Abstract
Despite the fact that colorectal cancer (CRC) is a highly treatable form of cancer if detected early, a very low proportion of the eligible population undergoes screening for this form of cancer. Integrating a genomic screening profile as a component of existing screening programs for CRC could potentially improve the effectiveness of population screening by allowing the assignment of individuals to different types and intensities of screening and also by potentially increasing the uptake of existing screening programs. We evaluated the utility and predictive value of genomic profiling as applied to CRC, and as a potential component of a population-based cancer screening program. We generated simulated data representing a typical North American population including a variety of genetic profiles, with a range of relative risks and prevalences for individual risk genes. We then used these data to estimate parameters characterizing the predictive value of a logistic regression model built on genetic markers for CRC. Meta-analyses of genetic associations with CRC were used in building science to inform the simulation work, and to select genetic variants to include in logistic regression model-building using data from the ARCTIC study in Ontario, which included 1,200 CRC cases and a similar number of cancer-free population-based controls. Our simulations demonstrate that for reasonable assumptions involving modest relative risks for individual genetic variants, that substantial predictive power can be achieved when risk variants are common (e.g., prevalence > 20%) and data for enough risk variants are available (e.g., ~140–160). Pilot work in population data shows modest, but statistically significant predictive utility for a small collection of risk variants, smaller in effect than age and gender alone in predicting an individual’s CRC risk. Further genotyping and many more samples will be required, and indeed the discovery of many more risk loci associated with CRC before the question of the potential utility of germline genomic profiling can be definitively answered.
Introduction
Colorectal cancer (CRC) screening in Canada
CRC is the second leading cause of cancer death in Canadians, with an estimated 22,000 new cases and 9,100 deaths in 2009 (Canadian Cancer Society Steering Committee 2009). Although incidence and mortality rates for CRC in Canada are among the highest in the world, a very low proportion of the population undergoes screening for this form of cancer (Rabeneck and Paszat 2004). In fact, more men undergo prostate cancer screening than CRC screening, whereas the weight of evidence as to efficacy and effectiveness is greater for the latter (Sirovich et al. 2003). Recommendations put forward in 2002 by the Canadian National Committee on Colorectal Cancer Screening to introduce population-based fecal occult blood (FOB) screening of average risk individuals 50 years and older (National Committee on Colorectal Cancer Screening 2002) are now being implemented in several Canadian provinces. However, there are concerns about the likely population impact of FOB screening. First, considerable variation (46–92%) in the sensitivity of FOB screening has been reported (Hewitson et al. 2007), leading to uncertainty about the extent to which the findings of the RCTs will translate into screening outcomes in practice, although recent advances in immunochemical-based FOB testing suggests the latest FOB assays will detect 60–94% of cancers and 20–67% of advanced neoplasia (Allison et al. 2007). Second, it has been suggested that about a quarter of the decrease in colorectal mortality attributable to FOB screening reported in the Funen and Nottingham trials may have been due to better disease awareness on the part of patients who developed interval CRC who had been assigned to receive screening, and therefore that the reduction in CRC mortality attributed to FOB screening had been over-estimated (Autier et al. 2003). Third, a recent review casts doubt on the overall efficacy of FOB screening in prolonging the lives of those who are screened, on the basis of an examination of all-cause mortality in data from the 2000 Cochrane review (Moayyedi and Achkar 2006). Evidence on the effectiveness of screening modalities other than FOB testing is limited. Case–control and cohort studies suggest that screening by sigmoidoscopy reduces incidence and mortality due to distal CRC, but randomized-control trial (RCT) evidence is not available (UK Trial of Early Detection of Breast Cancer Group 1988; Selby et al. 1992; Newcomb et al. 1992; Müller and Sonnenberg 1995). Participation rates may be similar to FOB screening (Bampton et al. 2000; UK Flexible Sigmoidoscopy Screening Trial Investigators 2002), or lower (Multicentre Australian Colorectal-neoplasia Screening (MACS) Group 2006. For colonoscopy screening, it is not clear that any potential gain in benefit compared with flexible sigmoidoscopy screening outweighs the higher risk of perforation and complications associated with the use of sedatives, particularly if this form of screening is repeated over time; participation rates are lower for colonoscopy than sigmoidoscopy-based programs (Rex et al. 1997; Simon 2000; Bampton et al. 2000; Atkin 2003), and there is concern about the capacity of the Canadian health system to provide greater access to colonoscopy-based screening (Schabas 2003).
Potential utility of genomic information in CRC screening
Given that about 30% of the risk of CRC is attributable to heritable factors (Lichtenstein et al. 2000), and less than 5% to high penetrance variants (Aaltonen et al. 2007), the balance of the genetic risk is likely explained by numerous low penetrance variants, many of which may be common. It is well established that genetic testing for low penetrance alleles one at a time is not useful in a screening or diagnostic context (Vineis et al. 2001; Khoury et al. 2004; Madlensky et al. 2005; Janssens et al. 2008) and may even cause unintended psychosocial harm. However, because a moderate number of common, low penetrance variants, in combination, may account for a substantial proportion of the disease (Yang et al. 2003; Khoury et al. 2004; Yang et al. 2005), it seems reasonable to postulate that information on a combination of genetic variants may be useful in screening. There are several ways in which inclusion of an adjunctive genomic screening profile might improve population screening for CRC. First, genomic information might be useful in triaging individuals to different types and intensities of screening. Second, it is possible that an adjunctive profiling tool based on genomic information might increase the uptake of existing screening methods. For example, population-based studies have shown that the likelihood of participating in CRC screening is positively associated with having a family history of the disease (Slattery et al. 2000, 2004; Mandelson et al. 2000; Madlensky et al. 2003; Ramji et al. 2005). In a study of patient and physician preferences for various types of tests to detect CRC, both patients and physicians stated that they considered accuracy as the most important feature in deciding on which test they would choose; thus, improved efficacy through an adjunctive test might also lead to higher uptake (Ling et al. 2001). Third, since the magnitude of the effect of population-based cancer screening on cancer-specific mortality depends on the extent of repeated compliance (Gertig et al. 1998; Kronborg et al. 2004), it is possible that inclusion of an adjunctive profile at the initial screening might help identify, and target effort towards, those at greatest need of repeated screening. Finally, genomic information may assist in decisions about preventive interventions or subsequent screening modality or intensity in individuals after the result of the first screening test is known and any related interventions have been carried out. For example, genomic information could assist in decisions about subsequent management (including surveillance) of individuals in whom polyps were detected (Ransohoff 2002).
Evidence for the potential value of genomic information in CRC risk profiling
Although a substantial proportion of candidate gene studies of common variants have failed to replicate, a number of variants have been associated with the risk for CRC (Houlston and Tomlinson 2001; de Jong et al. 2002; Chapelle 2004; Kemp et al. 2004; Sharp and Little 2004; Chen et al. 2005; Little and Sharp 2007). In addition, recent results from a number of genome-wide association (GWA) studies have implicated at least ten loci as being associated with CRC (Broderick et al. 2007; Tomlinson et al. 2007, 2008; Houlston et al. 2008; Jaeger et al. 2008; Tenesa et al. 2008). Combining the information for robust candidate gene associations and GWA studies may not only explain a substantial share of the as yet unexplained genetic component of CRC etiology, but may also shed light on the pathways and mechanisms at play. Previous authors have published simulation studies looking at the number of hypothetical low risk variants that would be necessary to achieve various benchmarks of population attributable risk and predictive utility (Yang et al. 2003, 2005; Janssens et al. 2007). What is unclear is how such a genetic test might perform as an adjunct to established CRC screening modalities such as FOB testing, colonoscopy and sigmoidoscopy.
Aims of our study
In this paper we evaluate a hypothetical genetic test including increasing numbers of simulated genetic variants, and evaluate the degree to which such tests might be useful in enhancing the efficacy of established screening modalities. Particularly, we have focused on a very low relative risk range given that the most recently characterized risk variants from GWA studies have typically had marginal relative risks in the range of 1.1–1.3. In addition, we develop risk prediction models in population-based case–control data from the ARCTIC study (Zanke et al. 2007) to evaluate the utility and predictive value of available genomic information from an already completed study of risk factors for CRC. The variants chosen for inclusion in risk prediction models were informed by an extensive literature review of genetic risk factors for CRC completed by the authors (Campbell et al. 2009).
Methods
Statistical methods: simulation studies
For all simulations, a simple genetic model was assumed, whereby the risk allele was either present or absent for each of multiple loci. Using this approach, the risk status for each individual could be simulated using a binomial distribution with n = number of genes, x = number of genes with the risk allele, and p = prevalence of risk genotype for each individual gene such that,
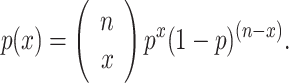 |
This binomial distribution was used to create a table of probabilities and combined odds ratios for genotypes with increasing numbers of risk variants from 0 to n, the number of genes specified in the simulation scenario. From this table, a simulated population was generated whereby the genotype for each individual was assigned based on a random draw from a uniform [0, 1] distribution. The odds ratio corresponding to the assigned genotype was calculated by exponentiating the per allele odds ratio according to the number of risk alleles. This corresponds to an additive (on the log scale) risk model. Although clearly important, we ignored the potential effects of gene–gene and gene–environment interactions in our simulations. We assumed that risk variants were either present or absent, and conferred the stated risk independent of the presence or absence of other genetic or environmental effects. Although multiplicative effects may be important in multi-locus genetic effects, we chose to report on the more conservative additive model that assumes no synergy among risk loci. All simulations were carried out in the R statistical package. Inputs to the simulation were based on existing empirical evidence. We used odds ratio estimates of 1.1–1.5 to demonstrate the range of possible outcomes for a variety of scenarios, but we focus on odds ratio = 1.2 and prevalence of 20% because these values are consistent with recent GWA findings for CRC and several other chronic diseases.
Genetic variance
Under the polygenic model, the distribution of risk approximately follows a lognormal distribution, thus log(R) = Y ~ N(μ, σ
2). The relative risk of disease in monozygous twins (which we denote λmonozygotic) and dizygotous twins/siblings (which we denote λsibling) are related to each other, and to the genetic variance (i.e., variance of the polygenic risk model on the log scale) by equation: 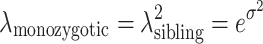 (Pharoah et al. 2002, 2008). Previous studies have reported estimates in the range of 2–4 for λ
sibling for common cancers including breast and colorectal (Pharoah et al. 2002, 2008; Lichtenstein et al. 2000; Ahlbom 1997), hence we have conservatively assumed a λ
sibling = 2, which corresponds to a standard deviation of 1.2 for the polygenic lognormal risk model (Pharoah et al. 2002, 2008). Based on the above assertions, we have used SD = 1.2 to define the ceiling at which all of the hereditary variance has been explained in our simulation work.
(Pharoah et al. 2002, 2008). Previous studies have reported estimates in the range of 2–4 for λ
sibling for common cancers including breast and colorectal (Pharoah et al. 2002, 2008; Lichtenstein et al. 2000; Ahlbom 1997), hence we have conservatively assumed a λ
sibling = 2, which corresponds to a standard deviation of 1.2 for the polygenic lognormal risk model (Pharoah et al. 2002, 2008). Based on the above assertions, we have used SD = 1.2 to define the ceiling at which all of the hereditary variance has been explained in our simulation work.
Sensitivity, specificity and area under the ROC curve (AUC)
Receiver operator characteristic (ROC) curves which plot sensitivity versus (1-specificity) were used in assessing the performance of our fitted logistic regression models (Hanley and McNeil 1982). Areas under the ROC curve (AUC) were estimated using the concordance index statistic c (Harrell et al. 1982). The c-statistic was calculated using the somers2 function from the Hmisc package in R (Harrell et al., http://cran.rproject.org/web/packages/). Sensitivity, specificity, positive and negative predictive values were also reported, requiring that a cutoff be specified as a classification rule. An effective screening test generally has a very high sensitivity (i.e., identifies a high proportion of disease cases) with the trade-off of accepting a certain proportion of false-positives. Our objective is not the development of a stand-alone screening tool, but rather an adjunctive test, to enrich the screening pool for conventional screening methods. Hence, we have focused on maximizing AUC, and have chosen a somewhat naïve cutoff, such that the sum of sensitivity and specificity is maximized, and in the results we report the sensitivity, specificity, and positive predictive value at this cutpoint. As another measure of predictive power, we also calculated the proportion of CRC cases that would be captured in the top 50% of ranked risk scores. If the test was used to calculate a risk score for a large number of individuals, and these scores were ranked from highest to lowest, then the top 50% based on rank were selected as positive tests. This is another somewhat arbitrary rule; however, screening is currently recommended in 100% of individuals over the age of 50 in North America, so our aim is to illustrate the properties of an adjunctive test that could focus effort on the half of the population at the highest genetic risk. A test that provides no more predictive ability than a coin toss will capture about 50% of cases in the top 50% of ranked risk scores. A highly predictive test will capture much more than 50% of cases thus creating an enriched screening pool. We proposed as a benchmark, a test that could capture 80% of all future CRC cases in the top 50% of ranked test scores. Such a test would enrich the screening pool enough to be useful as an adjunctive screening test.
Empirical confidence intervals
In order to estimate directly the variance associated with each of our statistics of interest, we repeated the Monte Carlo approach used to generate the simulated populations 500 times. Using these 500 samples of size of 100,000 each, we calculated all of the statistics of interest described above for each sample. We calculated point estimates using the median of the 500 estimates, and calculated non-parametric 95% confidence intervals by ranking the point estimates from the 500 samples and computing the 2.5th and 97.5th percentiles. This approach does not require distributional assumptions (e.g., normality) and the confidence limits are not constrained to be symmetrical. This allows the generation of more representative plausible ranges for each statistic of interest.
Simulating the impact of varying scenarios on screening
We simulated the impact of increasing predictive power for an adjunctive screening test on enrichment of the screening pool. For example, a genetic test that could capture 80% of subjects will develop CRC within their lifetime, within the top 50% of ranked scores of those tested would allow screening to be focused on those at the highest genetic risk of disease. Ten-year age-specific cancer incidence rates and population structure data were obtained from National Cancer Institute of Canada data (Canadian Cancer Society Steering Committee 2009), and Statistics Canada 2006 Census data (Statistics Canada 2006), respectively. We also simulated the effect of increasing levels of screening compliance concomitantly with the enriching of the screening pool. Simulations were carried out varying the degree of enrichment of the screening pool from 50% (no enrichment) up to 90% of CRC cases being captured in the top 50% of scores in the genetic test. For illustrative purposes, it was assumed that resources were limited for conventional screening and that 1,000,000 conventional screening tests were to be performed. Therefore, if 2,000,000 genetic tests were administered then the top 1,000,000 scores (or any other arbitrary cutoff) could be flagged as candidates for conventional screening. We classified simulated subjects into 10-year age and sex risk groups and estimated the number of subjects who would develop CRC in the next 10-year window, who would be flagged to undergo screening under varying genetic test predictive power, screening compliance and age and sex categories. We calculated these numbers first within age and sex subgroups, and then a weighted overall average based on Canadian population structure.
Statistical methods: analyses in a population-based case–control study of CRC, the Assessment of Risk for Colorectal Tumors in Canada (ARCTIC) Study
Multivariate modeling was carried out using available case control data and genotyping from the Assessment of Risk for Colorectal Tumors in Canada (ARCTIC) Study. The ARCTIC case–control study has been described in detail elsewhere (Zanke et al. 2007). Briefly, the ARCTIC study involved 1,257 CRC cases and 1,336 matched community controls from the Ontario Familial Colorectal Cancer Registry. Cases with known germline APC, MSH2, MLH1, MSH6 or biallelic MUTYH mutations were excluded. Extensive genotyping was performed for each subject, including several large genotyping arrays involving markers for over 600,000 single nucleotide polymorphisms (SNPs). Through a meta-analysis and systematic review, we identified an extensive list of polymorphisms which had been studied in relation to CRC (Campbell et al. 2009). Using the results of this review as a starting point, we selected about 80 polymorphisms, spanning over 30 genes and nearly all chromosomes. These variants were selected for inclusion based on the strength of association, consistency and quality of the evidence and validation of findings. These included genetic polymorphisms affecting xenobiotic metabolism (GSTM1, GSTT1, GSTA1, NAT1, NAT2); related to folate and one-carbon metabolism (MTHFR, MTRR, MTR); associated with alcohol metabolism (ADH1C, CYP2E1); influencing iron metabolism (HFE); identified via GWA studies [8q24, 8q23.3 (EIFH), 10p14, 11q23, 15q13, 18q21(SMAD7), 14q22(BMP4), 16q22(CDH1),19q13(RHPN2), and 20p12]; influencing vitamin D and calcium metabolism (VDR); influencing lipid or insulin metabolism (APOE, IRS1), affecting inflammation and immune response (HRAS1, IGF1, IL6, IL8, PPARG, TNF-α, PTGS1), tumor growth factors (TGFBR1, CCND1, CDH1), and highly penetrant variants with large effects (APC, MLH1, MLH3, MSH2). To address missing values for SNP covariates in multivariate modeling, we imputed missing genotypes using the haplotype clustering method implemented in the open-source software BEAGLE (Browning and Browning 2007). Genotype frequencies for each bi-allelic genotype were compared to that expected to arise from random mating. A χ 2 test was used to test whether any departures from HWE were more extreme than would be expected by chance (Thomas 2004). Multivariate logistic regression analyses and model validation procedures were carried out in SAS version 9.1 (SAS Institute, Cary, NC) and R version 2.9 (The R Foundation for Statistical Computing, Vienna, Austria).
Results
Genomic profiling simulations
In Table 1, the results for selected simulations are presented, for 20–400 putative risk alleles. For simulations with 20 putative genes, odds ratio of 1.2 for each risk variant, and risk variant prevalence of 20%, AUC was 59.2% and 61.8% of cases were captured in the top 50% of ranked risk scores (abbreviated as CCNT for case concentration). At the maximum observed sensitivity + specificity, sensitivity was 49.5%, specificity was 63.9% and positive and negative predictive values were 9.3 and 94.4%, respectively. Hence, only 9 out of every 100 positive tests were true positives under this prediction model. If a test threshold was selected such that CCNT = 0.80, then for the same allele frequency and average relative risk, roughly 140–160 risk variants would be required. At this level, positive predictive value is still only about 13%. Hence even with a highly predictive test, where 80% of cases are being captured in the “test positive” pool, 87 out of every 100 positive tests are false-positives. We also ran simulations to evaluate the relative contribution of a small number of rare variants with higher penetrance, with varying prevalence and odds ratios. For example, from Table 1, with 20 risk variants, odds ratios of 1.2 per variant, and risk variant prevalence of 20%, AUC is 59.2%, and CCNT is 61.8%. Adding the effect of five higher risk variants with odds ratios of 20 per variant, and rare prevalence of 0.1%, raises AUC to 0.607, and CCNT to 63.3%. However, adjusting the parameters such that the five rare variants have a prevalence of 1% but much lower odds ratios of 5.0 per rare variant leads to an AUC of 0.64, and CCNT of 66.3%. These results are compatible with the view that a collection of common low penetrance variants could together explain a much larger proportion of the genetic variance than a small number of rare variants with extremely high relative risks. However, a substantial number of such variants would be necessary. Figure 1 summarizes results for the simulations for numbers of risk alleles ranging from 20 to 400 risk variants, for odds ratios of 1.1–1.5. The plot of number of risk variants versus CCNT in Fig. 1 suggests that with average relative risk of 1.2, at least 140–160 risk alleles would be required for the combined genomic information to really be useful in a screening or risk triage setting, based on the benchmark of requiring a test to be able to correctly identify 80% of future CRC cases in the top 50% of ranked test scores (horizontal reference line denotes this benchmark). To reach the upper limit of genetic variance (explaining all possible hereditary CRC) would require in excess of 300 risk alleles (horizontal reference line of SD = 1.2 in Fig. 2). At this threshold, close to 90% of cases would be contained in the top 50% (CCNT > 87.3%).
Table 1.
Simulation results for 20–400 risk alleles: statistics and empirical confidence intervals
| Simulation no. | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Number of loci | 20 | 40 | 60 | 80 |
| AUC | 0.592 (0.585, 1.598) | 0.629 (0.622, 0.636) | 0.656 (0.649, 0.663) | 0.678 (0.672, 0.685) |
| CCNT | 0.618 (0.607, 0.629) | 0.666 (0.654, 0.677) | 0.700 (0.690, 0.711) | 0.728 (0.718, 0.739) |
| SD | 0.302 (0.301, 0.303) | 0.428 (0.426, 0.429) | 0.524 (0.522, 0.526) | 0.605 (0.602, 0.607) |
| Sensitivity | 0.495 (0.484, 0.609) | 0.582 (0.569, 0.593) | 0.633 (0.512, 0.645) | 0.662 (0.560, 0.683) |
| Specificity | 0.639 (0.523, 0.642) | 0.606 (0.603, 0.610) | 0.592 (0.589, 0.710) | 0.587 (0.582, 0.691) |
| PPV | 0.093 (0.088, 0.096) | 0.100 (0.097, 0.103) | 0.105 (0.102, 0.118) | 0.111 (0.106, 0.124) |
| NPV | 0.944 (0.942, 0.948) | 0.950 (0.949, 0.952) | 0.955 (0.951, 0.957) | 0.958 (0.954, 0.961) |
| Simulation no. | 5 | 6 | 7 | 8 |
|---|---|---|---|---|
| Number of loci | 100 | 120 | 140 | 160 |
| AUC | 0.697 (0.690, 0.702) | 0.713 (0.707, 0.719) | 0.727 (0.721, 0.733) | 0.740 (0.734, 0.746) |
| CCNT | 0.750 (0.740, 0.760) | 0.770 (0.761, 0.780) | 0.787 (0.778, 0.796) | 0.802 (0.792, 0.810) |
| SD | 0.676 (0.674, 0.679) | 0.741 (0.738, 0.744) | 0.801 (0.797, 0.804) | 0.856 (0.852, 0.860) |
| Sensitivity | 0.613 (0.602, 0.709) | 0.648 (0.635, 0.661) | 0.677 (0.598, 0.689) | 0.700 (0.624, 0.714) |
| Specificity | 0.674 (0.578, 0.677) | 0.663 (0.659, 0.667) | 0.655 (0.652, 0.730) | 0.649 (0.645, 0.721) |
| PPV | 0.123 (0.110, 0.127) | 0.126 (0.122, 0.131) | 0.129 (0.125, 0.145) | 0.132 (0.128, 0.147) |
| NPV | 0.962 (0.960, 0.963) | 0.964 (0.960, 0.966) | 0.966 (0.962, 0.968) | 0.966 (0.964, 0.970) |
| Simulation no. | 9 | 10 | 11 | 12 |
|---|---|---|---|---|
| Number of loci | 180 | 200 | 250 | 300 |
| AUC | 0.751 (0.746, 0.757) | 0.762 (0.756, 0.767) | 0.785 (0.779, 0.790) | 0.803 (0.798, 0.808) |
| CCNT | 0.815 (0.806, 0.824) | 0.827 (0.819, 0.836) | 0.853 (0.844, 0.860) | 0.873 (0.866, 0.880) |
| SD | 0.908 (0.905, 0.912) | 0.957 (0.953, 0.961) | 1.071 (1.066, 1.075) | 1.173 (1.169, 1.178) |
| Sensitivity | 0.663 (0.650, 0.732) | 0.685 (0.673, 0.748) | 0.731 (0.673, 0.743) | 0.727 (0.683, 0.776) |
| Specificity | 0.710 (0.640, 0.713) | 0.703 (0.638, 0.706) | 0.689 (0.686, 0.745) | 0.729 (0.677, 0.775) |
| PPV | 0.146 (0.130, 0.150) | 0.148 (0.133, 0.152) | 0.152 (0.148, 0.169) | 0.168 (0.153, 0.184) |
| NPV | 0.966 (0.964, 0.970) | 0.967 (0.966, 0.971) | 0.971 (0.968, 0.973) | 0.973 (0.970, 0.976) |
| Simulation no. | 13 | 14 | ||
|---|---|---|---|---|
| Number of loci | 350 | 400 | ||
| AUC | 0.819 (0.814, 0.823) | 0.830 (0.826, 0.835) | ||
| CCNT | 0.889 (0.882, 0.895) | 0.901 (0.895, 0.908) | ||
| SD | 1.268 (1.263, 1.273) | 1.353 (1.348, 1.359) | ||
| Sensitivity | 0.758 (0.711, 0.771) | 0.753 (0.740, 0.756) | ||
| Specificity | 0.719 (0.715, 0.765) | 0.751 (0.707, 0.755) | ||
| PPV | 0.171 (0.166, 0.190) | 0.185 (0.168, 0.191) | ||
| NPV | 0.975 (0.972, 0977) | 0.976 (0.974, 0.979) |
AUC area under receiver–operator curve, CCNT % cases captured in top 50% of ranked model scores, SD standard deviation, PPV positive predictive value, NPV negative predictive value
SD of the genetic risk distribution: 1.2 is the theoretical threshold where all of the hereditary component of CRC has been explained
Fig. 1.

Simulation results: 20–500 risk alleles and OR = 1.1–1.5, for area under the ROC curve (AUC), case concentration (CCNT) and genetic risk standard deviation (SD). #Horizontal reference line indicates threshold of 80% of CRC cases being captured in top 50% of ranked test scores. *Horizontal reference line indicates theoretical maximum standard deviation of the genetic risk distribution (SD) explained by heritable factors (SD = 1.2)
Fig. 2.

Univariate frequencies and odds ratios for candidate risk genes in ARCTIC data. MM/mm major/minor allele homozygote, Mm heterozygote
Application of simulated genomic profiles in the context of population screening
Table 2 shows the number of cases of cancer that would be screened within varying test predictive power, and compliance levels. These are expressed as CRC cases per 1,000,000 subjects counseled to participate in FOB/endoscopy based on being in the top 50% of the 2,000,000 subjects in whom the simulated genetic profile is evaluated, who then may or may not present themselves for screening based on the stated compliance rates ranging from 25 to 100%. Numbers are presented for individual age groups and then an overall weighted population total is estimated. For example, for a situation of having an equivocal genetic test (50/50, no predictive utility), and 25% compliance with conventional screening, 2,746 cancer patients would present themselves for the FOB/endoscopic screening test. If a strongly predictive profiling test was deployed that concentrated 80% of cases into the top 50% of the sample, and screening compliance remained at 25%, 4,398 cancers would be brought into screening. If, as hypothesized, the screening compliance of the subjects was to improve to say to 40%, this would result in 7,039 cancers being brought to screening. Hence, the combination of a strongly predictive genetic test and the improved screening compliance leads to an additional 4,293 CRC cases being brought into the screening pool. The majority of these additional cancers would be detected by the latest FOB/endoscopic tests. In Table 3, the impact of a genomic test that captures 80% of cases in the top 50% of test scores is explored in individual age and gender categories.
Table 2.
Future cancers potentially screened and detected early out of 1,000,000 triaged screenees based on predictive power of the genetic profile, and projected increase in screening compliance (weighted by Canadian population structure from 2006 Canadian Census)
| CCNT | Compliance ratea | |||||
|---|---|---|---|---|---|---|
| 25 | 30 | 40 | 50 | 60 | 100 | |
| 0.50 | 2,746 | 3,297 | 4,398 | 5,500 | 6,599 | 11,004 |
| 0.60 | 3,297 | 3,958 | 5,279 | 6,599 | 7,920 | 13,204 |
| 0.70 | 3,847 | 4,617 | 6,157 | 7,700 | 9,240 | 15,405 |
| 0.80 | 4,398 | 5,279 | 7,039 | 8,801 | 10,561 | 17,608 |
| 0.90 | 4,949 | 5,940 | 7,920 | 9,901 | 11,882 | 19,808 |
CCNT % of all CRC cases in top 50% of ranked genomic profile scores
aCompliance refers to the proportion of subjects counseled to undergo screening who actually attend screening
Table 3.
Future cancers potentially screened and detected early for a genomic profile that detects 80% of cancers in the top 50% of ranked profile scores in age and sex groups and overall, with the impact of increasing levels of compliance across columns
| Age and sex | Compliance ratea | |||||
|---|---|---|---|---|---|---|
| 25 | 30 | 40 | 50 | 60 | 100 | |
| Male 40–49 | 133 | 160 | 213 | 267 | 320 | 534 |
| Male 50–59 | 453 | 544 | 725 | 907 | 1,088 | 1,814 |
| Male 60–69 | 712 | 855 | 1,140 | 1,425 | 1,710 | 2,851 |
| Male 70–79 | 730 | 876 | 1,168 | 1,460 | 1,753 | 2,921 |
| Male 80–89 | 313 | 375 | 501 | 626 | 751 | 1,253 |
| Female 40–49 | 137 | 165 | 220 | 275 | 330 | 551 |
| Female 50–59 | 351 | 421 | 561 | 702 | 842 | 1,404 |
| Female 60–69 | 489 | 586 | 782 | 978 | 1,173 | 1,956 |
| Female 70–79 | 628 | 754 | 1,005 | 1,256 | 1,508 | 2,513 |
| Female 80–89 | 452 | 543 | 724 | 905 | 1,086 | 1,811 |
| Total | 4,398 | 5,279 | 7,039 | 8,801 | 10,561 | 17,608 |
aCompliance refers to the proportion of subjects counseled to undergo screening who actually attend screening
Risk models via logistic regression in ARCTIC case–control data
Less than one-third of the targeted candidate polymorphisms were available for study based on completed genotyping in ARCTIC data. Univariate analyses of all available candidate SNPs from ARCTIC study genotyping data were conducted. Allele frequencies were calculated and checked for departures from Hardy–Weinberg Equilibrium (HWE). No serious departures from HWE were noted. Frequencies and univariate odds ratios were calculated for each SNP. Odds ratios were calculated for three contrasts of interest: (1) heterozygotes versus reference allele homozygotes, (2) variant homozygotes versus reference allele homozygotes and (3) “per allele” odds ratios where individuals were scored as 0, 1 or 2 corresponding to the number of variant alleles they carried. The results of univariate analyses are reported in Fig. 2. SNPs demonstrating nominal significance in univariate analyses were included in multivariate modeling procedures. A number of environmental risk factors were also included in order to explore relevant gene × environment interactions. These biologically relevant gene–environment interactions were considered in individual logistic models. SNP–environment interactions that achieved nominal significance (P < 0.10) in individual models were included as candidates in multivariate modeling. Initial models included only SNPs and SNP × SNP interactions. Stepwise variable selection was used, with P < 0.10 the cutoff to enter the model, and P < 0.20 to be removed from the model once included. Model hierarchy was respected in the modeling process, meaning in order for an interaction to be included in the model, both of the main effects were forced into the model. To address overfitting, tenfold cross-validation was used, and all reported statistics are from cross-validated models. The final model from this procedure included variants: rs1801282-PPARG P10A, rs2289046-IRS2, rs2472300-CYP1A2 C-164A tag snp, rs3099844-TNF-alpha G-308A, rs4779584-CRAC1, rs10505477-ARCTIC 8q24, rs10735810-VDR FokI T > C and interactions: (rs1801282 × rs4779584), (rs1801282 × rs10505477) and (rs1801282 × rs10735810). AUC as measured by the c-statistic (concordance index) was 0.54. Models additionally including age and gender gave an AUC of 63%. In contrast, a model with only age and sex gave an AUC of 60%. Stepwise variable selection resulted in a model that included age, sex, rs1801282, rs2289046, rs2472300, rs4779584, rs10505477, rs7296651, tertile of red meat consumption (rmtert), history of colitis (coli), aspirin use (asp), ibuprofen use (ibp), and interactions: age × sex, rs1801282 × rs4779584, rs1801282 × rs7296651, rs4779584 × rs7296651, rs2289046 × rmtert, rs7296651 × rmtert and rs2289046 × asp. The AUC for this model was 0.66. Given that most candidate polymorphisms available in ARCTIC failed to demonstrate nominal statistical significance individually, an aggregate model was constructed to explore pooled effects of increasing numbers of variant alleles. All SNPs were re-parameterized so that the reference allele conferred low risk and the variant allele conferred elevated risk. Considering all 26 variants collectively by scoring each locus as having 0, 1 or 2 risk alleles and pooling and grouping the data showed a highly statistically significant trend of increasing risk with increasing numbers of risk alleles; however, the effect size was small in magnitude. For every additional risk allele, risk increased by about 8% [odds ratio 1.08 (95% CI 1.05–1.11)] (see Fig. 3a). Grouping subjects into quintiles, the odds ratio for subjects with 27+ risk alleles versus 0–21 risk alleles was 1.67 (95% CI 1.28–2.18) (see Fig. 3b). More detailed grouping demonstrated that subjects with 30 or more variant alleles have an odds ratio of 2.62 (95% CI 1.27–4.04) versus subjects with 20 or fewer (see Fig. 3c). Although modest in magnitude, the effects showed a strong and consistent linear trend.
Fig. 3.
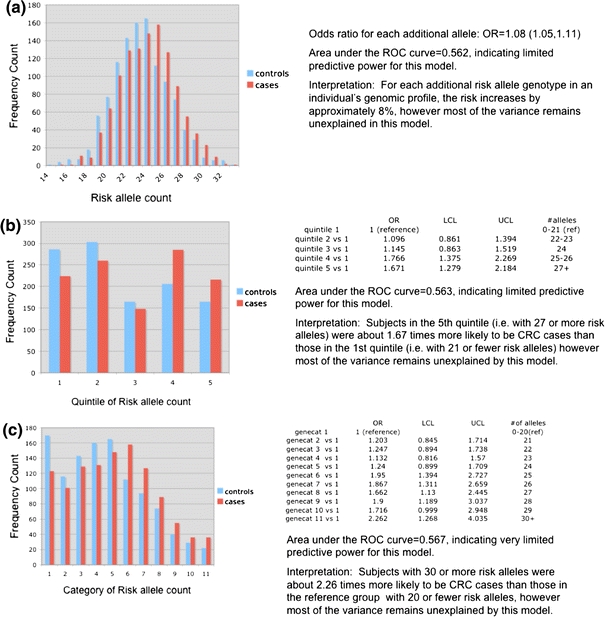
a Combining all alleles into a risk allele score (risk allele count = 0, 1 or 2 per locus), maximum of possible 52 alleles for 26 variants. b Combining all alleles into a risk allele score (risk allele count = 0, 1 or 2 per locus), risk allele count grouped into quintiles. c Combining all alleles into a risk allele score (risk allele count = 0, 1 or 2 per locus), risk allele counts grouped into detailed categories
Discussion
We have performed a variety of analyses using both simulated and empirical population data to illustrate how a genetic profile with moderate predictive power, while not a viable screening test in isolation, could be an important tool when employed as an adjunct to conventional CRC screening modalities such as FOB testing and endoscopy (colonoscopy/flexible sigmoidoscopy). We developed risk prediction models in data from the ARCTIC case–control data using a collection of polymorphisms identified through an extensive literature search for risk variants identified in genetic association studies (both candidate gene and GWA). Our list consisted of approximately 80 polymorphisms, of which less than 30 were available in the existing ARCTIC data. Model building in this limited subset led to models with significant effects from a statistical perspective, but which failed to demonstrate sufficient predictive power to be useful. Only a handful of polymorphisms reached statistical significance individually. When all the loci were pooled together, subjects with 30 or more risk alleles had more than a twofold risk increase over subjects with fewer than 20 risk alleles (OR 2.26 95% CI 1.27–4.04).The odds ratio per additional allele was 1.08 (95% CI 1.05–1.11). Although there have been a number of risk loci identified in candidate gene studies, and many have been replicated in more than one study, only a handful of these candidate gene effects were replicated in ARCTIC data. These included variants Cyp1A1 T461N (rs1799814), VDR Fok1T > C(rs10735810), Cyp1a2 C-164A (rs2472300 tag SNP) and PPARγ P10A (rs1801282). This is consistent with another recent report that found that very few positive findings from previous association studies could be replicated in a European population (Küry et al. 2008). A panel of 52 polymorphisms in 35 genes were tested in 1,023 patients with sporadic CRC and 1,121 controls from France. The polymorphisms were from inflammation, xenobiotic detoxification, one-carbon, insulin signaling, and DNA repair pathways. Logistic regression was used to model the risk of CRC associated with the panel of variants both individually in univariate analyses and in multivariate models. The researchers were able to replicate the association for only five of the polymorphisms. Three SNPs were shown to increase CRC risk: PTGS1 C639A, IL8 T-352A, and MTHFR A1298C. Two other SNPs, PLA2G2A C230T and PPARG C1431T, were associated with a decrease in CRC risk. Combinations of risk variants were also identified that led up to a twofold increase in risk (OR 1.97, 95% CI 1.31–2.97) (Küry et al. 2008). None of the replicated SNPs overlapped with those replicated in our work thus far, and no other similar attempts at replicating candidate gene study findings in large, independent cohorts have been reported. In another recently published report, a combined analysis of the impact of all ten risk loci implicated in GWA studies together demonstrated a per allele odds ratio of 1.16 (95% OR 1.13–1.18), and a nearly tenfold increased risk for subjects with ≥15 risk alleles versus subjects with ≤4 risk alleles (Houlston et al. 2008). When combined, the cumulative findings from replicated candidate gene studies could significantly compliment the accruing evidence from GWA studies to eventually comprise a useful risk model if all the variants could be measured on a single, easy to collect and analyze genomic panel. In our simulation work we attempted to extrapolate from the findings in population data to look at the hypothetical impact of an expanded genomic risk profile. For models including a plausible number of risk variants with risk allele frequency of approximately 20% and odds ratios in 1.1–1.5 range, it is possible to achieve meaningful predictive power and substantial population attributable fraction (PAF). Our results suggest it would take at least 300 independent risk alleles to explain all of the genetic component of CRC, and between 140 and 160 risk alleles to provide a genomic profile that could capture 80% of future cancers in the 50% of subjects at highest genetic risk. Similar reports have been published for other common cancers. A recent study estimates that given current knowledge of breast cancer susceptibility loci, the half of the population at highest genetic risk based on current knowledge, account for 60% of all cases of breast cancer (Pharoah et al. 2008). Estimates that give current knowledge of breast cancer susceptibility loci, the half of the population at highest genetic risk based on current knowledge, account for 60% of all cases of breast cancer. If all possible susceptibility loci were known, they estimate that the top 50% of the population in terms of genetic risk would represent about 88% of breast cancer cases. These estimates are consistent with those reported in similar investigations of the hereditary component of risk for sporadic CRC.
Despite the fact that germline genomic profiling in disease prevention has not as yet been adequately assessed (Janssens et al. 2004; Khoury et al. 2004; Yang et al. 2003), several companies have begun offering personalized genomic testing that includes a very small number of variants identified mostly in GWA studies that have shown association with CRC among other diseases. Some go as far as to provide a composite risk score based on the clients’ genotypes at these loci. The variants included in these tests are often not supported by adequate evidence (i.e., systematic review, meta-analysis repeated replication of findings) (Caulfield et al. 2001; Human Genetics Commision, UK 2003; Gollust et al. 2003; Gray and Olopade 2003; Williams-Jones 2003; American College of Medicine Genetics. Board of Directors 2004; McCabe and McCabe 2004; Kutz 2006; Janssens et al., 2008).
The trajectory and rate of discovery of novel risk loci in GWA studies and meta-analysis of GWAs suggest it is possible that within a few years enough risk alleles will have been characterized to allow the formulation of a useful genomic profile. However, at some point, there will inevitably be diminished returns with regard to the magnitude of effect sizes for novel risk loci. In the near future, while the coverage of the genome is still improving, it is possible that newly discovered risk variants will be comparable in effect size to those described before. However, once all of the overt risk loci have been described, then new alleles will only be detected through the increased power inherent in pooled analysis of multiple GWA studies or larger studies conducted via investigator networks. So, some argue it is inevitable that the relative risk for newly described loci will trend consistently downward (Goldstein 2009). Should the per-allele relative risk keep dropping well below 1.1, this will hamper the likelihood of a useful predictive model emerging that includes exclusively genomic factors. There are a number of established non-genetic risk factors that could easily be incorporated into future genomic profiles. Robust gene–gene and gene–environment interactions could also contribute valuable information as they become better characterized. Hence, the evolution of genomic profiling need not be limited to inclusion of solely genetic risk factors.
Assuming an adequate number of alleles are identified and have sufficient effect size, our simulations have demonstrated that a predictive genetic model as an adjunct test within a population screening program could potentially lead to better targeting of screening services to those at highest risk, and better adherence in those to whom screening is recommended. Overfitting is an issue that must be addressed in building predictive models to be used to risk-stratify new individuals—models need to be appropriately validated and calibrated to account for this model uncertainty. Much larger samples are required to detect small effects—especially for interactions, and multivariate modeling. Although our motivating example has been CRC, our results could easily be extended to other chronic diseases with a hereditary component. Indeed, future directions will undoubtedly involve a genetic profile that encompasses a spectrum of diseases. In many cases, the most important genetic variants act in the etiology of more than one disease. Although more low penetrance risk alleles will need to be identified before genomic information can be utilized in an impactful way, our results provide evidence that the concept is promising given a reasonable and realistic set of conditions. Although the ARCTIC study was well powered to look at associations between common SNPS and CRC, power diminishes rapidly for evaluation of gene–environment and gene–gene interactions, as well as for polymorphisms with much smaller minor allele frequencies, and investigations in subgroups of interest. Pooled analysis among several large CRC case–control studies would allow for a much more powerful statistical evaluation of these questions. This will be facilitated via existing networks of investigators (e.g., HUGEnet) and existing research collaborations.
Acknowledgments
This study was supported by the Cancer Risk Evaluation (CaRE) Program Grant from the Canadian Cancer Society Research Instituted, the Canadian Institutes of Health Research Team in Interdisciplinary Research on Colorectal Cancer, CIHR pilot project grant in colorectal cancer screening. TJH and BWZ are recipients of Senior Investigator Awards from the Ontario Institute for Cancer Research, through generous support from the Ontario Ministry of Research and Innovation. JL holds a Tier 1 Canada Research Chair in Human Genome Epidemiology.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Steven J. Hawken, Email: shawken@ohri.ca
Julian Little, Email: jlittle@uottawa.ca.
References
- Aaltonen L, Johns L, Järvinen H, Mecklin JP, Houlston R. Explaining the familial colorectal cancer risk associated with mismatch repair (mmr)-deficient and mmr-stable tumors. Clin Cancer Res. 2007;13(1):356–361. doi: 10.1158/1078-0432.CCR-06-1256. [DOI] [PubMed] [Google Scholar]
- Ahlbom A. Cancer in twins: genetic and nongenetic familial risk factors. J Natl Cancer Inst. 1997;89:287–293. doi: 10.1093/jnci/89.4.287. [DOI] [PubMed] [Google Scholar]
- Allison JE, Sakoda LC, Levin TR, Tucker JP, Tekawa IS, Cuff T, Pauly MP, Shlager L, Palitz AM, Zhao WK, Schwartz JS, Ransohoff DF, Selby JV. Screening for colorectal neoplasms with new fecal occult blood tests: update on performance characteristics. J Natl Cancer Inst. 2007;99(19):1462–1470. doi: 10.1093/jnci/djm150. [DOI] [PubMed] [Google Scholar]
- American College of Medicine Genetics. Board of Directors Acmg statement on direct-to-consumer genetic testing. Genet Med. 2004;6(1):60. doi: 10.1097/01.GIM.0000106164.59722.CE. [DOI] [PubMed] [Google Scholar]
- Atkin W. Options for screening for colorectal cancer. Scand J Gastroenterol Suppl. 2003;237:13–16. doi: 10.1080/00855910310001421. [DOI] [PubMed] [Google Scholar]
- Autier P, Boyle P, Buyse M, Bleiberg H. Is FOB screening really the answer for lowering mortality in colorectal cancer? Recent Results Cancer Res. 2003;163:254–263. doi: 10.1007/978-3-642-55647-0_23. [DOI] [PubMed] [Google Scholar]
- Bampton PA, Young GP, John JS. Screening for colorectal cancer will save lives. Med J Aust. 2000;172(12):576–577. doi: 10.5694/j.1326-5377.2000.tb124121.x. [DOI] [PubMed] [Google Scholar]
- Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K, Fielding S, Jaeger E, Vijayakrishnan J, Kemp Z, Gorman M, Chandler I, Papaemmanuil E, Penegar S, Wood W, Sellick G, Qureshi M, Teixeira A, Domingo E, Barclay E, Martin L, Sieber O, Consortium C, Kerr D, Gray R, Peto J, Cazier JB, Tomlinson I, Houlston RS. A genome-wide association study shows that common alleles of smad7 influence colorectal cancer risk. Nat Genet. 2007;39(11):1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- Browning S, Browning B. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell H, Hawken S, Theodoratou E, Demarsh A, Hutchings K, Johnson CY, Masson L, Sharp L, Tait V, Little J (2009) Cumulative assessment of the role of human genome variation in colorectal cancer. In: Human genome epidemiology, 2nd edn. Oxford University Press, Oxford
- Canadian Cancer Society Steering Committee (2009) Canadian cancer statistics 2009. http://www.cancer.ca/canada-wide/aboutcancer/cancerstatistics/canadiancancerstatistics.aspx
- Caulfield T, Burgess M, Williams-Jones B. Providing genetic testing through the private sector: a view from Canada. Can J Policy Res. 2001;2(3):72–81. [Google Scholar]
- Chapelle ADL. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4(10):769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- Chen K, Jiang QT, He HQ. Relationship between metabolic enzyme polymorphism and colorectal cancer. World J Gastroenterol. 2005;11(3):331–335. doi: 10.3748/wjg.v11.i3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong MM, Nolte IM, te Meerman GJ, van der Graaf WTA, de Vries EGE, Sijmons RH, Hofstra RMW, Kleibeuker JH. Low-penetrance genes and their involvement in colorectal cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2002;11(11):1332–1352. [PubMed] [Google Scholar]
- Gertig DM, Stampfer M, Haiman C, Hennekens CH, Kelsey K, Hunter DJ. Glutathione s-transferase GSTM1 and GSTT1 polymorphisms and colorectal cancer risk: a prospective study. Cancer Epidemiol Biomarkers Prev. 1998;7(11):1001–1005. [PubMed] [Google Scholar]
- Goldstein D. Common genetic variation and human traits. N Engl J Med. 2009;360(17):1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- Gollust SE, Wilfond BS, Hull SC. Direct-to-consumer sales of genetic services on the internet. Genet Med. 2003;5(4):332–337. doi: 10.1097/01.GIM.0000076972.83711.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Olopade OI. Direct-to-consumer marketing of genetic tests for cancer: buyer beware. J Clin Oncol. 2003;21(17):3191–3193. doi: 10.1200/JCO.2003.12.069. [DOI] [PubMed] [Google Scholar]
- Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- Harrell FE, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247(18):2543–2546. doi: 10.1001/jama.247.18.2543. [DOI] [PubMed] [Google Scholar]
- Hewitson P, Glasziou P, Irwig L, Towler B, Watson E (2007) Screening for colorectal cancer using the faecal occult blood test, hemoccult. Cochrane Database Syst Rev 1:CD001,216 (online) [DOI] [PMC free article] [PubMed]
- Houlston RS, Tomlinson IP. Polymorphisms and colorectal tumor risk. Gastroenterology. 2001;121(2):282–301. doi: 10.1053/gast.2001.26265. [DOI] [PubMed] [Google Scholar]
- Houlston RS, Webb E, Broderick P, Pittman AM, Bernardo MCD, Lubbe S, Chandler I, Vijayakrishnan J, Sullivan K, Penegar S, Carvajal-Carmona L, Howarth K, Jaeger E, Spain SL, Walther A, Barclay E, Martin L, Gorman M, Domingo E, Teixeira AS, Kerr D, Cazier JB, Niittymäki I, Tuupanen S, Karhu A, Aaltonen LA, Tomlinson IPM, Farrington SM, Tenesa A, Prendergast JGD, Barnetson RA, Cetnarskyj R, Porteous ME, Pharoah PDP, Koessler T, Hampe J, Buch S, Schafmayer C, Tepel J, Schreiber S, Völzke H, Chang-Claude J, Hoffmeister M, Brenner H, Zanke BW, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40(12):1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Genetics Commission UK (2003) Genes direct: ensuring the effective oversight of genetic tests directly to the public. http://www.dh.gov.uk/en/Publicationsandstatistics/Publications/PublicationsPolicyAndGuidance/DH4084423
- Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, Walther A, Spain S, Pittman A, Kemp Z, Sullivan K, Heinimann K, Lubbe S, Domingo E, Barclay E, Martin L, Gorman M, Chandler I, Vijayakrishnan J, Wood W, Papaemmanuil E, Penegar S, Qureshi M, Consortium C, Farrington S, Tenesa A, Cazier JB, Kerr D, Gray R, Peto J, Dunlop M, Campbell H, Thomas H, Houlston R, Tomlinson I. Common genetic variants at the crac1 (hmps) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40(1):26–28. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- Janssens ACJW, Pardo MC, Steyerberg EW, Duijn CMV. Revisiting the clinical validity of multiplex genetic testing in complex diseases. Am J Hum Genet. 2004;74(3):585–588. doi: 10.1086/382052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens ACJW, Moonesinghe R, Yang Q, Steyerberg EW, Duijn CMV, Khoury MJ. The impact of genotype frequencies on the clinical validity of genomic profiling for predicting common chronic diseases. Genet Med. 2007;9(8):528–535. doi: 10.1097/GIM.0b013e31812eece0. [DOI] [PubMed] [Google Scholar]
- Janssens ACJW, Gwinn M, Bradley LA, Oostra BA, Duijn CMV, Khoury MJ. A critical appraisal of the scientific basis of commercial genomic profiles used to assess health risks and personalize health interventions. Am J Hum Genet. 2008;82(3):593–599. doi: 10.1016/j.ajhg.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp Z, Thirlwell C, Sieber O, Silver A, Tomlinson I. An update on the genetics of colorectal cancer. Hum Mol Genet. 2004;13(2):R177–R185. doi: 10.1093/hmg/ddh247. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Yang Q, Gwinn M, Little J, Flanders WD. An epidemiologic assessment of genomic profiling for measuring susceptibility to common diseases and targeting interventions. Genet Med. 2004;6(1):38–47. doi: 10.1097/01.GIM.0000105751.71430.79. [DOI] [PubMed] [Google Scholar]
- Kronborg O, Jørgensen OD, Fenger C, Rasmussen M. Randomized study of biennial screening with a faecal occult blood test: results after nine screening rounds. Scand J Gastroenterol. 2004;39(9):846–851. doi: 10.1080/00365520410003182. [DOI] [PubMed] [Google Scholar]
- Küry S, Buecher B, du Pont SR, Scoul C, Colman H, Neel TL, Houérou CL, Faroux R, Ollivry J, Lafraise B, Chupin LD, Sébille V, Bézieau S. Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer. 2008;8:326. doi: 10.1186/1471-2407-8-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz G (2006) Nutrigenetic testing: tests purchased from four web sites mislead consumers: testimony before the special committee on aging, US senate. http://purl.access.gpo.gov/GPO/LPS73075
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Ling BS, Moskowitz MA, Wachs D, Pearson B, Schroy PC. Attitudes toward colorectal cancer screening tests. J Gen Intern Med. 2001;16(12):822–830. doi: 10.1046/j.1525-1497.2001.10337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J, Sharp L (2007) Colorectal cancer. In: Epidemiology of colorectal cancer. Informa Healthcare, New York, pp 43–75
- Madlensky L, Esplen MJ, Gallinger S, McLaughlin JR, Goel V. Relatives of colorectal cancer patients: factors associated with screening behavior. Am J Prev Med. 2003;25(3):187–194. doi: 10.1016/S0749-3797(03)00202-2. [DOI] [PubMed] [Google Scholar]
- Madlensky L, McLaughlin JR, Carroll JC, Goel V, Frank JW. Risks and benefits of population-based genetic testing for Mendelian subsets of common diseases were examined using the example of colorectal cancer risk. J Clin Epidemiol. 2005;58(9):934–941. doi: 10.1016/j.jclinepi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Mandelson MT, Curry SJ, Anderson LA, Nadel MR, Lee NC, Rutter CM, LaCroix AZ. Colorectal cancer screening participation by older women. Am J Prev Med. 2000;19(3):149–154. doi: 10.1016/S0749-3797(00)00193-8. [DOI] [PubMed] [Google Scholar]
- McCabe LL, McCabe ERB (2004) Direct-to-consumer genetic testing: access and marketing. Genet Med 6(1):60 [DOI] [PubMed]
- Moayyedi P, Achkar E. Does fecal occult blood testing really reduce mortality? A reanalysis of systematic review data. Am J Gastroenterol. 2006;101(2):380–384. doi: 10.1111/j.1572-0241.2006.00537.x. [DOI] [PubMed] [Google Scholar]
- Müller AD, Sonnenberg A. Protection by endoscopy against death from colorectal cancer. A case–control study among veterans. Arch Intern Med. 1995;155(16):1741–1748. doi: 10.1001/archinte.155.16.1741. [DOI] [PubMed] [Google Scholar]
- Multicentre Australian Colorectal-neoplasia Screening (MACS) Group A comparison of colorectal neoplasia screening tests: a multicentre community-based study of the impact of consumer choice. Med J Aust. 2006;184(11):546–550. doi: 10.5694/j.1326-5377.2006.tb00377.x. [DOI] [PubMed] [Google Scholar]
- National Committee on Colorectal Cancer Screening (2002) Recommendations for population-based colorectal cancer screening. http://www.phac-aspc.gc.ca/publicat/ncccs-cndcc/techrep-eng.php
- Newcomb PA, Norfleet RG, Storer BE, Surawicz TS, Marcus PM. Screening sigmoidoscopy and colorectal cancer mortality. J Natl Cancer Inst. 1992;84(20):1572–1575. doi: 10.1093/jnci/84.20.1572. [DOI] [PubMed] [Google Scholar]
- Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31(1):33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- Pharoah PDP, Antoniou AC, Easton DF, Ponder BAJ. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358(26):2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- Rabeneck L, Paszat LF. A population-based estimate of the extent of colorectal cancer screening in Ontario. Am J Gastroenterol. 2004;99(6):1141–1144. doi: 10.1111/j.1572-0241.2004.30623.x. [DOI] [PubMed] [Google Scholar]
- Ramji F, Cotterchio M, Manno M, Rabeneck L, Gallinger S. Association between subject factors and colorectal cancer screening participation in Ontario, Canada. Cancer Detect Prev. 2005;29(3):221–226. doi: 10.1016/j.cdp.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Ransohoff DF. Lessons from the UK sigmoidoscopy screening trial. Lancet. 2002;359(9314):1266–1267. doi: 10.1016/S0140-6736(02)08284-3. [DOI] [PubMed] [Google Scholar]
- Rex DK, Rahmani EY, Haseman JH, Lemmel GT, Kaster S, Buckley JS. Relative sensitivity of colonoscopy and barium enema for detection of colorectal cancer in clinical practice. Gastroenterology. 1997;112(1):17–23. doi: 10.1016/S0016-5085(97)70213-0. [DOI] [PubMed] [Google Scholar]
- Schabas RE. Colorectal cancer screening in Canada: it’s time to act. Can Med Assoc J. 2003;68(2):178–179. [PMC free article] [PubMed] [Google Scholar]
- Selby JV, Friedman GD, Quesenberry CP, Weiss NS. A case–control study of screening sigmoidoscopy and mortality from colorectal cancer. N Engl J Med. 1992;326(10):653–657. doi: 10.1056/NEJM199203053261001. [DOI] [PubMed] [Google Scholar]
- Sharp L, Little J. Polymorphisms in genes involved in folate metabolism and colorectal neoplasia: a huge review. Am J Epidemiol. 2004;159(5):423–443. doi: 10.1093/aje/kwh066. [DOI] [PubMed] [Google Scholar]
- Simon JB. Screening colonoscopy: is it time? Can Med Assoc J. 2000;163(10):1277–1278. [PMC free article] [PubMed] [Google Scholar]
- Sirovich BE, Schwartz LM, Woloshin S. Screening men for prostate and colorectal cancer in the United States: does practice reflect the evidence? JAMA. 2003;289(11):1414–1420. doi: 10.1001/jama.289.11.1414. [DOI] [PubMed] [Google Scholar]
- Slattery ML, Edwards SL, Ma KN, Friedman GD. Colon cancer screening, lifestyle, and risk of colon cancer. Cancer Causes Control. 2000;11(6):555–563. doi: 10.1023/A:1008924115604. [DOI] [PubMed] [Google Scholar]
- Slattery ML, Kinney AY, Levin TR. Factors associated with colorectal cancer screening in a population-based study: the impact of gender, health care source, and time. Prev Med. 2004;38(3):276–283. doi: 10.1016/j.ypmed.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Statistics Canada (2006) Age and sex distribution of the Canadian population. http://www12.statcan.gc.ca/english/census06/data/topics/Index.cfm
- Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, Barnetson RA, Theodoratou E, Cetnarskyj R, Cartwright N, Semple C, Clark AJ, Reid FJ, Smith LA, Kavoussanakis K, Koessler T, Pharoah PD, Buch S, Schafmayer C, Tepel J, Schreiber S, Volzke H, Schmidt CO, Hampe J, Chang-Claude J, Hoffmeister M, Brenner H, Wilkening S, Canzian F, Capella G, Moreno V, Deary IJ, Starr JM, Tomlinson IP, Kemp Z, Howarth K, Carvajal-Carmona L, Webb E, Broderick P, Vijayakrishnan J, Houlston RS, Rennert G, Ballinger D, Rozek L, Gruber SB, Matsuda K, Kidokoro T, Nakamura Y, Zanke BW, Greenwood CM, Rangrej J, Kustra R, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40(5):631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC. Statistical methods in genetic epidemiology. New York: Oxford University Press; 2004. [Google Scholar]
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, Corgi C, Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier JB, Houlston R. A genome-wide association scan of tag snps identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39(8):984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- Tomlinson IPM, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, Jaeger E, Fielding S, Rowan A, Vijayakrishnan J, Domingo E, Chandler I, Kemp Z, Qureshi M, Farrington SM, Tenesa A, Prendergast JGD, Barnetson RA, Penegar S, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop DT, Gray R, Maher ER, Lucassen A, Kerr D, Evans DGR, Consortium C, Schafmayer C, Buch S, Völzke H, Hampe J, Schreiber S, John U, Koessler T, Pharoah P, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellví-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, Consortium E, Försti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JWC, Cheng KK, Sham PC, Luk J, Agúndez JAG, Ladero JM, de la Hoya M, Caldés T, Niittymäki I, Tuupanen S, Karhu A, Aaltonen L, Cazier JB, Campbell H, Dunlop MG, Houlston RS (2008) A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet 40(5):623–630 [DOI] [PubMed]
- UK Flexible Sigmoidoscopy Screening Trial Investigators Single flexible sigmoidoscopy screening to prevent colorectal cancer: baseline findings of a UK multicentre randomised trial. Lancet. 2002;359(9314):1291–1300. doi: 10.1016/S0140-6736(02)08268-5. [DOI] [PubMed] [Google Scholar]
- UK Trial of Early Detection of Breast Cancer Group First results on mortality reduction in the UK trial of early detection of breast cancer. Lancet. 1988;2(8608):411–416. [PubMed] [Google Scholar]
- Vineis P, Schulte P, McMichael AJ. Misconceptions about the use of genetic tests in populations. Lancet. 2001;357(9257):709–712. doi: 10.1016/S0140-6736(00)04136-2. [DOI] [PubMed] [Google Scholar]
- Williams-Jones B. Where there’s a web, there’s a way: commercial genetic testing and the internet. Community genetics. 2003;6(1):46–57. doi: 10.1159/000069538. [DOI] [PubMed] [Google Scholar]
- Yang Q, Khoury MJ, Botto L, Friedman JM, Flanders WD. Improving the prediction of complex diseases by testing for multiple disease-susceptibility genes. Am J Hum Genet. 2003;72(3):636–649. doi: 10.1086/367923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Khoury MJ, Friedman J, Little J, Flanders WD. How many genes underlie the occurrence of common complex diseases in the population? Int J Epidemiol. 2005;34(5):1129–1137. doi: 10.1093/ije/dyi130. [DOI] [PubMed] [Google Scholar]
- Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, Sundararajan S, Roumy S, Olivier JF, Robidoux F, Sladek R, Montpetit A, Campbell P, Bezieau S, O’Shea AM, Zogopoulos G, Cotterchio M, Newcomb P, McLaughlin J, Younghusband B, Green R, Green J, Porteous ME, Campbell H, Blanche H, Sahbatou M, Tubacher E, Bonaiti-Pellie C, Buecher B, Riboli E, Kury S, Chanock SJ, Potter J, Thomas G, Gallinger S, Hudson TJ, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]


