Abstract
Background
Maltose-1-phosphate was detected in Mycobacterium bovis BCG extracts in the 1960's but a maltose-1-phosphate synthetase (maltokinase, Mak) was only much later purified from Actinoplanes missouriensis, allowing the identification of the mak gene. Recently, this metabolite was proposed to be the intermediate in a pathway linking trehalose with the synthesis of glycogen in M. smegmatis. Although the M. tuberculosis H37Rv mak gene (Rv0127) was considered essential for growth, no mycobacterial Mak has, to date, been characterized.
Results
The sequence of the Mak from M. bovis BCG was identical to that from M. tuberculosis strains (99-100% amino acid identity). The enzyme was dependent on maltose and ATP, although GTP and UTP could be used to produce maltose-1-phosphate, which we identified by TLC and characterized by NMR. The Km for maltose was 2.52 ± 0.40 mM and 0.74 ± 0.12 mM for ATP; the Vmax was 21.05 ± 0.89 μmol/min.mg-1. Divalent cations were required for activity and Mg2+ was the best activator. The enzyme was a monomer in solution, had maximal activity at 60°C, between pH 7 and 9 (at 37°C) and was unstable on ice and upon freeze/thawing. The addition of 50 mM NaCl markedly enhanced Mak stability.
Conclusions
The unknown role of maltokinases in mycobacterial metabolism and the lack of biochemical data led us to express the mak gene from M. bovis BCG for biochemical characterization. This is the first mycobacterial Mak to be characterized and its properties represent essential knowledge towards deeper understanding of mycobacterial physiology. Since Mak may be a potential drug target in M. tuberculosis, its high-level production and purification in bioactive form provide important tools for further functional and structural studies.
Background
The loss of human lives for tuberculosis (TB) remains unhampered as a result of the synergy with the AIDS pandemic and the emergence of drug resistant strains. Each year, there are almost 2 million TB-related deaths worldwide and nearly 9 million people become infected. Moreover, one third of the world's population is estimated to be infected with TB [1]. The available therapies target only a few bacterial functions and the growing numbers of extensively drug-resistant strains urges for the identification of novel pathways and new drug targets. While promising antimycobacterials have been recently synthesized, they are still far from being available for inclusion in TB therapies [2-4].
Maltose-1-phosphate was initially identified in Mycobacterium bovis BCG cell extracts [5]. An enzyme synthesizing maltose-1-phosphate from maltose and ATP was later identified in Actinoplanes missouriensis and named maltokinase (Mak) [6]. Apparently, it was constitutively expressed, regardless of the sugars present in the growth medium, thus unlikely to play an essential role in maltose catabolism [7]. A recombinant Mak from Streptomyces coelicolor was also partially characterized [8]. Maltose-1-phosphate could also be synthesized without the requirement of ATP in E. coli, where it was proposed to be involved in the regulation of maltose metabolism [9]. The maltokinase gene (mak) is present in most of the available mycobacterial genomes and it was considered essential for the growth of M. tuberculosis H37Rv [10]. Moreover, mak is always linked with the trehalose synthase gene (treS) and in some organisms fused into one gene. This highly conserved sequential arrangement is found in many bacterial groups and suggests a shared metabolic route.
Trehalose is a ubiquitous disaccharide found in bacteria, archaea, fungi, plants and in some invertebrates, where it plays a multitude of biological roles [11,12]. In mycobacteria and related organisms, trehalose is synthesized by three different pathways and serves a functional role as a compatible solute under osmotic or thermal stress, and a structural role in cell wall components [13-15]. Mycobacterium smegmatis mutants defective in the three pathways are unable to grow unless trehalose in supplemented to the growth medium [14]. However, each pathway seems to have a specific role and hierarchy in closely related actinobacterial species. While the OtsA/OtsB pathway seems to be essential for the survival of M. tuberculosis in infected mice, the inactivation of the TreY/TreZ did not cause obvious phenotypic effects under the examined conditions [16]. However, the inactivation of TreS promoted increased survival of the mutant in mice, when compared to the wild-type strain [16]. In Corynebacterium glutamicum, the TreY/TreZ pathway dominates, the OtsA/OtsB plays only a minor accessory role and the TreS pathway seems to be involved in trehalose degradation [17,18]. Single mutations targeting each of the pathways in M. smegmatis resulted in no apparent phenotypic defects indicating that the three pathways are redundant in this organism [14].
The TreS from M. smegmatis is capable of interconverting trehalose and maltose [19]. TreS was also shown to catalyze the formation of trehalose from glycogen via its amylase activity and with maltose as the intermediate [20]. Moreover, the synthesis of glycogen from trehalose has also recently been reported in M. smegmatis [21]. In this proposed three-step pathway, TreS and Mak sequentially convert trehalose into maltose and further into maltose-1-phosphate while a final maltosyltransferase transfers maltose from maltose-1-phosphate to glycogen. Although M. smegmatis has an essential trehalase to degrade trehalose directly into glucose [22], it has been suggested that it is energetically advantageous to convert trehalose to glycogen rather than to hydrolyze it to glucose [21], which cannot accumulate in cells.
Despite of the increasing numbers of genome sequences showing that mak genes are widely distributed throughout the bacterial world, there is still a tremendous lack of biochemical data on the corresponding enzymes as only the Mak from A. missouriensis and Streptomyces coelicolor have been biochemically characterized [7,8]. Therefore, we cloned and successfully expressed in E. coli the mak gene from M. bovis BCG, the organism where maltose-1-phosphate was firstly detected. We purified the recombinant bioactive enzyme and determined its biochemical and kinetic properties to further understand its role in mycobacterial physiology.
Results
Identification of mycobacterial maltokinase (Mak) and sequence analysis
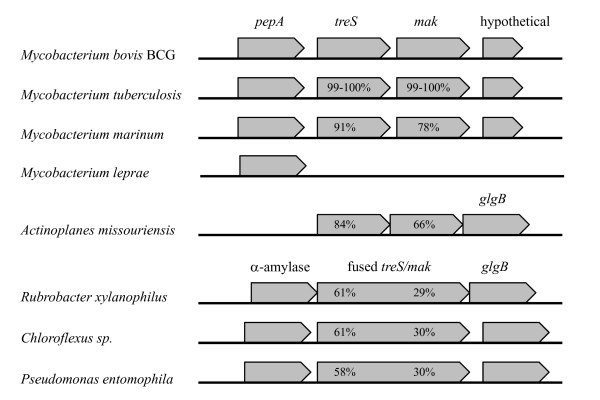
BLAST analysis with the Actinoplanes missouriensis and Streptomyces coelicolor maltokinase sequences within mycobacterial genome databases showed that the mak gene was annotated as a hypothetical protein/trehalose synthase fused maltokinase gene with homologues in most of these genomes. The amino acid identities of these proteins to the A. missouriensis sequence were between 62% and 67%, and 39% to 41% to the S. coelicolor Mak sequence. The maltokinase gene (mak) from M. bovis BCG contained 1368 bp coding for a polypeptide with 455 amino acids with a calculated molecular mass of 49.9 kDa and a calculated isoelectric point of 5.2. The Mak protein was 99 to 100% identical to the homologues from all M. tuberculosis and M. bovis strains with the genomes available (Fig. 1). Additional Mak homologues were found in M. avium (79% amino acid identity), M. kansasii (79%), M. marinum (79%), M. intracellulare (78%), M. ulcerans (78%), M. smegmatis (64%), M. gilvum (60%) and M. vanbaalenii (59%), but not in M. leprae nor in M. abscessus, which also lack treS genes. In other organisms like Rubrobacter xylanophilus or Pseudomonas fluorescens the mak gene is fused with the treS gene (Fig. 1). In M. bovis, M. tuberculosis and M. marinum the mak and treS genes have intergenic regions of about 72 to 102 bp that could accommodate a promoter sequence, implying independent regulation of mak expression. Our attempts to detect consensus promoter motifs in this region indicated about 69 to 75% probability of these elements within the 3' end of the treS gene (Fig. 1).
Figure 1.
Genetic environment of the mak gene in different organisms. Organization of the region containing the mak gene in Mycobacterium bovis BCG, M. tuberculosis, M. marinum, M. leprae, Actinoplanes missouriensis, Rubrobacter xylanophilus, Chloroflexus sp. and Pseudomonas entomophila. Arrows represent genes and their orientation. pepA, serine protease; treS, trehalose synthase; mak, maltokinase; glgB, glycogen branching enzyme; fused treS/mak, probable fused trehalose synthase/maltokinase.
The Mak protein had significant sequence similarity with putative aminoglycoside phosphotransferases from several organisms of the phylum Actinobacteria, namely Kribbella flavida (NCBI accession number YP_003382767, 49% amino acid identity), Actinosynnema mirum (YP_003103727, 45% identity) and Nakamurella multipartita (YP_003202415, 45% identity).
Expression and properties of the recombinant maltokinase
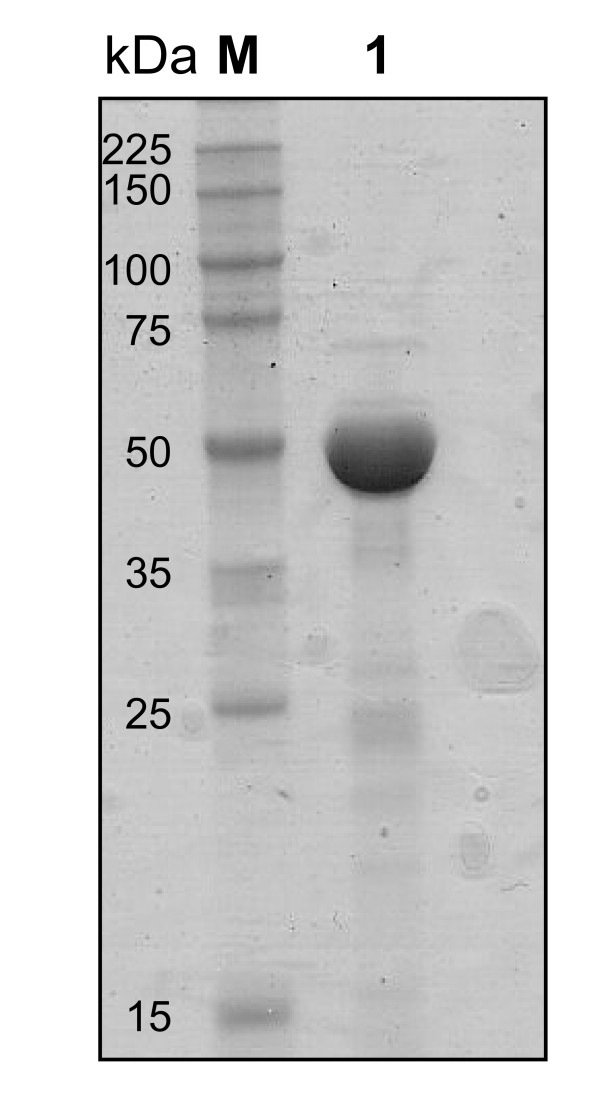
Expression of the M. bovis BCG mak gene in E. coli resulted in the high level production of recombinant His-tagged protein (Fig 2). The Mak identity was confirmed by peptide mass fingerprinting. Gel filtration experiments indicated that the recombinant His-tagged Mak behaved as a monomeric protein in solution, with a molecular mass of about 50.7 ± 4.2 kDa.
Figure 2.
SDS-PAGE showing the recombinant maltokinase (Mak) from M. bovis BCG. Lane 1 - Purified recombinant Mak. M - Protein molecular weight markers.
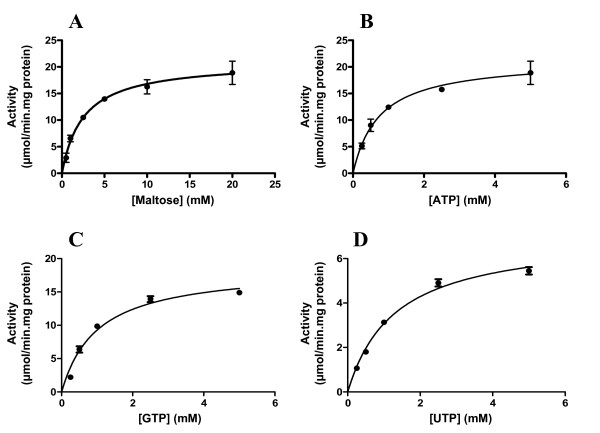
The recombinant Mak used ATP, GTP and UTP as phosphate donors with comparable but decreasing efficiency (Table 1). Residual activity was also detected with ADP and GDP (< 7%). Among the sugar substrates tested, maltose was, by far, the preferred acceptor. Residual activity was also detected with maltotriose, maltoetraose, maltopentaose and maltoheptaose (< 2%), which may be due to maltose contamination. Due to the sequence similarities of Mak with putative aminoglycoside phosphotransferases, the aminoglycoside antibiotics gentamicin, hygromycin B, kanamycin and streptomycin were also tested as possible acceptors of phosphate but no activity was detected. Kinetic experiments showed that the recombinant Mak exhibited Michaelis-Menten kinetics at 37°C with ATP, GTP and UTP up to 5 mM (Table 1 and Fig. 3). Higher concentrations of these phosphate donors were progressively inhibitory, in the reaction mixture containing 20 mM maltose and 10 mM MgCl2. While maximum Mak activity was observed with 5 mM ATP, the activity dropped to 75% with 10 mM ATP and to about 50% with 20 mM ATP.
Table 1.
Kinetic parameters of the recombinant maltokinase from Mycobacterium bovis BCG
| Substrate | Maltose | ATP | GTP | UTP |
|---|---|---|---|---|
| Km (mM) | 2.52 ± 0.40 | 0.74 ± 0.12 | 1.01 ± 0.17 | 1.30 ± 0.13 |
| Vmax(μmol/min.mg-1) | 21.05 ± 0.89 | 21.42 ± 1.03 | 18.65 ± 1.10 | 7.07 ± 0.28 |
Figure 3.
Kinetic studies of Mak activity. Dependence of Mak activity on the concentrations of maltose (A), ATP (B), GTP (C) and UTP (D).
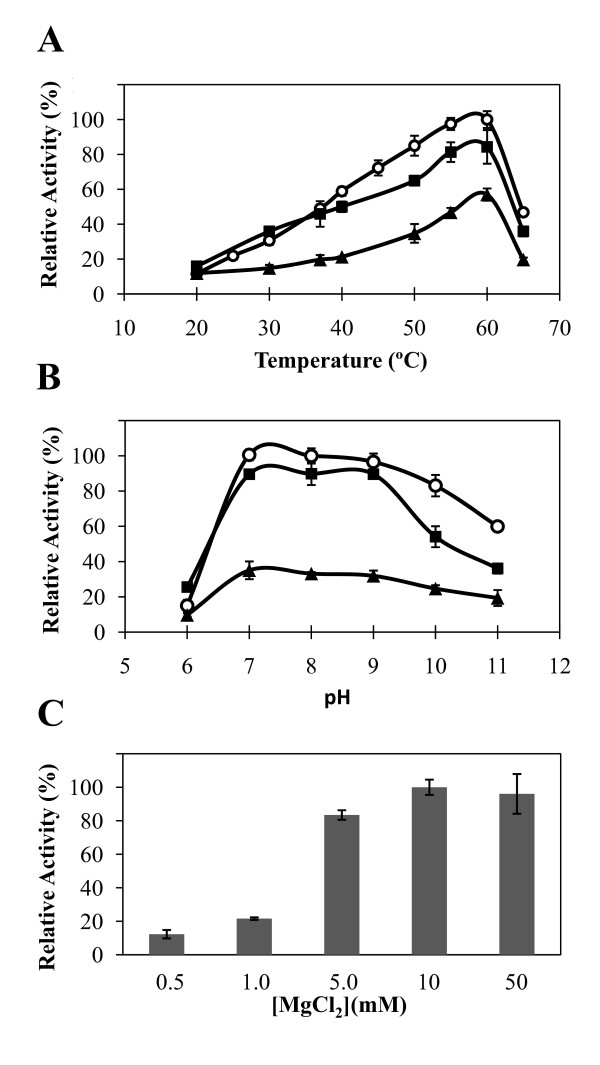
The enzyme was active between 20 and 65°C, with maximal activity at about 60°C (Fig. 4A). At 37°C, the enzyme was active between pH 6 and 11, optimally between pH 7 and 9 with maltose and ATP, GTP or UTP (Fig. 4B). The recombinant Mak was strictly dependent on divalent cations with Mg2+ (10 mM) having the most pronounced stimulatory effect (Fig. 4C). Other divalent cations like Co2+ and Mn2+ also activated Mak, but to lower extent (~41% and ~14% of maximal activity, respectively). The enzyme retained only about 40% of activity after 7 days storage at 4°C in 50 mM BTP (pH 7.5). Most of the activity was also lost after freeze/thawing the enzyme at -20°C in the same buffer. Glycerol strongly inhibited Mak activity, even at very low concentrations (1%). The addition of 10 mM maltose (final concentration) only slightly improved the stability at 4°C to about 50% of maximal activity. However, the addition of 50 mM NaCl dramatically improved the stability of the enzyme, as the residual activity after 1 week at 4°C was still above 90% of maximal activity.
Figure 4.
Effects of temperature, pH and Mg2+ concentration on Mak activity. Temperature profile (A) and pH dependence (B) using ATP (white circle), GTP (black circle) and UTP (black triangle) as phosphate donors. (C) Effect of Mg2+ concentration.
Identification of maltose-1-phosphate by NMR
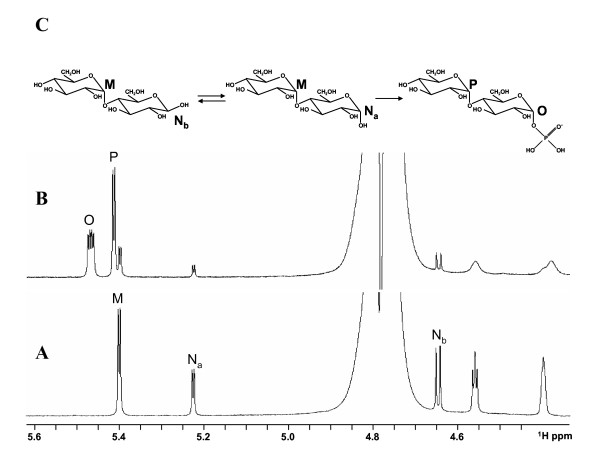
Maltose is composed of two glucose residues linked via an alpha-1,4-glycosidic bond. This means that one of the residues is blocked in the alpha configuration, while the other has a free aldehyde group at position one and therefore can adopt either the alpha or the beta configuration (Fig. 5C). This explains the presence of the three signals in the 1H-NMR spectrum of maltose (Fig. 5A), in the region where protons 1 of hexoses resonate; signal M corresponding to proton 1 of the blocked residue and signals Na and Nb corresponding to proton 1 of unblocked residue in the alpha and beta configurations, respectively. After incubating the reaction mixture with the enzyme (Fig 5B), signals M, Na, and Nb almost disappear and only two new signals arise (signals O and P). These signals present H-H coupling constants 3J1,2 around 3 Hz, meaning that the respective residues are in the alpha configuration. Additionally, resonance O presents a second 7 Hz splitting of the signal, consistent with a H-P coupling constant. This phosphorylation was confirmed by a 1H-31P HSQC spectrum that presents a correlation between signal O and a phosphorus resonance at 1.64 ppm (the region where phosphomonoesters resonate). This means that the reaction product has two hexose residues blocked in the alpha configuration and that one of them is phosphorylated at position 1, thus we conclude to be in the presence of alpha-D-maltose-1-phosphate. This result was further confirmed by the acquisition of a 1H-13C HSQC spectrum and comparison of the 1H and 13C resonances with the literature [6].
Figure 5.
1H-NMR spectra of the Mak reaction. Mixture contained 5 mM each ATP and maltose, 5 mM MgCl2 in 10 mM BTP at pH 8, prior (A) and after (B) the addition of 15 μg of Mak and incubation for 10 min at 37°C. The top panel (C) represents the structures of maltose and maltose-1-phosphate and is labeled according to their assigned resonances.
Discussion
Maltose-1-phosphate was identified for the first time in Mycobacterium bovis BCG [5] but a maltose-1-phosphate synthesizing enzyme (maltokinase, Mak) was only identified much later in Actinoplanes missouriensis [6]. Although the corresponding gene (mak) can now be identified in many bacterial genomes, including those of most mycobacteria, only the Mak from A. missouriensis and S. coelicolor have, so far, been characterized [7,8]. Interestingly, the M. tuberculosis H37Rv mak gene (Rv0127) was considered essential for the pathogen's growth [10]. This information and the absence of biochemical data on mycobacterial maltokinases prompted us to study the enzyme from the slow-growing species M. bovis BCG.
Our results show that, in contrast to the A. missouriensis and S. coelicolor enzymes, which use ATP as the sole phosphate donor (GTP and UTP are used at less than 3% by the A. missouriensis Mak and not used by the S. coelicolor enzyme), the M. bovis BCG maltokinase was able to use ATP, GTP and UTP with comparable efficiency. This substrate flexibility may reflect an absolute requirement for maltose-1-phosphate, corroborating the proposed essentiality of the mak gene in M. tuberculosis [10]. However, lower flexibility of the Mak for NTP substrates may occur in vivo. Similar behavior has been detected between native and recombinant forms of enzymes involved in trehalose metabolism [23,24]. Interestingly, some unprecedented residual activity (< 7%) could also be detected with ADP and GDP as phosphate donors. Like the enzymes from A. missouriensis and S. coelicolor, only maltose served as the acceptor substrate. Although the Km values for maltose were similar for the A. missouriensis and M. bovis BCG Mak (no kinetic data is available for the S. coelicolor Mak), the latter had a Km value for ATP (0.74 mM) slightly higher than that measured for the A. missouriensis Mak (0.54 mM).
A major obstacle for the characterization of the M. bovis BCG maltokinase was its very low stability. Our attempts to stabilize the enzyme with glycerol failed as it severely inhibited the activity. Unlike the native Mak from A. missouriensis, which could be stabilized by maltose, this disaccharide only moderately stabilized the mycobacterial Mak while trehalose was ineffective. Only the addition of 50 mM NaCl to the preparation stabilized the enzyme for characterization. The mycobacterial Mak was, like the A. missouriensis and the S. coelicolor enzymes, a monomer in solution. While no data is available for the cation dependence of the S. coelicolor Mak, both the A. missouriensis and the M. bovis BCG enzymes were dependent on Mg2+ ions for maximal activity. The cations Co2+ or Mn2+ could partially replace Mg2+ while Zn2+ was inhibitory. Divalent cations seem to play an essential role in the activity of kinases, namely in the stabilization of the negatively charged phosphate donor groups [7]. The temperature profile for activity of the recombinant Mak from M. bovis BCG was similar to that of the native Mak from A. missouriensis, with maximum near 60°C. The S. coelicolor recombinant Mak had maximal activity at about 45°C. All three enzymes had optimal activity between pH 7 and 9 [7,8], which is also the optimum pH range for the activity of other kinases [7]. The Mak from M. bovis BCG had no detectable aminoglycoside phosphotransferase activity, which is in agreement with the data obtained with the S. coelicolor Mak [8]. It is likely that many of the putative aminoglycoside phosphotransferases with significant amino acid identity with maltokinases (>40%) are incorrectly annotated and may instead have maltokinase activity.
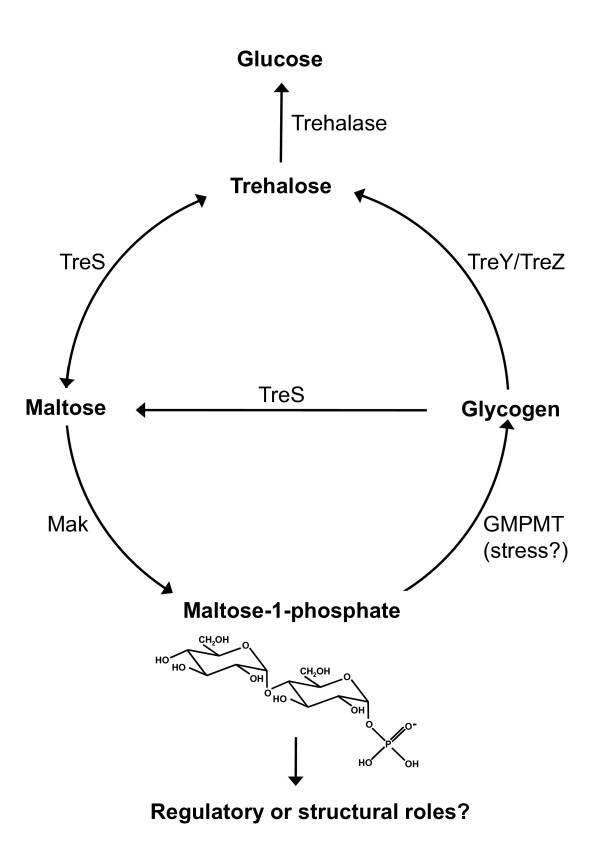
The frequent association of the mak gene with the trehalose synthase (treS) gene, either as a bicystronic unit or as a fused gene, strongly suggests a synergistic action and a common biochemical pathway. While such a pathway has been previously proposed [8], very recent research [21] corroborate this hypothesis by demonstrating that M. smegmatis converts excess trehalose into glycogen through the sequential action of TreS, Mak and of a maltosyltransferase (Fig. 6). The latter enzyme was shown to use maltose-1-phosphate as the substrate for the transfer of maltose to glycogen [21]. The genetic organization in A. missouriensis lend additional support to this hypothesis since a glycogen-branching enzyme gene (glgB) is located immediately downstream the contiguous treS and mak genes. In Rubrobacter xylanophilus, a bacterium that constitutively accumulates high levels of trehalose, a glgB gene is also located immediately downstream a fused treS-mak bifunctional gene and a similar function may be anticipated. Interestingly, the M. smegmatis TreS was also found to possess amylase activity and capable to catalyze the synthesis of trehalose from glycogen, with maltose as intermediate [20]. Curiously, Mycobacterium leprae, in which glycogen has not been reported, lacks both treS and mak homologues [25]. However, the reason for the missing glycogen may be explained by the absence of other genes involved in glycogen synthesis like glgB or glgP, which are pseudogenes in M. leprae. On the other hand, both M. gilvum and M. vanbaalenii possess a mak but lack a treS gene. Unfortunately no data on trehalose or glycogen synthesis is available for these organisms. Nevertheless, among the mycobacterial enzymes involved in this metabolic circuit (trehalose → maltose → maltose-1-phosphate → glycogen → maltose → trehalose) (Fig. 6), all enzymes but the maltokinase have been biochemically studied [13,19,21].
Figure 6.
Metabolic circuit between trehalose, maltose and glycogen and proposal of alternative roles for maltose-1-phosphate. Data adapted from references [13,20,21]. Mak, maltokinase, TreS, trehalose synthase, TreY/TreZ, maltooligosyltrehalose synthase/trehalohydrolase, GMPMT, α-1,4-glucan: maltose-1-P maltosyltransferase.
Mycobacteria possess three different pathways for the synthesis of trehalose [13,16]. This disaccharide is constitutively present in the cytoplasm of mycobacterial cells, an essential structural component of cell walls and its levels increase during osmotic and thermal stress [13-15]. Although the TreS has much higher affinity for maltose than for trehalose [19], in most cases it seems to be involved in trehalose degradation [16] and has only been implicated in trehalose biosynthesis in M. smegmatis [14]. M. smegmatis possesses an essential trehalase that converts trehalose into glucose [22] (Fig. 6). Elbein and coworkers argue that it is energetically favorable to convert trehalose to glycogen by the recently discovered TreS-Mak-maltosyltransferase pathway (Fig. 6) [21]. However, normal ATP intracellular concentrations within the 1 to 10 mM range [26] are inhibitory of the maltosyltransferase (GMPMT) activity [21] but not of the Mak activity. These authors also propose that the role of this pathway is to convert excess trehalose into glycogen during stress conditions. Therefore, under non-stressing conditions, in which the ATP levels rise, maltose-1-phosphate cannot be the substrate for glycogen, which may be synthesized via ADP-glucose [21]. Since mycobacteria contain other alpha-1,4-glucans and polysaccharides [25], it is tempting to speculate that maltose-1-phosphate may also be used as donor of maltose units for those macromolecules, under specific conditions (Fig. 6). Moreover, the constitutive expression of Mak in A. missouriensis [7] and the suggested essentiality of the M. tuberculosis mak gene (Rv0127) indicate alternative roles for maltose-1-phosphate in mycobacterial physiology. This is also supported by the fact that only mak, but not the treS gene (Rv0126), was considered essential for growth. Moreover, the presence of a putative promoter within the terminal part of the treS gene indicating independent regulation of mak expression, also suggests an alternative role for the Mak and maltose-1-phosphate in mycobacteria. Since trehalose-6-phosphate is an important signaling molecule in yeast and plant metabolism [12,27], maltose-1-phosphate could, hypothetically, play a similar role in mycobacteria. In fact, the involvement of maltose-1-phosphate in the regulation of sugar metabolism in E. coli has been proposed earlier but not confirmed experimentally [9].
Several questions remain unanswered and the role(s) of maltose-1-phosphate in mycobacterial metabolism only now begins to be unraveled. While further studies are required to answer these questions, the biochemical properties of the maltokinase from M. bovis BCG, the first of this bacterial group to be characterized, provide an important step towards the clarification of its role in mycobacterial metabolism.
Conclusions
Although the maltokinase gene was considered essential for M. tuberculosis growth [10], implying that the corresponding enzyme might be a suitable target for the design/development of anti-mycobacterial drugs, no mycobacterial maltokinase had been characterized to date. The fast and straightforward method for high-level production and purification of bioactive maltokinase from M. bovis BCG, a major bottleneck in mycobacterial research, and the determination of its biochemical properties, some of which are unique among these type of enzymes, contribute to the knowledge of these enzyme's abilities and provide insights about its unexplored role in mycobacterial physiology.
Methods
Identification, sequence analysis, cloning and functional overexpression of the maltokinase gene (mak) from Mycobacterium bovis BCG, strains and growth conditions
To identify the mak gene in mycobacterial genomes we used the amino acid sequences of the Actinoplanes missouriensis and Streptomyces coelicolor maltokinases (GenPept accession number AAQ01690 and CAA04602, respectively) in BLAST searches at the National Center for Biotechnology Information (NCBI, http://blast.ncbi.nlm.nih.gov/Blast.cgi) database. Promoter identification was carried out using the prokaryotic promoter prediction software from Berkeley University, available at http://www.fruitfly.org. The mak gene was amplified from the chromosomal DNA from M. bovis BCG (DSM 43990) obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany), with a pair of primers based on the gene sequence retrieved from the Institut Pasteur database http://genolist.pasteur.fr. An NdeI restriction site (highlighted in bold) was added to the forward primer MtuNde (5'-CTTACATATGACTCGGTCGGACACGC-3'). A HindIII restriction site (highlighted in bold) was added to the reverse primer MtuHind (5'-ATTAAGCTTGCTAGCGGTCAGGCGGG-3'). The stop codon was removed from the reverse primer to allow the translation of a C-terminal 6 × His-tag encoded by the expression vector pET30a (Novagen). PCR was carried out with the AccuPrime GC-Rich DNA Polymerase (Invitrogen). DNA (200 ng) was denatured by 95°C for 5 min followed by 30 cycles of 1 min denaturation step at 95°C, 1 min annealing step at 62°C, and 1.5 min extension step at 72°C. The PCR product was purified from agarose gel (NZYTech, Portugal), digested and cloned into pET30a. The construct was sequenced to confirm the identity of the insert (AGOWA, Berlin, Germany), and transformed into E. coli BL-21, which was used as host for expression of the mak gene. Recombinant E. coli was grown in a 5L fermentor, with continuous aeration and stirred at 180 rpm at 37°C, pH 7.0 in LB medium with kanamycin (30 μg/ml), to mid-exponential phase of growth (OD610 = 0.8). IPTG was added at a final concentration of 0.5 mM to induce gene expression, and temperature was reduced to 20°C. The cells were harvested 18 h later by centrifugation (9000 × g, 10 min, 4°C).
Preparation of cell-free extracts
Escherichia coli cells carrying the recombinant maltokinase (Mak) from M. bovis BCG were suspended in 25 mM Bis-tris propane buffer (BTP) at pH 7.5 with 50 mM NaCl for enzyme assays, or in 20 mM sodium phosphate buffer at pH 7.4 with 0.5 M NaCl and 20 mM imidazole, for protein purification. A protease inhibitor cocktail (Roche), 10 μg/ml DNAse I and 5 mM MgCl2 were added to the suspension. Cells were disrupted twice in a French-press cell followed by centrifugation (15000 × g, 4°C, 30 min).
Enzyme assays
The activity of the recombinant Mak from M. bovis BCG in E. coli extracts and during purification was detected after 15 min at 37°C in reaction mixtures (50 μl) containing 25 μl of cell-free extract, 3.0 mM (each) of ATP and maltose, and 10 mM MgCl2 in 50 mM BTP, pH 8.0. The synthesis of maltose-1-phosphate was monitored by thin-layer chromatography (TLC) with solvent systems composed by acetic acid/ethyl acetate/water/ammonia 25% (6:6:2:1, v/v) and butanol/ethanol/water (5:3:2, v/v). Trehalose-6-phosphate, ATP, GTP, UTP, ADP, GDP, AMP and maltose standards were used for comparative purposes. The maltose-1-phosphate formed was identified by NMR as described below. E. coli cell-free extracts carrying an empty vector were used as negative controls. Protein concentration was determined by the Bradford method [28].
Purification of recombinant Mak from M. bovis BCG
The His-tagged recombinant Mak from M. bovis BCG was purified in a prepacked Ni-Sepharose high-performance column (His-Prep FF 16/10) equilibrated with 20 mM sodium phosphate, pH 7.4, 0.5 M NaCl, and 20 mM imidazole. Elution was carried out with 500 mM imidazole and the purity of the fractions was determined by SDS-PAGE. The purest active fractions were pooled, diluted ten times with 25 mM BTP at pH 7.4 and loaded into a Q-Sepharose fast-flow column (Hi-Load FF 16/10), equilibrated with 25 mM BTP at pH 7.4 with 50 mM NaCl, and eluted by a linear gradient of NaCl (50 to 500 mM). The purity of the fractions was determined by SDS-PAGE and the purest active fractions were pooled, concentrated by ultracentrifugation in 30 kDa cutoff centricons (Amicon), equilibrated with 25 mM BTP at pH 7.4 with 200 mM NaCl, and loaded into a Superdex 200 fast-flow column equilibrated with the same buffer. After SDS-PAGE analysis the active pure fractions were concentrated and equilibrated with 50 mM BTP at pH 7.4 with 50 mM NaCl. Protein content of the samples was determined by the Bradford assay [28]. The identity of the purified maltokinase from M. bovis BCG was confirmed by Peptide Mass Fingerprinting (IPATIMUP Proteomics Unit, Porto, Portugal).
Characterization of the recombinant Mak from M. bovis BCG
The substrate specificity of the recombinant Mak from M. bovis BCG was determined using glucose, galactose, mannose, maltose, isomaltose, trehalose, maltotriose, maltotetraose, maltopentaose and maltoheptaose as possible acceptors and with ADP, CDP, GDP, TDP, UDP, ATP, CTP, GTP, TTP and UTP as possible phosphate donors (all from Sigma-Aldrich). Since the Mak protein had high sequence identity (>40%) with putative aminoglycoside phosphotransferases, this activity was also tested using the aminoglycoside antibiotics gentamicin, kanamycin, streptomycin and hygromycin B as possible phosphate acceptors (all from Sigma-Aldrich). The reaction mixtures (50 μl) containing (0.5 μg) pure recombinant Mak, 3 mM of each substrate, and 10 mM MgCl2 in 50 mM BTP at pH 8, were incubated at 37°C for 10 min. The products were visualized by TLC as described above.
Temperature and pH profile, effect of cations and thermal stability of Mak were determined by the addition of 0.5 μg of Mak to 50 μl reaction mixtures containing the appropriate buffer, 3 mM maltose and 3 mM NTP and stopped at different times by cooling on ethanol-ice. The Mak was inactivated by the addition of 5 μl of 1N HCl and neutralized by 5 μl of 1N NaOH. Controls were performed to account for possible NTP degradation following acid treatment. The amount of NDP released was determined at 340 nm after incubation of the sample with 3 U of pyruvate kinase and lactate dehydrogenase, 0.3 mM NADH and 2.5 mM phosphoenolpyruvate (all from Sigma-Aldrich) in 1 ml mixture (total volume) for 10 min at 30°C [29]. The temperature profile was determined between 20 and 65°C in 50 mM BTP at pH 7.0, with 10 mM MgCl2. The effect of pH was determined at 37°C in 50 mM BTP (pH 6.0 to 9.0) and in 50 mM CAPS (pH 9.0 to 11.0), with 10 mM MgCl2. The effect of cations was examined by incubating reaction mixture containing the appropriate substrates, with the chloride salts of Mg2+, Mn2+, Co2+, Zn2+ (0.5 to 50 mM) or without cations, at 37°C.
The kinetic parameters for the recombinant Mak were determined by measuring the amount of NDP released, as described above. The Km values for the substrates ATP, GTP, UTP and maltose were determined at 37°C, from Lineweaver-Burk plots. All experiments were performed in triplicate.
The molecular mass of the recombinant Mak from M. bovis BCG was estimated by gel filtration on a Superdex 200 column and the molecular mass standards were aprotinin (6.5 kDa), ribonuclease (13.7 kDa), carbonic anhydrase (29 kDa), ovalbumine (43 kDa), conalbumine (75 kDa), aldolase (158 kDa). Blue Dextran 2000 was used to determine the void volume (Amersham).
Enzyme stabilization assays
Pure recombinant Mak was stored in several aliquots containing 50 mM BTP at pH 8.0, and one of the following substances (final concentration) was added: 20% glycerol, 50% glycerol, 10 mM maltose, 10 mM trehalose or 50 mM NaCl. The initial Mak activity was measured at 37°C in 50 mM BTP at pH 8.0, with 10 mM MgCl2 and 3 mM (each) of ATP and maltose. Aliquots were stored at 4 and -20°C for seven days. Control aliquots were kept at 4° and -20°C only in 50 mM BTP at pH 8.0. After seven days storage the residual activity was measured for each of the conditions described above.
NMR spectroscopy
For the NMR experiments, 10% (v/v) of D2O was added to the reaction mixture containing 5 mM (each) of ATP and maltose, 5 mM MgCl2 in 10 mM BTP at pH 8.0. After spectral acquisition, 15 μg of enzyme were added to the NMR tube and incubated for 10 min at 37°C. All spectra were acquired on a Bruker AVANCE III 800 spectrometer (Bruker, Rheinstetten, Germany) working at a proton operating frequency of 800.33 MHz, equipped with a four channel 5 mm inverse detection probe head with pulse-field gradients along the Z axis.
Spectra were run at 25°C using standard Bruker pulse programs. 1H and 13C chemical shifts are referenced to 3-(trimethylsilyl)propane sulfonic acid, and 31P resonances to external 85% phosphoric acid. In the 1H-13C and the 1H-31P heteronuclear two-dimensional single quantum coherence (HSQC) spectra, delays of 3.44 and 71.4 ms were used for evolution of the JH, C and JH, P couplings, respectively; proton decoupling was, in both cases, achieved using the GARP4 sequence [30].
Abbreviations
Mak: maltokinase; TreS: trehalose synthase; GMPMT: α-1,4-glucan:maltose-1-P maltosyltransferase; TreY/TreZ: maltooligosyltrehalose synthase/trehalohydrolase; OtsA/OtsB: trehalose-phosphate synthase/phosphatase; glgB: gene for glycogen branching enzyme; glgP: glycogen phosphorylase gene; TLC: thin layer chromatography; NMR: nuclear magnetic resonance; SDS-PAGE: sodium dodecyl sulfate polyacrylamide gel electrophoresis; BTP: Bis-Tris propane buffer.
Authors' contributions
VM and NE designed the concept and experiments of this study. VM cloned, expressed and purified the maltokinase. VM and AM performed the experiments for the determination of the biochemical and kinetic data. PL designed the NMR experiments and acquired the data. VM, PL and NE were involved in the analysis and interpretation of the data. VM, NE and MSC drafted the manuscript, whereas the other authors helped to draft the manuscript. All authors have approved the final manuscript.
Contributor Information
Vítor Mendes, Email: vitor_g_mendes@yahoo.com.
Ana Maranha, Email: anamaranha@gmail.com.
Pedro Lamosa, Email: lamosa@itqb.unl.pt.
Milton S da Costa, Email: milton@ci.uc.pt.
Nuno Empadinhas, Email: numenius@cnc.uc.pt.
Acknowledgements
V. Mendes acknowledges a scholarship from FCT (SFRH/BD/36373/2007). We thank Hugo Osório (IPATIMUP Proteomics Unit, Porto, Portugal) for the mass spectrometry. The NMR spectrometers are part of The National NMR Network (REDE/1517/RMN/2005), supported by "Programa Operacional Ciência e Inovação (POCTI) 2010" and Fundação para a Ciência e a Tecnologia (FCT).
References
- Dye C. Doomsday postponed? Preventing and reversing epidemics of drug-resistant tuberculosis. Nat Rev Microbiol. 2009;7(1):81–87. doi: 10.1038/nrmicro2048. [DOI] [PubMed] [Google Scholar]
- Makarov V, Manina G, Mikusova K, Mollmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science. 2009;324(5928):801–804. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras-Dominguez M, Kim J, Genovesio A, Carralot JP, Ewann F, Kim EH. High content screening identifies decaprenyl-phosphoribose 2' epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathog. 2009;5(10):e1000645. doi: 10.1371/journal.ppat.1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balganesh TS, Alzari PM, Cole ST. Rising standards for tuberculosis drug development. Trends Pharmacol Sci. 2008;29(11):576–581. doi: 10.1016/j.tips.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Narumi K, Tsumita T. Identification of alpha, alpha-trehalose 6,6'-dimannosylphosphate and alpha-maltose 1-phosphate of Mycobacteria. J Biol Chem. 1967;242(9):2233–2239. [PubMed] [Google Scholar]
- Drepper A, Peitzmann R, Pape H. Maltokinase (ATP:maltose 1-phosphotransferase) from Actinoplanes sp.: demonstration of enzyme activity and characterization of the reaction product. FEBS Lett. 1996;388(2-3):177–179. doi: 10.1016/0014-5793(96)00554-6. [DOI] [PubMed] [Google Scholar]
- Niehues B, Jossek R, Kramer U, Koch A, Jarling M, Schroder W, Pape H. Isolation and characterization of maltokinase (ATP: maltose 1-phosphotransferase) from Actinoplanes missouriensis. Arch Microbiol. 2003;180(4):233–239. doi: 10.1007/s00203-003-0575-y. [DOI] [PubMed] [Google Scholar]
- Jarling M, Cauvet T, Grundmeier M, Kuhnert K, Pape H. Isolation of mak1 from Actinoplanes missouriensis and evidence that Pep2 from Streptomyces coelicolor is a maltokinase. J Basic Microbiol. 2004;44(5):360–373. doi: 10.1002/jobm.200410403. [DOI] [PubMed] [Google Scholar]
- Decker K, Gerhardt F, Boos W. The role of the trehalose system in regulating the maltose regulon of Escherichia coli. Mol Microbiol. 1999;32(4):777–788. doi: 10.1046/j.1365-2958.1999.01395.x. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48(1):77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Iturriaga G, Suarez R, Nova-Franco B. Trehalose metabolism: from osmoprotection to signaling. Int J Mol Sci. 2009;10(9):3793–3810. doi: 10.3390/ijms10093793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Primavesi LF, Jhurreea D, Zhang Y. Trehalose metabolism and signaling. Annu Rev Plant Biol. 2008;59:417–441. doi: 10.1146/annurev.arplant.59.032607.092945. [DOI] [PubMed] [Google Scholar]
- De Smet KA, Weston A, Brown IN, Young DB, Robertson BD. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology. 2000;146(Pt 1):199–208. doi: 10.1099/00221287-146-1-199. [DOI] [PubMed] [Google Scholar]
- Woodruff PJ, Carlson BL, Siridechadilok B, Pratt MR, Senaratne RH, Mougous JD, Riley LW, Williams SJ, Bertozzi CR. Trehalose is required for growth of Mycobacterium smegmatis. J Biol Chem. 2004;279(28):28835–28843. doi: 10.1074/jbc.M313103200. [DOI] [PubMed] [Google Scholar]
- Takayama K, Wang C, Besra GS. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev. 2005;18(1):81–101. doi: 10.1128/CMR.18.1.81-101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy HN, Stewart GR, Mischenko VV, Apt AS, Harris R, McAlister MS, Driscoll PC, Young DB, Robertson BD. The OtsAB pathway is essential for trehalose biosynthesis in Mycobacterium tuberculosis. J Biol Chem. 2005;280(15):14524–14529. doi: 10.1074/jbc.M414232200. [DOI] [PubMed] [Google Scholar]
- Wolf A, Kramer R, Morbach S. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Mol Microbiol. 2003;49(4):1119–1134. doi: 10.1046/j.1365-2958.2003.03625.x. [DOI] [PubMed] [Google Scholar]
- Tzvetkov M, Klopprogge C, Zelder O, Liebl W. Genetic dissection of trehalose biosynthesis in Corynebacterium glutamicum: inactivation of trehalose production leads to impaired growth and an altered cell wall lipid composition. Microbiology. 2003;149(Pt 7):1659–1673. doi: 10.1099/mic.0.26205-0. [DOI] [PubMed] [Google Scholar]
- Pan YT, Koroth Edavana V, Jourdian WJ, Edmondson R, Carroll JD, Pastuszak I, Elbein AD. Trehalose synthase of Mycobacterium smegmatis: purification, cloning, expression, and properties of the enzyme. Eur J Biochem. 2004;271(21):4259–4269. doi: 10.1111/j.1432-1033.2004.04365.x. [DOI] [PubMed] [Google Scholar]
- Pan YT, Carroll JD, Asano N, Pastuszak I, Edavana VK, Elbein AD. Trehalose synthase converts glycogen to trehalose. FEBS J. 2008;275(13):3408–3420. doi: 10.1111/j.1742-4658.2008.06491.x. [DOI] [PubMed] [Google Scholar]
- Elbein AD, Pastuszak I, Tackett AJ, Wilson T, Pan YT. The last step in the conversion of trehalose to glycogen: A mycobacterial enzyme that transfers maltose from maltose-1-phosphate to glycogen. J Biol Chem. 2010;285(13):9803–9812. doi: 10.1074/jbc.M109.033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JD, Pastuszak I, Edavana VK, Pan YT, Elbein AD. A novel trehalase from Mycobacterium smegmatis - purification, properties, requirements. FEBS J. 2007;274(7):1701–1714. doi: 10.1111/j.1742-4658.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- Cardoso FS, Castro RF, Borges N, Santos H. Biochemical and genetic characterization of the pathways for trehalose metabolism in Propionibacterium freudenreichii, and their role in stress response. Microbiology. 2007;153(Pt 1):270–280. doi: 10.1099/mic.0.29262-0. [DOI] [PubMed] [Google Scholar]
- Nobre A, Alarico S, Fernandes C, Empadinhas N, da Costa MS. A unique combination of genetic systems for the synthesis of trehalose in Rubrobacter xylanophilus: properties of a rare actinobacterial TreT. J Bacteriol. 2008;190(24):7939–7946. doi: 10.1128/JB.01055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg S, Kaur D, Jackson M, Brennan PJ. The glycosyltransferases of Mycobacterium tuberculosis - roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology. 2007;17(6):35–56R. doi: 10.1093/glycob/cwm010. [DOI] [PubMed] [Google Scholar]
- Veech RL, Lawson JW, Cornell NW, Krebs HA. Cytosolic phosphorylation potential. J Biol Chem. 1979;254(14):6538–6547. [PubMed] [Google Scholar]
- Blazquez MA, Lagunas R, Gancedo C, Gancedo JM. Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett. 1993;329(1-2):51–54. doi: 10.1016/0014-5793(93)80191-V. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Qu Q, Lee SJ, Boos W. TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem. 2004;279(46):47890–47897. doi: 10.1074/jbc.M404955200. [DOI] [PubMed] [Google Scholar]
- Shaka AJ, Barker PB, Freeman R. Computer-Optimized Decoupling Scheme for Wideband Applications and Low-Level Operation. J Magn Reson. 1985;64(3):547–552. [Google Scholar]