the large N-terminal extension of myosin XVA is required for hearing. Hum Mutat. 2007;28(10):1014–1019. doi: 10.1002/humu.20556. [DOI] [PubMed] [Google Scholar]
- 7.Kalay E, Uzumcu A, Krieger E, et al. MYO15A (DFNB3) mutations in Turkish hearing loss families and functional modeling of a novel motor domain mutation. Am J Med Genet A. 2007;143A(20):2382–2389. doi: 10.1002/ajmg.a.31937. [DOI] [PubMed] [Google Scholar]
- 8.Lezirovitz K, Pardono E, de Mello Auricchio MT, et al. Unexpected genetic heterogeneity in a large consanguineous Brazilian pedigree presenting deafness. Eur J Hum Genet. 2008;16(1):89–96. doi: 10.1038/sj.ejhg.5201917. [DOI] [PubMed] [Google Scholar]
- 9.Probst FJ, Fridell RA, Raphael Y, et al. Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science. 1998;280(5368):1444–1447. doi: 10.1126/science.280.5368.1444. [DOI] [PubMed] [Google Scholar]
- 10.Liang Y, Wang A, Belyantseva IA, et al. Characterization of the human and mouse unconventional myosin XV genes responsible for hereditary deafness DFNB3 and shaker 2. Genomics. 1999;61(3):243–258. doi: 10.1006/geno.1999.5976. [DOI] [PubMed] [Google Scholar]
- 11.Belyantseva IA, Boger ET, Friedman TB. Myosin XVa localizes to the tips of inner ear sensory cell stereocilia and is essential for staircase formation of the hair bundle. Proceedings of the National Academy of Sciences. 2003;100(24):13958–13963. doi: 10.1073/pnas.2334417100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krendel M, Mooseker MS. Myosins: tails (and heads) of functional diversity. Physiology (Bethesda) 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- 13.Di X, Matsuzaki H, Webster TA, et al. Dynamic model based algorithms for screening and genotyping over 100 K SNPs on oligonucleotide microarrays. Bioinformatics. 2005;21(9):1958–1963. doi: 10.1093/bioinformatics/bti275. [DOI] [PubMed] [Google Scholar]
- 14.Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22(1):7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- 15.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21(16):3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 16.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 17.International HapMap Consortium. Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiele H, Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21(8):1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 19.Frolenkov GI, Belyantseva IA, Friedman TB, Griffith AJ. Genetic insights into the morphogenesis of inner ear hair cells. Nat Rev Genet. 2004;5(7):489–498. doi: 10.1038/nrg1377. [DOI] [PubMed] [Google Scholar]
- 20.Delprat B, Michel V, Goodyear R, et al. Myosin XVa and whirlin, two deafness gene products required for hair bundle growth, are located at the stereocilia tips and interact directly. Hum Mol Genet. 2005;14(3):401–410. doi: 10.1093/hmg/ddi036. [DOI] [PubMed] [Google Scholar]
- 21.Belyantseva IA, Boger ET, Naz S, et al. Myosin-XVa is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nat Cell Biol. 2005;7(2):148–156. doi: 10.1038/ncb1219. [DOI] [PubMed] [Google Scholar]
- 22.Stepanyan R, Belyantseva IA, Griffith AJ, Friedman TB, Frolenkov GI. Auditory mechanotransduction in the absence of functional myosin-XVa. J Physiol. 2006;576(3):801–808. doi: 10.1113/jphysiol.2006.118547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown SD, Hardisty-Hughes RE, Mburu P. Quiet as a mouse: dissecting the molecular and genetic basis of hearing. Nat Rev Genet. 2008;9(4):277–290. doi: 10.1038/nrg2309. [DOI] [PubMed] [Google Scholar]
- 24.Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431(7006):325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- 25.Boger ET, Folenkov GI, Friedman TB, Belyantseva IA. Myosin XVA. In: Coluccio LM, editor. Myosins. Springer; Netherlands: 2007. pp. 441–467. [Google Scholar]
- 26.Holme RH, Kiernan BW, Brown SDM, Steel KP. Elongation of hair cell stereocilia is defective in the mouse mutant whirler. J. Comp. Neur. 2002;450(1):94–102. doi: 10.1002/cne.10301. [DOI] [PubMed] [Google Scholar]
- 27.Hildebrand MS, Newton SS, Gubbels SP, et al. Advances in molecular and cellular therapies for hearing loss. Mol Ther. 2008;16(2):224–236. doi: 10.1038/sj.mt.6300351. [DOI] [PubMed] [Google Scholar]