Abstract
Mitogen-Activated Protein Kinase Kinase Kinases (MAPKKKs) are important components of MAPK cascades, which are universal signal transduction modules and play important role in plant growth and development. In the sequenced Arabidopsis genome 80 MAPKKKs were identified and currently being analysed for its role in different stress. In rice, economically important monocot cereal crop only five MAPKKKs were identified so far. In this study using computational analysis of sequenced rice genome we have identified 75 MAPKKKs. EST hits and full-length cDNA sequences (from KOME or Genbank database) of 75 MAPKKKs supported their existence. Phylogenetic analyses of MAPKKKs from rice and Arabidopsis have classified them into three subgroups, which include Raf, ZIK and MEKK. Conserved motifs in the deduced amino acid sequences of rice MAPKKKs strongly supported their identity as members of Raf, ZIK and MEKK subfamilies. Further expression analysis of the MAPKKKs in MPSS database revealed that their transcripts were differentially regulated in various stress and tissue-specific libraries.
Keywords: MAPK cascade, MAPKKK, gene family, rice
1. Introduction
Mitogen-Activated Protein Kinase (MAPK) cascade plays an important role in plant growth and development, transferring the extracellular stimuli into intracellular response. MAPK cascades are evolutionarily conserved signalling modules in eukaryotes including animals, yeasts and plants.1,2 MAPK cascades are composed of three protein kinases: MAPKs, MAPK Kinases (MAPKKs/MKKs) and MAPKK Kinases (MAPKKKs/MEKKs). MAPKs are activated when both tyrosine and threonine residues in the TXY motif are phosphorylated by MAPKKs. MAPKKs are activated when serine and serine/threonine residues in the S/TXXXXXS/T motif are phosphorylated by MAPKKKs.3
By sequence comparison and signature motif searches, putative orthologue of MAPK cascade members have been identified in rice, Medicago sativa, Zea mays, tobacco and tomato. Southern blot analysis have also revealed that genomes of monocotyledonous plants such as rice and maize possesses sequences that are homologous to the NPKl gene (MAPKKK related gene in tobacco), an indication that NPKl-related genes are present in a number of plant species.4 During the past decade, incredible progress has been made towards the functional understanding of all genes in the model dicot Arabidopsis. In the sequenced Arabidopsis genome, 20 MAPKs, 10 MAPKKs and 80 MAPKKKs were identified.5,6 However, little is known about the MAPK gene family and their function and regulation in rice (Oryza sativa) and other economically important cereal crops.7 After the completion rice genome project 16 MAPKs and 8 MAPKKs are reported in rice.8 However, reports on the presence of MAPKKKs in rice have been very slow and so far a total of only five MAPKKKs have been reported. MAPKKK, which gets activated by upstream signals, forms a very important component of MAPK cascade. In plants the roles MAPKKK has been identified in various stresses,9,10 plant cytokinesis,11,12 ethylene signalling,13 innate immunity14 and defence responses,15,16,17 among many others.
However, the poor information about MAPKKK gene family in rice is proving to be bottleneck in elucidating MAPK cascade in this very important monocot crop. In the present study, an in silico search of rice genome databases was conducted to identify members of the rice MAPKKK gene family. A total of 75 genes were identified and among which 70 were novel. A phylogenetic tree was constructed and MAPKKKs were grouped into three different subfamilies. Conserved consensus motifs were analysed in all the subfamilies to support their association. To further validate the findings, expression analysis of all the 75 MAPKKKs in different stresses and tissue-specific libraries were carried out using MPSS database.
2. Materials and methods
2.1. Sequence and database search for OsMAPKKKs
In an attempt to obtain all the MAPKKKs, rice protein sequences available in Rice Genome Annotation Project (ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/ O_sativa/annotation_dbs/pseudomolecules/version_6.0) were downloaded to construct a local protein database. It comprised of 67393 sequences. This database was searched with MAPKKK query sequences downloaded from NCBI (ncbi.nlm.nih.gov), TAIR (arabidopsis.org), Royal Holloway University of London (www.rhul.ac.uk./Biological-Sciences/AcademicStaff/Bogre/mapkkk.htm), MIPS (mips.gsf.de), Uniprot (uniprot.org), BrassicaDB (brassica.bbsrc.ac.uk/BrassicaDB), MPSS (mpss.udel.edu/rice) and Maizegdb (maizegdb.org). The query consisted of 100 sequences from 13 different plant species which included Arabidopsis thaliana, Brassica napus, Cucumis melo, Cucumis sativus, Lotus japonicus, Lycopersicon esculentum, M. sativa, Nicotiana tabacum, O. sativa, Solanum chacoense, Triticum aestivum, Vitis vinifera and Z. mays. The search was carried out using BLASTP and 50% identity was taken as the threshold for the sequences obtained from BLAST analysis. Further all the MAPKKK sequences were aligned using CLUSTAL 2.0.3 (http://www.ebi.ac.uk/clustalw/) multiple sequence alignment and used to construct HMM profile. Using HMMER 2.3.2 (ftp://ftp.genetics.wustl.edu/pub/eddy/hmmer), the local protein database was searched on the basis of the HMM profile. HMMER and BLAST hits were compared and parsed by manual editing as well as using small PERL scripts. In addition self BLAST of the sequences was carried out to remove the redundancy and then they were analysed for the presence of domains. Subcellular localization prediction of each of the rice MAPKK kinases was carried out using the CELLO v2.5 server (http://cello.life.nctu.edu.tw/).
2.2. Phylogenetic tree construction
The Kinase domain of all the sequences (including both query and hits obtained) were analysed using ScanProsite program from Expasy server (au.expasy.org/prosite/). All the kinase domains were aligned using ClustalW. To investigate the evolutionary relationship among MAPKKK proteins, a phylogenetic tree was constructed by employing the minimal evolution (ME) method and the neighbour-joining (NJ) method wrapped in MEGA4 software suite.
2.3. Multiple alignment and conserved signature detection
Multiple sequence alignments of related proteins belonging to each group from both Arabidopsis and rice were performed using Multalin, which creates a multiple sequence alignment from a group of related sequences using progressive pairwise alignments (http://bioinfo.genotoul.fr/multalin/multalin). Programs INTERPROSCAN, SMART and MOTIF SCAN were employed to detect conserved domains.
2.4. Analysis of MPSS database for expression profiles
To gain insight into expression profiles of OsMAPKKK members in O. sativa in different stress and tissues, the MPSS database (http://mpss.udel.edu/rice/) was searched (opting 20-nt signature sequences) using the locus ID given in the TIGR database. The data thus obtained have been analysed and grouped based on stress and tissue specificity.
3. Results and discussion
3.1. In silico search and identification of novel MAPKKK genes
Availability of complete rice genome sequences18 has made it possible for the first time to identify all the MAPKKK family members in this plant species. In order to identify the MAPKKK genes, 100 query sequences of MAPKKKs from different plant species were analysed by BLASTP against 67393 sequences of the local protein database of rice from RGP, which resulted in 122 hits as subject sequences. These hits were passed through several layers of filters which primarily included 50% identity with the query sequence which further reduced to 107 hits as subject sequences. This list included all the five MAPKKKs from rice, which were already reported in NCBI and MPSS database. Simultaneously an HMM profile (HMMER version 2.3.2) was created with the 100 query sequences and a profile search was carried out against the local rice proteome database which resulted in 1574 hits. After the comparison of the sequences obtained from BLAST hits and HMM searches, the number of MAPKKKs was further reduced to 102. A self BLAST of these sequences followed by manual editing to remove the redundancy finally resulted in identification of 75 MAPKKK genes. This analysis has revealed that the O. sativa genome has 75 putative MAPKKK genes including five already known MAPKKKs. However, in Arabidopsis 80 putative MAPKKKs were predicted.6 Most of the MAPKKKs existence was supported by EST hits and full-length cDNA sequences from KOME or GenBank database (Table 1) except in 14 MAPKKKs where no such data were available. It indicates either all 14 are not expressed in all the conditions used for analysis or that they are expressed in very low quantity, which cannot be detected. Expression of other members was found in either of the two conditions mentioned above. MPSS database measures the absolute expression level of most genes in the sample and provide information about potentially novel transcripts.19 Out of the 14 MAPKKKs which were not reported in EST or cDNA database, eight of them were represented in MPSS database with their expression levels in different tissues and stresses and in the remaining six MAPKKKs, five were without any expression in the tested conditions additionally OsMAPKKK53 was not represented in MPSS database. Since there was no standard nomenclature followed for MAPKKKs neither in Arabidopsis nor in rice, we named the MAPKKKs sequentially based on the HMM search output. All the 75 MAPKKKs were having conserved protein kinase domain which is backbone for MAPK family. The predicted protein localization of most of the MAPKKKs varied from cytoplasm, mitochondria, chloroplast to nucleus except in MAPKKK33 and MAPKKK47 where it was present in cytoskeleton and peroxisomes, respectively.
Table 1.
List of MAPKKKs from rice
| Sl. No. | MAPKKKs | Score | TIGR/MSU ID | Amino acid length | Total number of mapped ESTs | ID's of cDNAs | Subcellular localization | TIGR predicted function (from MPSS Db.) |
|---|---|---|---|---|---|---|---|---|
| 1 | MAPKKK1 | 529.3 | LOC_Os03g06410 | 1017 | 74 | AY167575 AK111595 | Nuclear | EDR1, putative, expressed |
| 2 | MAPKKK2 | 522 | LOC_Os10g29540 | 972 | 30 | AK121718 | Nuclear | EDR1, putative, expressed |
| 3 | MAPKKK3* | 515.1 | LOC_Os02g32610 | 781 | 25 | CT835420 | Chloroplast | Protein kinase domain containing protein, expressed |
| 4 | MAPKKK4 | 513.1 | LOC_Os02g12810 | 864 | 31 | N/A | Chloroplast | MAP3K delta-1 protein kinase, putative, expressed |
| 5 | MAPKKK5 | 512.9 | LOC_Os12g37570 | 758 | 24 | N/A | Nuclear | ATP binding protein, putative, expressed |
| 6 | MAPKKK6 | 497.4 | LOC_Os02g50970 | 1111 | 42 | AK102767 | Nuclear | Protein kinase domain containing protein, expressed |
| 7 | MAPKKK7 | 491.2 | LOC_Os06g12590 | 1078 | 17 | AK099500 AK105681 | Plastid | ATP binding protein, putative, expressed |
| 8 | MAPKKK8 | 489.3 | LOC_Os11g10100 | 653 | 35 | N/A | Nuclear | Mitogen-activated kinase kinase kinase alpha, putative, expressed |
| 9 | MAPKKK9 | 489.1 | LOC_Os02g44642 | 894 | 22 | AK073040 | Nuclear | YDA, putative, expressed |
| 10 | MAPKKK10 | 485.3 | LOC_Os04g47240 | 894 | 38 | N/A | Nuclear | YDA, putative, expressed |
| 11 | MAPKKK11# | 482.4 | LOC_Os07g02 780 | 753 | 7 | AK069889 | Chloroplast | MAPKKK5, putative, expressed |
| 12 | MAPKKK12 | 482 | LOC_Os09g39320 | 1220 | 6 | N/A | Cytoplasm | CTR1-like protein kinase, putative, expressed |
| 13 | MAPKKK13 | 479.6 | LOC_Os09g21510 | 848 | 0 | N/A | Nuclear | Mitogen-activated protein kinase kinase kinase 2, putative, expressed |
| 14 | MAPKKK14 | 464.8 | LOC_Os04g52 140 | 778 | 48 | AK120898 | Chloroplast | Protein kinase domain containing protein, expressed |
| 15 | MAPKKK15 | 463.4 | LOC_Os08g32600 | 690 | 9 | AK103087 | Nuclear | Protein kinase domain containing protein, expressed |
| 16 | MAPKKK16 | 462.9 | LOC_Os04g35700 | 708 | 38 | AK061622 AK068725 | Nuclear | Mitogen-activated kinase kinase kinase alpha, putative, expressed |
| 17 | MAPKKK17 | 461.8 | LOC_Os09g37230 | 603 | 57 | AK072690 | Cytoplasmic | ATP binding protein, putative, expressed |
| 18 | MAPKKK18 | 461.6 | LOC_Os03g55560 | 777 | 112 | N/A | Chloroplast | MAPKKK5, putative, expressed |
| 19 | MAPKKK19 | 457.5 | LOC_Os02g35010 | 690 | 9 | AK287889 | Mitochondria | MAP3KA, putative, expressed |
| 20 | MAPKKK20 | 453.9 | LOC_Os07g38530 | 704 | 32 | DQ837532 AK100426 AF080436 | Nuclear | Protein kinase domain containing protein, expressed |
| 21 | MAPKKK21 | 446.7 | LOC_Os07g25680 | 1219 | 6 | AK241123 | Nuclear | Protein kinase domain containing protein, expressed |
| 22 | MAPKKK22* | 443.1 | LOC_Os03g49640 | 654 | 15 | N/A | Nuclear | Mitogen-activated protein kinase, putative, expressed |
| 23 | MAPKKK23 | 440.3 | LOC_Os12g40279 | 4262 | 1 | N/A | Plastid | Protein kinase domain containing protein, expressed |
| 24 | MAPKKK24# | 440.1 | LOC_Os04g56530 | 1357 | 54 | AK099839 | Cytoplasm | MAPKKK7, putative, expressed |
| 25 | MAPKKK25 | 438 | LOC_Os02g38080 | 352 | 4 | N/A | Cytoplasm | Mitogen-activated protein kinase kinase kinase 7, putative, expressed |
| 26 | MAPKKK26 | 437.6 | LOC_Os07g29330 | 439 | 27 | AK066198 | Cytoplasm | Serine/threonine-protein kinase CTR1, putative, expressed |
| 27 | MAPKKK27 | 436.5 | LOC_Os03g43760 | 379 | 98 | AK243690 | Cytoplasm | ATP binding protein, putative, expressed |
| 28 | MAPKKK28 | 436.1 | LOC_Os03g15570 | 597 | 51 | AK242766 CT835152 CT835203 | Nuclear | Mitogen-activated protein kinase 1, putative, expressed |
| 29 | MAPKKK29 | 434.4 | LOC_Os02g45130 | 612 | 42 | AK073845 | Chloroplast | WNK6, putative, expressed |
| 30 | MAPKKK30 | 434.3 | LOC_Os02g02780 | 583 | 49 | AK060220 AK103704 | Cytoplasm | ATP binding protein, putative, expressed |
| 31 | MAPKKK31 | 429.9 | LOC_Os01g45380 | 388 | 1 | AK108130 | Cytoplasm | ATP binding protein, putative, expressed |
| 32 | MAPKKK32 | 426.2 | LOC_Os08g12750 | 418 | 103 | AY156510 AK099003 AK111601 AK111800 AK121704 AY646225 | Cytoplasm | Serine/threonine-protein kinase, putative, expressed |
| 33 | MAPKKK33 | 423.9 | LOC_Os02g07790 | 421 | 83 | AY156512 AK101327 | Cytoskeleton | Serine/threonine-protein kinase CTR1, putative, expressed |
| 34 | MAPKKK34 | 423.4 | LOC_Os05g50190 | 381 | 23 | AY224453 AK107217 | Nuclear | ATP binding protein, putative, expressed |
| 35 | MAPKKK35 | 419.3 | LOC_Os02g54510 | 1083 | 2 | N/A | Nuclear | Protein kinase domain containing protein, expressed |
| 36 | MAPKKK36 | 418.6 | LOC_Os05g01780 | 621 | 40 | AK070061 | Nuclear | ZIK1 protein, putative, expressed |
| 37 | MAPKKK37 | 418.5 | LOC_Os04g51950 | 422 | 54 | AY156511 AK111698 | Cytoplasm | Serine/threonine-protein kinase, putative, expressed |
| 38 | MAPKKK38 | 416.7 | LOC_Os06g45300 | 428 | 49 | AY224431 AK112024 | Cytoplasm | Serine/threonine-protein kinase, putative, expressed |
| 39 | MAPKKK39 | 416.2 | LOC_Os06g08280 | 1273 | 27 | AK067771 | Nuclear | Protein kinase domain containing protein, expressed |
| 40 | MAPKKK40 | 415.7 | LOC_Os01g48330 | 801 | 66 | AK070808 AK102209 AK111983 | Nuclear | ATP binding protein, putative, expressed |
| 41 | MAPKKK41 | 408.5 | LOC_Os06g43840 | 1112 | 49 | AK243413 | Nuclear | Protein kinase domain containing protein, expressed |
| 42 | MAPKKK42 | 405.8 | LOC_Os03g60150 | 383 | 67 | CT830832 | Cytoplasm | Tyrosine-protein kinase 2, putative, expressed |
| 43 | MAPKKK43 | 400 | LOC_Os06g50920 | 564 | 16 | AK073747 | Cytoplasm | ATP binding protein, putative, expressed |
| 44 | MAPKKK44 | 399.5 | LOC_Os02g14530 | 790 | 50 | AK111618 | Nuclear | ATP binding protein, putative, expressed |
| 45 | MAPKKK45 | 399 | LOC_Os06g43030 | 398 | 0 | N/A | Chloroplast | HT1 protein kinase, putative, expressed |
| 46 | MAPKKK46 | 394.2 | LOC_Os11g06140 | 439 | 3 | AK070490 | Nucleus | Serine/threonine-protein kinase WNK4, putative, expressed |
| 47 | MAPKKK47 | 380.6 | LOC_Os07g08750 | 601 | 37 | AK060552 AK100930 | Peroxisome | WNK1, putative, expressed |
| 48 | MAPKKK48 | 371.7 | LOC_Os01g01740 | 376 | 32 | AK059460 | Nuclear | HT1 protein kinase, putative, expressed |
| 49 | MAPKKK49 | 366.1 | LOC_Os05g44290 | 604 | 3 | N/A | Nuclear | Protein kinase, putative, expressed |
| 50 | MAPKKK50 | 363.3 | LOC_Os12g02250 | 619 | 48 | AK067447 AK072172 | Nuclear | Mitogen-activated protein kinase, putative, expressed |
| 51 | MAPKKK51 | 361.1 | LOC_Os01g54350 | 637 | 38 | AK102467 | Nuclear | Protein kinase, putative, expressed |
| 52 | MAPKKK52 | 357.4 | LOC_Os12g06490 | 418 | 71 | AK062812 AK073772 | Cytoplasm | Serine/threonine-protein kinase WNK3, putative, expressed |
| 53 | MAPKKK53 | 355.8 | LOC_Os11g02305 | 622 | 0 | N/A | Nuclear | – |
| 54 | MAPKKK54 | 355.5 | LOC_Os03g28300 | 859 | 63 | CT828499 | Nuclear | ATP binding protein, putative, expressed |
| 55 | MAPKKK55 | 354.1 | LOC_Os01g50400 | 418 | 0 | N/A | Chloroplast | Mitogen-activated protein kinase kinase kinase 1, putative, expressed |
| 56 | MAPKKK56 | 349.8 | LOC_Os05g01780 | 621 | 20 | AY336987 | Mitochondria | ZIK1 protein, putative, expressed |
| 57 | MAPKKK57# | 349.4 | LOC_Os05g46750 | 591 | 0 | N/A | Chloroplast | MAPKKK16, putative, expressed |
| 58 | MAPKKK58 | 348.8 | LOC_Os03g39150 | 351 | 0 | N/A | Cytoplasm | Serine/threonine-protein kinase CTR1, putative |
| 59 | MAPKKK59 | 346.4 | LOC_Os12g41260 | 400 | 1 | AK109696 | Cytoplasm | ATMRK1, putative, expressed |
| 60 | MAPKKK60 | 341.8 | LOC_Os03g53410 | 407 | 11 | N/A | Chloroplast | ATMRK1, putative, expressed |
| 61 | MAPKKK61 | 334.7 | LOC_Os01g10450 | 563 | 25 | AK069537 | Nuclear | Protein kinase, putative, expressed |
| 62 | MAPKKK62 | 329.7 | LOC_Os01g50420 | 541 | 0 | N/A | Mitochondria | Mitogen-activated protein kinase kinase kinase 2, putative, expressed |
| 63 | MAPKKK63 | 328.8 | LOC_Os01g50370 | 484 | 0 | N/A | Chloroplast | Mitogen-activated protein kinase kinase kinase 1, putative, expressed |
| 64 | MAPKKK64 | 325.1 | LOC_Os07g39520 | 327 | 42 | CT832200 AK072014 AK072176 AK105268 | Mitochondria | Serine/threonine-protein kinase WNK2, putative, expressed |
| 65 | MAPKKK65 | 324 | LOC_Os07g43900 | 321 | 12 | AK241519 | Cytoplasmic | Mitogen-activated protein kinase kinase kinase 10, putative, expressed |
| 66 | MAPKKK66 | 320.8 | LOC_Os10g04010 | 525 | 0 | N/A | Chloroplast | Mitogen-activated protein kinase kinase kinase 1, putative |
| 67 | MAPKKK67 | 318.7 | LOC_Os10g04000 | 526 | 0 | N/A | Cytoplasm | Mitogen-activated protein kinase kinase kinase 1, putative |
| 68 | MAPKKK68 | 315.8 | LOC_Os12g30570 | 384 | 0 | N/A | Nuclear | ATMRK1, putative |
| 69 | MAPKKK69 | 305.8 | LOC_Os05g46760 | 441 | 8 | AK105946 | Chloroplast | Mitogen-activated protein kinase kinase kinase 1, putative, expressed |
| 70 | MAPKKK70 | 300.2 | LOC_Os01g50410 | 451 | 0 | N/A | Mitochondria | Mitogen-activated protein kinase kinase kinase 1, putative, expressed |
| 71 | MAPKKK71 | 278.4 | LOC_Os02g21700 | 413 | 0 | N/A | Cytoplasm | Mitogen-activated protein kinase kinase kinase 2, putative, expressed |
| 72 | MAPKKK72 | 265.4 | LOC_Os01g54480 | 468 | 29 | AK104674 | Mitochondria | Serine/threonine-protein kinase TNNI3K, putative, expressed |
| 73 | MAPKKK73 | 258.8 | LOC_Os03g18170 | 511 | 0 | N/A | Chloroplast | Mitogen-activated protein kinase kinase kinase 1, putative, expressed |
| 74 | MAPKKK74 | 225 | LOC_Os01g66860 | 500 | 13 | AK070097 | Chloroplast | Ankyrin-kinase, putative, expressed |
| 75 | MAPKKK75 | 212.5 | LOC_Os02g39560 | 502 | 13 | AK071922 AK099756 | Chloroplast | Non-receptor tyrosine kinase spore lysis A, putative, expressed |
*From NCBI database (MAPKKK3 and MAPKKK22).
#From MPSS database (MAPKKK11, MAPKKK24 and MAPKKK57).
3.2. Phylogenetic analysis of MAPKKKS
The family of MAPKKKs forms the largest group of MAPK pathway components. Arabidopsis contains 80 MAPKKKs,6 which can be subdivided into three major subtypes, Raf, MEKK and ZIK.20 Among these two groups, MEKK group is most similar to animal MEKKs and yeast MAPKKKs. In total, it consists of 21 MEKK-like and 11 ZIK kinases along with Arabidopsis ANP1-3 (Arabidopsis NPK1-like protein kinases). The other group consists of 48 genes encoding Raf-like protein kinases including Arabidopsis CTR1 and EDR1.6 To further characterize the MAPKKKs from rice and to evaluate the phylogenetic relationships with Arabidopsis MAPKKKs, the kinase domains of rice and Arabidopsis were aligned using ClustalW and analysed using MEGA4. Phylogenetic tree was constructed by employing the NJ method (Fig. 1) and ME method have shown similar topologies, with only minor modifications at deep nodes. On the basis of phylogenetic analysis, MAPKKKs in rice were classified in to three categories, which include Raf, ZIK and MEKK subfamilies. There were 43 MAPKKKs from rice and 48 from Arabidopsis grouped under Raf subfamily, 22 MAPKKKs from rice and 21 from Arabidopsis were grouped in to MEKK subfamily where as only 10 MAPKKKs from rice and 11 from Arabidopsis were grouped under ZIK family. As the results indicate, rice and Arabidopsis show similarity in the number of MAPKKKs in each subgroup.
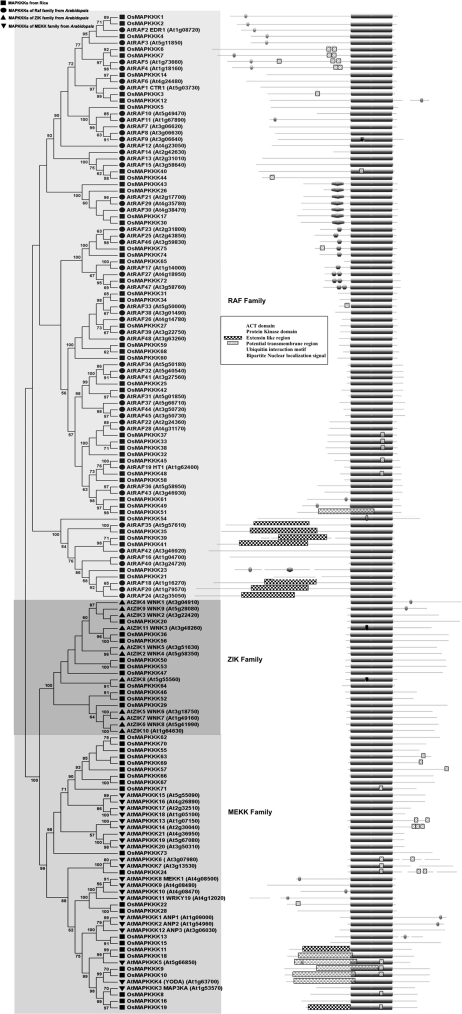
Figure 1.
Phylogenetic tree and domain organization of MAPKKKs from rice and Arabidopsis. Kinase domains of putative MAPKKKs from Arabidopsis and rice were aligned using CLUSTALW program. The NJ phylogenetic tree was created using MEGA4 software suite. Bootstrap value above 50% was shown. The domain organization was depicted on the right, gaps in lines were introduced while depicting the protein sequences of bigger size for convenience. Scanning of the protein sequences for the presence of known motifs and domains was performed using PlantsP. To identify the species of origin for each MAPKKK, a species acronym is included before the protein name: At, Arabidopsis thaliana; Os, Oryza sativa.
Domain architecture of MAPKKKs from Arabidopsis and rice revealed that most of the Raf family proteins have a C-terminal kinase domain and a long N-terminal regulatory domain. In contrast, majority of the ZIK family members have N-terminal kinase domain whereas members of MEKK family has less conserved protein structure with kinase domain located either at N- or C-terminal or central part of the protein. Bipartite NLS and transmembrane regions are distributed across the members of all the subfamilies whereas extensin like region is observed in Raf and MEKK members. Ubiquitin-interaction motif and ACT domain which is known to play a role in the regulation of a wide range of metabolic enzymes by responding to amino acid concentration are present only in the members of Raf family from rice and Arabidopsis (Fig. 1).
Extrapolating the phylogenetic tree shown in Fig. 1 along with 14 additional sequences from 11 different plants revealed that most of the known MAPKKK from other plants are grouped with MEKK subfamily (Supplementary Fig. S1). Comparing MPK and MKK gene families in rice, Arabidopsis and Populus genomes it was reported that the recent duplication events in amplifying the respective gene families are more evident in eudicots compared with monocot rice.8 We also observed comparing the MAPKKK gene family between rice and Arabidopsis that the eudicot has undergone more recent duplication events, more so in the Raf subfamily (Supplementary Fig. 1).
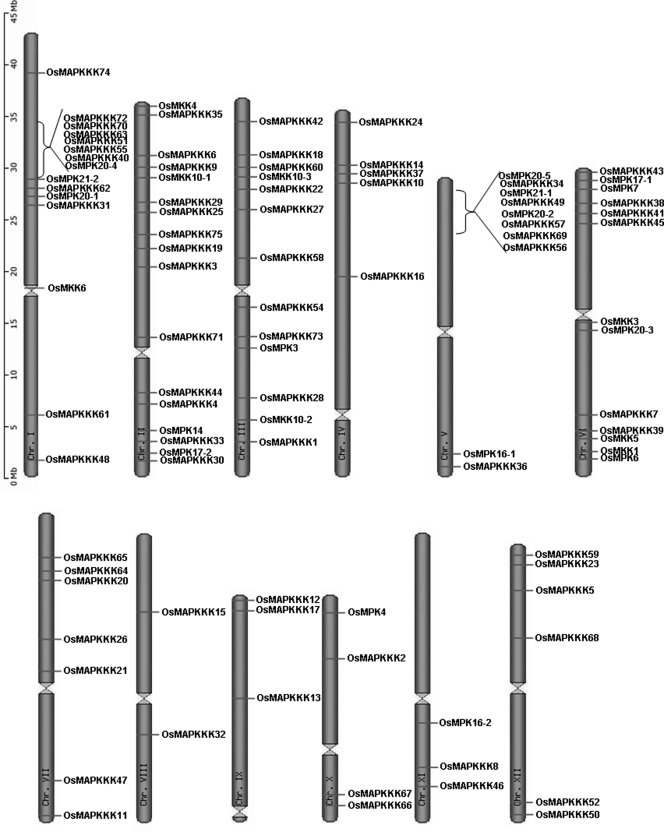
In silico localization of MAPKKKs on chromosomes indicated that all the 75 MAPKKKs are distributed on all 12 chromosomes of rice and half of them were present in first three chromosomes, which include chromosome 1, 2 and 3. Among all, chromosomes 8 and 11 were sharing only two MAPKKKs each, whereas chromosome 2 was having as many as 13 MAPKKKs (Fig. 2). Although similar analysis for other two components of MAPK cascade namely, MAPKK/MKK and MAPK/MPK revealed that eight MKKs are present in four chromosomes and 16 MPKs are distributed on seven chromosomes.
Figure 2.
Graphical (scaled) representation of location of MAPK, MAPKK and MAPKKK genes on rice chromosomes.
3.3. Analysis of conserved motifs among MAPKKKs
The pattern of amino acid residues found in many subdomains is conserved among the family members. All the rice MAPKKKs that were grouped under Raf, ZIK and MEKK subfamilies were further analysed for the presence of specific signatures. Raf subfamily consists of the largest number of MAPKKKs in both Arabidopsis and rice. Experimental data from Drosophila and Caenorhabditis elegans have provided much evidence that the Rafs sensu stricto stimulate MAP2K and MAPK activation.21 Human Rafs are involved in a signalling network that controls cell proliferation, cell differentiation and apoptosis. Many of their effects are transmitted through the ERK/MAPK pathway.22,23
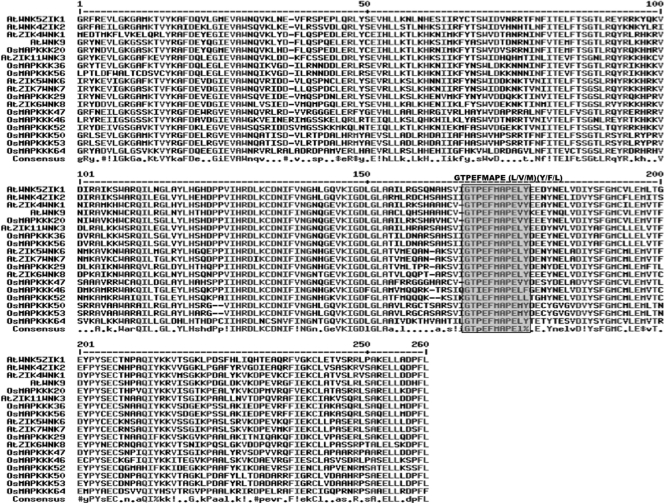
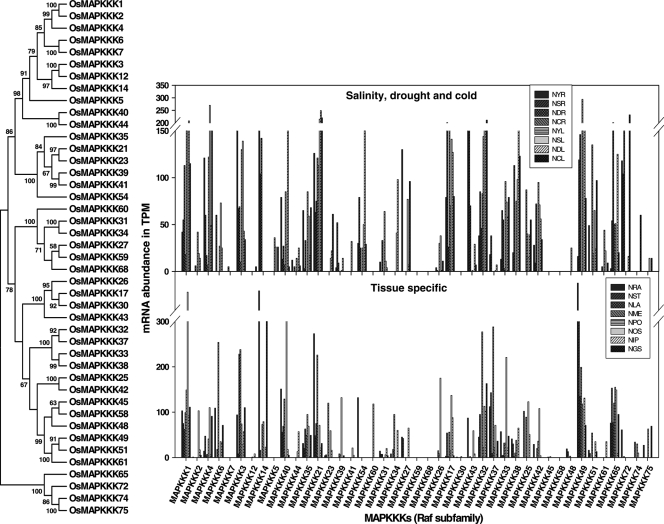
Raf family has a conserved signature in its kinase domain across the members. Analysis of the rice MAPKKKs along with the Arabidopsis for Raf specific signature GTXX (W/Y) MAPE was carried out by multiple alignments of kinase domains. The data revealed the presence of signature in all the members of Raf family in rice (Fig. 3) and strongly supported their identity as members of Raf subfamily. Around 43 MAPKKKs were grouped under Raf subfamily in rice where as in Arabidopsis this number is higher and consists of 48 members. The kinase domain of human B-Raf is a strong activator of MEK and has a high affinity for MEK.24,25 In Arabidopsis, members of Raf subfamily CTR1 and EDR1 act as negative regulators in ethylene signalling13 and in response to powdery mildew attack,26 respectively. Further CTR1 has been found to interact with the histidine kinase domain of ETR1 and the ethylene response sensor (ERS1) in vitro.27
Figure 3.
Alignment of MAPKKKs of Raf subfamily from rice and Arabidopsis. The highlighted part shows the conserved signature motif.
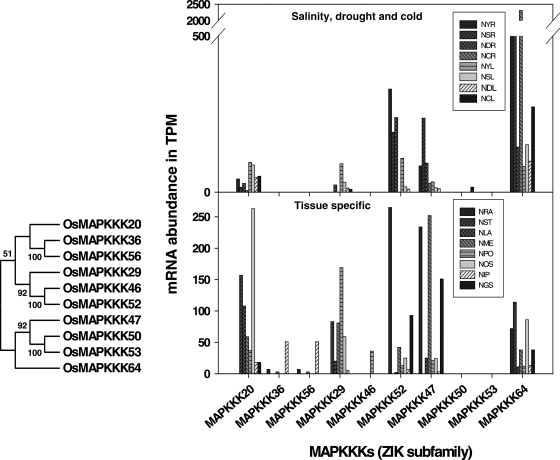
The ZIK subfamily consists of 10 putative MAPKKKs in rice and 11 in Arabidopsis. Recently, the Arabidopsis ZIK protein WNK1 (At3g04910) was demonstrated to phosphorylate a protein involved in the control of circadian rhythms,28 suggesting a function different from that of other MAPKKKs. The characteristic feature of this family consists of a conserved signature GTPEFMAPE (L/V/M) (Y/F/L) across the members. Rice putative MAPKKKs were analysed for the presence of the above signature, which reassured that 10 MAPKKKs out of 75 have ZIK specific signatures (Fig. 4) and eventually grouped under this family.
Figure 4.
Alignment of MAPKKKs of ZIK subfamily from rice and Arabidopsis. The highlighted part shows the conserved signature motif.
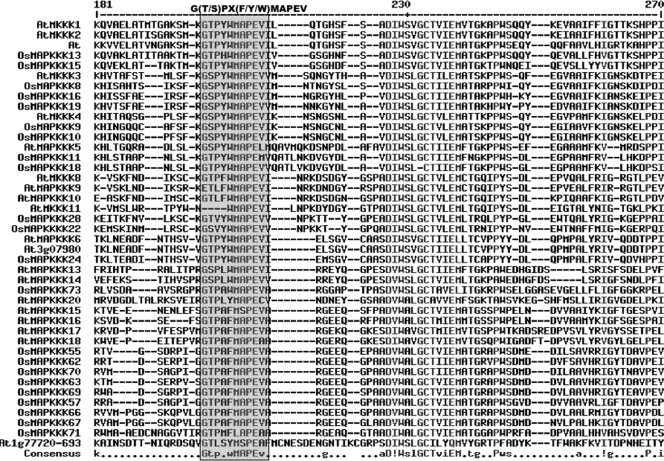
Among the three families MEKK subfamily is relatively well characterized. NPK1 gene from tobacco whose role in cytokinesis have been established,11,29 ScFRK2, a MAPKKK from S. chacoense involved in fertilization and embryogenesis,30 OMTK1, MAPKKK from M. sativa which channels oxidative stress signalling31 were some of the members of MEKK subfamily. Further ANP1 in Arabidopsis was found responsive to oxidative stress and is involved in negative regulation of auxin signal transduction pathway.32,33 It was also reported that Arabidopsis ANP1, ANP2, ANP3 are involved in plant cytokinesis11,34 and MAPKKKα in relation to defence response.15 Role of YODA a MAPKKK from Arabidopsis was characterized in stomatal development35 with its targeting downstream MAPKKs.36 Another member of this family MEKK1 functions in integrating ROS homeostasis with plant development and hormone signalling.37,38 Twenty-two MAPKKKs from rice and 21 from Arabidopsis belong to this subfamily. Relationship analysis based on the amino acid sequences of the protein kinase catalytic domain shows that the sequence, G (T/S) PX (F/Y/W) MAPEV forms a conserved signature of this family (Fig. 5). Presence of this signature in 18 putative rice MAPKKKs further confirmed their association with MEKK family. Moreover analysis of the transcript abundance of MAPKKK subfamilies from rice in different tissue and stress specific libraries revealed that they are differentially expressed and regulated.
Figure 5.
Alignment of MAPKKKs of MEKK subfamily from rice and Arabidopsis. The highlighted part shows the conserved signature motif.
3.4. In silico analysis of expression of MAPKKKs based on MPSS database
MPSS is a valuable tool to have an insight into gene expression.39 It has been used previously for genome-level expression analysis in several systems including Arabidopsis.40 To extract information about the relative abundance of transcripts of O. sativa MAPKKK members, we have carried out the analysis in the available MPSS database (http://mpss.udel.edu/rice/). This database is derived from the TIGR O. sativa genome sequence and the search has been performed employing the 20 nucleotides long signatures in tissue-specific and stress-related libraries. Our analysis has revealed that among the different tissue-specific libraries, maximum numbers of MAPKKKs were expressed and their transcript abundance was found in crown vegetative meristematic tissue (NME), ovary and mature stigma (NOS) tissue libraries. Libraries of salinity and cold stress in rice roots have shown maximum transcript abundance of OsMAPKKK64 which is a member of ZIK subfamily (Fig. 6) and with drought stress elevated transcripts of OsMAPKKK28 was observed (Fig. 7). OsMAPKKK28 is a member of MEKK subfamily and an orthologue of AtMEKK1 from Arabidopsis. Similarly in leaves highest Transcripts Per Million (TPM) of OsMAPKKK4 a member of Raf subfamily in salinity stress (Fig. 8), OsMAPKKK63 in cold, OsMAPKKK8 in drought, all members of MEKK subfamily, were observed. This suggested the involvement of MAPKKK members from all the subfamilies and more precisely MEKK subfamily in the regulation of abiotic stress. These observations further strengthen their prediction and the differential regulation in various stress conditions suggests their active participation in stress signalling.
Figure 6.
Transcript abundance of MAPKKK members of ZIK subfamily in salinity, drought, cold and tissue-specific libraries from MPSS database along with their dendrogram. Different libraries in MPSS database were analysed for the expression level of MAPKs. NYR, 14 days young roots; NSR, 14 days young roots stressed in 250 mM NaCl for 24 h; NDR, 14 days young roots stressed in drought for 5 days; NCR, 14 days young roots stressed in 4°C cold for 24 h; NYL, 14 days young leaves; NSL, 14 days young leaves stressed in 250 mM NaCl for 24 h; NDL, 14 days young leaves stressed in drought for 5 days; NCL, 14 days young leaves stressed in 4°C Cold for 24 h; NRA, mature roots (60 days) replicate A; NST, mature stem (60 days); NLA, mature leaves (60 days) replicate A; NME, Crown vegetative meristematic tissue (60 days); NPO, mature pollen; NOS, ovary and mature stigma; NIP, Immature panicle; NGS, 3 days germinating seed.
Figure 7.
Transcript abundance of MAPKKK members of MEKK subfamily in salinity, drought, cold and tissue-specific libraries from MPSS database along with their dendrogram. Different libraries in MPSS database were analysed for the expression level of MAPKs. NYR, 14 days young roots; NSR, 14 days young roots stressed in 250 mM NaCl for 24 h; NDR, 14 days young roots stressed in drought for 5 days; NCR, 14 days young roots stressed in 4°C cold for 24 h; NYL, 14 days young leaves; NSL, 14 days young leaves stressed in 250 mM NaCl for 24 h; NDL, 14 days young leaves stressed in drought for 5 days; NCL, 14 days young leaves stressed in 4°C cold for 24 h; NRA, mature roots (60 days) replicate A; NST, mature stem (60 days); NLA, mature leaves (60 days) replicate A; NME, crown vegetative meristematic tissue (60 days); NPO, mature pollen; NOS, ovary and mature stigma; NIP, immature panicle; NGS, 3 days germinating seed.
Figure 8.
Transcript abundance of MAPKKK members of Raf subfamily in salinity, drought, cold and tissue-specific libraries from MPSS database along with their dendrogram. Different libraries in MPSS database were analysed for the expression level of MAPKs. NYR, 14 days young roots; NSR, 14 days young roots stressed in 250 mM NaCl for 24 h; NDR, 14 days young roots stressed in drought for 5 days; NCR, 14 days young roots stressed in 4°C cold for 24 h; NYL, 14 days young leaves; NSL, 14 days young leaves stressed in 250 mM NaCl for 24 h; NDL, 14 days young leaves stressed in drought for 5 days; NCL, 14 days young leaves stressed in 4°C cold for 24 h; NRA, mature roots (60 days) replicate A; NST, mature stem (60 days); NLA, mature leaves (60 days) replicate A; NME, crown vegetative meristematic tissue (60 days); NPO, mature pollen; NOS, ovary and mature stigma; NIP, immature panicle; NGS, 3 days germinating seed.
3.5. Conclusion
The present study has provided the full list of MAPKKKs present in rice for the first time. In silico search of various rice protein databases using BLASTP and HMM profile resulted in identification of 75 MAPKKK genes from rice among these 70 were novel. EST hits and full-length cDNA sequences (from KOME or Genbank database) of 75 MAPKKKs supported their existence. Phylogenetic analysis of MAPKKKs from rice and Arabidopsis has classified them in to three subgroups that include Raf, ZIK and MEKK. Conserved motifs in the deduced amino acid sequences of rice MAPKKKs strongly supported their identity as members of Raf, ZIK and MEKK subfamilies. Expression analysis of the MAPKKKs in MPSS database revealed that maximum number of MAPKKK transcripts was represented in crown vegetative meristematic tissue (NME), ovary and mature stigma (NOS) tissue libraries. Libraries of salinity and cold stress in rice roots have shown maximum transcript abundance of OsMAPKKK64 which is a member of ZIK subfamily and with drought stress elevated transcripts of OsMAPKKK28 was observed. Similarly in leaves highest TPM of OsMAPKKK4 a member of Raf subfamily in salinity stress, OsMAPKKK63 in cold, OsMAPKKK8 in drought, which are members of MEKK subfamily were observed. The information generated will be very significant for further investigating the regulation mechanism of MAPKKKs and eventually MAPK cascade in response to extracellular stimuli and their central roles in various biological functions. Additionally, the information generated will serve the purpose in elucidating still very poorly characterized MAP kinase cascade in plants in general and in rice in particular.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
The work is financially supported by the core grant of National Institute of Plant Genome Research, New Delhi, India.
Supplementary Material
Acknowledgements
K.P.R. and K.K. acknowledge the fellowship provided by University Grant Commission, India, while B.R. thanks Department of Biotechnology, India for fellowship.
Footnotes
Edited by Kazuo Shinozaki
References
- 1.Tena G., Asai T., Chiu W.L., Sheen J. Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 2001;4:392–401. doi: 10.1016/s1369-5266(00)00191-6. doi:10.1016/S1369-5266(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 2.Mishra N.S., Tuteja R., Tuteja N. Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 2006;452:55–68. doi: 10.1016/j.abb.2006.05.001. doi:10.1016/j.abb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Ichimura K., Mizoguchi T., Irie K., et al. Isolation of ATMEKK1 (a MAP kinase kinase kinase) interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem. Biophys. Res. Commun. 1998;253:532–43. doi: 10.1006/bbrc.1998.9796. doi:10.1006/bbrc.1998.9796. [DOI] [PubMed] [Google Scholar]
- 4.Machida Y., Nishihama R., Kitakura S. Progress in studies of plant homologs of mitogen-activated protein (MAP) kinase and potential upstream components in kinase cascades. Crit. Rev. Plant Sci. 1997;16:481–96. doi:10.1080/713608155. [Google Scholar]
- 5.MAPK Group. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7:301–8. doi: 10.1016/s1360-1385(02)02302-6. doi:10.1016/S1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- 6.Jonak C., Okresz L., Bogre L., Hirt H. Complexity, cross talk and integration of plant MAP kinase signaling. Curr. Opin. Plant Biol. 2002;5:415–24. doi: 10.1016/s1369-5266(02)00285-6. doi:10.1016/S1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 7.Reyna N.S., Yang Y. Molecular Analysis of the Rice MAP Kinase Gene family in relation to Magnaporthe grisea infection. Mol. Plant Microbe Interact. 2006;19:530–40. doi: 10.1094/MPMI-19-0530. doi:10.1094/MPMI-19-0530. [DOI] [PubMed] [Google Scholar]
- 8.Hamel L.P., Nicole M.C., Sritubtim S., et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006;11:192–8. doi: 10.1016/j.tplants.2006.02.007. doi:10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Mizoguchi T., Ichimura K., Irie K., et al. Identification of a possible MAP kinase cascade in Arabidopsis thaliana based on pairwise yeast two hybrid analysis and functional complementation tests of yeast mutants. FEBS Lett. 1998;437:56–60. doi: 10.1016/s0014-5793(98)01197-1. doi:10.1016/S0014-5793(98)01197-1. [DOI] [PubMed] [Google Scholar]
- 10.Gao L., Xiang C.B. The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Mol. Biol. 2008;67:125–34. doi: 10.1007/s11103-008-9306-8. doi:10.1007/s11103-008-9306-8. [DOI] [PubMed] [Google Scholar]
- 11.Krysan P.J., Jester P.J., Gottwald J.R., Sussman M.R. An Arabidopsis mitogen-activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell. 2002;14:1109–20. doi: 10.1105/tpc.001164. doi:10.1105/tpc.001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishihama R., Ishikawa M., Araki S., Soyano T., Asada T., Machida Y. The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev. 2001;15:352–63. doi: 10.1101/gad.863701. doi:10.1101/gad.863701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–41. doi: 10.1016/0092-8674(93)90119-b. doi:10.1016/0092-8674(93)90119-B. [DOI] [PubMed] [Google Scholar]
- 14.Asai T., Tena G., Plotnikova J., et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–83. doi: 10.1038/415977a. doi:10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 15.del Pozo O., Pedley K.F., Martin G.B. MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 2004;23:3072–82. doi: 10.1038/sj.emboj.7600283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., et al. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007;143:661–9. doi: 10.1104/pp.106.091389. doi:10.1104/pp.106.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frye C.A., Tang D., Innes R.W. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA. 2001;98:373–8. doi: 10.1073/pnas.98.1.373. doi:10.1073/pnas.011405198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. doi:10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 19.Nobuta K., Vemaraju K., Meyers B.C. Methods for Analysis of Gene Expression in Plants Using MPSS. Methods Mol. Biol. 2007;406:387–407. doi: 10.1007/978-1-59745-535-0_19. doi:10.1007/978-1-59745-535-0_19. [DOI] [PubMed] [Google Scholar]
- 20.Wrzaczek M., Hirt H. Plant MAP kinase pathways: how many and what for. Biol. Cell. 2001;93:81–7. doi: 10.1016/s0248-4900(01)01121-2. doi:10.1016/S0248-4900(01)01121-2. [DOI] [PubMed] [Google Scholar]
- 21.Dickson B., Hafen E. Genetics of signal transduction in invertebrates. Curr. Opin. Genet. Dev. 1994;4:64–70. doi: 10.1016/0959-437x(94)90092-2. doi:10.1016/0959-437X(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 22.Dhillon A.S., Meikle S., Peyssonnaux C., et al. Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 2002;l21:64–71. doi: 10.1093/emboj/21.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzivion G., Luo Z., Avruch J. A dimeric 14–3-3 protein is an essential cofactor for Raf kinase activity. Nature. 1998;394:88–92. doi: 10.1038/27938. [DOI] [PubMed] [Google Scholar]
- 24.Marais R., Light Y., Paterson H.F., Mason C.S., Marshall C.J. Differential regulation of Raf-1, A-Raf and B-Raf by oncogenic Ras and tyrosine kinases. J. Biol. Chem. 1997;272:4378–83. doi: 10.1074/jbc.272.7.4378. [DOI] [PubMed] [Google Scholar]
- 25.Papin C., Denouel-Galy A., Laugier D., Georges C., Eychène A. Modulation of kinase activity and oncogenic properties by alternative splicing reveals a novel regulatory mechanism for B-Raf. J. Biol. Chem. 1998;273:24939–47. doi: 10.1074/jbc.273.38.24939. doi:10.1074/jbc.273.38.24939. [DOI] [PubMed] [Google Scholar]
- 26.Frye C.A., Innes R.W. An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell. 1998;10:947–56. doi: 10.1105/tpc.10.6.947. doi:10.1105/tpc.10.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark K.L., Larsen P.B., Wang X., Chang C. Association of the Arabidopsis CTR1 Raf like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. USA. 1998;95:5401–6. doi: 10.1073/pnas.95.9.5401. doi:10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami-Kojima M., Nakamichi N., Yamashino T., Mizuno T. The APRR3 component of the clock associated APRR1/TOC1 quintet is phosphorylated by a novel protein kinase belonging to the WNK family, the gene for which is also transcribed rhythmically in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:675–83. doi: 10.1093/pcp/pcf084. doi:10.1093/pcp/pcf084. [DOI] [PubMed] [Google Scholar]
- 29.Soyano T., Nishihama R., Morikiyo K., Ishikawa M., Machida Y. NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev. 2003;17:1055–67. doi: 10.1101/gad.1071103. doi:10.1101/gad.1071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsumune M.G., Brien M.O., Bertrand C., Tebbji F., Nantel A., Matton D.P. Loss of ovule identity induced by overexpression of the fertilization-related kinase 2 (ScFRK2), a MAPKKK from Solanum chacoense. J. Exp. Bot. 2006;57:4171–87. doi: 10.1093/jxb/erl194. [DOI] [PubMed] [Google Scholar]
- 31.Nakagami H., Kiegerl S., Hirt H. OMTK1, a novel MAPKKK, channels oxidative stress signaling through direct MAPK interaction. J. Biol. Chem. 2004;279:28959–66. doi: 10.1074/jbc.M312662200. [DOI] [PubMed] [Google Scholar]
- 32.Kovtun Y., Chiu W.L., Zeng W., Sheen J. Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature. 1998;395:716–20. doi: 10.1038/27240. [DOI] [PubMed] [Google Scholar]
- 33.Kovtun Y., Chiu W.L., Tena G., Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA. 2000;97:2940–5. doi: 10.1073/pnas.97.6.2940. doi:10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukowitz W., Roeder A., Parmenter D., Somerville C. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell. 2004;116:109–19. doi: 10.1016/s0092-8674(03)01067-5. doi:10.1016/S0092-8674(03)01067-5. [DOI] [PubMed] [Google Scholar]
- 35.Bergmann D.C., Lukowitz W., Somerville C.R. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304:1494–7. doi: 10.1126/science.1096014. doi:10.1126/science.1096014. [DOI] [PubMed] [Google Scholar]
- 36.Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007;19:63–73. doi: 10.1105/tpc.106.048298. doi:10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichimura K., Casais C., Peck S.C., Shinozaki K., Shirasu K. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 2006;281:36969–76. doi: 10.1074/jbc.M605319200. doi:10.1074/jbc.M605319200. [DOI] [PubMed] [Google Scholar]
- 38.Nakagami H., Soukupova H., Schikora A., Zarsky V., Hirt H. A Mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 2006;281:38697–704. doi: 10.1074/jbc.M605293200. doi:10.1074/jbc.M605293200. [DOI] [PubMed] [Google Scholar]
- 39.Brenner S., Johnson M., Bridgham J., et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–4. doi: 10.1038/76469. doi:10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 40.Meyers B.C., Tej S.S., Vu T.H., et al. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 2004;14:1641–53. doi: 10.1101/gr.2275604. doi:10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.