Abstract
Background
The study of P transposable element repression in Drosophila melanogaster led to the discovery of the Trans-Silencing Effect (TSE), a homology-dependent repression mechanism by which a P-transgene inserted in subtelomeric heterochromatin (Telomeric Associated Sequences, “TAS”) has the capacity to repress in trans, in the female germline, a homologous P-lacZ transgene located in euchromatin. Phenotypic and genetic analysis have shown that TSE exhibits variegation in ovaries, displays a maternal effect as well as epigenetic transmission through meiosis and involves heterochromatin (including HP1) and RNA silencing.
Principal Findings
Here, we show that mutations in squash and zucchini, which are involved in the piwi-interacting RNA (piRNA) silencing pathway, strongly affect TSE. In addition, we carried out a molecular analysis of TSE and show that silencing is correlated to the accumulation of lacZ small RNAs in ovaries. Finally, we show that the production of these small RNAs is sensitive to mutations affecting squash and zucchini, as well as to the dose of HP1.
Conclusions and Significance
Thus, our results indicate that the TSE represents a bona fide piRNA-based repression. In addition, the sensitivity of TSE to HP1 dose suggests that in Drosophila, as previously shown in Schizosaccharomyces pombe, a RNA silencing pathway can depend on heterochromatin components.
Introduction
Mobilization of transposable elements (TEs) is regulated by complex mechanisms involving proteins encoded by the TEs themselves, as well as heterochromatin formation and small RNA silencing mechanisms [1]–[11]. Genomic sites containing full-length or defective copies of TEs have been identified which are sufficient to establish complete repression of the other copies of the same family scattered throughout the genome. For example in Drosophila, the flam/COM locus, located in pericentromeric heterochromatin, represses various families of “Type I” TEs (retrotransposons which transpose via an RNA intermediate) [12]–[15] and the TAS (Telomeric Associated Sequence) region of sub-telomeric heterochromatin houses strong regulatory P elements (“Type II” TEs whose transposition occurs via a DNA intermediate) [2], [16]–[18]. The flam/COM locus represses expression of gypsy, Zam, and Idefix in somatic follicle cells, thereby preventing transfer of these retrotransposons to the oocyte [19], [20]. By contrast, P element repression by telomeric P copies takes place in the germline of both sexes [17], [18], [21], [22] and it is in this tissue that all P element transposition steps take place [3], [23], [24]. It has been shown recently that the RNA silencing pathways implicated in both the germline and somatic follicle cells of the ovary rely on the piwi-interacting RNA (piRNAs) silencing pathway [8], although the mechanisms at work in these two tissues differ since some actors of the piRNA machinery are present only in the germline [25]–[28].
The study of the mechanism of P element repression in the germline, elicited by telomeric P copies, has been facilitated by the use of P-transgenes instead of natural P transposons. The P-lacZ transgene carries an in-frame fusion of the N-terminal region of the transposase with the E. coli lacZ gene and can be used as an enhancer-trap [29]. It has been shown that the presence of one or two copies of P-lacZ in TAS, can repress another P-lacZ copy in trans, irrespective of the genomic location of the latter copy [30]–[32]. This repression occurs in the female germline (nurse cells and oocytes), but not in the somatic follicle cells [32]. This phenomenon, termed “Trans-Silencing Effect” (TSE) [30], thus allows the precise study of the genetic and phenotypic properties of piRNA-based repression in the context of the germline. It has been shown that TSE displays a maternal effect, epigenetic transmission through meiosis and variegation between egg chambers when repression is incomplete [31], [33]. TSE was also shown to be affected by mutations in genes involved in heterochromatin formation (including HP1) and the piRNA silencing pathway [33]. In particular, TSE was shown to be completely abolished by mutations affecting aubergine, armitage, homeless (spindle-E) and a partial dose effect of piwi was also found [33]. All these genes have been shown to be necessary for the production of piRNAs in the germline [25].
In the present study, we explore further the genetic and molecular properties of TSE with regard to the piRNA-based mechanism of repression. We first tested the effect on TSE of mutations in squash (squ) and zucchini (zuc), encoding two putative nucleases which have been shown recently to be involved in the piRNA pathway [25], [34]. SQUASH and ZUCCHINI both interact with AUBERGINE and mutants exhibit dorso-ventral patterning defects similar to those associated with aub mutations. Mutations in squ and zuc induce the transcription upregulation of Het-A and TART telomeric retrotransposons and result in the loss of piRNAs in the germline [25], [34]. We first show that the loss of function of squ and zuc has a very strong negative effect on TSE. Second, we provide the first molecular support of the mechanism of TSE showing that trans-silencing is correlated to the presence of lacZ small RNAs in ovaries, the levels of these small RNAs being strongly affected by mutations in squ and zuc. Third, we show that accumulation of these small RNAs in ovaries is also sensitive to a mutation affecting HP1 levels. These results open the possibility of a functional reciprocal dependence between heterochromatin formation and RNA silencing in Drosophila. Thus TSE in the fly could parallel the “self-reinforcing loop” of RNA silencing and heterochromatin previously shown to occur in Schizosaccharomyces pombe [35]–[37].
Materials and Methods
Experimental conditions
All crosses were performed at 25°C and involved 3–5 couples in most of the cases. All ovary lacZ expression assays were carried out using X-gal overnight staining as described in Lemaitre et al. 1993 [21], except that ovaries were fixed for 6 min [33].
Transgenes and strains
P-lacZ fusion enhancer trap transgenes (P-1152, BQ16) contain an in-frame translational fusion of the E. coli lacZ gene to the second exon of the P transposase gene and contain rosy + as a transformation marker [38]. The P-1152 insertion (FBti0005700) comes from stock #11152 in the Bloomington Stock Center and was mapped at the telomere of the X chromosome (site 1A); this stock was previously described to carry a single P-lacZ insertion in TAS [30]. However, in our #11152 stock, we have mapped two P-lacZ insertions in the same TAS unit and in the same orientation which might have resulted from an unequal recombination event duplicating the P-lacZ transgene [33]. P-1152 is homozygous viable and fertile. BQ16 is located at 64C in euchromatin of the third chromosome [32] and is homozygous viable and fertile. P-1152 shows no lacZ expression in the ovary, whereas BQ16 is strongly expressed in the nurse cells and in the oocyte.
Lines carrying transgenes have M genetic backgrounds (devoid of P transposable elements), as do the multi-marked balancer stocks used in genetic experiments and the strains carrying mutations used for the candidate gene analysis. The Cantony line was used as a control line, completely devoid of any P element or transgene (true “M” line).
Mutations used for the candidate gene analysis
Su(var)205, squash (squ) and zucchini (zuc) are located on chromosome 2. Loss of function is lethal in the case of Su(var)205, female sterile in the case of squash and zucchini.
Su(var)2–5 05 (or Su(var)205 05) was X-ray induced and corresponds to a null allele of Su(var)205 since it only encodes the first ten amino acids of the HP1 protein [39]. zuc and squ alleles were isolated from an EMS screen [40]. zucHM27 contains a stop codon at residue 5, zucSG63 a substitution of Histidine 169 with a Tyrosine in the conserved HKD domain presumably involved in nuclease activity. squHE47 and squPP32 are generated by insertion of stop codons at residues 100 and 111, respectively [34]. Lines carrying mutations of squash and zucchini were kindly provided by Attilio Pane and Trudi Schüpbach and the line carrying the Su(var)2–5 05 allele was kindly provided by Gunter Reuter. All the alleles described above are maintained over a Cy balancer chromosome. Cy balancer chromosomes have been shown not to affect TSE (unpublished results). Additional information about mutants and stocks are available at flybase: http://flybase.bio.indiana.edu/.
Quantification of TSE
When TSE is incomplete, variegation is observed since “on/off” lacZ expression is seen between egg chambers: that is, egg chambers can show strong expression (dark blue) or no expression, but intermediate expression levels are rarely found. TSE was quantified as previously described [33] by determining the percentage of egg chambers with no expression. We scored the number of these repressed chambers among the first five egg chambers of a given ovariole for ten ovarioles chosen at random per ovary. For a given genotype more than 1000 egg chambers were counted.
Statistical analysis
The levels of TSE produced in flies of different genotypes were compared using the non-parametric Mann-Whitney test, conducted on TSE percentages per ovary.
RNase protection assays (RPA)
Small RNAs from adult flies were extracted using the Ambion mirVana™ miRNA isolation kit. Per each condition, 400 ovaries were used for RNA extraction. Aliquots of 4 µg of small RNAs were used in RPA experiments. The radiolabelled RNA probe homologous to the 5′ region of P-lacZ was 150 nt long (position 600 to 750 of the P{1ArB} transgene (FBtp0000160)). After purification, probes with a specific activity of 5×104 cpm were used. We used the Ambion mirVana™ miRNA detection kit for RPA experiments. Hybridization was performed overnight at 42°C and digestion of single-stranded RNA was carried out for 45 minutes at 37°C with RNase A/RNase T1. After RNase inactivation, protected fragments were precipitated and separated on a 15% acrylamide/polyacrylamide (19∶1) gel running in 0.5×TBE. Protected fragments were detected by autoradiography after 4 weeks of exposure.
Results
Functional assay for the Trans-Silencing Effect in zuc and squ mutants
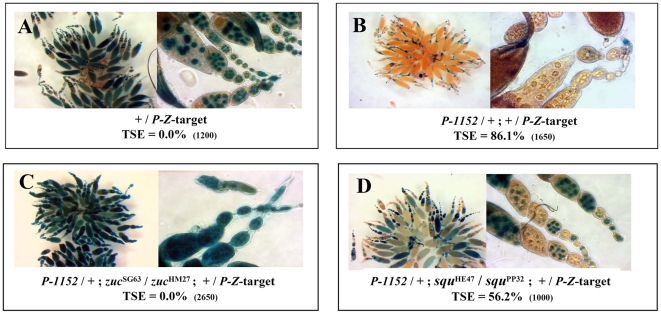
Given the role of squash and zucchini in the piRNA pathway [25], [34], the effect of mutant alleles of these genes on TSE was tested (Figure 1). For a given assay, a P-1152 telomeric silencer was combined with a P-lacZ target expressed in the female germline, in the absence (TSE positive control), or presence of mutant alleles of the candidate gene. The P-1152 silencer was inherited, in each case from a homozygous P-1152 female. The first gene tested was zucchini (Figure 1C). The TSE positive control produced a strong repression (Fig. 1B, TSE = 86.1%, n = 1650), whereas females having a heteroallelic zuc SG63/zuc HM27 genotype showed a complete loss of repression (Fig. 1C, TSE = 0.0%, n = 2650). The same result was found for females having the reciprocally inherited heteroallelic zuc HM27/zuc SG63 combination (i.e. the mutant alleles were inherited by the reciprocal parent: TSE = 0.0%, n = 1600, data not shown). The same analysis was performed for squash and the heteroallelic squ HE47/squ PP32 genotype showed reduced TSE (Fig. 1D, TSE = 56.2%, n = 1000). The reciprocally-inherited heteroallelic genotype, squ PP32/squ HE47, showed a very similar result (TSE = 56.6%, n = 2200, data not shown). The percentage of TSE observed for each of the two kinds of heteroallelic squash mutant females (squ HE47/squ PP32 and squ PP32/squ HE47) was compared to that observed for the TSE positive control (Fig. 1B), using the non-parametric Mann-Whitney test: in both cases the difference is highly significant (P<0.001). By contrast, for both zuc and squ, no significant effect on TSE was detected for the heterozygous mutants (zuc: TSE = 79.4%, n = 1850; squ: TSE = 86.8%, n = 2600, data not shown). These levels do not differ from the TSE positive control level (Fig. 1B), as tested with the non-parametric Mann-Whitney test. In conclusion, the loss of function of either of these two genes affects TSE, zuc having a more severe effect than squ, a result consistent with data reported by Pane et al. [34] and Malone et al. [25] showing that the zuc mutant context has a more severe effect than the squ mutant context on the production of piRNAs.
Figure 1. TSE is sensitive to mutations affecting squash and zucchini.
(A) Expression control in ovaries of the P-lacZ transgene used as a TSE target (BQ16, located on chromosome 3). (B) G1 females produced from the cross between P-1152 females and BQ16 males. (C–D) Heteroallelic females for mutant alleles of zuc or squ: these females have inherited the BQ16 target paternally and the P-1152 telomeric silencer from a homozygous P-1152 female. The maternally-introduced zuc or squ mutant allele is written first. In each case, the percentage of TSE is given with the total number of egg chambers assayed in parenthesis.
Silencing is correlated to the accumulation of lacZ small RNAs in ovaries whose production is sensitive to squash and zucchini mutations
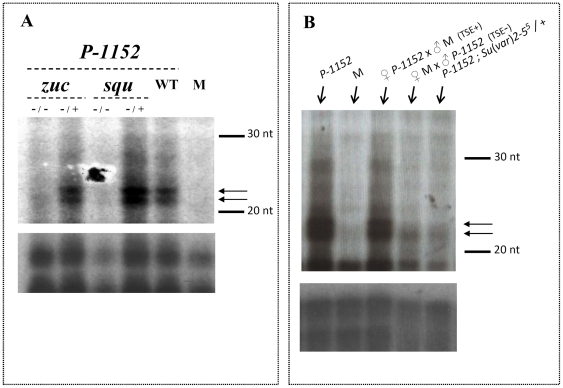
Since TSE is highly sensitive to mutations in genes involved in the piRNA silencing pathway, we tested whether lacZ small RNAs were present in ovaries of females which carry the P-1152 telomeric silencer locus and, if so, whether the production of these small RNAs requires the squ and zuc functions. We used an RNAse protection assay to detect lacZ small RNAs in ovaries from females carrying two copies of P-1152 and otherwise wild-type, heterozygous or heteroallelic mutants for squ and zuc. Ovaries from the M line Cantony were also analyzed as a negative control. RNAse protection analysis allowed detection of two abundant small RNAs in ovaries from homozygous P-1152 females (Fig. 2A, lane 5 and Fig. 2B, lane 1 – arrows to the right of the autoradiography), which were not detected in M females (Fig. 2A, lane 6 and Fig. 2B, lane 2). Females heterozygous for squ or zuc mutant alleles, also exhibited abundant accumulation of the lacZ small RNAs of the same size in ovaries (Fig. 2A, lanes 2 and 4, respectively). By contrast, these RNAs were almost undetectable for females heteroallelic for mutant alleles of squ or zuc (Fig. 2A, lanes 1 and 3, respectively). This analysis shows that telomeric P-lacZ silencer transgenes produce lacZ small RNAs in the ovary and that loss of function of squ and zuc has a strong negative effect on the accumulation of these lacZ small RNAs. In addition, as for the TSE assay, no dose effects for squ or zuc were observed on lacZ small RNAs accumulation.
Figure 2. TSE is correlated with the presence of small RNAs whose production depends on the piRNA pathway and HP1.
(A–B) RNAse protection was carried out using a lacZ sense riboprobe (150 nt) hybridized to RNAs extracted from ovaries from 3–6 day-old females. Data concerning the 20–30 nt region are shown together with aspecific bands used as a loading control (shown below). Cantony was used as an M strain, (devoid of any P element or P transgene). (A) Small RNA detection and effect of mutations in squash and zucchini. WT corresponds to P-1152 females which are wild-type for both squ and zuc. Two small RNAs (arrows) are highly abundant in females carrying the P-1152 telomeric TSE silencer at the homozygous state (WT), but are not detected in ovaries of females devoid of the P-1152 transgene (M). Females carrying the P-1152 telomeric silencer at the homozygous state and mutant for squash and zucchini were analyzed. The same two abundant small RNAs found in P-1152 (WT) can be detected in females carrying one functional allele of squ and zuc, but are undetectable in squ or zuc heteroallelic mutant females. Thus, accumulation of lacZ small RNAs occurring in P-1152 ovaries requires squ and zuc functions. (B) TSE maternal effect and effect of mutations affecting HP1. TSE+ indicates that this cross allows a strong TSE in G1 females due to the maternal transmission of the telomeric P-1152 silencer, whereas TSE- means that only a weak TSE is recovered from this cross in which P-1152 is inherited paternally. P-1152 homozygous females and M females were analyzed as positive and negative controls, respectively. The two most abundant small RNAs are indicated by arrows. A strong signal for these small RNAs is obtained for P-1152 homozygous females and for females having inherited a P-1152 transgene maternally (TSE+), but is undetectable in negative control M females. The signal for the small RNAs is significantly reduced for females having inherited P-1152 paternally (TSE-), as well as for P-1152 homozygous females carrying one null allele of Su(var)205 which encodes HP1. Therefore, accumulation of lacZ small RNAs is correlated to the maternal effect of TSE and depends on HP1 dose.
Accumulation of lacZ small RNAs in ovaries is correlated with the maternal effect of TSE
TSE was shown to exhibit a maternal effect: crossing females carrying a telomeric transgene with males carrying a target transgene produces G1 females which show strong TSE, whereas the reciprocal cross produces G1 females showing only weak TSE [31]–[33]. TSE also shows maternal inheritance since this maternal effect presents a remanence which can extend through six generations following the reciprocal G0 crosses [33]. TSE is therefore, at least in part, epigenetically transmitted through meiosis. TSE maternal inheritance can also be observed in the presence of a telomeric silencer alone, i.e. in the absence of the target transgene [32]. We thus tested if lacZ small RNA detection in ovaries parallels the maternal effect of TSE. RNAse protection analysis allowed detection of the two abundant small RNAs in G1 females produced by the two (P-1152 x M) reciprocal crosses, but the intensity of the signal obtained with the progeny of the (female P-1152 x male M) cross (cross TSE+) was higher than that of the progeny of the reciprocal cross which induces only a weak level of TSE (cross TSE-) (Fig. 2B, lanes 3 and 4). This difference becomes particularly clear, when a comparison is made between the signal intensities of the aspecific bands shown below the lacZ small RNAs. However, the signal from TSE- females is not null, a result which is consistent with the weak but non null level of TSE (around 10%) which can be induced in this cross [32], [33]. In conclusion, the presence of lacZ small RNAs in ovaries is detected in a manner which is correlated to the maternal effect of TSE.
Accumulation of lacZ small RNAs is sensitive to HP1 dose
TSE was shown previously to be sensitive not only to mutations in genes involved in the piRNA pathway, but also to mutations in genes involved in heterochromatin formation, such as Su(var)205 which encodes HP1 [33]. For Su(var)205, a particularly clear dose effect on TSE was observed. We thus tested if the presence of lacZ small RNAs in ovaries is affected in P-1152 females having only one dose of the Su(var)205 gene compared to wild-type. RNAse protection was performed as previously on females carrying two copies of P-1152 and heterozygous for Su(var)2–5 05, an amorphic allele of Su(var)205. Figure 2B (lane 5) shows that the level of small RNAs detected for females having two copies of P-1152 and only one dose of Su(var)205 is strongly reduced when compared to P-1152 wild-type females (Fig. 2B, lane 1). Indeed, with one dose of HP1, the level of these small RNAs is comparable to that of females carrying a single paternally-inherited P-1152 copy (Fig. 2B, lane 4). Under these two latter conditions, comparable low levels of TSE were also found [33]. Therefore, the effect of mutations affecting HP1 on TSE [33], as for squ and zuc mutations, can be correlated to a significant reduction in the accumulation of small RNAs in ovaries produced by the telomeric P-1152 silencer locus.
Discussion
Trans-Silencing Effect, a typical piRNA germline repression mechanism
Trans-silencing was previously shown to be strongly impaired by mutations affecting several components of the piRNA silencing pathway (AUBERGINE, ARMITAGE, HOMELESS, PIWI) [8], [33]. By contrast, TSE was not impaired by mutations affecting R2D2, a component of the siRNA pathway [33], [41], or LOQUACIOUS, a component of both the miRNA and endo-siRNA pathways [33], [42]–[44]. This indicates that TSE likely involves the piRNA silencing pathway, a hypothesis which is consistent with the fact that TSE is restricted to the germline [32], the tissue in which the “canonical” piRNA pathway functions [25], [26]. Further, SQUASH and ZUCCHINI were found to interact with AUBERGINE and to localize to the nuage, a cytoplasmic organelle surrounding the nurse cell nuclei, which also contains AUBERGINE and ARMITAGE and appears to be involved in RNA silencing [34]. squ and zuc mutations were also shown to affect piRNA production in ovaries at the cytological 42AB repetitive sequence cluster, a typical piRNA-producing genomic region [25]. Regarding TE repression in the germline, squ and zuc mutants were found to derepress transcription of the telomeric retrotransposons Het-A and TART [34] and of the I factor, a retrotransposon involved in a Drosophila system of hybrid dysgenesis [45], [46]. It is noteworthy that the I factor and the Het-A retrotransposons have also been found to be sensitive to aub, armi and hls (spn-E) [5], [45], [47]. The genetic analysis reported here shows that TSE is also highly sensitive to zuc and squ mutations (Figure 1). TSE is therefore sensitive to mutations affecting all the genes of the germline piRNA pathway tested and thus appears to represents a bona fide piRNA-based repression.
The presence of lacZ small RNAs in ovaries of females carrying a TSE silencer was therefore investigated using RNase protection analysis. In addition, paternal vs maternal transmission of the telomeric silencer was compared. Indeed, TSE was previously shown to have a maternal effect, i.e. strong repression occurs only when the telomeric silencer is maternally inherited, whereas a paternally-inherited telomeric silencer has weak or null repression capacities [32], [33], [48]. More precisely, it was shown genetically that TSE requires inheritance of two components, a maternal cytoplasmic component plus a chromosomal copy of the transgene, but these two components can be transmitted separately [33]. Indeed, a paternally-inherited telomeric transgene can be “potentiated” by a maternally-inherited cytoplasm from a female bearing a silencer. This interaction also functions between telomeric silencers located on different chromosomal arms [32]. The RNase protection analysis reported here shows that: 1- P-1152, a telomeric P-lacZ silencer produces small lacZ RNAs in ovaries (Figure 2A–B); 2- P-1152 lacZ small RNA accumulation is negatively affected in squ and zuc mutants (Figure 2A); 3- maternal transmission of P-1152 leads to accumulation of higher levels of these small RNAs than that observed upon paternal P-1152 transmission (Figure 2B). We have reproduced these results with independent RNAse protection assays (two experiments for the effect of each mutant and three experiments for the maternal effect). The size of the small RNAs detected here appears smaller (around 22–23 nt) than that corresponding to piRNAs as characterized by deep sequencing (23–28 nt, [8]), but they are consistent with piRNAs as detected by RNAse protection assays in other studies [49]: this can result from the RNAse protection protocol which tends to reduce the size of the RNAs detected. In conclusion, our results strongly suggest that the lacZ small RNAs in P-1152 oocytes may correspond to cytoplasmically-transmitted piRNAs mediating the maternal effect of TSE, as well potentiating a paternally-inherited telomeric silencer [33].
Towards a mutual dependence between RNA silencing and heterochromatin formation
TSE was previously shown to be sensitive to mutations affecting HP1 since a negative, dose-dependent, effect on TSE was found with two loss of function alleles of Su(var)205 (including Su(var)2–5 05) [33]. RNase protection analysis shows here that lacZ small RNA accumulation is also negatively affected by the dose of HP1 (Figure 2B). Although we cannot exclude that this effect may be indirect, this opens the possibility that some piRNA-producing loci depend on the presence of HP1 itself at the locus to produce piRNAs. A similar model was recently proposed for rhino, a HP1 homolog, mutations of which strongly reduce the production of piRNAs by dual strand piRNA- producing loci [28]. The authors propose that rhino is required for the production of the long precursor RNAs which are further processed to produce primary piRNAs. Note that in their study, rhino mutants were shown to have a drastic effect on the production of piRNAs by the X-chromosome TAS locus [28]. A similar situation may therefore exist for HP1 at this locus and, if so, it would be interesting to characterize more precisely the function of HP1 in the production of piRNAs at the TAS locus.
HP1 was shown to be present at TAS [50], [51]. A first possibility would be that HP1 stimulates transcription of the TAS locus as a classical transcription factor, independent of any heterochromatic role at this locus. Consistent with this, it was shown that PIWI, a partner of HP1 [52], promotes euchromatin histone modification and piRNA transcription at the third chromosome TAS [49]. The precise status of TAS, however, remains complex since some studies have shown that TAS exhibit some of the properties attributed to heterochromatin [53]–[55] and carry primarily heterochromatic histone tags [56]. Therefore, a second possibility would be that HP1 enhances the heterochromatic status of TAS in the germline, such that production of aberrant transcripts being processed into piRNAs is enhanced. This would result in a “heterochromatin-dependent RNA silencing pathway”. Examples of heterochromatin formation that depends on RNA silencing (“RNA-dependent heterochromatin formation”) have been described in numerous species including yeast [37], ciliates [57] and plants [58]. In Drosophila, this type of interaction has been described for variegation of pigment production in the eye linked to the insertion of the white gene in different types of heterochromatin structures [59], [60], as well as for heterochromatin formation at telomeres in the germline [51]. Therefore, telomeric regions in fly may be submitted to both RNA-dependent heterochromatin formation [47], [51] and heterochromatin-dependent RNA silencing. RNA silencing may favor heterochromatin formation that in turn potentiates RNA silencing, resulting in a functional positive loop between transcriptional gene silencing and post-transcriptional gene silencing. In such cases, RNA silencing and heterochromatin may not only reinforce each other but may also be functionally interdependent. Such bidirectional reinforcement between RNA silencing and heterochromatin formation was demonstrated in S. pombe since: 1- deletion of genes involved in RNA silencing were shown to derepress transcriptional silencing from centromeric heterochromatic repeats and was accompanied by loss of Histone 3 Lysine 9 methylation and Swi6 (a HP1 homolog) delocalization [37]; 2 - Swi6 was found to be required for the propagation and the maintenance of the RNA Induced Transcriptional Silencing (RITS) complex at the mat locus, a complex involved in amplification of RNA silencing [35], [61]. A positive loop between RNA silencing and heterochromatin formation may therefore also be at play in the Drosophila germline. According to this model, the epigenetic transmission of TSE through meiosis, (i.e. six generations of maternal transmission of the silencer are required to elicit a strong TSE following maternal inheritance of a cytoplasm devoid of lacZ piRNAs [33]) would underlie progressive establishment of this loop. Note that RNAi-dependent DNA methylation in Arabidopsis thaliana was shown to occur progressively over several consecutive generations [62].
Since TSE can be considered as a sub-phenomenon within P regulation, it may underlie epigenetic transmission of the P element repression. P element mobilization is responsible for a syndrome of germline abnormalities, known as the “P-M” system of hybrid dysgenesis which includes a high mutation rate, chromosomal rearrangements, male recombination and an agametic temperature-sensitive sterility called GD sterility (Gonadal Dysgenesis) [63]. P-induced hybrid dysgenesis is repressed by a maternally inherited cellular state called the“ P cytotype” [3], [23], [64], [65]. The absence of P-repression is called M cytotype. G1 females produced from the cross (P cytotype females × M cytotype males) present a strong capacity for repression, whereas females produced from the reciprocal cross present a weak capacity for repression [64]. In the subsequent generations, cytotype is progressively determined by the chromosomal P elements but the influence of the initial maternal inheritance can be detected for up to five generations [64], [66]. Therefore, P cytotype exhibits partial epigenetic transmission through meiosis. Furthermore, the identification and use of telomeric P elements as P cytotype determinants [2], [16]–[18], has made it possible to show that P cytotype (like TSE) involves a strictly-maternally inherited component (called the pre-P cytotype) [67], is sensitive to mutations affecting HP1 [2], [68] and aubergine [1], [69] and is correlated to maternal deposition of piRNAs [70]. Some of these properties are also found for the I factor which is responsible for the occurrence of another system of hybrid dysgenesis (“I-R” system) [45], [46], [71]–. TSE therefore parallels germline regulation of TEs (P, I), and does not resemble regulation of TEs in the somatic follicle cells (gypsy, ZAM, Idefix [19], [20]) for which no epigenetic transmission of repression capacities through meiosis has been described so far. It will be interesting to test if previously described cases of RNA-dependent heterochromatin formation show the reciprocal dependence, thus being able to form a positive loop.
Acknowledgments
We thank Paula Graça for technical assistance. We thank Anne-Marie Pret, Augustin de Vanssay and Neel Randsholt for their help in the preparation of the manuscript. We thank Jean-Louis Couderc, Frank Laski, Gunter Reuter, Attilio Pane and Trudi Schüpbach for the gift of strains. We thank the Bloomington stock center (http://flystocks.bio.indiana.edu/) and Flybase (http://flybase.bio.indiana.edu/).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Centre National de la Recherche Scientifique (Laboratoire de Biologie du Développement - Unité Mixte de Recherche 7622 and Institut Jacques Monod - Unité Mixte de Recherche 7592), Université Pierre & Marie Curie - Paris 6, Université Paris-Diderot - Paris 7, Action Concertée Incitative - Biologie Moléculaire et Structurale programme and the Association pour la Recherche sur le Cancer. A.L.T. was supported by a post-doctoral fellowship from the Fondation pour la Recherche Médicale. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Reiss D, Josse T, Anxolabehere D, Ronsseray S. aubergine mutations in Drosophila melanogaster impair P cytotype determination by telomeric P elements inserted in heterochromatin. Mol Genet Genomics. 2004;272:336–343. doi: 10.1007/s00438-004-1061-1. [DOI] [PubMed] [Google Scholar]
- 2.Ronsseray S, Lehmann M, Nouaud D, Anxolabehere D. The regulatory properties of autonomous subtelomeric P elements are sensitive to a Suppressor of variegation in Drosophila melanogaster. Genetics. 1996;143:1663–1674. doi: 10.1093/genetics/143.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rio DC. P transposable elements elements in Drosophila melanogaster. 2002. pp. 484–518. Mobile DNA II, edited by NL Craig et al American Society for Microbiology, Washington, DC.
- 4.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 6.Vastenhouw NL, Fischer SE, Robert VJ, Thijssen KL, Fraser AG, et al. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr Biol. 2003;13:1311–1316. doi: 10.1016/s0960-9822(03)00539-6. [DOI] [PubMed] [Google Scholar]
- 7.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 9.Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 11.Blumenstiel JP, Hartl DL. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci U S A. 2005;102:15965–15970. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desset S, Buchon N, Meignin C, Coiffet M, Vaury C. In Drosophila melanogaster the COM Locus Directs the Somatic Silencing of Two Retrotransposons through both Piwi-Dependent and -Independent Pathways. PLoS ONE. 2008;3:e1526. doi: 10.1371/journal.pone.0001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desset S, Meignin C, Dastugue B, Vaury C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics. 2003;164:501–509. doi: 10.1093/genetics/164.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelisson A, Song SU, Prud'homme N, Smith PA, Bucheton A, et al. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. Embo J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmons MJ, Raymond JD, Niemi JB, Stuart JR, Merriman PJ. The P cytotype in Drosophila melanogaster: a maternally transmitted regulatory state of the germ line associated with telomeric P elements. Genetics. 2004;166:243–254. doi: 10.1534/genetics.166.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronsseray S, Lehmann M, Anxolabehere D. The maternally inherited regulation of P elements in Drosophila melanogaster can be elicited by two P copies at cytological site 1A on the X chromosome. Genetics. 1991;129:501–512. doi: 10.1093/genetics/129.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marin L, Lehmann M, Nouaud D, Izaabel H, Anxolabehere D, et al. P-Element repression in Drosophila melanogaster by a naturally occurring defective telomeric P copy. Genetics. 2000;155:1841–1854. doi: 10.1093/genetics/155.4.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chalvet F, Teysset L, Terzian C, Prud'homme N, Santamaria P, et al. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. Embo J. 1999;18:2659–2669. doi: 10.1093/emboj/18.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brasset E, Taddei AR, Arnaud F, Faye B, Fausto AM, et al. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology. 2006;3:25. doi: 10.1186/1742-4690-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemaitre B, Ronsseray S, Coen D. Maternal repression of the P element promoter in the germline of Drosophila melanogaster: a model for the P cytotype. Genetics. 1993;135:149–160. doi: 10.1093/genetics/135.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuart JR, Haley KJ, Swedzinski D, Lockner S, Kocian PE, et al. Telomeric P elements associated with cytotype regulation of the P transposon family in Drosophila melanogaster. Genetics. 2002;162:1641–1654. doi: 10.1093/genetics/162.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engels WR. P elements in Drosophila . In: Berg DE, Howe M. M., editors. Washington, D.C.: American Society for Microbiology; 1989. pp. 437–484. [Google Scholar]
- 24.Laski FA, Rio DC, Rubin GM. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- 25.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Vagin VV, Lee S, Xu J, Ma S, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tushir JS, Zamore PD, Zhang Z. SnapShot: Fly piRNAs, PIWI proteins, and the ping-pong cycle. Cell. 2009;139:634, 634 e631. doi: 10.1016/j.cell.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Klattenhoff C, Xi H, Li C, Lee S, Xu J, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellen HJ, O'Kane CJ, Wilson C, Grossniklaus U, Pearson RK, et al. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 30.Roche SE, Rio DC. Trans-silencing by P elements inserted in subtelomeric heterochromatin involves the Drosophila Polycomb group gene, Enhancer of zeste. Genetics. 1998;149:1839–1855. doi: 10.1093/genetics/149.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronsseray S, Josse T, Boivin A, Anxolabehere D. Telomeric transgenes and trans-silencing in Drosophila. Genetica. 2003;117:327–335. doi: 10.1023/a:1022929121828. [DOI] [PubMed] [Google Scholar]
- 32.Josse T, Maurel-Zaffran C, de Vanssay A, Teysset L, Todeschini AL, et al. Telomeric trans-silencing in Drosophila melanogaster: tissue specificity, development and functional interactions between non-homologous telomeres. PLoS ONE. 2008;3:e3249. doi: 10.1371/journal.pone.0003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Josse T, Teysset L, Todeschini AL, Sidor CM, Anxolabehere D, et al. Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet. 2007;3:1633–1643. doi: 10.1371/journal.pgen.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 36.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci U S A. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 38.O'Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eissenberg JC, Morris GD, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 42.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, et al. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, et al. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci U S A. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bucheton A, Paro R, Sang HM, Pelisson A, Finnegan DJ. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell. 1984;38:153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- 47.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ronsseray S, Boivin A, Anxolabehere D. P-Element repression in Drosophila melanogaster by variegating clusters of P-lacZ-white transgenes. Genetics. 2001;159:1631–1642. doi: 10.1093/genetics/159.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 50.Frydrychova RC, Mason JM, Archer TK. HP1 is distributed within distinct chromatin domains at Drosophila telomeres. Genetics. 2008;180:121–131. doi: 10.1534/genetics.108.090647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, et al. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karpen GH, Spradling AC. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cryderman DE, Morris EJ, Biessmann H, Elgin SC, Wallrath LL. Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. Embo J. 1999;18:3724–3735. doi: 10.1093/emboj/18.13.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Biessmann H, Prasad S, Semeshin VF, Andreyeva EN, Nguyen Q, et al. Two distinct domains in Drosophila melanogaster telomeres. Genetics. 2005;171:1767–1777. doi: 10.1534/genetics.105.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andreyeva EN, Belyaeva ES, Semeshin VF, Pokholkova GV, Zhimulev IF. Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J Cell Sci. 2005;118:5465–5477. doi: 10.1242/jcs.02654. [DOI] [PubMed] [Google Scholar]
- 57.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 58.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 59.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 60.Fagegaltier D, Bouge AL, Berry B, Poisot E, Sismeiro O, et al. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0809208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 63.Kidwell MG, Kidwell JF, Sved JA. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility, and male recombination. Genetics. 1977;86:813–833. doi: 10.1093/genetics/86.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engels WR. Hybrid dysgenesis in Drosophila melanogaster: rules of inheritance of female sterility. Genet Research. 1979;33:219–236. doi: 10.1017/S0016672308009592. [DOI] [PubMed] [Google Scholar]
- 65.Engels WR. Extrachromosomal control of mutability in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1979;76:4011–4015. doi: 10.1073/pnas.76.8.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coen D, Lemaitre B, Delattre M, Quesneville H, Ronsseray S, et al. Drosophila P element: transposition, regulation and evolution. Genetica. 1994;93:61–78. doi: 10.1007/BF01435240. [DOI] [PubMed] [Google Scholar]
- 67.Ronsseray S, Lemaitre B, Coen D. Maternal inheritance of P cytotype in Drosophila melanogaster: a “pre-P cytotype” is strictly extra-chromosomally transmitted. Mol Gen Genet. 1993;241:115–123. doi: 10.1007/BF00280208. [DOI] [PubMed] [Google Scholar]
- 68.Haley KJ, Stuart JR, Raymond JD, Niemi JB, Simmons MJ. Impairment of cytotype regulation of P-element activity in Drosophila melanogaster by mutations in the Su(var)205 gene. Genetics. 2005;171:583–595. doi: 10.1534/genetics.102.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simmons MJ, Ryzek DF, Lamour C, Goodman JW, Kummer NE, et al. Cytotype regulation by telomeric P elements in Drosophila melanogaster: evidence for involvement of an RNA interference gene. Genetics. 2007;176:1945–1955. doi: 10.1534/genetics.106.066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jensen S, Gassama MP, Heidmann T. Taming of transposable elements by homology-dependent gene silencing. Nat Genet. 1999;21:209–212. doi: 10.1038/5997. [DOI] [PubMed] [Google Scholar]
- 72.Dramard X, Heidmann T, Jensen S. Natural Epigenetic Protection against the I-factor, a Drosophila LINE Retrotransposon, by Remnants of Ancestral Invasions. PLoS ONE. 2007;2:e304. doi: 10.1371/journal.pone.0000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Picard G, Bregliano JC, Bucheton A, Lavige JM, Pelisson A, et al. Non-mendelian female sterility and hybrid dysgenesis in Drosophila melanogaster. Genet Res. 1978;32:275–287. doi: 10.1017/s0016672300018772. [DOI] [PubMed] [Google Scholar]