Abstract
The GTPases Rac1, RhoA and Cdc42 act in concert to control cytoskeleton dynamics1-3. Recent biosensor studies have shown that all three GTPases are activated at the front of migrating cells4-7 and biochemical evidence suggests that they may regulate one another: Cdc42 can activate Rac18, and Rac1 and RhoA are mutually inhibitory9-12. However, their spatiotemporal coordination, at the seconds and single micron dimensions typical of individual protrusion events, remains unknown. Here, we examine GTPase coordination both through simultaneous visualization of two GTPase biosensors and using a “computational multiplexing” approach capable of defining the relationships between multiple protein activities visualized in separate experiments. We found that RhoA is activated at the cell edge synchronous with edge advancement, whereas Cdc42 and Rac1 are activated 2 μm behind the edge with a delay of 40 sec. This indicates that Rac1 and RhoA operate antagonistically through spatial separation and precise timing, and that RhoA plays a role in the initial events of protrusion, while Rac1 and Cdc42 activate pathways implicated in reinforcement and stabilization of newly expanded protrusions.
Our computational multiplexing approach makes the critical assumption that the relationship between GTPase activation and the movements of the cell edge during constitutive protrusion and retraction cycles is preserved within a cell, and among cells. Thus, the initiation of protrusion and retraction can be used as a timing reference to indirectly determine the activation dynamics of multiple Rho GTPases: First, for each GTPase, the timing of activation relative to protrusion/retraction events is determined in separate experiments. Then, the activation timings of different GTPases are aligned, using protrusion/retraction events as a common reference. GTPase activities were imaged in Mouse Embryo Fibroblasts (MEF) using biosensors for Rac14, Cdc425, or RhoA6 (Fig 1a-c; Supplementary Figs S1 and S2). The Rac1 biosensor was improved over the previously published version4, using a fluorescent protein rather than a covalently attached dye. Images were captured at 10 s intervals, sufficient to sample the protrusion-retraction cycles below Nyquist frequency (Supplementary Fig. S3).
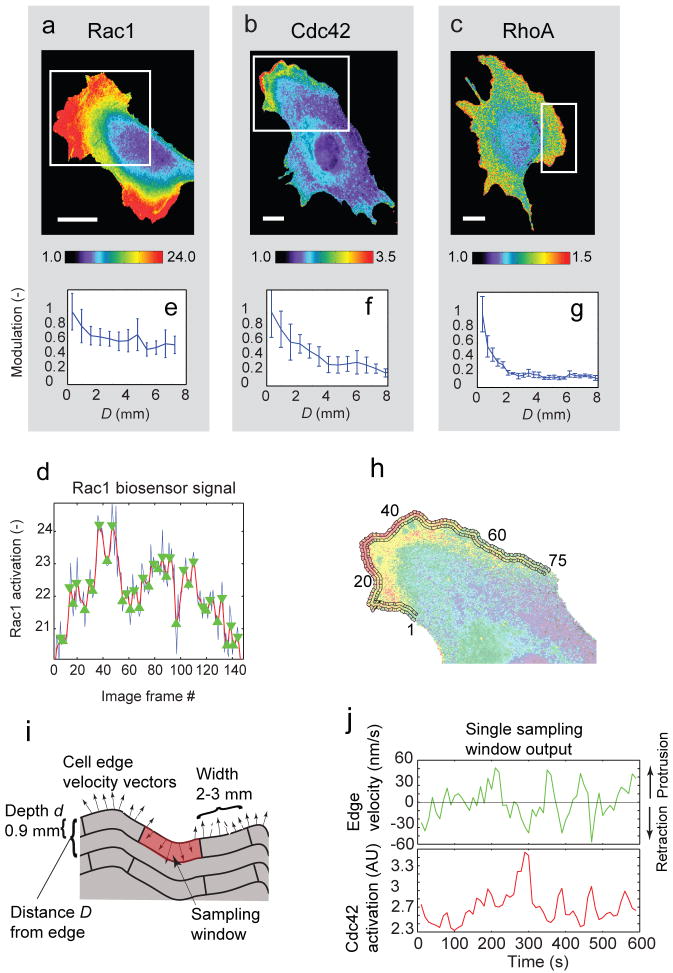
Figure 1. Activation of Rho GTPases in migrating mouse embryonic fibroblasts.
(a) Rac1, (b) Cdc42, and (c) RhoA activation reported by biosensors. White rectangle: Region of interest selected for analysis. Scale bar: 20 μm. (d) Time course (blue line) of Rac1 activation within 0.6 μm from the cell edge, averaged over a 5 μm long portion of the cell edge. Red line: filtered time-course (Gaussian filter, σ=5 frames). Green triangles: local maxima and minima of filtered time-course. (e-g) Modulation of GTPase activation (mean absolute difference between consecutive local extrema of the time-course) as a function of the distance D from the leading edge. Values are normalized to the modulation at the cell edge. Error bars: s.e.m. of n = 6 (Rac1); n = 4 (Cdc42); n = 4 (RhoA) cells. (h) Sampling windows of 0.9 μm depth placed at the cell edge and at D = 1.8 μm from the cell edge. (i) Parameters to define the position and size of a sampling window. In each window, the time course of GTPase activation was recorded (average of ∼10 pixels). For a sampling window at D = 0, a time course of edge velocity was recorded (mean of 6 – 8 measurements). (j) Time courses of edge velocity (green) and GTPase activation (Cdc42, red) recorded in one sampling window.
As reported before4-6, all GTPases, including RhoA, were maximally activated proximal to the leading edge. Visual inspection of time lapse sequences indicated substantial fluctuations in GTPase activity as the cell edges protruded and retracted (Movies 1-6). To quantify the magnitude and location of the fluctuations we extracted time courses of GTPase activity at multiple distances from the cell edge and quantified the extent of signal modulation (Fig 1d). For all three GTPases the modulation was highest at the cell edge, decreased monotonically within ∼2 – 4 μm, and then reached a plateau of baseline activity (Fig 1e-g). Rac1 displayed the least significant decay, mainly because of cell-to-cell variations in the extent of the region with high activation. RhoA, on the other hand, decayed tightly over 2 μm. Thus, GTPase activities are regulated most prominently within a few micrometers from the leading edge, supporting our hypothesis that these fluctuations are linked to cell edge movements.
To investigate how GTPase activation relates to edge movement spatially and temporally, we tracked the position of the cell edge13 and compared edge velocities with the biosensor signal in forty to eighty sampling windows moving with the leading edge (Fig. 1h; Movie7). The window width was set to 1.8 - 3 μm, the distance over which edge movements were correlated along the cell boundary, and 0.9 μm in depth, the distance over which signaling molecules bound to the plasma membrane would be expected to diffuse between consecutive movie frames14. Edge velocities were sampled every 400 nm (see Methods), so that 5 - 7 velocity measurements would fall within a window (Fig. 1i). Thus each window yielded time courses of both edge velocity and GTPase activation level (Fig. 1j), with independent measurements in each adjacent window. Changes in edge velocity appeared to parallel changes in GTPase activity (shown for the example of a Cdc42 data set), but with a time lag.
To study more systematically the potential relationships between edge dynamics (Fig. 2a, d, g) and GTPase activation we copied the data from the individual sampling windows along the cell boundary into activity maps13 (Fig. 2b, c, e, f, h, i). This data representation revealed tight correlation of morphological dynamics and GTPase activation. Protrusion and retraction occurred in a quasi-cyclic fashion with a periodicity of ∼100 s (Supplementary Fig. S3). As reported previously13, protrusion cycles propagated transversally along the leading cell edge (Fig. 2b, e, h). Both Cdc42 and RhoA cycled with similar periodicity (Supplementary Fig. S3) and their activity maps visually resembled those of the morphological dynamics (Fig. 2f, i). Rac1 exhibited a broader time response (Supplementary Fig. S3), consistent with a spatiotemporally more diffuse activation (Fig. 2c). Quiescent regions of the cell edge (i.e. regions Q) exhibited low GTPase activity compared to protruding and retracting regions. Transitions between active and quiescent portions of the cell edge were sharp (2 – 3 μm) and were accompanied by a steep gradient in GTPase activity. Thus, this data suggests an immediate coupling of GTPase activation and cell morphological dynamics, in both time and space.
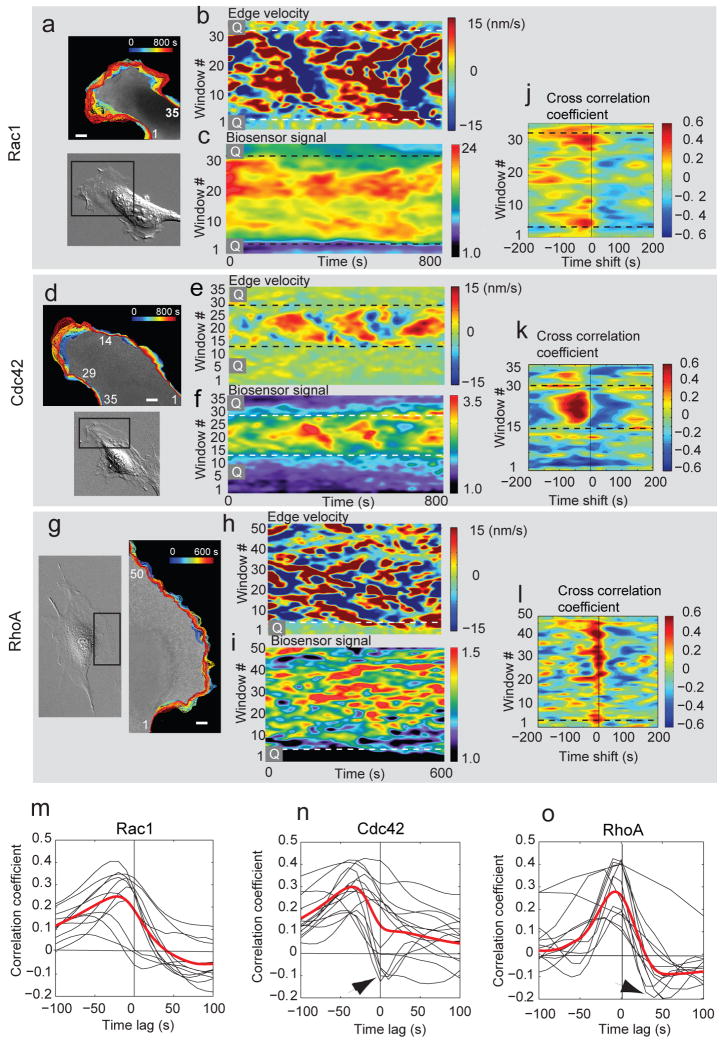
Figure 2. Dynamics of cell edge morphology and GTPase activation.
(a, d, g) Overview of cell morphology (in DIC) and evolution of cell edge positions (color-encoded from blue (early) to red (late) time points). Scale bar: 5 μm. (b, e, h) Activity maps of edge movement: Velocity values along the edge are copied time point by time point into the columns of the map. Color coding: red = protrusion; blue = retraction. Quiescent regions of the cell edge are marked by ‘Q’. (c, f, i) Activity maps of Rho GTPase activation: Colors designate the activation level of the Rho GTPase recorded in a sampling window at D = 0 μm. (j, k, l) Cross-correlation coefficients (color-coded) between edge velocity and Rac1 (j), Cdc42 (k) and RhoA (l) as a function of the sampling window and time shift. (m, n, o) Temporal cross-correlation functions for individual cells (black). Red: Average cross-correlation functions per GTPase resulting from a spline fit to the combined single cell cross-correlation functions; n = 11 cells (Rac1); n = 12 cells (Cdc42); n = 12 cells (RhoA). Arrowheads: some cells show negative correlation coefficients at positive time lags, likely associated with ruffling (see text).
To quantify the coupling we computed, for every sampling window, Pearson's correlation coefficient between edge velocity and the GTPase activity, as a function of the time lag between the two variables. High correlation coefficients indicated a tight coupling of GTPase activation and cell protrusion/retraction events, whereas low correlation coefficients suggested a more remote relationship. Moreover, if a significant maximum correlation coefficient was obtained at a non-zero time lag, it indicated the relative timing of the two processes. For actively protruding regions, we observed significant correlation coefficients for all three GTPases. For Cdc42 and Rac1, high correlation values occurred predominantly at negative time lags (Fig. 2j, k), i.e. the activation of these two GTPases was delayed relative to cell edge advancements. In contrast, for RhoA high correlation values were narrowly distributed around zero time lags (Fig. 2l). Thus, RhoA activation appeared to be synchronous with forward movements of the cell edge. These results show that GTPases are activated over a fixed time interval relative to the dynamics of the leading edge.
To more precisely estimate the activation of the three GTPases relative to protrusion/retraction events, we averaged the correlation coefficients over all sampling windows of a cell (Supplementary Fig. S4). A comparison between cells showed that despite substantial variation in shape and morphodynamic behavior, the relationships between GTPase activation and edge movements were preserved (Fig. 2m - o). To estimate the average timing between protrusion/retraction and GTPase activation across cells the correlation coefficients of individual cells were pooled and the significant maximum of a spline function fitted through the data determined (Fig. 2m–o, red curves; Supplementary Methods). A few cells displayed correlation minima at positive time lags, particularly with Cdc42 and RhoA (Fig. 2n, o, arrowheads). These were likely due to ruffling events in which GTPases reach a high activation level a few seconds before the lamellipodium is lifted off the substrate and the edge retracts (see Movies 3 – 4). Edge retraction generates a negative velocity in the velocity map, and hence a negative cross correlation with elevated GTPase activation. These ruffling events did not affect our finding that both Rac1 and Cdc42 activation lag edge protrusion, whereas RhoA is activated during cell protrusion.
We speculated that the GTPases might be organized not only temporally, but also spatially, within the band of modulated signaling activity (see Fig. 1e-g). Therefore, we repeated the correlation analysis between protrusion and retraction events and activation of Rac1, Cdc42, and RhoA in sampling windows at various distances from the cell edge (Fig. 3a-c, see also Movie 7). Maxima of the correlation coefficients CM and the corresponding time lags Δt were determined for each distance and fitted with a smoothing spline (Fig. 3d-f). For RhoA the correlation coefficient was highest at the leading edge and monotonically decreased at larger distances from the edge (Fig. 3f, blue line). At D > 2 μm the correlation coefficient was smaller than 0.2 (Fig. 3d-f, black dashed line), the 95% confidence level for correlation coefficients of individual cells (Supplementary Methods). Thus, beyond 2 μm RhoA activity is no longer related to protrusion events (Fig. 3f, arrow). Rac1 and Cdc42 activities were correlated with edge movements over a wider region (4.5 μm and 3.2 μm, respectively; Fig 3d, e, blue curve). Interestingly, the highest correlation coefficients were found at a distance D = 1.8±0.7 μm for Rac1 and at D = 1.3±0.7 μm for Cdc42. The same distance D was obtained when identifying the distance with the shortest time lag between edge velocity fluctuations and GTPase activation (Fig 3d, e, red curve), which indicates the location of initial GTPase activation. These results indicate that both Rac1 and Cdc42 are activated at ∼1.8 μm from the cell edge. The larger time lags and lower correlation coefficients at distances other than 1.8 μm suggest that the activation of Rac1 and Cdc42 then propagates in anterior and posterior directions loosing the close coordination with edge movements.
Figure 3. Correlation of Rho GTPase activation and cell edge velocity as a function of time and space.
(a-c) Cross-correlation functions between edge velocity and GTPase activation at D = 0 (red line), 0.6, 1.3, 2.5, 4.4 and 6.3 μm. Dashed line: 95% confidence level for correlation values. (d-f) Blue line: magnitude of cross-correlation maximum as a function of D (blue bracket in b). Red line: time lag of cross-correlation maxima as a function of D (red bracket in b). Dashed lines: 95% confidence interval estimated by bootstrap sampling of residual distributions to the spline fits (see Supplementary Methods). Grey area: region with correlation values below 95% confidence. Of note, time lags of cross-correlation maxima in this area are no longer meaningful. Data derived from n = 6 (Rac1), n=5 (Cdc42) and n = 5 (RhoA) cells.
Next, we estimated the between-cell variation of the cross-correlation time lag (Fig 4a) by bootstrapping 2000 samples from the residual distribution of the spline fit to the individual correlation functions in Fig. 2m–o. The time lags were consistently negative for all three GTPases, i.e. the peak in their activation was delayed relative to the protrusion of the cell edge (time lag to protrusion for Rac1: -41 [-61 -13]; Cdc42: -46 [-51 -39]; RhoA -6 [-8 -4] s; mean, ± 95% confidence interval as determined by bootstrap analysis). Hence, Rac1, Cdc42 and RhoA each have a different timing of activation, which is well conserved between cells.
Figure 4. Spatiotemporal coordination of Rac1, Cdc42, and RhoA activation.
(a) Timing of Rho GTPase activation relative to edge velocity, as determined by temporal cross-correlation functions. The variable ‘edge movement’ is being used as the reference signal. Thus, correlation maxima in the sector labeled “After” indicate that the GTPase reaches the activation maximum after the protrusion event (time point of fastest edge advancement). Dashed lines: 95% confidence intervals. Data originate from n = 11 cells (Rac1), n = 12 cells (Cdc42), and n=12 cells (RhoA). Confidence intervals were computed by bootstrap sampling of 2000 residuals to the spline fits. (b) Timing of RhoA activity relative to Cdc42 activity, both monitored in the same cell, as determined by the temporal cross-correlation function. The variable Cdc42 is being used as the reference signal. Thus, the correlation maximum in the sector labeled “Before” indicates that RhoA reaches the activation maximum before Cdc42 (n = 7 cells). (c) Model of GTPase activation during protrusion and retraction. Green: Rac1 activation. Black: RhoA activation. Blue: Cdc42 activation.
We were concerned that the measured spatiotemporal shifts of Rac1, Cdc42, and RhoA activation might be attributed to the different biosensor designs we used for each of these proteins (Supplementary Fig. S1). Some designs might be more susceptible to competition from native ligands, affecting readouts of activation kinetics. Therefore, we titrated the concentrations of biosensor components (Supplementary Figs S8 and S9) and repeated the timing experiments using different biosensor designs for the same GTPase (Supplementary Fig. S10). These control experiments confirmed that neither the relative activation kinetics nor the localization were a function of biosensor design (see Supplementary Materials).
We also sought to validate the time shifts between GTPase activations predicted by computational multiplexing by directly observing two signaling activities in the same cell. To this end, we modified the Cdc42 biosensor with new fluorophores (Supplementary Fig. S6), enabling simultaneous imaging of Cdc42 and RhoA activation in one cell (Movie 9). The resulting four-channel image data provided unprecedented temporal and spatial resolution for the study of the coordination between two signaling molecules, without using the cell edge velocity as a reference. Here, fluctuation correlation analysis was valuable to harness the full potential of the new simultaneous imaging capability. The data from multiplex biosensor imaging was again analyzed locally in sampling windows at the leading edge. Averaging the correlation coefficients between the Cdc42 and RhoA time courses within one cell and over n=7 cells we obtained a time lag of -34 [-30, -38] s, i.e. Cdc42 was activated after RhoA (Fig 4b). This was within the confidence band of the computationally predicted difference between Cdc42 and RhoA activation (cf. Fig 4a), confirming that direct visualization and indirect inference of signaling relationships yield the same result.
Together, these data indicate the following dynamics of Rho family GTPase activation in one protrusion–retraction cycle (Fig 4c): RhoA activation increases and decreases in synchrony with protrusion and retraction. RhoA activation that is correlated with leading edge dynamics is confined to a band 2 μm from the leading edge. Cdc42 and Rac1 reach their peak activation with a 40 s delay relative to protrusion, and their activation is initiated 1.8 μm from the leading edge. Rac1 and Cdc42 are temporally as well as spatially less coupled to protrusion. Cdc42 and even more so Rac1 maintain significant levels of activity during the retraction phase, such that lower levels of Rac1 activation overlap the onset of the next protrusion cycle. Rac1 and Cdc42 signals propagate from their location of initial activation in anterograde and retrograde directions via as yet unidentified mechanisms.
The delayed activation of Rac1 and Cdc42 suggests that in spontaneous cell motility, i.e. without active stimulation by growth factors, their primary role may not be in initiating protrusion, as is currently supposed15. While the early, low activation level of Rac1 we observe may still be sufficient for this role, the peak activities for Rac1 and Cdc42 are localized closer to the sites of maturating adhesions (∼1.8 μm behind the protruding leading edge16). Rac1 and Cdc42 appear to be affecting protrusion by regulating adhesion dynamics17-19, likely by reinforcing adhesion sites that are formed to balance increasing protrusive forces at the leading edge20. In addition, our data supports the notion that Rac1 operates as an antagonist to RhoA12. RhoA activity is rapidly suppressed as Rac1 reaches its maximum activation. RhoA is usually thought to be a mediator of contractility. Here, we propose that the synchronized activation of RhoA with protrusion relates to a second function of RhoA as an initiator of actin polymerization at the onset of the protrusion-retraction cycle, possibly via its ability to activate members of the formin family such as mDia21,22. In addition, the RhoA – mDia pathway is known to stabilize microtubules specifically in leading edge adhesions23. This may contribute to the activation of Rac1 we observed at adhesion sites, perhaps via positive feedback between microtubule growth and Rac1 activation24 and/or via the engagement of integrins that regulate the coupling of Rac1 to its effector Pak125, which is known to exhibit mutually positive feedback18.
We present two complementary approaches to in situ analysis of cellular pathways. Simultaneous imaging presents unprecedented spatial and temporal resolution to explore the relationship between two pathways, while “computational multiplexing” enables correlation of many signaling activities. Computational multiplexing makes use of the spontaneous activation of pathways by random local changes in signaling molecules due to low level stochastic stimulation of receptors, variations in concentration, etc.. Thus it is potentially less perturbing than methods that rely on acute cell stimulation to initiate a signaling cascade. The sensitivity required to capture the coupling of spontaneous molecular activities is achieved for two reasons: First, by very local image measurement, below the diffusion radius of signaling molecules, our analysis captures the immediate relationship between the states of a signaling pathway and its morphological outputs. While the activity levels of pathway components and hence the pathway outputs may vary between cellular locations and between cells, the hierarchy and timing between pathway components are conserved. Second, measurement noise is greatly reduced by averaging thousands of local relationships between activities. Together, these properties permit the pathway to be reconstructed despite significant cell-to-cell heterogeneity.
We chose to illustrate computational multiplexing for a pathway controlling cell morphology, enabling us to use cell morphological dynamics as a reference to align distinct signaling events observed in different experiments. However, the method is readily scalable to more and diverse activities. Relationships between signals can also be inferred in pathways without spatial cues. In this case, one of the biosensors may itself be used as the common reference in a series of experiments where it is imaged simultaneously in pairings with other biosensors. Again, the statistical approaches described for computational multiplexing will be useful to integrate into a pathway model the heterogeneous and complex responses made visible by simultaneous imaging of different biosensor combinations.
Methods Summary
Biosensors
The three biosensors were made and used as originally described 4-6, with modifications where noted in the text. The Cdc42 biosensor was modified for multiplex imaging by removing EGFP and replacing it with an Alexa750 dye. This produced a biosensor with wavelengths orthogonal to those of the RhoA biosensor. The new Cdc42 biosensor was fused to maltose binding protein to enhance solubility. For control studies a new dual chain RhoA biosensor was constructed by eliminating the linker in the published RhoA biosensor (see Supplementary Material).
Live cell imaging
Biosensors were imaged as previously described26,27. In order to obtain sufficient signal/noise for multiplexing, cells with biosensor were filmed at 10 s intervals, using a 40×/1.3 NA objective and binning, resulting in an effective pixel size in object space of 330 nm.
Ratiometric corrections
Raw images were aligned and noise-filtered to allow ratiometric correction of volume effects, bleed-through and variation in sensor concentration.
Image analysis
Corrected biosensor images were segmented and cell edge displacements tracked as in13. Sampling windows of 0.9 μm depth and 1.8 - 3μm width were constructed to follow morphological changes at a fixed distance from the cell edge. For each window, biosensor activation time courses were recorded. For windows placed at the cell edge, a time course of protrusion/retraction velocity was recorded additionally. Coupling of two activity time courses was analyzed per window by Pearson's cross-correlation function. Subsequently, for a cell the per-window correlation functions were averaged over all windows following the edge at a specific distance D (Fig. 1i). Per-cell correlation functions were averaged over multiple cells and statistically analyzed by bootstrap sampling to determine the significance and time lag of the coupling between two activities. All procedures are detailed in Supplementary Methods.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding from the Swiss National Science Foundation and the Novartis Foundation, formerly the Ciba-Geigy Jubilee Foundation (M.M.), NIH R01 GM57464 (K.M.H.), NIH R01 GM71868 (G.D.), and the Cell Migration Consortium, Grant No U54 GM064346 from NIGMS (G.D. and K.M.H.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.M. initiated the project, conceptualized the idea of computational multiplexing, wrote all image analysis software pertinent to multiplexing, and contributed to the writing of the manuscript; L.H. developed simultaneous imaging of RhoA and Cdc42, including the modification and validation of the meroCBD probe, developed the intermolecular RhoA sensor, including controls and validation, studying the effects of biosensor stoichiometry and expression level, developed the new version of the Rac biosensor, and contributed to writing of the manuscript. C.W. produced stable cell lines of the intermolecular RhoA biosensor and conducted imaging experiments for the comparison of intra- and intermolecular biosensor designs; H.E. contributed simulations for validation of the correlation analysis and assisted with image processing; O.P. and P.N. contributed image data of RhoA and Cdc42 activity, respectively; A.A. and G.L.J. contributed valuable advice and unpublished reagents. K.M.H. and G.D. coordinated the study and wrote the final version of the manuscript and supplement.
References
- 1.Jaffe AB, Hall A. Rho GTPases: Biochemistry and Biology. Annual Review of Cell and Developmental Biology. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 2.Burridge K. Wennerberg, K., Rho and Rac Take Center Stage. Cell. 2004;116(2):167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 3.Ridley AJ, et al. Cell Migration: Integrating Signals from Front to Back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 4.Kraynov V, et al. Localized Rac Activation Dynamics Visualized in Living Cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 5.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of Endogenous Cdc42 Visualized in Living Cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 6.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006 doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 7.Kurokawa K, Matsuda M. Localized RhoA Activation as a Requirement for the Induction of Membrane Ruffling. Mol Biol Cell. 2005;16(9):4294–4303. doi: 10.1091/mbc.E04-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 9.Rottner K, Hall A, Small JV. Interplay between Rac and Rho in the control of substrate contact dynamics. Current Biology. 1999;9(12):640–649. doi: 10.1016/s0960-9822(99)80286-3. [DOI] [PubMed] [Google Scholar]
- 10.Nimnual AS, Taylor LJ, Bar-Sagi D. Redox-dependent downregulation of Rho by Rac. Nature Cell Biology. 2003;5:236–241. doi: 10.1038/ncb938. [DOI] [PubMed] [Google Scholar]
- 11.Arthur WT, Burridge K. RhoA Inactivation by p190RhoGAP Regulates Cell Spreading and Migration by Promoting Membrane Protrusion and Polarity. Mol Biol Cell. 2001;12(9):2711–2720. doi: 10.1091/mbc.12.9.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nature Cell Biology. 2006;8(8):803–U835. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- 13.Machacek M, Danuser G. Morphodynamic Profiling of Protrusion Phenotypes. Biophys J. 2006;90(4):1–14. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marguet D, Lenne PF, Rigneault H, He HT. Dynamics in the plasma membrane: how to combine fluidity and order. Embo Journal. 2006;25(15):3446–3457. doi: 10.1038/sj.emboj.7601204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Developmental Biology. 2004;265(1):23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci. 2003;116(22):4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 17.Del Pozo MA, et al. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat Cell Biol. 2002;4:232–239. doi: 10.1038/ncb759. [DOI] [PubMed] [Google Scholar]
- 18.Nayal A, et al. Paxillin phosphorylation at ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. Journal of Cell Biology. 2006;173(4):587–599. doi: 10.1083/jcb.200509075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ten Klooster JP, Jaffer ZM, Chernoff J, Hordijk PL. Targeting and activation of Rac1 are mediated by the exchange factor ß-Pix. Journal of Cell Biology. 2006;172(5):759–769. doi: 10.1083/jcb.200509096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat Cell Bio. 2008;10(12):1393–1400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narumiya S, Ishizaki T, Watanabe N. Rho effectors and reorganization of actin cytoskeleton. FEBS Lett. 1997;410:68–72. doi: 10.1016/s0014-5793(97)00317-7. [DOI] [PubMed] [Google Scholar]
- 22.Yamana N, et al. The Rho-mDia1 Pathway Regulates Cell Polarity and Focal Adhesion Turnover in Migrating Cells through Mobilizing Apc and c-Src. Mol Cell Biol. 2006;26:6844–6858. doi: 10.1128/MCB.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palazzo A, Cook T, Alberts A, Gundersen G. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nature Cell Biology. 2001;3(8):723–729. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez OC, et al. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nature Cell Biology. 2003;5(7):599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- 25.Del Pozo MA, Price LS, Alderson NB, Ren XD, Schwartz MA. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. Embo Journal. 2000;19(9):2008–2014. doi: 10.1093/emboj/19.9.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodgson L, Nalbant P, Shen F, Hahn K. Imaging and photobleach correction of Mero-CBD, sensor of endogenous Cdc42 activation in Methods in Enzymology, Vol 406, Regulators and Effectors of Small Gtpases: Rho Family. 2006;406:140–156. doi: 10.1016/S0076-6879(06)06012-5. [DOI] [PubMed] [Google Scholar]
- 27.Hodgson L, Shen F, Hahn K. Biosensors for characterizing the dynamics of Rho family GTPases in living cells. Curr Protocols Cell Biol In Press. 2009 doi: 10.1002/0471143030.cb1411s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.