Abstract
The complete genome sequence of the thermophilic sulphur-reducing bacterium, Deferribacter desulfuricans SMM1, isolated from a hydrothermal vent chimney has been determined. The genome comprises a single circular chromosome of 2 234 389 bp and a megaplasmid of 308 544 bp. Many genes encoded in the genome are most similar to the genes of sulphur- or sulphate-reducing bacterial species within Deltaproteobacteria. The reconstructed central metabolisms showed a heterotrophic lifestyle primarily driven by C1 to C3 organics, e.g. formate, acetate, and pyruvate, and also suggested that the inability of autotrophy via a reductive tricarboxylic acid cycle may be due to the lack of ATP-dependent citrate lyase. In addition, the genome encodes numerous genes for chemoreceptors, chemotaxis-like systems, and signal transduction machineries. These signalling networks may be linked to this bacterium's versatile energy metabolisms and may provide ecophysiological advantages for D. desulfuricans SSM1 thriving in the physically and chemically fluctuating environments near hydrothermal vents. This is the first genome sequence from the phylum Deferribacteres.
Keywords: Deferribacter desulfuricans, whole genome sequence, hydrothermal vent, chemolithotroph
1. Introduction
Generally, the greatest challenge to thermophilic microorganisms living on hydrothermal vents may be posed by the risk of being swept out of the range of the vent and thereby losing the temperature range and necessary chemical supplies. They solve this problem by clinging to rocks in communal mats or swimming with a whip-like flagellum as sensing temperature or chemical stimuli to guide their directional movements.1 Deferribacter desulfuricans SMM1T (DSM 14783T) has been isolated from a deep-sea hydrothermal vent chimney at the Suiyo Seamount in the Izu-Bonin Arc, Japan.2 The strain SMM1 is thermophilic (optimal temperature, 60–65°C) and a strictly anaerobic heterotroph capable of using complex organic compounds such as yeast extract and tryptone, ethanol, and various organic acids as sources of energy and carbon. There are three other species in the genus Deferribacter—D. abyssi and D. autotrophicus, which have been isolated from deep-sea hydrothermal vent environments, and D. thermophilus, which has been isolated from a subseafloor petroleum reservoir.3–5 These bacteria are strictly anaerobic chemolithotrophs utilizing various organic compounds and H2 as electron donors and nitrate, S0 (D. desulfuricans, D. abyssi, and D. autotrophicus), and Fe (III) and Mn (IV) (D. thermophilus, D. abyssi, and D. autotrophicus) as electron acceptors. Such versatility for energy generation may provide an ecological advantage for deep-sea vent-dominating chemolithotrophs as has been proposed for members of Aquificales and Epsilonproteobacteria.6 It has been shown, depending on the hydrothermal vent chimney environment, that Deferribacter-related species are the dominant species in these locations.7,8 Thus, it is intriguing to compare the genomes of Deferribacter-related species with other deep-sea vent chemolithotrophic species such as Thiomicrospira crunogena,9 Nautilia profundicola,10 Sulfurovum sp. NBC37-1,11 Nitratiruptor sp. SB155-2,11 and Persephonella marina12 in order to highlight the genomic features that reflect their lifestyle in the environment of deep-sea hydrothermal vent chimneys. Here, we report the complete genome sequence of thermophilic D. desulfuricans SMM1 determined as the first published bacterial genome sequence from the phylum Deferribacteres. We provide a comparative analysis of the genome of D. desulfuricans SMM1 with those of five other chemolithotrophs isolated from deep-sea hydrothermal vent chimneys.
2. Materials and methods
2.1. Genome sequencing and assembly
Deferribacter desulfuricans SSM1 was grown heterotrophically as described previously,2 and total DNA was isolated using proteinase K treatment followed by phenol extraction. The DNA was fragmented by HydroShear (Genomic Solutions), and three shotgun libraries were generated: small- and medium-insert (1.5 and 5 kb) plasmid libraries and a large-insert fosmid (35 kb) library as described previously.13 All three libraries provided 15 times coverage of the genome. Assemblies were accomplished using the PHRED/PHRAP/CONSED suite (http://www.pharap.org).14 Gaps between the assembled sequences were primarily closed by primer walking on gap-spanning library clones or with PCR products from genomic DNA. Quality assessment of the final assembly was performed as described previously.13
2.2. Gene identification and annotation
Putative non-translated genes were identified by using the Rfam15 and tRNAscan-SE16 programs, whereas rRNA genes were identified using the BLASTN program.17 Protein-coding sequences (CDSs) were predicted using a combination of GLIMMER18 and GeneMarkS.19 The remaining parts of the genome were screened further to find missed CDSs by a BLASTX homology search against protein databases. The predicted CDSs were translated and submitted for BLAST analysis against the NCBI non-redundant (nr),20 UniProt,21 and Kyoto Encyclopedia of Genes and Genomes (KEGG) GENES22 databases. The results were collated and presented via a home-made browser (GBROWN) for manual verification. The start sites were manually inspected and altered in an alignment comparison with the best match. The revised CDS set was searched against Pfam23 in addition to the previously mentioned databases. Specific functional assignments suggested by matches with these databases were only accepted when they had a minimum 30% amino acid sequence identity over 70% of the gene length and were supported by Pfam domain assignments, or were consistent with gene context in the genome (e.g. membership in a potential operon with other genes with convincing matches to curated databases). The KEGG PATHWAY and MetaCyc24 databases were used for pathway reconstruction. Signal peptides were predicted with SignalP,25 whereas transmembrane helices were predicted with SOSUI.26 Clusters of regularly interspaced repeats [clustered regularly interspaced short palindromic repeats (CRISPR)] were identified using the CRISPRFinder.27
2.3. Comparative genomics analyses
Protein sequence data for bacterial genomes were obtained from the KEGG database.22 Pairwise ortholog families were identified with the InParanoid program.28 In order to examine genome-wide relationships of D. desulfuricans in terms of gene repertories, the ordination of bacterial genomes was generated using the non-metric multidimensional scaling (NMDS) method, which is a major branch of multivariate analysis. NMDS ordinations attempt to place all samples in an arbitrary two-dimensional space, in which the relative distances between the samples indicate the corresponding pairwise similarity. Hence, closely related organisms in the NMDS ordination would have similar gene repertoires. The gene context of each genome, based on clusters of orthologous groups (COGs),29 was constructed with the following procedure. Each complete proteome was compared with the COG profile database (4450 families) using the RPSBLAST program.17 A significant match (E-value of 10−5) was determined for each COG, so that any outlier detected by the Smirnov–Grubbs test at the 0.01 significance level was excluded from the COG profile. Any proteome was thus converted into a binary character matrix indicating the presence (1) or absence (0) of the COG families. The genome distance between species (A and B) was calculated by the formula: DAB = 1 − JAB, where JAB is the Jaccard's coefficient, which reflects the similarity of gene content between A and B.30 NMDS ordination was calculated using metaMDS in the vegan library running with the R programming language and environment (R package version 2.7.2: R Development Core Team, 2007, http://www.R.project.org).
2.4. Nucleotide sequence accession numbers
The sequences of the D. desulfuricans genome, consisting of the chromosome and plasmid sequences, are available at GenBank/EMBL/DDBJ under accession numbers AP011529 and AP011530, respectively.
3. Results and discussion
3.1. General genome features
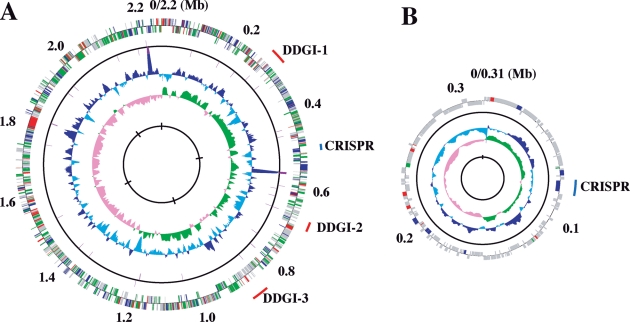
The genome of Deferribacter desulfuricans SSM1 contains a single circular chromosome of 2 234 389 bp and a megaplasmid (pDF308) of 308 544 bp (Table 1). Their average G + C contents are 31.1% and 24.5%, respectively. The chromosome displays two clear GC skew transitions that likely correspond to the DNA replication origin and terminus (Fig. 1). Annotation of the chromosomal sequence reveals 2117 CDSs, of which 1404 (66%) can be functionally assigned. The megaplasmid encodes 257 CDSs, whereas more than two-thirds are unique and exhibit no apparent similarity with any of the CDSs present in the database. Interestingly, 17 copies of gene cluster encoding two transposases belonging to the IS200 and IS605 family present in the megaplasmid but none were found in the chromosome. The chromosome contains two ribosomal RNA operons with a 16S–23S–5S rRNA gene alignment. In all, 43 tRNA genes were identified (Table 1). In addition to the regular tRNA genes, the D. desulfuricans genome also contains selC for the selenocysteine tRNA. The other components necessary for the selenocysteine system, e.g. selenocysteine synthetase (SelA), the specific elongation factor (SelB), selenophosphate synthetase (SelD), and seryl-tRNA synthetase (SerS), are also present in the genome. As for small RNA-encoding genes, potential genes for RNase P RNA (RnpB) and 6S RNA (SsrS) were assigned by using the Rfam database.
Table 1.
Summary of genome features of D. desulfuricans SSM1
| Characteristics | Chromosome | Plasmid |
|---|---|---|
| Size (bp) | 2 234 389 | 308 544 |
| G + C content (%) | 31.1 | 24.5 |
| No. of identified protein-coding genes | ||
| Total | 2117 | 257 |
| Functionally assigned | 1404 | 63 |
| Conserved hypothetical | 494 | 20 |
| Hypothetical | 219 | 174 |
| Pseudogenes | 17 | 9 |
| Average gene length (bp) | 976 | 993 |
| Coding density (%) | 93.7 | 88.2 |
| No. of identified RNA genes | ||
| rRNA operons | 2 | — |
| tRNA | 43 | — |
| Small RNAa | 2 | — |
| Size (bp) of genomic islands (% G + C content) | ||
| DDGI-1 | 35 072 (31.4%) | — |
| DDGI-2 | 20 995 (28.2%) | |
| DDGI-3 | 36 900 (31.0%) | |
| CRISPR elements | 2 | 2 |
aIncludes genes for RNase P RNA and 6S RNA.
Figure 1.
Circular representation of the D. desulfuricans SSM1 genome. (A) Chromosome. (B) Megaplasmid pDF308. From the inside, the first and second circles show the GC skew (values greater than or less than zero are indicated in green and pink, respectively) and the G + C percent content (values greater or smaller than the average percentage in the overall chromosome or plasmid are shown in blue and sky blue, respectively) in a 10-kb window with 100-bp step, respectively. The third and fourth circles show the presence of RNAs (rRNA, tRNA, and small RNA genes); CDSs aligned in the clockwise and counterclockwise directions are indicated in the upper and lower sides of the circle, respectively. Different colours indicate different functional categories: red for information storage and processing; green for metabolism; blue for cellular processes and signalling; grey for poorly characterized function; and purple for RNA genes. The outermost circle shows the location of genomic islands (red) and CRISPR/Cas systems (blue). The ‘0’ marked on the outmost circles corresponds to the putative replication origin, and the putative replication termination site of the chromosome is at 1.23 Mb.
The D. desulfuricans chromosome contains three genomic islands, termed DDGI-1 (coordinates 278 054–313 125), DDGI-2 (coordinates 687 244–708 238), and DDGI-3 (coordinates 864 851–901 750), possibly acquired via horizontal gene transfer (Fig. 1 and Table 1). They have many specific features of genomic island, such as the tRNA gene locus at junctions and the presence of direct repeats and phage integrases (DEFDS_0277, DEFDS_0708, and DEFDS_0871), but no anomalous GC content.31 Most of the genes in the genomic islands encode hypothetical proteins, but some of those appeared to encode the functional proteins related to the adaptation mechanisms for the hydrothermal vent environments. For example, the DDGI-1 includes two heavy-metal transporting P-type ATPases (DEFDS_0300 and DEFDS_0301). The heavy-metal transporters could be responsible for heavy metal tolerance in the hydrothermal vent environment. The DDGI-2 has a toxin–antitoxin system (DEFDS_0716 and DEFDS_0717) involved in phage defence and the stress response.32
Similar to most other thermophiles, D. desulfuricans has the CRISPR elements together with their associated genes (cas), which would serve as immunity against phages, possibly by an RNA-interference-like mechanism.33 The CRISPR/Cas systems were identified in the chromosome (coordinates 77 878– 86 387) and the plasmid (coordinates 267 868–276 377). Both systems have an identical repeat, and the cas genes also show a high relevance with each other. Regarding the repeat sequence and structure of the cas genes, the systems of D. desulfuricans were closely related with those observed in other thermophiles, such as Sulfurihydrogenibium azorense, Thermoanaerobacter pseudethanolicus, and the homo-acetogen Clostridium thermocellum. Therefore, the D. desulfuricans CRISPR/Cas systems may have been acquired horizontally via the megaplasmid.
3.2. Orthologous relationships among the bacterial species
Among the proteins identified in the D. desulfuricans genome, ∼30% of them showed the highest similarity to those species from Deltaproteobacteria, especially Geobacter, Pelobacter, Desulfovibrio, and Syntrophobacter spp., and ∼14% were found to be most similar to those from clostridial species. The remaining proteins showed the highest similarities to several species from Aquificae (5.3%), Gammaproteobacteria (4.8%), and Epsilonproteobacteria (3.7%). Approximately half of the proteins were shared between D. desulfuricans and deltaproteobacterial species, such as Geobacter sulfurreducens PCA (1106 orthologs), Pelobacter carbinolicus (1046 orthologs), Syntrophobacter fumaroxidans (985 orthologs), and Desulfovibrio vulgaris subsp. vulgaris Hildenborough (936 orthologs), although D. desulfuricans is phylogenetically distant from Deltaproteobacteria.
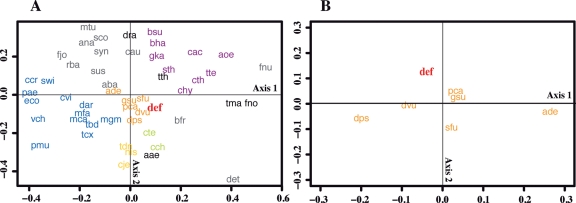
For the next step of orthologous analysis, we performed multivariate analyses on the basis of the gene repertories from 51 bacterial species in order to schematically express the orthologous relationships of D. desulfuricans among other bacterial species (Fig. 2A). As expected, the correlation map indicated that D. desulfuricans and the deltaproteobacterial species, which have strong orthologous relationships, gathered around the centre of map. The same analysis to focus on six species of the class Deltaproteobacteria with D. desulfuricans showed that D. desulfuricans is especially related to three species, G. sulfurreducens (gsu), P. carbinolicus (pca), and D. vulgaris (dvu), possessing sulphur- or sulphate-reducing properties (Fig. 2B). This result seems to attribute that these four species share many genes involved in physiological and metabolic properties such as anaerobiosis and assimilations system for small organic molecules (e.g. acetate, pyruvate, and lactate) as carbon and energy source.
Figure 2.
Ordination plot of bacterial genomes using NMDS. (A) Analysis with 50 species belonging to 12 phyla and 5 classes, and D. desulfuricans SSM1. (B) Analysis with six species within Deltaproteobacteria and D. desulfuricans SSM1. Distances were calculated from gene profiles based on COG families. The abbreviation corresponding to the KEGG organism code is used as the label for the species name (detailed explanations are described in Supplementary Table S1). The labels are colour-coded according to their taxonomic groups (phylum/class): red, D. desulfuricans SSM1 (def); orange, Deltaproteobacteria; yellow, Epsilonproteobacteria; blue, other Proteobacteria; purple, Firmicutes; green, Chlorobi; black, Aquificae, Thermotogae, and Deinococcus-Thermus; and grey, other bacteria.
3.3. Central metabolism
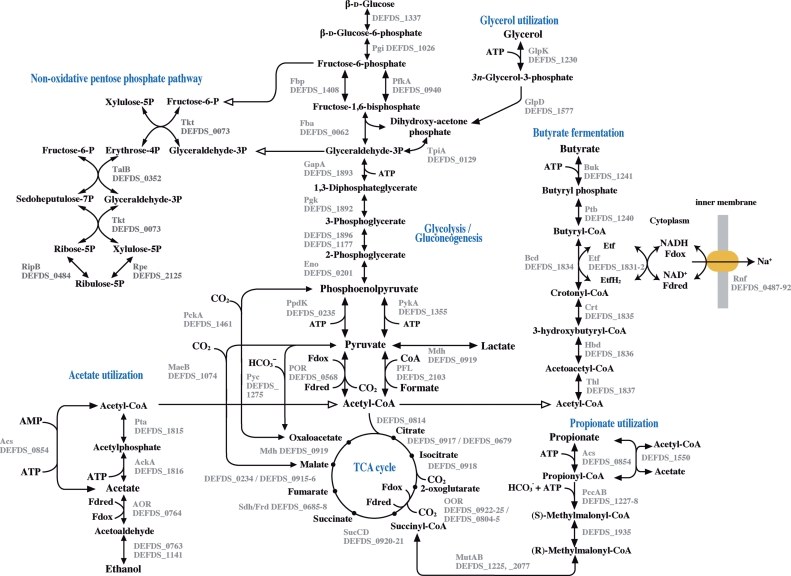
Deferribacter desulfuricans grows heterotrophically using a variety of organic acids (formate, acetate, propionate, pyruvate, and lactate) with nitrate or S0 as a primary electron acceptor.2 As shown in Fig. 3, the reconstructed central metabolic pathways from the D. desulfuricans genome certainly showed that these organic acids could be utilized as energy and carbon sources via the oxidative tricarboxylic acid (TCA) cycle and various anaplerotic pathways. Acetate used as a D. desulfuricans SSM1 growth substrate can be activated to acetyl-CoA either via a single-step reaction by an acetyl-CoA synthetase (Acs: DEFDS_0854) or a two-step reaction by acetate kinase (AckA: DEFDS_1816) and phosphate acetyltransferase (Pta: DEFDS_1815). The AckA–Pta reaction can also operate in acetate production from acetyl-CoA in a fermentative metabolism of pyruvate, and the enzymatic action of AckA results in ATP production by substrate-level phosphorylation. Lactate may be oxidized to pyruvate by a malate dehydrogenase with a broad substrate specificity. In contrast to acetate and lactate, propionate is fed directly into the TCA cycle via succinyl-CoA. Propionyl-CoA, produced by the activation of propionate by acetyl-CoA synthetase, is converted to succinyl-CoA through a methylmalonyl pathway, including a propionyl-CoA carboxylase (DEFDS_1227-8), a methylmalonyl-CoA epimerase (DEFDS_1935), and a methylmalonyl-CoA mutase (MutA: DEFDS_2077 and MutB: DEFDS_1225). The catabolism of pyruvate reflects the anaerobic nature of D. desulfuricans. Conversion of pyruvate to acetyl-CoA is performed by either pyruvate ferredoxin oxidoreductase (POR: DEFDS_0568) or pyruvate-formate lyase (PFL: DEFDS_2103).
Figure 3.
Central metabolism based on potential growth substrates and metabolic capacities reconstructed from the D. desulfuricans genome. This figure displays the flow of carbon in the metabolism of various organic acids (acetate, propionate, butyrate, lactate, and glycerol) predicted from the genome information of D. desulfuricans SSM1. The reversible and irreversible reactions catalysed by enzymes are indicated with both and single arrowhead, respectively. POR, pyruvate ferredoxin oxidoreductase; PFL, pyruvate formate-lyase; Pyc, pyruvate carboxylase; Pck, phosphoenolpyruvate carboxykinase; PykA, pyruvate kinase; Ppd, pyruvate phosphate dikinase; MaeB, malate dehydrogenase (oxaloacetate-decarboxylating); Mdh, malate dehydrogenase; SucCD, succinyl-CoA synthase; Sdh, succinate dehydrogenase; OOR, 2-oxogultarate ferredoxin oxidoreductase; Fba, fructose-bisphosphatase; Pfk, 6-phosphofructokinase; Acs, acetyl-CoA synthetase; Ack, acetate kinase; Pta, phosphate acetyltransferase; AOR, aldehyde ferredoxin oxidoreductase; GlpK, glycerol kinase; GlpD, glycerol-3-phosphate dehydrogenase; Thl, acetoacetyl-CoA thiolase; Hbd, 3-hydroxybutyryl-CoA dehydrogenase; Crt, 3-hydroxybutyryl-CoA dehydratase (crotonase); Bcd, butyryl-CoA dehydrogenase; Ptb, phosphate butyryltransferase; Buk, butyrate kinase; Rnf, Rnf-type ion-translocating electron transport complex; Etf, electron transfer flavoprotein complex; PccAB, propionyl-CoA carboxylase; MutAB, methylmalonyl-CoA mutase; 2Pi, diphosphate; and Fdox/Fdred, ferredoxin, oxidized and reduced forms respectively.
Deferribacter desulfuricans is capable of growing using formate as its sole source of carbon and energy. According to the reconstructed metabolic pathways from the D. desulfuricans genome, formate is oxidized to CO2 by membrane-bound formate dehydrogenase (Fdh: DEFDS_1329-31), which is energetically coupled with the respiratory nitrate or S0 reduction. Methanogens and homo-acetogens are known to assimilate formate to acetyl-CoA via the Wood–Ljungdahl pathway (reductive acetyl-CoA pathway).34 Since the reductive acetyl-CoA pathway is absent in the D. desulfuricans genome, formate must be assimilated by another pathway. A potential reaction for formate assimilation is the direct or indirect conversion of formate and acetyl-CoA to pyruvate, the reverse reaction of pyruvate-formate lyase or the POR-catalysing carboxylation of CO2 coupled with the formate oxidation by the Fdh protein. The pyruvate can then be converted to various biosynthetic intermediates by the TCA cycle and anaplerotic pathways (Fig. 3).
The D. desulfuricans genome has a variety of anaplerotic pathways, which include the three enzymes, malic enzyme (MaeB: DEFDS_1074), pyruvate carboxylase (Pyc: DEFDS_1275), and phosphoenolpyruvate carboxykinase (PckA: DEFDS_1461). These pathways replenish the intermediates of the TCA cycle for gluconeogenesis and amino acid biosynthesis, and their anaplerotic CO2 fixation may be an important function for D. desulfuricans living heterotrophically in a limited organic carbon such as hydrothermal vent environment. In the gluconeogenesis pathway, pyruvate may be dominantly produced by POR from acetyl-CoA and further converted to phosphoenolpyruvate by pyruvate phosphate dikinase (PpdK: DEFDS_0235).
The genome possesses genes for the Embden–Meyerhof–Parnas (EMP) pathway and the non-oxidative branch of the pentose phosphate pathway (Fig. 3). The EMP pathway includes fructose-1,6-bisphosphatase (DEFDS_1408), which is a key enzyme in gluconeogenesis. The TCA cycle of D. desulfuricans includes 2-oxoglutarate ferredoxin oxidoreductases (OOR: DEFDS_0922-25 and DEFDS_0804-5) instead of 2-oxoglutarate dehydrogenases that are typically found in aerobic bacteria. These enzymes catalyse the reversible oxidative decarboxylation of 2-oxoglutarate to form succinyl-CoA, while they are also key enzymes of the reductive TCA (rTCA) cycle. Therefore, the TCA cycle has the potential to proceed in the reverse direction. Although no ATP-dependent citrate lyase (AclAB) is present, the presence of other key enzymes of the rTCA cycle suggests that the rTCA cycle could be partially operative in D. desulfuricans. Actually, oxaloacetate is produced from pyruvate and phosphoenolpyruvate by the anaplerotic enzymes, Pyc and PckA, respectively, and then oxaloacetate is converted to 2-oxoglutarate by operating the TCA cycle in the reductive direction. Among the previously characterized members of the genus Deferribacter, D. abyssi and D. autotrophicus grow autotrophically.3,5 We have also isolated several strictly autotrophic strains of D. desulfuricans from different deep-sea hydrothermal environments (data not shown). Thus, the autotrophy may not be an unusual feature within the genus Deferribacter. The lack of an autotrophic phenotype in the strain SMM1 is presumably caused by the lack of the aclAB genes.
Genome analysis suggested the capability of butyrate-fermentation in D. desulfuricans, although it has not been confirmed experimentally. In this pathway, as shown in Fig. 3, butyryl-CoA is converted from acetyl-CoA by the four enzymes—acetoacetyl-CoA thiolase (Thl; DEFDS_1837), 3-hydroxybutyryl-CoA dehydrogenase (Hbd; DEFDS_1836), 3-hydroxybutyryl-CoA dehydratase (Crt; DEFDS_1835), and butyryl-CoA dehydrogenase (Bcd; DEFDS_1834)—and the electron transfer flavoprotein complex (Etf; DEFDS_1831-1832). The order of this gene cluster is conserved in the genomes of Geobacter metallireducens, Geobacter uraniireducens, and a butyrate-producing clostridial species, Butyrivibrio fibrisolvens.35 Butyryl-CoA is converted to butyrate by phosphate butyryltransferase (DEFDS_1240) and butyrate kinase (DEFDS_1241) resulting in the generation of ATP by the substrate-level phosphorylation. This process can be reversible under a certain condition36 and may thus serve for the butyrate utilization.
As shown in Fig. 3, the D. desulfuricans genome also contains a set of six genes (rnfCDGEAB, DEFDS_0487-92) related to the potential membrane-bound electron transport complex (Rnf) recently found in various bacteria and archaea.37–39 In nitrogen-fixing bacteria, the Rnf complex transports electrons from NADH to ferredoxin, which donates electrons to nitrogenase.40 It is further proposed that the same enzyme apparently runs in the reverse direction: electron transfer from reduced ferredoxin to NAD+ driving the electrogenic pumping of Na+ out of the cell.39 Regional synteny for the rnf genes in the D. desulfuricans genome was found in several clostridial genomes, such as Clostridium tetani, Clostridium kluyveri, and Halothermothrix orenii. It is supposed that the Rnf complex is involved in the regeneration of NADH for the fermentation of butyrate.41 Since the D. desulfuricans genome indicated the possible fermentation of butyrate and the requirement of reduced ferredoxin for the anabolic reactions, the Rnf complex may be operative in either the forward or reverse flow of electrons, or both. In addition, it is suggested that glycerol can be utilized by glycerol kinase (GlpK, DEFDS_1230), sn-glycerol-3-phosphate dehydrogenase (GlpD, DEFDS_1577), and triosephosphate isomerase (DEFDS_0129). However, no growth was observed when glycerol was used as the sole source of energy and carbon,2 and this phenomenon is presumably explained by the lack of a glycerol-uptake-facilitator protein.
Supporting the heterotrophic ability of D. desulfuricans, a variety of transporters for mono-, di-, and tricarboxylates are encoded within the genome. Actually, the genes encoding two sodium:solute symporter (SSS) family proteins (DEFDS_1547 and DEFDS_1574) and a YaaH family protein, which are presumably involved in acetate uptake, were identified in the genome. In addition, the genes encoding five transporter systems of tripartite ATP-independent periplasmic (TRAP) family (DEFDS_0682-4, DEFDS_0770-2, DEFDS_1399-1400, DEFDS_1846-7, and DEFDS_2070-1) may be involved in uptake of various dicarboxylates such as fumarate, malate, and succinate. In terms of tricarboxylate transporters, the genes for two citrate transporters (DEFDS_1180 and DEFDS_2162) were identified in the genome. On the other hand, the genome encodes two phosphotransferase system sugar transporters for fructose and mannose, although D. desulfuricans could not grow by using sugar.
3.4. Respiration
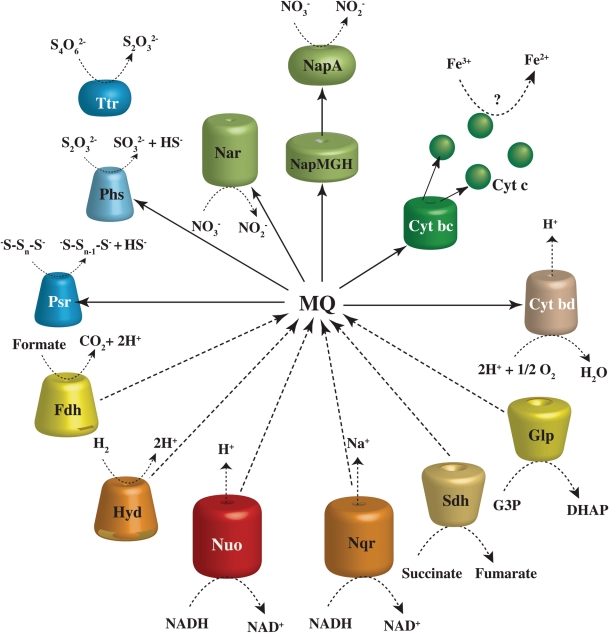
Since D. desulfuricans SSM1 is capable of using molecular hydrogen (H2) as an energy source,2 a membrane-bound NiFe-hydrogenase (Hyd) (DEFDS_0075-77) would function as an H2-uptake hydrogenase. As shown in Fig. 4, the oxidation of molecular hydrogen is coupled to the reduction of menaquinone. The genome has two distinctive nitrate-reducing enzymes for respiration, which are coupled to the electron transfer chains (Fig. 4). One is a membrane-bound (Nar: DEFDS_2086-89) and the other is a periplasmic (Nap: DEFDS_1819-23) nitrate reductase, which does not directly contribute to the generation of a proton motive force but contributes to redox balancing.42 It is generally known that bacteria have Nar-type nitrate reductases, but some bacterial species have a Nap-type or both types of nitrate reductase. Nap-type nitrate reductase usually functions as a two-subunit enzyme comprising a catalytic subunit (NapA) that binds a bis-MGD cofactor and a [4Fe-4S] cluster, and an electron transfer subunit (NapB) that binds two c-type haems. However, since D. desulfuricans does not possess the napB gene, the nitrate reductase of D. desulfuricans may be monomeric, which is differentiated from the heterodimeric NapAB structure typically found in bacteria. Actually, NapB-independent periplasmic nitrate reductases have been reported for some species in Deltaproteobacteria and Clostridia.43 The organization of Nap genes in the D. desulfuricans genome is similar to that of Desulfovibrio desulfuricans. Although nitrate is sequentially reduced to N2 (denitrification) or NH4 (ammonification) in many cases, no enzyme for nitrite reduction was identified in the D. desulfuricans genome. This is consistent with experimental results, because the accumulation of nitrite is observed during culture of this organism.2
Figure 4.
Genome-based models for the energy-conserving electron-transport pathways of D. desulfuricans SSM1. Reducing power acquired by catabolic metabolism (NADH, succinate, and sn-glycerol-3-phosphate) or by oxidation of hydrogen and/or formate is used to reduce sulphur compounds and nitrate, and potentially iron ions via the quinol pool for energy conservation or dissipation. Membrane-binding components are indicated with cylinders or cones, where the upper and the lower reactions are catalysed on the periplasmic side and the cytoplasmic side, respectively. Periplasmic components are indicated with ellipsoids or spheres. Hyd, membrane-binding NiFe-hydrogenase; Nar, respiratory membrane-bound nitrate reductase; Nap, periplasmic nitrate reductase; Psr, polysulphide reductase; Phs, thiosulphate reductase; Ttr, tetrathionate reductase; Fdh, formate dehydrogenase; Nuo, proton-pumping NADH dehydrogenase; Nqr, sodium-translocating NADH:quinone oxidoreductase; Sdh, succinate dehydrogenase; Cyt bd, cytochrome bd quinol oxidase; Cyt bc, cytochrome bc complex; Glp, sn-glycerol-3-phosphate dehydrogenase; G3P, sn-glycerol-3-phosphate; and DHAP, dihydroxyacetone phosphate.
It has been reported that the reduction of energy-yielding sulphur compounds (sulphur-respiration) is catalysed by some key enzymes: a polysulphide reductase (Psr) in Wolinella succinogenes (Epsilonproteobacteria),44 a thiosulphate reductase (Phs) in Salmonella enterica (Gammaproteobacteria),45 and a hydrogenase-sulphur reductase multienzyme (Sre) in Acidianus ambivalens (Crenarchaeota) or Aquifex aerolicus (Aquificae).46,47 Psr and Phs reduce soluble polysulphide derived from S0 to sulphide, whereas Sre is considered to reduce S0 directly to sulphide. Genome analysis revealed that D. desulfuricans has three gene clusters encoding Psr/Phs enzymes, DEFDS_0670-72, DEFDS_1691-93, and DEFDS_1697-99. The genes in each cluster encode three subunits that construct the following membrane-bound complex enzyme: a catalytic subunit with molybdopterin (PsrA/PhsA), an electron transfer subunit with the [Fe–S] cluster (PsrB/PhsB), and a membrane anchor subunit (PsrC/PhsC). The amino acid sequences of the PsrA/PhsA subunits contain the motif necessary for translocation of the protein towards the periplasmic space via the twin arginine-translocation systems, suggesting that the polysulphide reduction should occur in the periplasmic space. Moreover, an alternative molybdopterin-containing enzyme, tetrathionate reductase (Ttr), is encoded by DEFDS_1446-47, but it is unclear whether this enzyme is involved in energy conservation because of the absence of a membrane anchor subunit. Deferribacter desulfuricans SSM1 presumably uses the polysulphides chemically formed from the reaction of S0 and sulphide, which readily occurs in hydrothermal vents,48 and therefore it needs to transfer polysulphide to the periplasmic space across the outer membrane (Supplementary Fig. S1A). The periplasmic Sud protein in W. succinogenes has been proposed to serve as a polysulphide-binding protein and to transfer polysulphide-sulphur to the active site of polysulphide reductase.49 A Psr/Phs gene cluster (DEFDS_0670-72) in the D. desulfuricans genome is associated with the genes coding an outer membrane porin and lipoprotein (DEFDS_0667-68). The amino acid sequence of DEFDS_0667 represents the functional domain of porin_O_P (phosphate-selective porins O and P) that are conserved in anion-specific porins.50 The lipoprotein DEFDS_0668 contains a rhodanese (thiosulphate sulphurtransferase) domain as well as the Sud protein, although there is no homology between the two proteins. Thus, these two proteins may facilitate sulphur respiration in D. desulfuricans as a polysulphide-specific porin in the outer membrane and a polysulphide-binding protein in the periplasmic space, respectively. This locus is adjacent to other genes related to respiration, two multihaem c-type cytochromes (DEFDS_0665-66) and a cytochrome bc complex (DEFDS_0674-77). Interestingly, a similar locus without the Psr/Phs genes was found in the genomes of the iron- and sulphur-reducing bacteria, Geobacter spp. and Desulfuromonas acetoxidans DSM 684 (Supplementary Fig. S1B).
The D. desulfuricans genome has a gene cluster (DEFDS_0741-60) that includes many genes encoding membrane-bound or periplasmic multihaem c-type cytochromes, which are potentially distributed in the inner or outer membranes or in the periplasmic space (Supplementary Fig. S2A). As demonstrated in G. sulfurreducens and Shewanella oneidensis, multihaem c-type cytochromes may be involved in the potential dissimilatory reduction of metal ions.51 Although D. desulfuricans SSM1 cannot grow by iron reduction, the capability of iron reduction is a symbolic physiological trait of the genus Deferribacter.3–5 On the basis of cultivation-dependent estimations, the members of the genus Deferribacter have been predicted to be the most numerically abundant iron-reducing prokaryotes in deep-sea vents.7,8,52 Therefore, the multihaem c-type cytochromes in D. desulfuricans may be a remnant of the metal reduction system.
As shown in Fig. 4, reductants produced from the oxidation of organic compounds would be oxidized by a proton-pumping NADH dehydrogenase (Nuo: DEFDS_1972-85) and succinate dehydrogenase (Sdh: DEFDS_0685-88) to provide reducing equivalents for the nitrate- and polysulphide reductions. The D. desulfuricans genome has an alternative complex for NADH oxidation, sodium-translocating NADH:quinone oxidoreductase (Nqr: DEFDS_1913-18). This complex possibly allows the generation of a sodium gradient, which could energize an Na+-dependent symporter of the organic substrates. This organism found to possess many sodium gradient-dependent symporters such as two solute:sodium symporters (SSS family), a nucleobase:cation symporter-2 (NCS2) family protein (DEFDS_1105), an alanine or glycine:cation symporter (AGCS) family protein (DEFDS_1601), and phosphate:sodium symporter (PNaS) family protein (DEFDS_0081).
3.5. Signal transduction, motility, and chemotaxis
Deferribacter desulfuricans SSM1 and its relatives inhabit the ever-changing steep physical and chemical gradients of hydrothermal vent chimney. The thermophilic microorganisms living on hydrothermal vents are generally sensing temperature or chemical stimuli to guide their directional movements by swimming with a whip-like flagellum. Thus, the D. desulfuricans genome should have the molecular machinery for sensing and responding to environmental changes. Indeed, D. desulfuricans has genes for various signalling systems, as other vent chemolithotrophs do9–11 (Table 2). More than 70 genes for two-component signal transduction (TCS) systems, including 23 histidine kinases, 41 response regulator genes, and 13 hybrid histidine kinases with regulator domains, were found in the D. desulfuricans genome. The frequency of TCS systems found in the D. desulfuricans genome is the highest (34 genes per megabase) among vent bacteria (ranging from 11 to 22 genes per megabase). Although most of them could not be functionally assigned due to very low homologies with well-defined systems, some functions were assumed to be phosphate regulation (PhoBR) and ammonium assimilation (NtrBC).
Table 2.
Abundance of genetic components for signal transduction systems in the genomes of representative deep-sea vent chemolithotrophs
| Signal transduction systems | Domains (Pfam) | Deferribacter desulfuricans | Thiomicrospira crunogena | Nautilia profundicola | Nitratiruptor sp. SB155-2 | Sulfurovum sp. NBC37-1 | Persephonella marina |
|---|---|---|---|---|---|---|---|
| Genome size (bp) | 2 234 389 | 2 427 734 | 1 676 444 | 1 877 931 | 2 562 277 | 1 930 284 | |
| Two-component signal transduction systems | |||||||
| Sensor histidine kinase | PF00512/PF02518 | 23 | 13 | 11 | 15 | 20 | 8 |
| Response regulator | PF00072 | 41 | 24 | 16 | 19 | 28 | 14 |
| Hybrid | 13 | 7 | 1 | 1 | 9 | 0 | |
| Total | 77 | 44 | 28 | 35 | 57 | 22 | |
| Cyclic diGMP signalling systems | |||||||
| GGDEF | PF00990 | 12 | 20 | 17 | 11 | 5 | 12 |
| EAL | PF00563 | 1 | 5 | 3 | 2 | 1 | 4 |
| HD-GYP | PF01966 | 3 | 3 | 2 | 0 | 0 | 2 |
| GGDEF-EAL | 3 | 16 | 11 | 19 | 12 | 12 | |
| Total | 19 | 44 | 33 | 32 | 18 | 30 | |
| Chemotaxis | |||||||
| Methyl-accepting chemotaxis proteins | PF00015 | 14 | 14 | 11 | 8 | 0 | 5 |
| Chemotaxis proteins | 32 | 13 | 3 | 13 | 0 | 6 | |
| Total | 46 | 27 | 14 | 21 | 0 | 11 | |
Signal transduction proteins were identified by querying the predicted gene products to the Pfam and COG databases. Genes with significant matches (E-value of less than 1E−5) were assigned a product description and classified using a set of rules based on the domain architecture of the protein. The final results were manually verified.
As an alternative signalling system, the D. desulfuricans genome has many genes relevant to a cyclic diguanylate (c-diGMP) signalling system,53 although these were less abundant than in other deep-sea vent bacteria (Table 2). This system is characterized by either diguanylate cyclase (GGDEF domain) or phosphodiesterase (EAL and HD-GYP domains) activity and is relevant to biofilm formation, motility, and virulence.53 In addition, at least one PAS domain was found in seven histidine kinase proteins and two c-diGMP signalling proteins as additional input modules. PAS domains are known to sense changes in the redox conditions inside or outside cells.54 It is worthy to note that the D. desulfuricans genome has a higher proportion of intracellular signalling systems than those of other deep-sea vent chemolithotrophs (Supplementary Fig. S3). These sensors may be essential for the versatile energy acquisition, carbon metabolisms, and the chemotaxis and redoxtaxis of D. desulfuricans SSM1 that are necessary under highly variable environmental conditions.
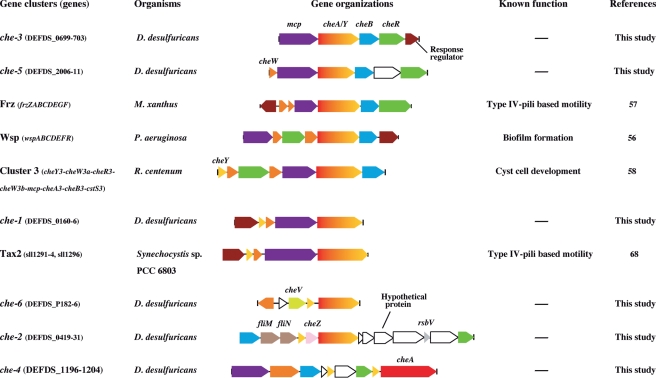
This genome encodes a large number of methyl-accepting chemotaxis proteins (14 MCPs) as observed in other deep-sea vent bacteria (5–14 MCPs), except for the immotile Sulfurovum sp. NBC37-1 (Table 2). These chemotaxis-specific receptors would, in part, contribute to the versatile sensing capabilities of D. desulfuricans. The MCP encoded by DEFDS_1855 possesses a Cache (Ca2+ channels and chemotaxis receptors) domain that is involved in the binding of amino acids and carbohydrates in the MCP of Bacillus subtilis.55 It is also notable that multiple che clusters (six clusters of che-1 to che-6) were identified in the genome of D. desulfuricans, whereas most deep-sea vent microorganisms usually have one or two che gene clusters (Table 2 and Fig. 5). The che-2 cluster is associated with genes for flagellar biosynthesis, suggesting that flagellar formation and motility may be linked with the chemotaxis pathway. Deferribacter desulfuricans SSM1 has almost all of the genes necessary for the flagellar apparatus although motility has not yet been observed in this strain.2 On the other hand, since motility has been confirmed in several strains of D. desulfuricans recently isolated by us, flagellar-based motility could be an intrinsic genetic potential in D. desulfuricans. Moreover, all che clusters, except for che-4, contain genes that encode a fusion protein (CheA/Y) consisting of histidine kinase (CheA) and response regulator (CheY) domains. In other bacteria, the gene clusters containing cheA/Y are involved in specific functions other than chemotaxis. For instances, the Pseudomonas aeruginosa Wsp cluster is involved in biofilm formation,56 and the Myxococcus xanthus Frz cluster functionally controls type IV pili-based motility for cellular aggregation.57 Also, the gene cluster 3 of Rhodospirillum centenum is involved in cyst cell development.58 As shown in Fig. 5, the che-3 and che-5 gene clusters of D. desulfuricans, in particular, have a similar gene organization to each other. There are significant homologies (more than 22% identity at the level of amino acid sequences) between CheA/Y proteins in D. desulfuricans and other organisms. Thus, various D. desulfuricans chemotaxis-like systems may be responsible not only for chemotaxis but also for other cellular functions involved in cell–cell interactions. In fact, this genome contains several gene clusters potentially involved in the formation of a type IV pilus that plays a role in the adhesion of bacteria to host cells and solid surfaces, and mediates bacterial twitching motility. Some genes encoding pilus-related proteins such as PilA of the major prepilin (DEFDS_1243), PilB of the assembly ATPase (DEFDS_1109 and DEFDS_1256), PilQ of secretin (DEFDS_1255), and PilD of prepilin peptidase (DEFDS_0112) were detected. Considering the presence of these genes, it is possible to propose a lifestyle of clinging to the chimney surface for survival under a constantly variable environment.59
Figure 5.
Gene arrangement of the representative chemotaxis-like gene clusters in the genomes of D. desulfuricans and related bacteria. Six chemotaxis-like gene clusters (che-1 to che-6) identified in the D. desulfuricans genome are compared with those CheA/Y-containing clusters that have been experimentally verified: M. xanthus Frz,57 P. aeruginosa Wsp,56 R. centenum Cluster 3,58 and Synechocystis sp. PCC 6803 Tax2.68
3.6. Adaptation to thermal environment
As in other thermophilic microbes, the proteome of D. desulfuricans, which can grow up to 70°C should be stable at least at the maximum growth temperature. Generally, it is known that a protein's amino acid composition has a great influence on its thermostability, and the proteins of thermophiles show a tendency to possess fewer non-charged amino acids and more charged amino acids than those of mesophiles.60 Actually, all of the proteins encoded in the D. desulfuricans genome were found to have fewer histidine, glutamine, and threonine residues, but more glutamic acid and lysine residues than mesophiles (Supplementary Fig. S4). Both charged amino acids presumably contribute to increase the thermostability of the protein by enhancing the occurrence of salt bridges and ion pairs.61
Also, the stabilization of DNA and RNA at a high temperature is indispensable for survival in the hydrothermal environment. It has been suggested that polyamines, RNA methyltransferases, and protamine P1 could contribute to the thermoadaptation of Geobacillus kaustophilus, which has a similar maximum temperature for growth (74°C) as D. desulfuricans.62 In Thermus thermophilus, which can grow up to 85°C, the inactivation of genes for polyamine biosynthesis or tRNA (adenine-N1)-methyltransferase (TrmI) results in a thermolabile phenotype.63 The D. desulfuricans genome encodes genes necessary for putrescine and spermidine synthesis, speA (DEFDS_1288) and speBDE (DEFDS_0967-9). In addition, we identified two genes for norspermidine synthesis—carboxynorspermidine dehydrogenase (DEFDS_1287) and carboxynorspermidine decarboxylase (DEFDS_1286). Norspermidine and norspermine are commonly found in hyperthermophilic bacteria.64 These polyamines stabilize DNA by binding to nucleic acids and can induce aggregation or conformational changes of DNA.65 On the other hand, a homolog (DEFDS_0605) of TrmI found in T. thermophilus, and four tRNA and three tRNA/rRNA methyltransferase genes in total were identified in D. desulfuricans. The genes described above appear to contribute to the thermoadaptation of D. desulfuricans as well as other thermophilic bacteria.
Deferribacter desulfuricans possesses many molecular chaperons used for protein folding and unfolding such as DnaJ-DnaK-GrpE (DEFDS_2113-4) and the HrcA repressor (DEFDS_2113-6), GroEL-GroES (DEFDS_0240-39), and small heat shock proteins categorized into Hsp20 family (DEFDS_1576 and DEFDS_1707). This organism also possesses ATP-dependent heat shock-responsive proteases, which is thought to be concerned in providing thermotolerance to cell on exposure to heat stress, such as HslVU (DEFDS_2155-6), ClpPX (DEFDS_0193-4), and Lon (DEFDS_0195 and DEFDS_1706).
3.7. Antioxidant system
Deferribacter desulfuricans SSM1 is a strict anaerobe: its genome possesses many genes potentially associated with resistance to oxidative stress. DEFDS_1019 and DEFDS_0573 encode a superoxide reductase (SOR) and a rubredoxin (Rd), respectively. The SOR–Rd proteins can catalyse the reduction of superoxide to hydrogen peroxide. Putative rubrerythrins (Rbr) encoded by DEFDS_0019, DEFDS_1568, and DEFDS_1971 further reduce hydrogen peroxide to water.66 DEFDS_1568 is located adjacent to the gene encoding the peroxide repressor (perR) (DEFDS_1567). A series of these genes presumably construct the antioxidant system of D. desulfuricans SSM1 inhabiting the oxic–anoxic transition zone in the deep-sea hydrothermal environment. Additionally, genes for peroxiredoxins (DEFDS_0018 and DEFDS_1350), thioredoxin (DEFDS_1167), and thioredoxin reductase (DEFDS_0503) may be involved in the antioxidant system.
On the other hand, D. desulfuricans possesses genes encoding two potential dioxygen scavenging systems. The first one is a cytochrome bd quinol oxidase (DEFDS_1619-1620), which is a respiratory terminal oxidase, and the second one is a rubredoxin-oxygen oxidoreductase (Roo), whose gene is located next to the rubredoxin gene (DEFDS_0573). In the sulphate-reducing Desulfovibrio-related species, the latter enzyme is proposed to be the cytoplasmic terminal oxidase for the non-energy-conserving respiratory reduction of O2 or nitric oxide.67 Thus, the bd-type oxidase and Roo of D. desulfuricans may also serve to relieve the environmental and intracellular oxidative stresses.
3.8. Conclusions
Genome analysis of D. desulfuricans SSM1 revealed its versatile energy and carbon metabolisms and its machineries for sensing and responding to the environmental changes in hydrothermal vent habitats. We showed that the molecular systems such as the multihaem c-type cytochrome clusters, two-component signal transducers, and abundant chemotaxis components could be tightly linked to the adaptation mechanisms required to adapt in physically and chemically variable environments. The multiple signal transduction systems for sensing dynamic changes in carbon source and temperature, and the type IV pili, which are likely to be useful for clinging to the chimney, allow D. desulfuricans to survive in such a harsh environment. The genome sequence of D. desulfuricans SSM1 should provide many clues for the better understanding of bacterial life in environments around hydrothermal vents from the ecological and evolutionary points of view. The sequences, as well as the gene information shown in this paper, are available in the web databases, ExtremoBase (http://www.jamstec.go.jp/gbrowser/cgi-bin/top.cgi) and DOGAN (http://www.bio.nite.g.o.jp/dogan/Top).
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Supplementary Material
Footnotes
Edited by Naotake Ogasawara
References
- 1.Gold T. In: The Deep Hot Biosphere. Gold T., editor. New York: Copernicus Books; 2001. pp. 19–22. [Google Scholar]
- 2.Takai K., Kobayashi H., Nealson K.H., Horikoshi K. Deferribacter desulfuricans sp. nov., a novel sulfur-, nitrate- and arsenate-reducing thermophile isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2003;53:839–46. doi: 10.1099/ijs.0.02479-0. [DOI] [PubMed] [Google Scholar]
- 3.Miroshnichenko M.L., Slobodkin A.I., Kostrikina N.A., et al. Deferribacter abyssi sp. nov., an anaerobic thermophile from deep-sea hydrothermal vents of the Mid-Atlantic Ridge. Int. J. Syst. Evol. Microbiol. 2003;53:1637–41. doi: 10.1099/ijs.0.02673-0. [DOI] [PubMed] [Google Scholar]
- 4.Greene A.C., Patel B.K., Sheehy A.J. Deferribacter thermophilus gen. nov., sp. nov., a novel thermophilic manganese- and iron-reducing bacterium isolated from a petroleum reservoir. Int. J. Syst. Bacteriol. 1997;47:505–9. doi: 10.1099/00207713-47-2-505. [DOI] [PubMed] [Google Scholar]
- 5.Slobodkina G.B., Kolganova T.V., Chernyh N.A., Querellou J., Bonch-Osmolovskaya E.A., Slobodkin A.I. Deferribacter autotrophicus sp. nov., an iron(III)-reducing bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2009;59:1508–12. doi: 10.1099/ijs.0.006767-0. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa S., Takai K., Inagaki F., et al. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 2005;7:1619–32. doi: 10.1111/j.1462-2920.2005.00856.x. [DOI] [PubMed] [Google Scholar]
- 7.Slobodkin A., Campbell B., Cary S.C., Bonch-Osmolovskaya E., Jeanthon C. Evidence for the presence of thermophilic Fe(III)-reducing microorganisms in deep-sea hydrothermal vents at 13 degrees N (East Pacific Rise) FEMS Microbiol. Ecol. 2001;36:235–43. doi: 10.1111/j.1574-6941.2001.tb00844.x. [DOI] [PubMed] [Google Scholar]
- 8.Takai K., Nunoura T., Ishibashi J.-I., et al. Variability in the microbial communities and hydrothermal fluid chemistry at the newly discovered Mariner hydrothermal field, southern Lau Basin. J. Geophys. Res.-Biogeosci. 2008;113:G02031. [Google Scholar]
- 9.Scott K.M., Sievert S.M., Abril F.N., et al. The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 2006;4:e383. doi: 10.1371/journal.pbio.0040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell B.J., Smith J.L., Hanson T.E., et al. Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 2009;5:e1000362. doi: 10.1371/journal.pgen.1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagawa S., Takaki Y., Shimamura S., Reysenbach A.-L., Takai K., Horikoshi K. Deep-sea vent ε-proteobacterial genomes provide insights into emergence of pathogens. Proc. Natl. Acad. Sci. USA. 2007;104:12146–50. doi: 10.1073/pnas.0700687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reysenbach A.-L., Hamamura N., Podar M., et al. Complete and draft genome sequences of six members of the Aquificales. J. Bacteriol. 2009;191:1992–3. doi: 10.1128/JB.01645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takarada H., Sekine M., Kosugi H., et al. Complete genome sequence of the soil actinomycete Kocuria rhizophila. J. Bacteriol. 2008;190:4139–46. doi: 10.1128/JB.01853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–85. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–41. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S.F., Madden T.L., Schäffer A.A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–41. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Besemer J., Lomsadze A., Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–18. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–8. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bairoch A., Apweiler R., Wu C.H., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–9. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–80. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn R.D., Tate J., Mistry J., et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caspi R., Foerster H., Fulcher C.A., et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–31. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 26.Hirokawa T., Boon-Chieng S., Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–9. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 27.Grissa I., Vergnaud G., Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–7. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remm M., Storm C.E., Sonnhammer E.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–52. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 29.Tatusov R.L., Fedorova N.D., Jackson J.D., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf Y.I., Rogozin I.B., Grishin N.V., Koonin E.V. Genome trees and the tree of life. Trends Genet. 2002;18:472–9. doi: 10.1016/s0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- 31.Hacker J., Kaper J.B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000;54:641–79. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 32.Gerdes K., Christensen S.K., Løbner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–82. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 33.Barrangou R., Fremaux C., Deveau H., et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 34.Ragsdale S.W. Enzymology of the Wood–Ljungdahl pathway of acetogenesis. Ann. N. Y. Acad. Sci. 2008;1125:129–36. doi: 10.1196/annals.1419.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asanuma N., Kawato M., Ohkawara S., Hino T. Characterization and transcription of the genes encoding enzymes involved in butyrate production in Butyrivibrio fibrisolvens. Curr. Microbiol. 2003;47:203–7. doi: 10.1007/s00284-002-3976-2. [DOI] [PubMed] [Google Scholar]
- 36.Wallrabenstein C., Schink B. Evidence of reversed electron transport in syntrophic butyrate or benzoate oxidation by Syntrophomonas wolfei and Syntrophus buswellii. Arch. Microbiol. 1994;162:136–42. [Google Scholar]
- 37.Curatti L., Brown C.S., Ludden P.W., Rubio L.M. Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii. Proc. Natl. Acad. Sci. USA. 2005;102:6291–6. doi: 10.1073/pnas.0501216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q., Li L., Rejtar T., Lessner D.J., Karger B.L., Ferry J.G. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 2006;188:702–10. doi: 10.1128/JB.188.2.702-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller V., Imkamp F., Biegel E., Schmidt S., Dilling S. Discovery of a ferredoxin:NAD + -oxidoreductase (Rnf) in Acetobacterium woodii: a novel potential coupling site in acetogens. Ann. N. Y. Acad. Sci. 2008;1125:137–46. doi: 10.1196/annals.1419.011. [DOI] [PubMed] [Google Scholar]
- 40.Kumagai H., Fujiwara T., Matsubara H., Saeki K. Membrane localization, topology, and mutual stabilization of the rnfABC gene products in Rhodobacter capsulatus and implications for a new family of energy-coupling NADH oxidoreductases. Biochemistry. 1997;36:5509–21. doi: 10.1021/bi970014q. [DOI] [PubMed] [Google Scholar]
- 41.Herrmann G., Jayamani E., Mai G., Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 2008;190:784–91. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno-Vivián C., Cabello P., Martínez-Luque M., Blasco R., Castillo F. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 1999;181:6573–84. doi: 10.1128/jb.181.21.6573-6584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jepson B.J.N., Marietou A., Mohan S., Cole J.A., Butler C.S., Richardson D.J. Evolution of the soluble nitrate reductase: defining the monomeric periplasmic nitrate reductase subgroup. Biochem. Soc. Trans. 2006;34:122–6. doi: 10.1042/BST0340122. [DOI] [PubMed] [Google Scholar]
- 44.Krafft T., Gross R., Kröger A. The function of Wolinella succinogenes psr genes in electron transport with polysulphide as the terminal electron acceptor. Eur. J. Biochem. 1995;230:601–6. doi: 10.1111/j.1432-1033.1995.0601h.x. [DOI] [PubMed] [Google Scholar]
- 45.Clark M.A., Barrett E.L. The phs gene and hydrogen sulfide production by Salmonella typhimurium. J. Bacteriol. 1987;169:2391–7. doi: 10.1128/jb.169.6.2391-2397.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guiral M., Tron P., Aubert C., Gloter A., Iobbi-Nivol C., Giudici-Orticoni T. A membrane-bound multienzyme, hydrogen-oxidizing, and sulfur-reducing complex from the hyperthermophilic bacterium Aquifex aeolicus. J. Biol. Chem. 2005;280:42004–15. doi: 10.1074/jbc.M508034200. [DOI] [PubMed] [Google Scholar]
- 47.Laska S., Lottspeich F., Kletzin A. Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology. 2003;149:2357–71. doi: 10.1099/mic.0.26455-0. [DOI] [PubMed] [Google Scholar]
- 48.Campbell B.J., Jeanthon C., Kostka J.E., Luther G.W., Cary S.C. Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 2001;67:4566–72. doi: 10.1128/AEM.67.10.4566-4572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klimmek O., Kreis V., Klein C., Simon J., Wittershagen A., Kröger A. The function of the periplasmic Sud protein in polysulfide respiration of Wolinella succinogenes. Eur. J. Biochem. 1998;253:263–9. doi: 10.1046/j.1432-1327.1998.2530263.x. [DOI] [PubMed] [Google Scholar]
- 50.Hancock R.E., Egli C., Benz R., Siehnel R.J. Overexpression in Escherichia coli and functional analysis of a novel PPi-selective porin, oprO, from Pseudomonas aeruginosa. J. Bacteriol. 1992;174:471–6. doi: 10.1128/jb.174.2.471-476.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi L., Squier T.C., Zachara J.M., Fredrickson J.K. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 2007;65:12–20. doi: 10.1111/j.1365-2958.2007.05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takai K., Nunoura T., Suzuki Y., et al. Variability in microbial communities in black smoker chimneys at the NW caldera vent field, Brothers volcano, Kermadec arc. Geomicrobiol. J. 2009;26:552–569. [Google Scholar]
- 53.Galperin M.Y. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 2004;6:552–67. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhulin I.B., Taylor B.L., Dixon R. PAS domain S-boxes in Archaea, bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 1997;22:331–3. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 55.Garrity L.F., Schiel S.L., Merrill R., Reizer J., Saier M.H., Ordal G.W. Unique regulation of carbohydrate chemotaxis in Bacillus subtilis by the phosphoenolpyruvate-dependent phosphotransferase system and the methyl-accepting chemotaxis protein McpC. J. Bacteriol. 1998;180:4475–80. doi: 10.1128/jb.180.17.4475-4480.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hickman J.W., Tifrea D.F., Harwood C.S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA. 2005;102:14422–7. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zusman D.R., Scott A.E., Yang Z., Kirby J.R. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 2007;5:862–72. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 58.Berleman J.E., Bauer C.E. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum. Mol. Microbiol. 2005;56:1457–66. doi: 10.1111/j.1365-2958.2005.04646.x. [DOI] [PubMed] [Google Scholar]
- 59.Reysenbach A.-L., Shock E. Merging genomes with geochemistry in hydrothermal ecosystems. Science. 2002;296:1077–82. doi: 10.1126/science.1072483. [DOI] [PubMed] [Google Scholar]
- 60.Singer G.A.C., Hickey D.A. Thermophilic prokaryotes have characteristic patterns of codon usage, amino acid composition and nucleotide content. Gene. 2003;317:39–47. doi: 10.1016/s0378-1119(03)00660-7. [DOI] [PubMed] [Google Scholar]
- 61.Zhou X.-X., Wang Y.-B., Pan Y.-J., Li F. Differences in amino acids composition and coupling patterns between mesophilic and thermophilic proteins. Amino Acids. 2008;34:25–33. doi: 10.1007/s00726-007-0589-x. [DOI] [PubMed] [Google Scholar]
- 62.Takami H., Takaki Y., Chee G.-J., et al. Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res. 2004;32:6292–303. doi: 10.1093/nar/gkh970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Droogmans L., Roovers M., Bujnicki J.M., et al. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 2003;31:2148–56. doi: 10.1093/nar/gkg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daniel R.M., Cowan D.A. Biomolecular stability and life at high temperatures. Cell. Mol. Life Sci. 2000;57:250–64. doi: 10.1007/PL00000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou M.H., Lin S.B., Yuann J.M., Lin W.C., Wang A.H., Kan Ls L. Effects of polyamines on the thermal stability and formation kinetics of DNA duplexes with abnormal structure. Nucleic Acids Res. 2001;29:5121–8. doi: 10.1093/nar/29.24.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weinberg M.V., Jenney F.E., Cui X., Adams M.W.W. Rubrerythrin from the hyperthermophilic archaeon Pyrococcus furiosus is a rubredoxin-dependent, iron-containing peroxidase. J. Bacteriol. 2004;186:7888–95. doi: 10.1128/JB.186.23.7888-7895.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wildschut J.D., Lang R.M., Voordouw J.K., Voordouw G. Rubredoxin:oxygen oxidoreductase enhances survival of Desulfovibrio vulgaris Hildenborough under microaerophilic conditions. J. Bacteriol. 2006;188:6253–60. doi: 10.1128/JB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhaya D., Takahashi A., Grossman A.R. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA. 2001;98:7540–5. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.