Abstract
The rich knowledge of morphological variation among organisms reported in the systematic literature has remained in free-text format, impractical for use in large-scale synthetic phylogenetic work. This noncomputable format has also precluded linkage to the large knowledgebase of genomic, genetic, developmental, and phenotype data in model organism databases. We have undertaken an effort to prototype a curated, ontology-based evolutionary morphology database that maps to these genetic databases (http://kb.phenoscape.org) to facilitate investigation into the mechanistic basis and evolution of phenotypic diversity. Among the first requirements in establishing this database was the development of a multispecies anatomy ontology with the goal of capturing anatomical data in a systematic and computable manner. An ontology is a formal representation of a set of concepts with defined relationships between those concepts. Multispecies anatomy ontologies in particular are an efficient way to represent the diversity of morphological structures in a clade of organisms, but they present challenges in their development relative to single-species anatomy ontologies. Here, we describe the Teleost Anatomy Ontology (TAO), a multispecies anatomy ontology for teleost fishes derived from the Zebrafish Anatomical Ontology (ZFA) for the purpose of annotating varying morphological features across species. To facilitate interoperability with other anatomy ontologies, TAO uses the Common Anatomy Reference Ontology as a template for its upper level nodes, and TAO and ZFA are synchronized, with zebrafish terms specified as subtypes of teleost terms. We found that the details of ontology architecture have ramifications for querying, and we present general challenges in developing a multispecies anatomy ontology, including refinement of definitions, taxon-specific relationships among terms, and representation of taxonomically variable developmental pathways.
Keywords: Bioinformatics, devo-evo, fish, morphology, ontology, Teleostei
Decades of comparative anatomical work in systematics have produced a rich, and still growing, body of data on the natural diversity of phenotypes. However, the noncomputable “free-text” format in which systematic data are published poses a challenge to the use of these data outside the narrow scope of the original study in which they were collected. The core of the problem is that, even where printed text is digitally rendered, the meaning of a text string such as “bone” is not interpretable to a computer; it is simply a string of letters. Thus, even a seemingly simple computational task, such as compiling a list of species possessing a similar structure, requires substantial human time investment and expert knowledge of the literature. A more complex investigation of morphology, such as a comparison of structures across a monophyletic group of species, requires processing such a large amount of data that it is rarely undertaken except by the most determined domain experts. Yet larger-scale analyses of the patterns of morphological evolution across multiple clades are simply not tractable. Testing, for example, whether sets of morphological characters vary in statistically similar ways across clades, or whether there is differential enrichment in the types of morphological qualities (shape, size, presence/absence) among sets of anatomical features or among clades, can only be done if morphology is represented in a format amenable to computation. The current moribund format of morphological data not only precludes expert analysis for a “big picture” view but also means that morphological data are inaccessible to scientists outside of the domain for use in interdisciplinary studies. The increasing rarity of morphology-based studies may be at least partially attributed to the perception that they are small, isolated, “old-fashioned,” essentially noncomputable data sets.
In contrast to the situation for morphological data, the data from genomics, genetics, development, and experimental assays are rapidly proliferating and simultaneously being databased and synthesized by model organism communities. Both genetic and phenotypic data are represented in these databases using ontologies. Ontologies are hierarchical vocabularies with well-defined relationships between terms that can be understood by humans and machines. Ontologies have been widely used in the model organism community to represent biological knowledge (e.g., genes, Blake and Harris 2008; anatomy, Rosse and Mejino 2003). Genetic data (along with genomic and developmental data) are curated using gene ontologies and are frequently associated with particular phenotypes. Anatomy ontologies that are specific to the model organism species have been developed for the purpose of annotating gene expression, mutant phenotypes, and associated data. These databases provide model organism communities with an extensive range of interconnected data, including links between genes and mutant phenotypes.
The application of ontologies to systematics has the potential to force clarification and improve communication about morphological character diversity across taxonomic domains. As a result, ontologies could extend the applicability and level of universality of characters for phylogenetic analysis and improve the knowledge of evolutionary transformations. These computable vocabularies could enable efficient computer processing of vast amounts of data and allow the exploration and aggregation of data across studies that is currently difficult to do in morphology-based phylogenetics. Finally, the use of ontologies to represent systematic data will also enable their connection to genetic and developmental data. Understanding the developmental genetic basis of evolutionary change in morphology, known as “devo-evo,” has become a major interdisciplinary focus in biology. The question of how to render comparative phenotype data computable and how to connect them to genetic and developmental data is critical not only to the field of devo-evo but also to all of the many areas of biology that depend on access to comparative phenotype data.
We have undertaken an effort to prototype a curated, ontology-based evolutionary morphology database that maps to genetic databases (http://kb.phenoscape.org; see also www.phenoscape.org). One of the first requirements for the Phenoscape system is an anatomy ontology that represents multiple species. Few ontologies have been developed with the explicit goal of representing anatomical diversity that is the result of evolutionary mechanisms like genetic drift, selection, and mutation. In contrast, the anatomical diversity represented in model organism databases reflects mutant phenotypes that are typically the result of experimentally induced mutations. We describe here the development of a multispecies anatomy ontology that we initiated for teleost fishes, named the Teleost Anatomy Ontology (TAO). The concepts and unique challenges that arise when constructing an ontology to represent such a high level of anatomical and developmental diversity, as well as some of our solutions, are likely general to all taxonomic groups and useful to other similar efforts aimed at representing morphological data with anatomy ontologies.
BACKGROUND
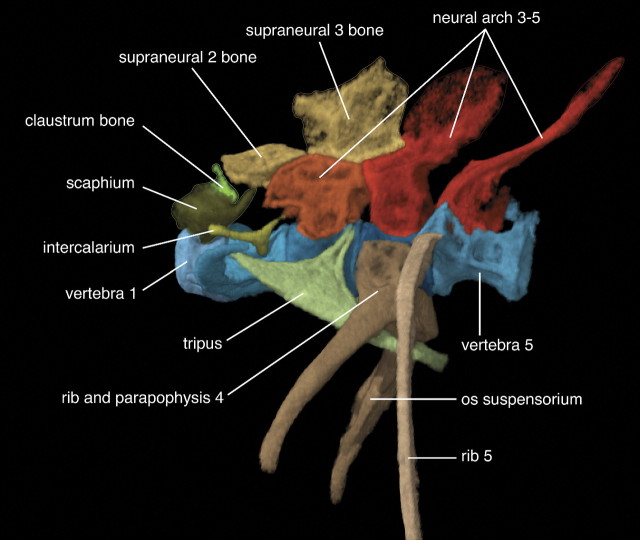
We illustrate many of the concepts and relations used in anatomy ontologies with an example from a set of bones in fishes termed the “Weberian apparatus” (Figs. 1 and 2; Tables 1 and 2). The Weberian apparatus is a complex of bones derived from the anterior vertebrae and associated skeletal structures that function in sound transmission from the air bladder to the inner ear. It is uniquely found in the Otophysi (Fink S.V. and Fink W.L. 1981), a diverse clade of freshwater fishes including the cypriniforms (zebrafish, minnows, carps, and loaches), characiforms (tetras), gymnotiforms (knifefishes), and siluriforms (catfishes).
FIGURE 1.
Three-dimensional model of the Weberian apparatus of the zebrafish Danio rerio in lateral view, anterior to left. The specimen is deposited at Academy of Natural Sciences, Philadelphia, ANSP 189304, standard length = 25.5 mm. The specimen was embedded in agarose gel and scanned at low voltage (30 kV) and high amperage (181 A) on the Skyscan 1172 Microfocus X-radiographic Scanner in laboratory of Dr. J. W. Hagadorn at Amherst College, Amherst, MA. The three-dimensional image processing was done with the software VGStudio MAX v.1.2.1 (Volume Graphics, Heidelberg, Germany) at the Department of Ichthyology, ANSP.
FIGURE 2.
Graphical representation of the Weberian apparatus and its parts in the TAO. Term names are enclosed in ovals; definitions shown in Table 1. Relationships between terms are shown as lines: is_a (red), part_of (blue), and develops_from (green). The dashed lines represent taxonomically variable is_a and part_of relationships (i.e., vertebra 1–4 are types of Weberian vertebra only in otophysan fishes, Fig. 1 and text). Not all relationships are illustrated.
TABLE 1.
TAO terms, identification numbers, and definitions and synonyms for the Weberian apparatus and its parts
| TAO term | TAO:ID | Definition | Synonym(s) | Comment |
| Weberian apparatus | TAO:0001188 | Anatomical cluster that consists of the anteriormost vertebrae and associated structures that connect the swim bladder to the inner ear | Vertebra 1–4, and sometimes vertebra 5 in some catfishes, are part of the Weberian apparatus. Weberian apparatus is present in Otophysi | |
| Neural complex | TAO:0001363 | Anatomical cluster that consists of supraneural 2 bone and supraneural 3 bone and their extensions and which is part of the Weberian apparatus | Weberian supraneural | |
| Supraneural bone | TAO:0000442 | Endochondral bone that is median and located in the dorsal skeletogenous septum between the cranium and the dorsal fin | ||
| Supraneural 2 cartilage | TAO:0001869 | Postcranial axial cartilage located dorsal to vertebra 2 | ||
| Supraneural 2 bone | TAO:0001191 | Supraneural bone located dorsal to vertebra 2 | Small supraneural, sn2 | In otophysans, supraneural 2 bone is part of the neural complex |
| Supraneural 3 cartilage | TAO:0001870 | Postcranial axial cartilage located dorsal to vertebrae 3 and 4 | ||
| Supraneural 3 bone | TAO:0001192 | Supraneural bone that is located dorsal to vertebrae 3 and 4 | Large supraneural, sn3 | In otophysans, supraneural 3 bone is part of the neural complex and fuses in development with neural arches 3 and 4 |
| Weberian ossicle | TAO:0000461 | Bone that is an element associated with a Weberian vertebra | ||
| Weberian ossicle set | TAO: 0001873 | Anatomical cluster that consists of the Weberian ossicles | ||
| Claustrum bone | TAO:0001592 | Endochondral bone, membrane bone, and Weberian ossicle that is located dorsal to the scaphium. The claustrum bone is bilaterally paired | First Weberian ossicle | |
| Claustrum cartilage | TAO:0000637 | Postcranial axial cartilage and Weberian ossicle that is located dorsal to the scaphium. The claustrum cartilage is bilaterally paired | First Weberian ossicle | |
| Scaphium | TAO:0000429 | Neural arch 1 and Weberian ossicle that is reduced in size relative to other neural arches. Scaphium has a process that articulates with the first centrum | Second Weberian ossicle, Weberian ossicle 2 | |
| Intercalarium | TAO:0000525 | Neural arch and Weberian ossicle that has been modified in shape relative to other neural arches. Intercalarium includes 3 parts: an anterolateral process (the manubrium) that is embedded in the interossicular ligament and the ascending and articulating processes | Third Weberian ossicle, Weberian ossicle 3 | |
| Tripus | TAO:0000698 | Endochondral bone, membrane bone, and Weberian ossicle that is an element of third vertebra. Anteriorly, it attaches to the interosseus ligament and posteriorly it extends as a transformator process embedded in the tunica externa of the swimbladder | Fourth Weberian ossicle, Weberian ossicle 4 | |
| Os suspensorium | TAO:0001171 | Endochondral bone and Weberian ossicle that is an element of the fourth vertebra that curves ventrally around the anterior head of the swim bladder | Fifth Weberian ossicle, Weberian ossicle 5 | |
| Weberian vertebra | TAO:0001190 | Vertebra that is part of the Weberian apparatus | Weberian vertebra are usually vertebra 1–4. Present in Otophysi | |
| Vertebra | TAO:0001189 | Endochondral bone that forms around the notochord and is part of the vertebral column |
Note: Relationships among these terms are shown in Figure 2.
TABLE 2.
Hypothesized homology relationships among the skeletal entities of the Weberian apparatus in TAO
| Entity 1 | Taxon 1 | Relationship | Entity 2 | Taxon 2 | Evidence | Reference |
| Scaphium | Otophysi | homologous_to | Neural arch 1 | Teleostei | IDS, IMS, IPS | Fink S.V. and Fink W.L. (1981);Rosen and Greenwood (1970) |
| Intercalarium | Otophysi | homologous_to | Neural arch 2 (ventral portion) | Teleostei | IDS, IMS, IPS | Rosen and Greenwood (1970) |
| Intercalarium | Otophysi | homologous_to | Neural arch 2 | Teleostei | NAS | Fink S.V. and Fink W.L. (1981) |
| Intercalarium | Otophysi | homologous_to | Neural arch 2 | Teleostei | IMS | Hora (1922) |
| Intercalarium | Otophysi | homologous_to | Rib of vertebra 2 | Teleostei | TAS | Hora (1922) |
| Tripus | Otophysi | homologous_to | Parapophysis + rib of vertebra 3 | Teleostei | IDS, IMS, IPS | Fink S.V. and Fink W.L. (1981); Rosen and Greenwood (1970) |
| Tripus | Otophysi | homologous_to | Parapophysis + rib of vertebra 3 + rib of vertebra 4 | Teleostei | IDS, NAS | Hora (1922) |
| Os suspensorium | Otophysi | homologous_to | Parapophysis + rib of vertebra 4 | Teleostei | IDS, IMS, IPS | Fink S.V. and Fink W.L. (1981); Rosen and Greenwood (1970) |
| Claustrum bone and claustrum cartilage | Otophysi | homologous_to | Accessory neural arch | Clupeomorphs, Salmoniforms | IDS, IMS, IPS | de Pinna and Grande (2003) |
| Claustrum bone and claustrum cartilage | Otophysi | homologous_to | Neural spine 1 | Teleostei | TAS | Chranilov (1927) |
| Claustrum bone and claustrum cartilage | Otophysi | not_homologous_to | Neural spine 1 | Teleostei | IPS | Hora (1922) |
| Claustrum bone and claustrum cartilage | Otophysi | homologous_to | Neural arch 1 (a piece of) | Teleostei | IPS | Hora (1922) |
| Claustrum bone and claustrum cartilage | Otophysi | homologous_to | Neural arch 1 (“dissociated upper part of the anterior neural arch”) | Teleostei | IMS | Fink S.V. and Fink W.L. (1981) |
| Claustrum bone and claustrum cartilage | Otophysi | homologous_to | Supradorsal | Teleostei | IMS | Rosen and Greenwood (1970) |
| Claustrum bone and claustrum cartilage | Otophysi | homologous_to | Supraneural 1 | Teleostei | IDS | Coburn and Futey (1996) |
| Claustrum bone and claustrum cartilage | Otophysi | homologous_to | Supraneural 1 | Teleostei | IMS | Gayet (1982) |
Notes: Entity 1 in Taxon 1 is homologous_to or not_homologous_to Entity 2 in Taxon 2 based on evidence. Terms from the ECO are defined in Table 4. IDS = inferred from developmental similarity; IMS = inferred from morphological similarity; IPS = inferred from positional similarity; NAS = nontraceable author statement; TAS = traceable author statement.
The standardization of terms and logical relationships in an ontology enables their use for both human and machine annotation and querying. Terms in ontologies are given textual definitions, assigned unique identifiers, and frequently associated with synonyms. For example, in TAO, the term supraneural 2 bone is defined as “supraneural bone located dorsal to vertebra 2” (Table 1) and assigned the identifier TAO:0001191. By including the synonyms “small supraneural” and “sn2,” which have been applied to this entity, a user can search on any of them and find all information associated with supraneural 2 bone.
The TAO, like other ontologies, is structured as a directed acyclic graph in which a term can have multiple relationships between child and parent terms. For example, Weberian apparatus is_a anatomical cluster and part_of the vertebral column and it has 3 part_of children (Weberian ossicle set, Weberian vertebra, and neural complex) (Fig. 2). The primary relations used in TAO (is_a, part_of, and develops_from) are those commonly used in anatomy ontologies (Burger et al. 2008). The formal definitions of these relations can be found in Smith et al. (2005) and in the Relations Ontology (http://obofoundry.org).
There are 32 anatomy ontologies (as of 25 February 2010) listed at the Open Biological and Biomedical Ontologies (OBO) Foundry (http://www.obofoundry.org; partial list shown in Table 3). The OBO Foundry (Smith et al. 2007) promotes common design principles in support of interoperability. Most of the anatomy ontologies in the OBO Foundry pertain to a single species and are linked to model organism databases. Single-species anatomy ontologies represent canonical anatomies in that they represent the generalized or prototypical structural composition of an idealized instance of the species (Haendel et al. 2008; Neuhaus and Smith 2008). Typically, these ontologies represent a chosen wild-type strain. Anatomy ontologies were first used by model organism communities to link anatomy to gene expression (Davidson et al. 1997) and mutant phenotypes (Gkoutos et al. 2004). Use of an ontology for these purposes allows one to query for genes (see Gene Ontology; The Gene Ontology Consortium 2000; The Gene Ontology Consortium 2001; The Gene Ontology Consortium 2008) that are expressed in a particular structure or for all genes that have an altered phenotype in a given structure. The significance of this is the reasoning that can be done across the ontologies to group annotations (Zhang et al. 2006; Keet et al. 2007). For example, if one queries for phenotypes that involve Weberian apparatus annotations to Weberian vertebra would also be retrieved due to the part _of relationship between these 2 terms (Fig. 2).
TABLE 3.
Selected multispecies (*) and single-species anatomy ontologies listed at the OBO Foundry (http://obofoundry.org/)
| Taxon | Ontology name (acronym) | Reference | Associated database or source |
| Teleosts* | Teleost Anatomy Ontology (TAO) | This study | http://kb.phenoscape.org/ |
| Zebrafish (Danio rerio) | Zebrafish Anatomical Ontology (ZFA) | Sprague et al. (2001, 2003, 2006) | http://zfin.org/ |
| Amphibians* | Amphibian Anatomical Ontology (AAO) | Maglia et al. (2007) | http://www.amphibanat.org/ |
| Xenopus | Xenopus Anatomy (XAO) | Segerdell et al. (2008) | http://xenbase.org/ |
| Mouse (Mus) | Mouse Adult Gross Anatomy (MA) | Bult et al. (2008) | http://www.informatics.jax.org/ |
| Human (Homo sapiens) | Foundational Model of Anatomy (FMA) | Rosse and Mejino (2003) | http://sig.biostr.washington.edu/projects/fm/index.html |
| Spiders* | Spider Ontology (SPD) | Ramirez et al. (2007) | http://research.amnh.org/atol/files/ |
| Fruit fly (Drosophila) | Drosophila Gross Anatomy (FBbt) | Wilson et al. (2008) | http://flybase.org/ |
| Hymenoptera* | Hymenoptera Anatomy | Deans et al. (2010) | http://purl.oclc.org/NET/hymontology |
| Ontology (HAO) | |||
| Plants* | Plant Ontology (PO) | Avraham et al. (2008); Ilic et al. (2007, 2008); Jaiswal et al. (2005) | http://www.plantontology.org/ |
| Fungi* | Fungal Gross Anatomy (FAO) | None | http://www.yeastgenome.org/fungi/fungal_anatomy_ontology/ |
To mine phenotypic variation across taxa, a multispecies anatomy ontology is required. The goal of such an ontology is to represent all the anatomical variation observed across species within a clade. Multispecies anatomy ontologies contain terms that represent generalizations from observations of museum voucher specimens that represent species and their higher taxa. Anatomy ontologies designed for multiple species are relatively few in number, with even fewer linked to databases. As of February 2010, there were 8 multispecies anatomy ontologies listed at the OBO Foundry (Table 3). The Plant Ontology, publicly released in July 2004 (Ilic et al. 2008), was the first anatomy ontology that applied to more than one species. Its purpose is to provide a set of terms that can be used for annotation of gene expression patterns, germplasm, and phenotypes of mutants and natural variants of multiple species from participating plant databases. The inception of 3 multispecies ontologies (teleosts, amphibians, and spiders) was associated with collaborative efforts of morphologists funded by the NSF Assembling the Tree of Life program. This work has required synthesis of disparate morphological data sets, which has driven standardization in terminology, a first step in creating an ontology.
Our approach of capturing the diversity of form among multiple species within a single ontology is motivated partly from the standpoint of practicality: there are simply too many species (e.g., 25,000 species of teleost fishes) to make a separate ontology for each one. A multispecies ontology provides a single source for terms to be used in annotating species variation and facilitates studies based upon interspecific comparisons. In addition, representation of the domain knowledge of systematic biology is most effectively done in a multispecies ontology because experts hold detailed knowledge about the phenotypic variation across versus within single species.
METHODS
Initialization from and Synchronization with the Zebrafish Anatomical Ontology
The TAO was developed and is being maintained according to OBO Foundry principles (Smith et al. 2007). In keeping with these principles, TAO is open (under CC0 license; http://creativecommons.org/license/zero), available to users, follows a community syntax (OBO), and employs a versioning system (Concurrent Versions System) to preserve previous versions of the ontology. Each term possesses a unique identifier consisting of the prefix “TAO,” which designates the unique ontology namespace relative to other OBO Foundry ontologies, followed by a 7-digit numerical code. The identifier is stable, and provided that the meaning of a term is maintained, the term name and textual definition can be modified. These identifiers are never reused or deleted. Instead, the identifier of an obsolete term is maintained in the ontology as a “paper trail” for references to that term.
The choice of maintaining TAO in the OBO format gave us direct access to several other relevant ontologies: a relations ontology (Relations Ontology, OBO_REL), a quality ontology (Phenotype and Trait Ontology, PATO), a spatial ontology used to describe anatomy (Spatial Ontology, BSPO), and an evidence code ontology (ECO), all available at OBO Foundry. Although other ontology languages have larger user bases than OBO, the OBO community is focused on representing biological and biomedical data, whereas other user communities are focused on different domains. The OBO language is also relatively simple for biologists to learn relative to OWL (Web Ontology Language; http://www.w3.org/2004/OWL/) or more logically expressive languages such as Common Logic or CycL (Matuszek et al. 2006). Another advantage of using OBO is that there is an ontology editor available, OBO-Edit (Day-Richter et al. 2007), which has been designed by and for biologists. Versions of the ontology in other ontology languages, such as OWL, are provided by the OBO Foundry Web site.
Because zebrafish (Danio rerio) is a subtype of the order Cypriniformes, and the yet more encompassing clades, Otophysi and Ostariophysi, and most broadly, the Teleostei (teleost fishes), we utilized the existing Zebrafish Anatomical Ontology (ZFA) to populate TAO with its first set of terms in September 2007. The initialization process of TAO used a Perl script, provided by Chris Mungall, which transformed a copy of the ZFA OBO file by changing the identifier prefix of each term from ZFA to TAO and added a cross-reference entry for each term that pointed back to its corresponding term in the ZFA. For example, the zebrafish term vertebra has the ID ZFA:0000323; this term was cloned into TAO as TAO:0000323 with a cross-reference back to the ZFA term ZFA:0000323. The namespace, which indicates which ontology the terms belong to, was set to teleost_anatomy. Both TAO and ZFA top level nodes are is_a children of corresponding higher level terms from the Common Anatomy Reference Ontology (CARO) (Haendel et al. 2008). CARO contains a framework of higher level anatomical terms that apply to all organisms and is designed to promote interoperability among anatomical ontologies.
After the initial cloning, TAO was generalized so as to be applicable to Teleostei. Specifically, we removed the ZFA adult anatomy and developmental stage terms that do not generalize across teleosts. ZFA terms are logically subtypes of TAO terms (e.g., ZFA:vertebra is_a TAO:vertebra), and thus ZFA has a subset of TAO terms, and additional information relevant specifically to the zebrafish model organism community. In contrast, TAO has many additional terms representative of the anatomical diversity within Teleostei.
We regularly synchronize TAO and ZFA to incorporate updates to either ontology. Cross references are added to each ontology to maintain logical interoperability. To automate this process, we have developed the synchronization tool, a plug-in for OBO-Edit that is publicly available from SourceForge (see https://www.phenoscape.org/wiki/Synchronization_Tool). The synchronization tool aids in keeping 2 ontologies aligned by checking for missing cross references between identically named terms, conflicting data between cross-referenced terms, terms present in one but missing from the other ontology, and structural differences such as differences in the parent of cross-referenced terms.
TAO was made publicly available in September 2007 within the OBO CVS repository (http://obofoundry.org) and from the National Center for Biomedical Ontology (NCBO) Bioportal (http://bioportal.bioontology.org/). Interested users can search and browse terms and visualize the relationships among terms using online ontology resources with a graphical user interface such as the NCBO Bioportal or the EBI-hosted Ontology Lookup Service (http://www.ebi.ac.uk/ontology-lookup/init.do).
Organization and Features
Terms in TAO represent anatomical structures found in teleost fishes. Some terms are subtypes of anatomical cluster (defined as “An anatomical group that has its parts adjacent to one another.”). For example, Weberian apparatus is an anatomical cluster because it consists of a group of entities closely associated in position with each other (Fig. 1). Term names are given in the singular because terms refer to single classes or types of entities and because singular names promote grammatical consistency with term definitions (Smith et al. 2006; Schober et al. 2009). Term definitions facilitate the consistent use of a term by curators for annotation of disparate data types. TAO follows the convention that each term should have a textual definition of the genus–differentia form (i.e., a subclass structure of A is an A that has properties X and Y that distinguish it from the other subclass structures of A) (Smith et al. 2007). Definitions for structures of the Weberian apparatus (Table 1) begin with the parent of the term being defined, followed by characteristics that distinguish the term from its siblings. These characteristics can include location, shape, and a list of the parts of the entity. For example, Weberian apparatus is defined as “Anatomical cluster [parent term] that consists of the anteriormost vertebrae and associated structures that connect the swim bladder to the inner ear [differentia].” Definitions are primarily based on structural criteria. Taxonomic statements relevant to a structure but that do not universally apply to all teleosts are recorded in the comment field for that term (e.g., comment for Weberian apparatus; Table 1). These statements are potentially helpful to the user for general understanding and identification of structures.
Given the broad taxonomic scope of TAO, term definitions are also necessarily broad to ensure applicability to all fishes in the clade. TAO definitions are typically suggested by a user such as a curator who requires the term for annotation. Definitions may be refined by community experts participating in the ontology term request mailing list (see Extending TAO section), or from discussion among experts at ontology development and annotation workshops.
In addition to is_a, part_of, and develops_from, TAO also uses the overlaps relation, which is used to represent the relationship between 2 entities that share a part (defined in draft form on Relations Ontology wiki page: http://www.bioontology.org/wiki/index.php/RO:Main_Page). All TAO terms have a relationship to a parent term, with the exception of the root term, which in the case of TAO is teleost anatomical entity. Even though a term in an ontology may have multiple parents with the same or different relationship types, the OBO Foundry principle of “single inheritance” recommends that each term only have a single-asserted is_a parent, which can help avoid errors in reasoning that occur in ontologies that contain terms with multiple is_a parents. We have strived to achieve this in TAO, but for searching convenience, a few concepts are currently represented with multiple is_a parentage. For example, tripus (Table 1; Fig. 2) develops from both endochondral and membrane ossifications, and it is therefore a subclass of both endochondral and membrane bone (in addition to being a subclass of Weberian ossicle). An alternative way to represent bone development without multiple parents would be to create a new class of bone, such as compound bone, which is defined as having both endochondral and membrane bone development. However, this classification has implications for database queries because terms that are children of compound bone will not appear in searches for endochondral bone or membrane bone as might be expected by a user. A representation whereby development is represented by a separate axis of classification is planned.
Extending TAO
TAO is a community resource for ichthyologists and vertebrate morphologists. To ensure that it represents high-quality domain knowledge for teleost fish morphology, its continued development must be a community exercise. We have made particular efforts to engage the community of ichthyologists in TAO development. A variety of experts in fish morphology are kept up to date with proposed additions and changes to TAO through a mailing list (Fig. 3). When a new term is required, a request is made through the SourceForge term request tracker, and an automated e-mail summarizing the request is sent to the list. Changes are discussed, amendments are made as required, and a record of the discussion is maintained by the TAO administrator.
FIGURE 3.
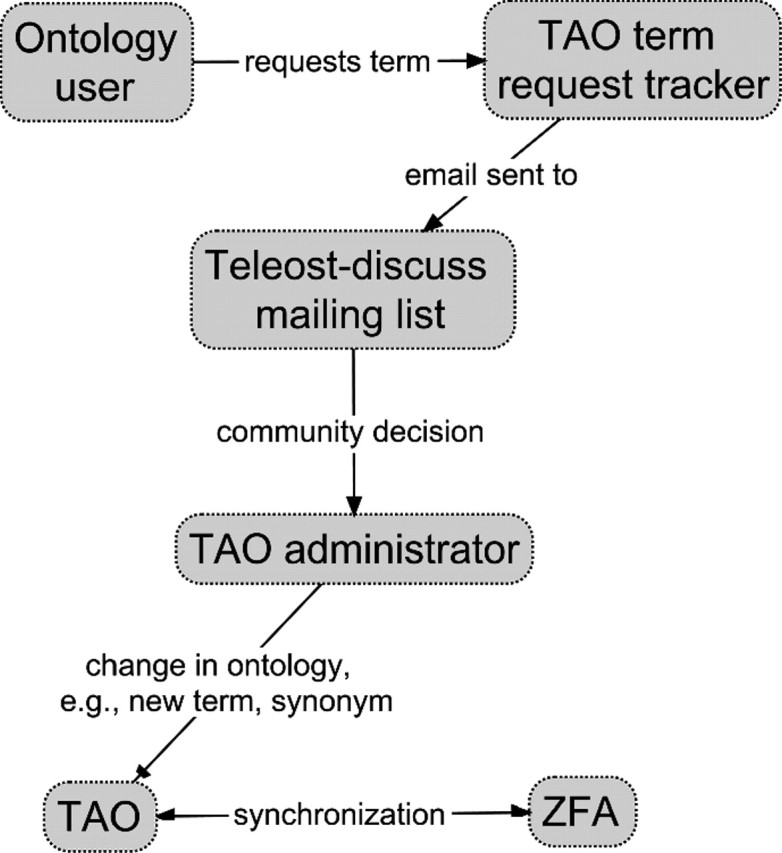
The process of updating TAO to include new terms, refined definitions, synonym additions, and structural changes begins with term requests from ontology users through the SourceForge Term Tracker (http://sourceforge.net/tracker/?group_id=76834&atid=994764). Requests trigger an automated e-mail to the ichthyological community through the teleost-discuss mailing list (https://lists.sourceforge.net/lists/listinfo/obo-teleost-discuss). Discussion ensues, a decision is reached, and the request is summarized by the TAO administrator on the tracker page. The request is then closed, and TAO is updated to include the requested change. Updates that also apply to the ZFA are made during synchronization.
Like other community-based ontologies, active extension of TAO is driven by user needs. Because the active use of ontologies is relatively new to evolutionary biology in general and systematics in particular, the biological research project currently driving development of TAO is the phenotype annotation of the systematic literature for teleost fishes by the Phenoscape project. The skeletal system is the focus of most systematic ichthyological studies, and thus, the development of TAO is focused on the skeletal system. Since cloning from ZFA in September 2007, TAO has grown from 1976 terms to 2662 terms (as of February 2010) in large part due to the greater number of terms required to represent the diversity of anatomical structures across species. Over half of these new terms (391 of 686) are skeletal.
Although TAO will continue to grow, very large ontologies become difficult for users to navigate and for curators and software tools to manage. An alternative to adding a new term to the ontology is to construct a postcomposition (Mungall et al. 2010). A postcomposition is a combination of existing terms that are drawn from an ontology or from multiple orthogonal ontologies to refer to a single entity. For example, a curator might need to refer to the process on vertebra 1 (“process” here refers to an anatomical structure rather than processes that are temporally unfolding entities). Rather than adding the new term vertebra 1 process to the ontology, the existing terms process and vertebra 1 can be joined on-the-fly using the part_of relation to create a new term. Although the postcomposed term does not have an identifier and is not added to the ontology, it provides the implied definition “process that is part of vertebra 1,” and it can be queried using the same reasoning principles as any other term. Postcompositional terms are ideal for structures that may not exist outside of a few species and thus are unlikely to be required for repeated annotation. Postcomposition can also prevent the ontology from being forced to grow solely to satisfy a need for combinatorial complexity.
Application of TAO
One common use of ontologies is the annotation or “tagging” of objects or observations, such as genetic sequences, gene expression patterns, or whole organism phenotypes, with ontology terms. The convention used by model organism databases to annotate mutant phenotypes is entity-quality (EQ) syntax in which an entity from an anatomical ontology is combined with a quality or nontaxon specific modifier (Gkoutos et al. 2004; Sprague et al. 2008). The Phenoscape project has adopted and extended EQ to annotate evolutionary phenotypes, specifically systematic characters for ostariophysan fishes. Using the Phenex software (Balhoff et al. 2010), entity terms are drawn from TAO and quality terms are provided by PATO (Mabee, Arratia, et al. 2007; Mabee, Ashburner, et al. 2007). For example, a character state might be “Antorbital, triangular.” This phenotype can be expressed using the TAO term antorbital and the PATO term triangular. Such phenotype annotations corresponding to systematic characters require the association of the anatomical term with a taxonomic name. Thus, TAO terms are associated with species or higher taxa through annotations to teleost scientific names from the Teleost Taxonomy Ontology (www.phenoscape.org; OBO CVS repository: http://obofoundry.org).
Another driving application that we anticipate to be of increasing importance is the annotation of images using anatomical ontology terms, which will increase the need for ontologies like TAO. TAO has been used for annotation of images by the Cypriniform Tree of Life (CToL) morphologists, and Morphbank (http://www.morphbank.net/) imported TAO-annotated images of the skeleton from the CToL portal. Other image and morphology databases are similarly beginning to experiment with the use of anatomical and phenotype ontologies for annotation (e.g., Morphster, MorphBank, Morphobank, CatfishBones) (Ramirez et al. 2007).
DISCUSSION
We have established the multispecies TAO by expanding a single-species ontology core to represent the anatomical diversity of many species. TAO was built at the outset of developing an evolutionary phenotype database because an appropriate anatomy ontology is a requirement for rendering morphological data in a computable format. The primary application of TAO currently is for curation of systematic characters and image annotation. Built primarily on a structural axis, TAO is being developed with the goal of maintaining interoperability with other ontologies. Here, we discuss the requirements and remaining challenges in the development and maintenance of this multispecies anatomy ontology, issues that are general to similar efforts.
Requirements of Multispecies Ontologies
Addition of many synonyms.—
Different research communities often use different terminologies to describe identical structures. Ontologies include synonyms to accommodate these differing nomenclature preferences and community traditions. Synonyms can also aid queries by allowing searches for these alternative names. The OBO format distinguishes among different types of synonyms, such as BROAD, NARROW, EXACT, and RELATED. TAO currently uses the latter 2 categories. EXACT synonyms include “true” synonyms that are alternative names for the same structure, such as “claustrum bone” and “first Weberian ossicle” (Table 1); misspellings in the literature such as “postemporal” instead of “posttemporal”; alternative spellings such as “hyomandibular” versus “hyomandibula”; and plural synonyms such as “lepidotrichia” versus “lepidotrichium.” RELATED synonyms are alternative names that stem from the incorrect usage of a term. For example, some ichthyologists use the term lacrimal, a tetrapod bone, to refer to the infraorbital 1 even though infraorbital 1 in fishes is not homologous to the lacrimal bone in tetrapods (see Homology section). Therefore, “lacrimal” is listed as a RELATED synonym of infraorbital 1 in TAO.
Coordination of anatomy ontologies across taxa.—
As multispecies anatomy ontologies are built, concepts within the single-species anatomy ontologies of the model species that are phylogenetically contained within the group will logically become subtypes. In the case of TAO, ZFA concepts became the subtypes. Frequent synchronization between subtype and parent is critical for interoperability, but by partially automating this with the synchronization tool, we have made this less time consuming and more consistent. Logically, both TAO and ZFA concepts would become subtypes of a vertebrate anatomy ontology, as would those concepts in, for example, the Xenopus, amphibian, human, and mouse anatomy ontologies. Limits to the cascade of synchronization required when a new term is added to a subtype ontology have yet to be determined. Coordination among ontologies in subtype relationships will be important for knowledgably exchanging information among databases separated by significant phylogenetic distance.
Representation of intermediate nodes.—
Multispecies ontologies can require the addition of a parent term (intermediate node) that is unnecessary for single-species ontologies. We added tooth to TAO, for example, as the parent term for the different types of teeth in fishes (e.g., premaxillary tooth, dentary tooth). It is not a term required for the ZFA, however, because zebrafish only have one type of tooth (ceratobranchial 5 tooth). On the other hand, a term that is an intermediate node in a single-species ontology may be represented as a leaf node in a multispecies ontology. For example, the ZFA terms precaudal vertebra and caudal vertebra refer to vertebrae that lack or possess hemal spines, respectively, and the count of these vertebral types is consistent in zebrafish: there are typically 10 non-Weberian precaudal vertebrae and 17 caudal vertebrae (Fig. 4a). The number of these vertebral types, however, is highly variable across teleost fishes, and it is not possible to universally associate a particular vertebra with one of these 2 regions (e.g., the assertion vertebra 7 is_a precaudal vertebra is not applicable to all teleosts). Thus, the TAO terms precaudal vertebra and caudal vertebra do not have child terms, and all individual vertebral terms have the same parent, vertebra (Fig. 4b). It is important to recognize that differences in the parent of the individual vertebra terms in the ZFA versus TAO does not preclude queries that require reasoning across them because they are related via cross references and standard relations.
FIGURE 4.
Representation of the subtypes of vertebra in a) ZFA and b) TAO. Danio rerio exhibits a modal number of 4 Weberian vertebra, 10 precaudal vertebra, and 17 caudal vertebra (Bird and Mabee 2003), and precaudal vertebra and caudal vertebra may be represented as intermediate nodes in the ZFA (a), whereas they are necessarily leaf nodes in TAO (b) because of variability in number of each type of vertebra among teleost species. Not all individual, numbered vertebra are represented in ZFA and TAO.
General Challenges
Homology.—
Homology, the similarity in cross-species characteristics due to common ancestry, is a central concept in evolutionary biology, and the relationship of homology to ontologies is important to consider. TAO primarily follows a structural definition of terms. The goal of this ontology is to contain all possible anatomical terms applicable to fishes, but it is not meant to imply homology between terms or when the same term is used for different taxa. Instead, we record homology statements between differently or identically named terms in different taxa outside of the ontology, with evidence and attribution. We use the homologous_to and not_homologous_to relations (working definitions available from the Relations Ontology wiki page: http://www.bioontology.org/wiki/index.php/RO:Main_Page) to record homology statements. Where a use case or query requires viewing competing hypotheses of homology, they can be drawn from these tables outside the ontology according to established criteria.
Opposing views of homology for particular structures are typically based on different lines of evidence or homology criteria. Standard lines of evidence that are used to assess homology a priori include similarity in shape and size, topographic position, complexity, and development (Remane 1952; Roth 1984; Patterson 1988). Homology is tested a posteriori by the distribution of character states on phylogenies resulting from character analysis (Mayden and Wiley 1992). In essence, these criteria represent different kinds of evidence for an homology assertion, and thus, the possible types of evidence can be codified similarly to the types of evidence for gene function annotation (http://www.geneontology.org/GO.evidence.shtml). We followed OBO community standards for establishing and using evidence codes to annotate evidence for homology of anatomical structures. Where possible, we used the same evidence codes that are used for the annotation of gene function, but we also proposed new evidence codes for inclusion in the ECO, and these were accepted in January 2008 (Table 4).
TABLE 4.
Terms used for homology annotation in the ECO (http://obofoundry.org/cgi-bin/detail.cgi?id=evidence_code)
| Evidence code | ID | Definition | Parent |
| Inferred from positional similarity | ECO:0000060 | Used when an annotation is made based on the similarity of the location and or arrangement of structures | Inferred from phenotypic similarity (parent: inferred from similarity) |
| Inferred from compositional similarity | ECO:0000063 | Used when an annotation is made based on the similarity of the histological makeup of structures | Inferred from phenotypic similarity (parent: inferred from similarity |
| Inferred from developmental similarity | ECO:0000067 | Used when an annotation is made based on the similarity of embryological and/or postembryonic origin of structures | Inferred from phenotypic similarity (parent: inferred from similarity) |
| Inferred from morphological similarity | ECO:0000071 | Used when an annotation is made based on the similarity of the shape, structure, or overall configuration of structures | Inferred from phenotypic similarity (parent: inferred from similarity) |
| Inferred from gene expression similarity | ECO:0000075 | Used when an annotation is made based on the similarity of expression of genes in structures | Inferred from phenotypic similarity (parent: inferred from similarity) |
| Inferred from phylogeny | ECO:0000080 | Used when an annotation is made based on the common ancestry of structures on a particular phylogenetic tree. Typically, other evidence (a type of similarity) support a prior hypothesis of homology for these structures | Parent: inferred from reviewed computational analysis |
| Nontraceable author statement | ECO:0000034 | An author states a homology relationship, but no evidence is provided or cited; that is, the statement not backed up with traceable data | Parent: evidence code |
| Traceable author statement | ECO:0000033 | An author states a homology relationship and cites a source for the data | Parent: evidence code |
An example of how these codes can be used to annotate differing evidence for homology comes from the Weberian apparatus. Intercalarium, for example, is a modified neural arch of vertebra 2 according to developmental, positional, and morphological criteria (Table 2). On the other hand, claustrum bone (and claustrum cartilage in which the bone forms) has been considered homologous to accessory neural arch, neural spine 1, neural arch 1, supradorsal, and supraneural 1, sometimes with specific evidence presented but other times simply stated by an author. In the latter case, the evidence is weaker. This type of evidence is given the code “traceable author statement” and linked to the source that the author references as providing evidence for a particular homology relationship or “non-traceable author statement” if the author asserts a homology without citing a source (Table 4).
Although TAO endeavors to maintain a clear separation of homology from its ontological representation of anatomy, an argument can be made that some structures are represented in the ontology according to a hypothesized homology with another structure. For example, TAO asserts that the intercalarium is a subtype of (stands in an is_a relationship to) neural arch (Fig. 2) because direct uncontested developmental observations indicate that the intercalarium develops from the neural arch of the second vertebra. Intercalarium is also hypothesized as homologous_to neural arch 2 (Table 2) to facilitate queries on intercalarium and its homologs.
Homologous structures in phylogenetically distant taxa may be referred to by different names or by the same name, and thus, it is critical to have a mechanism such as a homology table in place to enable the informed use of these terms. For example, the bone that is termed the frontal bone in teleost fishes is not the homolog of the frontal bone in tetrapods (Jollie 1962; Schultze and Arsenault 1985). Instead, the frontal bone in teleosts is the homolog of the parietal bone in tetrapods. If, for example, “parietal bone” was simply added as a synonym of frontal bone in TAO, then a query for all the genes expressed in the frontal bone would also return genes expressed in the parietal bone. Thus, although synonyms can be used to represent homologs in a multispecies ontology (e.g., Plant Structure Ontology; Ilic et al. 2008), refining these relationships using evidence codes and explicit homology links is a more conservative approach that avoids encoding controversial homology assertions within the anatomy ontology, while still enabling users and computer applications to take advantage of comparative evolutionary reasoning.
Representing serial homologs.—
Representation of serial homologs in a multispecies anatomy ontology requires accommodation of different naming methods used in the literature for elements of the series. Serial homologs are usually given names based on ordinal position (e.g., “vertebra 3”), but some elements, frequently the first or last of a series, may also be referred to by a structural name. Consequently, a term may have its name also designated as a synonym of another term. For example, the infraorbital series is an iterated set of bones that encircle the eye in fishes. The most posterior bone of the series is considered homologous across many fishes because of similarity in position and structure. It is either named the dermosphenotic or given a number according to its ordinal position. In some cypriniform fish species, for example, the dermosphenotic is the terminal of 5 bones and is also termed “infraorbital 5.” In some characiform fishes, however, there are 6 bones in the infraorbital series and the terminal bone is named the dermosphenotic or infraorbital 6. To represent these alternative names for dermosphenotic in TAO, the term dermosphe- notic is given the RELATED synonyms “infraorbital 5” and “infraorbital 6.” Likewise, the terms infraorbital 5 and infraorbital 6 are given the RELATED synonym “dermosphenotic.”
Representation of taxonomic differences in development of anatomical structures.—
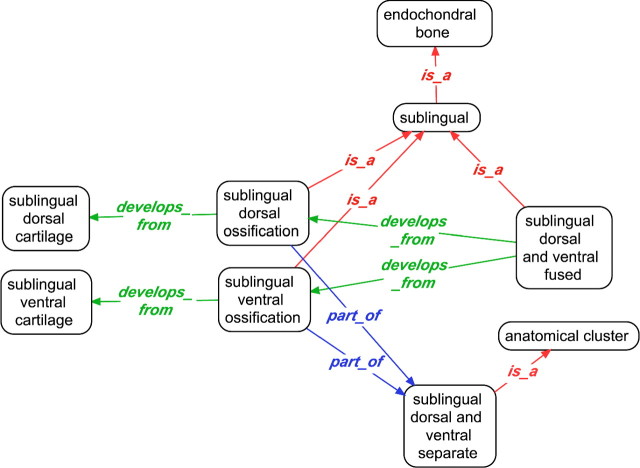
Because of differences in the patterns of development across species, even of homologous structures, multispecies anatomy ontologies must be able to accommodate multiple developmental pathways. In cypriniform fishes, for example, there are differences in the development of the sublingual, a median element(s) of the lower hyoid arch. There are 2 sublinguals (dorsal and ventral) in some species and 1 sublingual in others. The single sublingual of at least one species (Catostomus commersonii) is formed through developmental fusion of dorsal and ventral cartilage precursors (Engeman et al. 2009). Representation of the dual developmental origin of the single sublingual required creation of terms for the sublingual formed via fusion (sublingual dorsal and ventral fused) and the sublingual with separate parts (sublingual dorsal and ventral separate). These 2 terms have different relations (develops_from vs. part_of) to the dorsal and ventral components of the sublingual. In addition, the terms for fused and separate sublingual ossifications (“ossification” here refers to anatomical structures rather than the process of bone formation) have different is_a parents (Fig. 5): sublingual dorsal and ventral fused is a single structure (is_a sublingual), whereas sublingual dorsal and ventral separate is composed of 2 adjacent parts (is_a anatomical cluster). Searching on the term sublingual would not return data for both fused and separate sublinguals. However, if these terms are designated as homologous in a relational table, then data for both types would be returned together.
FIGURE 5.
Multiple developmental pathways of the sublingual represented in the TAO. The sublingual is a median element of the lower hyoid arch in cypriniform fishes. In at least one species, the ossified sublingual (sublingual dorsal and ventral fused) results from the fusion of 2 cartilage elements, whereas in other species, the sublingual exists as separate ossified elements (sublingual dorsal and ventral separate). These terms have different is_a parents due to structural differences.
Remaining challenges
Representing taxonomically variable relationships for a single entity.—
Different species may demonstrate variation in the relationship between 2 entities. For example, vertebrae 1–4 are considered subtypes of Weberian vertebra but only in otophysan fishes (Fig. 2, dashed lines indicate that this is_a relationship is only present in some fishes). To assert this subtype relationship between vertebra 1 and Weberian apparatus in TAO (which we have not done) incorrectly implies that every instance of vertebra 1 in all teleosts is a type of Weberian vertebra. One solution (not shown in Fig. 2) is to create a new term, Weberian vertebra 1, that is a subtype (child) of Weberian vertebra and has a part_of relationship to Weberian apparatus. Disadvantages to adding a new term are that it inflates the size of the ontology and it requires a user to know which term to select for annotation (e.g., vertebra 1 vs. Weberian vertebra 1).
An approach we are exploring to express these taxonomically variable relationships is to use postcomposition to create taxon-specific subtypes of anatomical terms. In the above example, vertebra 1 in D. rerio could be asserted as a subtype of Weberian vertebra either within the ontology or only within our database. Database queries for Weberian vertebra will then return annotations to vertebra 1 in taxon D. rerio. Another alternative would take advantage of phenotype annotations to infer a taxonomically variable relationship. In this case, a rule could be constructed that would allow a reasoner to infer from the (separately asserted) presence of Weberian apparatus in a taxon (e.g., Otophysi) that vertebra 1 is_a Weberian vertebra in that taxon. Because the presence of the Weberian apparatus is a morphological character state and as such a phenotype, this strategy would use phenotype assertions to inject additional and taxon-specific relationships into the anatomy ontology, without the need for an ontology curator to maintain those separately.
Aligning multiple ontologies.—
Ontology alignment is the process of determining correspondences between terms (and less often relations) between 2 or more ontologies. This process, an important step in any comparative analysis using ontologies, poses a significant challenge for both single- and multispecies anatomy ontologies that represent phylogenetically diverse organisms. That said, single-species ontologies used by model organism communities such as fly, zebrafish, and mouse are commonly developed in collaboration with each other to promote cross-species comparisons (Sprague et al. 2006). Still, given the rapid development of new anatomy ontologies and the growing importance of multispecies ontologies, alignment of terms and relations has become a significant problem.
Alignment is much more than just matching terms with the same name. Even semantically similar terms may be found at different levels in the class hierarchies of their respective ontologies. This problem, sometimes referred to as the terms being “out of phase,” would occur with naive matching of terms (e.g., the term “upper jaw” represents a cluster of different bones in teleosts vs. amphibian anatomy ontologies). Although tools for (semi-) automatic ontology alignment have been developed (see http://ontologymatching.org/projects.html), relatively little effort has been focused on alignment of biological ontologies. However, several such tools exist (e.g., COBRA, www.xspan.org; Homolontol, Bastian et al. 2008; and UBERON, described below).
The “Minimal Anatomy Terminology” (MAT) is a recent approach in which synonyms and identifiers from other anatomy ontologies (>20) are grouped into high-level categories (Bard et al. 2008). The MAT terminology (not a formal ontology) covers basic anatomy for all common taxa from fungi, plants, and animals. Its purpose is to provide primary search terms to access tissue-associated data, and it allows data integration and interoperation among many ontologies. Another approach has been to create a cross-species anatomy ontology that is based on alignment of terms and relations held in common across ontologies (UBERON; Mungall et al. 2010). UBERON was initially seeded from contributing ontologies using a string matching algorithm (Mungall 2004) and then curated for further alignment. UBERON therefore groups similar structures in different organism based on any kind of similarity, while maintaining cross references to the contributing anatomy ontologies. This approach is useful because it may uncover homology at different levels of granularity or instances of convergent evolution where genetic pathways are reused. One difficulty with an overarching ontology approach such as UBERON is that the number of shared terms and relations across ontologies may be few, and the terms in common may be mainly higher level parent terms in the ontology. Thus, queries enabled by this higher level ontology may not be granular enough to be informative.
Yet another alignment approach would be to represent the anatomy of the most recent common ancestor and thereby formalize homology between structures within the ontology. This would involve creating a separate higher level anatomy ontology for the common ancestor of the taxa represented in the anatomy ontologies and then use a derives_from relation, which is defined in terms of continuous genetic ancestry, to relate descendant structures to ancestral structures. Although this approach could be advantageous for model organism databases to link homologous structures (e.g., zebrafish heart and mouse heart), it would preclude testing alternative phylogenetic views if only a single derives_from relationship could be captured for any one structure. Phylogenetic analysis depends upon the iterative testing of homology using alternative phylogenies and/or different types of evidence for the homology assertion. Furthermore, no phylogenetic methods currently exist for computing ancestral anatomy (or character) ontologies. If nothing else, the variety of possibilities and lack of a commonly accepted methodology demonstrate that the alignment of separately growing anatomy ontologies will require new approaches, new software tools, and coordination across communities.
CONCLUSIONS
Aggregation of comparative data to address evolutionary questions requires tools like ontologies that accurately, flexibly, and in a computable manner represent a historical yet still growing body of knowledge. Multispecies anatomy ontologies like TAO can effectively represent the broad knowledgebase of comparative biology, and their development will necessarily be a community exercise as the need for shared resources grows (e.g., interaction between image repositories and morphology databases). In developing TAO by extending the core of a single-species ontology, we have encountered a number of issues that are likely general to other attempts at establishing multispecies ontologies. As the number of ontologies at various taxonomic levels continues to grow, the need to align them will present a challenge across phylogenetically distant clades. To be interoperable, these anatomical ontologies will need to conform to a common set of standards. Ultimately, these efforts will facilitate integration of these data with those contained in model organism and genomic databases, efforts that are simply not possible in the current paradigm of comparative biology. Anatomy ontologies are a first step in making computable the vast stores of evolutionary phenotypic data accumulated from decades of comparative and systematic work. It is our hope that lessons learned from TAO will be of use in other efforts to create multispecies anatomy ontologies that embrace ever more branches of the tree of life.
FUNDING
This work was supported by the National Science Foundation (NSF DBI 0641025), National Institutes of Health (HG002659), and the National Evolutionary Synthesis Center (NSF EF-0423641).
Acknowledgments
We are grateful to the following for their contributions to ontology development, mostly at NESCent working groups and Phenoscape data jamborees: G. Arratia, M. Ashburner, J. Blake, S. Blum, M. Coburn, K. Conway, Q. Cronk, M. de Pinna, J. Engeman, G.V. Gkoutos, T. Grande, B. Hall, E.J. Hilton, C. Kothari, S. Lewis, A. Maglia, R.L. Mayden, C. Mungall, S. Rhee, N. Rios, M. Sabaj Pérez, E. Segerdell, B. Sidlauskas, B. Smith, M. Wallinga, N. Washington, and J. Webb. We thank E. Jockusch, C. Mungall, J. Sullivan, and an anonymous referee for providing valuable comments. K. Luckenbill, Department of Ichthyology, Academy of Natural Sciences, Philadelphia, conducted the MicroCT scan and three-dimensional image processing for Figure 1. J.W. Hagadorn and D. Kelly generously provided access to and training in MicroCT use in Hagadorn's laboratory at Amherst College. J. Sprague at Zebrafish Information Network provided the zebrafish specimen used for the MicroCT scan.
References
- Avraham S, Tung C-W, Ilic K, Jaiswal P, Kellogg E, McCouch S, Pujar A, Reiser L, Rhee S, Sachs M, Schaeffer M, Stein L, Stevens P, Leszek V, Zapata F, Ware D. The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Res. 2008;36:D449–D454. doi: 10.1093/nar/gkm908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhoff JP, Dahdul WM, Kothari CR, Lapp H, Lundberg JG, Mabee PM, Midford PE, Westerfield M, Vision TJ. Phenex: Ontological annotation of phenotypic diversity. PLoS ONE. 2010 doi: 10.1371/journal.pone.0010500. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard JBL, Malone J, Rayner TF, Parkinson H. Proceedings of the 11th Annual Bio-Ontologies Meeting, 2008 July 20. Toronto: Canada; 2008. Minimal Anatomy Terminology (MAT): a species-independent terminology for anatomical mapping and retrieval. [Google Scholar]
- Bastian F, Parmentier G, Roux J, Moretti S, Laudet V, Robinson-Rechavi M. Bgee: integrating and comparing heterogeneous transcriptome data among species. In: Bairoch A, Cohen-Boulakia S, Froidevaux C, editors. Data Integration in the Life Sciences, 5th International Workshop, DILS 2008, Evry France. Berlin: Springer-Verlag; 2008. [Google Scholar]
- Bird NC, Mabee PM. The developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae) Dev. Dyn. 2003;228:337–357. doi: 10.1002/dvdy.10387. [DOI] [PubMed] [Google Scholar]
- Blake J, Harris M. The Gene Ontology (GO) project: structured vocabularies for molecular biology and their application to genome and expression analysis. Curr. Protoc. Bioinformatics. 2008;7.2:1–9. doi: 10.1002/0471250953.bi0702s23. [DOI] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. Members of the Mouse Genome Database Group. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A, Davidson D, Baldock R. Anatomy ontologies for bioinformatics: principles and practice. London: Springer; 2008. [Google Scholar]
- Chranilov NS. Beitrage zur kenntnis des Weber'schen apparates der Ostariophysi. I. Vergleichend-anatomishe ubersicht der knochenelemente der Weber'schen apparates bei Cypriniformes. Zool. Jahrb. 1927;49:501–597. [Google Scholar]
- Coburn MM, Futey LM. The ontogeny of supraneurals and neural arches in the cypriniform Weberian apparatus (Teleostei: Ostariophysi) Zool. J. Linn. Soc. 1996;116:333–346. [Google Scholar]
- Davidson D, Bard J, Brune R, Burger A, Dubreuil C, Hill W, Kaufman M, Quinn J, Stark M, Baldock R. The mouse atlas and graphical gene-expression database. Cell Dev. Biol. 1997;8:509–517. doi: 10.1006/scdb.1997.0174. [DOI] [PubMed] [Google Scholar]
- Day-Richter J, Harris MA, Haendel M The Gene Ontology OBO-Edit Working Group. Lewis S. OBO-Edit—an ontology editor for biologists. Bioinformatics. 2007;23:2198–2200. doi: 10.1093/bioinformatics/btm112. [DOI] [PubMed] [Google Scholar]
- de Pinna MCC, Grande T. Ontogeny of the accessory neural arch in pristigasteroid clupeomorphs and its bearing on the homology of the otophysan claustrum (Teleostei) Copeia. 2003;2003:838–845. [Google Scholar]
- Deans AR, Miko I, Yoder MJ, Seltmann KS The HAO Consortium. 2010 [Thu Feb 25 18:03:10–0600 2010]. Hymenoptera Glossary. Available from: http://purl.oclc.org/NET/hymontology. [Google Scholar]
- Engeman J, Aspinwall N, Mabee PM. Development of the pharyngeal arch skeleton in Catostomus commersonii (Teleostei: Cypriniformes) J. Morphol. 2009;270:291–305. doi: 10.1002/jmor.10688. [DOI] [PubMed] [Google Scholar]
- Fink SV, Fink WL. Interrelationships of the ostariophysan fishes (Teleostei) Zool. J. Linn. Soc. 1981;72:297–353. [Google Scholar]
- Gayet M. Considération sur la phylogénie et la paléobiogéographie des ostariophysaires. Geobios Mem. Spec. 1982;6:39–52. [Google Scholar]
- The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. The Gene Ontology Project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkoutos GV, Green EC, Mallon AM, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol. 2004;6:R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendel MA, Neuhaus F, Osumi-Sutherland DS, Mabee PM, Mejino JLV, Mungall CJ, Smith B. CARO—the common anatomy reference ontology. In: Burger A, Davidson D, Baldock R, editors. Anatomy ontologies for bioinformatics: principles and practice. London: Springer; 2008. pp. 327–349. [Google Scholar]
- Hora SL. The homology of the Weberian ossicles. J. Proc. Asiat. Soc. Beng. 1922;18:1–4. [Google Scholar]
- Ilic K, Kellogg EA, Jaiswal P, Zapata F, Stevens PF, Vincent LP, Avraham S, Reiser L, Pujar A, Sachs MM, Whitman NT, McCouch SR, Schaeffer ML, Ware DH, Stein LD, Rhee SY. The plant structure ontology, a unified vocabulary of anatomy and morphology of a flowering plant. Plant Physiol. 2007;143:587–599. doi: 10.1104/pp.106.092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic K, Rhee SY, Kellogg EA, Stevens PF. Plant Structure Ontology (PSO)—a morphological and anatomical ontology of flowering plants. In: Burger A, Davidson D, Baldock R, editors. Anatomy ontologies for bioinformatics: priniciples and practice. London: Springer; 2008. [Google Scholar]
- Jaiswal P, Avraham S, Ilic K, Kellogg E, McCouch S, Pujar A, Reiser L, Rhee S, Sachs M, Schaeffer M, Stein L, Stevens P, Vincent L, Ware D, Zapata F. Plant Ontology (PO): a controlled vocabulary of plant structures and growth stages. Comp. Funct. Genomics. 2005;6:388–397. doi: 10.1002/cfg.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jollie M. Chordate morphology. New York: Reinhold Books; 1962. [Google Scholar]
- Keet CM, Roos M, Marshall MS. A survey of requirements for automated reasoning services for bio-ontologies in OWL. Third International Workshop OWL: Experiences and Directions (OWLED 2007) 2007;Vol. 258 2007 June 6–7; Innsbruck, Austria. CEUR-WS. [Google Scholar]
- Mabee PM, Arratia G, Coburn M, Haendel M, Hilton EJ, Lundberg JG, Mayden RL, Rios N, Westerfield M. Connecting evolutionary morphology to genomics using ontologies: a case study from Cypriniformes including zebrafish. J. Exp. Zool. B Mol. Dev. Evol. 2007;308B:655–668. doi: 10.1002/jez.b.21181. [DOI] [PubMed] [Google Scholar]
- Mabee PM, Ashburner M, Cronk Q, Gkoutos G, Haendel M, Segerdell E, Mungall C, Westerfield M. Phenotype ontologies: the bridge between genomics and evolution. Trends Ecol. Evol. 2007;22:345–350. doi: 10.1016/j.tree.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Maglia AM, Leopold JL, Pugener LA, Gauchi S. An anatomical ontology for amphibians. Pac. Symp. Biocomput. 2007;12:367–378. [PubMed] [Google Scholar]
- Matuszek C, Cabral J, Witbrock M, Oliveira JD. Proceedings of the 2006 AAAI Spring Symposium on Formalizing and Compiling Background Knowledge and Its Applications to Knowledge Representation and Question Answering. 2006. An introduction to the syntax and content of Cyc. 2006 Mar. 27–29; Menlo Park, CA: The AAAI Press. [Google Scholar]
- Mayden RL, Wiley EO. The fundamentals of phylogenetic systematics. In: Mayden RL, editor. Systematics, historical ecology, and North American freshwater fishes. Stanford (CA): Stanford University Press; 1992. pp. 114–185. [Google Scholar]
- Mungall CJ. Obol: integrating language and meaning in bio-ontologies. Comp. Funct. Genomics. 2004;5:509–520. doi: 10.1002/cfg.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, Gkoutos GV, Smith CL, Haendel MA, Lewis SE, Ashburner M. Integrating phenotype ontologies across multiple species. Genome Biol. 2010;11:R2. doi: 10.1186/gb-2010-11-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus F, Smith B. Modelling principles and methodologies—relations in anatomical ontologies. In: Burger A, Davidson D, Baldock R, editors. Anatomy ontologies for bioinformatics: priniciples and practice. London: Springer; 2008. pp. 289–306. [Google Scholar]
- Patterson C. Homology in classical and molecular biology. Mol. Biol. Evol. 1988;5:603–625. doi: 10.1093/oxfordjournals.molbev.a040523. [DOI] [PubMed] [Google Scholar]
- Ramirez M, Coddington J, Maddison W, Midford P, Prendini L, Miller J, Griswold C, Hormiga G, Sierwald P, Scharff N, Benjamin S, Wheeler W. Linking of digital images to phylogenetic data matrices using a morphological ontology. Syst. Biol. 2007;56:283–294. doi: 10.1080/10635150701313848. [DOI] [PubMed] [Google Scholar]
- Remane A. Die Grundlagen des naturlichen Systems der vergleichenden Anatomie und der Phylogenetick. Leipzig: Geest und Portig; 1952. [Google Scholar]
- Rosen DE, Greenwood PH. Origin of the Weberian apparatus and the relationships of the ostariophysan fishes. Am. Mus. Novit. 1970;2428:1–25. [Google Scholar]
- Rosse C, Mejino J. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J. Biomed. Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Roth VL. On homology. Biol. J. Linn. Soc. 1984;22:13–29. [Google Scholar]
- Schober D, Smith B, Lewis S, Kusnierczyk W, Lomax J, Mungall C, Taylor C, Rocca-Serra P, Sansone S-A. Survey-based naming conventions for use in OBO Foundry ontology development. BMC Bioinformatics. 2009;10:125. doi: 10.1186/1471-2105-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze HP, Arsenault M. The panderichthyid fish Elpistostege: a close relative of tetrapods? Paleontology. 1985;28:293–309. [Google Scholar]
- Segerdell E, Bowes J, Pollet N, Vize P. An ontology for Xenopus anatomy and development. BMC Dev Biol. 2008;8:92. doi: 10.1186/1471-213X-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg L, Eilbeck K, Ireland A, Mungall C, Consortium TO, Leontis N, Rocca-Serra P, Ruttenberg A, Sasone S, Scheuermann R, Shah N, Whetzel P, Lewis S. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ceusters W, Klagges B, Köhler J, Kumar A, Lomax A, Mungall C, Neuhaus F, Rector A, Rosse C. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Kusnierczyk W, Schober D. Towards a reference terminology for ontology research and development in the biomedical domain. Proceedings of the KR-MED. 2006;2006 2006 Nov. 8; Baltimore, MD. p. 57–66. [Google Scholar]
- Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe D, Knight J, Mani P, Moxon S, Pich C, Ramachandran S, Schaper K, Segerdell E, Shao X, Singer A, Song P, Sprunger B, Van Slyke C, Westerfield M. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe D, Mani P, Ramachandran S, Schaper K, Segerdell E, Song P, Sprunger B, Taylor S, Van Slyke C, Westerfield M. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Clements D, Conlin T, Edwards P, Frazer K, Schaper K, Segerdell E, Song P, Sprunger B, Westerfield M. The Zebrafish Information Network (ZFIN): the zebrafish model organism database. Nucleic Acids Res. 2003;31:241–243. doi: 10.1093/nar/gkg027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Doerry E, Douglas S, Westerfield M. The Zebrafish Information Network (ZFIN): a resource for genetic, genomic and developmental research. Nucleic Acids Res. 2001;29:87–90. doi: 10.1093/nar/29.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RJ, Goodman JL, Strelets VB the FlyBase Consortium. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Bodenreider O, Goldbreich C. Experience in reasoning with the Foundational Model of Anatomy in OWL DL. Pac. Symp. Biocomput. 2006;11:200–211. [PMC free article] [PubMed] [Google Scholar]