Abstract
The 2.1 Å crystal structure of tryptophanyl-tRNA synthetase (TrpRS) from the diplomonad Giardia lamblia reveals that the N-terminus of this class I aminoacyl-tRNA synthetase forms a 16-residue α-helix. This helix replaces a β-hairpin that is required by human TrpRS for normal activity and has been inferred to play a similar role in all eukaryotic TrpRS. The primary sequences of TrpRS homologs from several basal eukaryotes including Giardia lack a set of three residues observed to stabilize interactions with this β-hairpin in the human TrpRS. Thus the present structure suggests that the activation reaction mechanism of TrpRS from the basal eukaryote G. lamblia differs from that of higher eukaryotes. Furthermore, the protein as observed in the crystal forms an (α2)2 homotetramer. The canonical dimer interface observed in all previous structures of tryptophanyl-tRNA synthetases is maintained, but in addition each N-terminal α-helix reciprocally interlocks with the equivalent helix from a second dimer to form a dimer of dimers. Although we have no evidence for tetramer formation in vivo, modeling indicates that the crystallographically observed tetrameric structure would be compatible with the tRNA binding mode used by dimeric TrpRS and TyrRS.
Keywords: aminoacyl-tRNA synthetase, protozoa, structural genomics
1. Introduction
The intestinal parasite Giardia lamblia (Giardia intestinalis) is a eukaryote from the class Diplomonadida. The biology of these organisms presents puzzles that were deepened by analysis of the completed G. lamblia genome (Morrison et al., 2007). For example, diplomonads possess two transcriptionally active diploid nuclei, but lack both mitochondria and peroxisomes. These odd features are reflected in the set of aminoacyl-tRNA synthetases (aaRS) found in the G. lamblia genome. Eukaryotes in general use two different aaRS for each amino acid, one mitochondrial and one cytosolic. Consistent with its lack of mitochondria, the G. lamblia genome contains only a single aaRS for each amino acid. Many of these are more closely related to archaeal homologs than to aaRS sequences from higher eukaryotics, but the sequence of the tryptophanyl-tRNA synthetase (TrpRS) from G. lamblia is close to that seen for cytosolic homologs in higher eukaryotes. TrpRS is a class I aminoacyl-tRNA synthetase, characterized by an α-helical anticodon binding domain and a separate canonical Rossman-fold catalytic domain. Class I aaRS contain two characteristic sequence motifs, HIGH and KMSKS, near the active site. The latter motif is KMSAS in G. lamblia TrpRS, as it is in mammals and many other eukaryotic TrpRS. The G. lamblia TrpRS sequence contains a 49-residue insertion in the catalytic domain relative to homologous sequences in higher eukaryotes. Such insertions are typical for G. lamblia genomic sequences (Morrison et al., 2007). Disregarding this insertion, the Giardia core domain sequence is ~ 50% identical to that of mammalian cytosolic TrpRS, a slightly higher identity than it shares with the core domain in archaeal sequences. All TrpRS structures reported to date are α2 homodimers. A single cognate tRNATrp molecule spans both monomers on binding. The tRNA anticodon loop binds to the α-helical domain of one monomer, while the tRNA acceptor arm extends into the active site in the catalytic domain of the other monomer (Shen et al., 2006; Yang et al., 2006). This mode of tRNA recognition and binding to an α2 dimer is shared by bacterial, eukaryotic, and archaeal TrpRS and TyrRS (Yaremchuk et al., 2002; Tsunoda et al., 2007). Eukaryotic and archaeal TrpRS sequences also contain a weakly conserved N-terminal extension from the core domain consisting of up to several hundred additional residues. Schimmel and coworkers have structurally and biochemically characterized the 154 residue extension in human TrpRS as being comprised of several functionally important components (Yang et al., 2003, 2007). In Giardia TrpRS, this extension contains 68 residues (Figure 1).
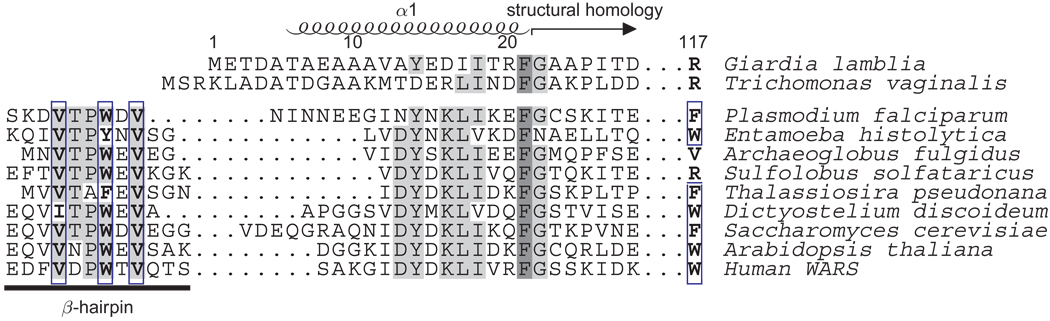
Figure 1. Sequence alignment of the G. lamblia TrpRS N-terminus with other eukaryotic and archaeal sequences.
The Giardia and human TrpRS are structurally homologous beginning at residue 20. In the Giardia structure, residues N-terminal to this point form part of an initial α-helix comprising residues 6–22. In the human TrpRS these residues have been observed instead to form a β-hairpin involved in ATP binding. A triad Val-Trp-Val of hydrophobic residues, shown boxed in the figure, stabilizes association of the hairpin with a Trp residue at the positions equivalent to Arg 117 in the Giardia sequence, and with a highly conserved Phe residue equivalent to Phe 301 in the Giardia sequence. This hydrophobic triad and the corresponding tryptophan are recognizably present in most eukaryotic TrpRS sequences, but are missing from some protozoan homologs.
We report here a crystal structure of the TrpRS from Giardia lamblia determined at 2.1 Å resolution. Unlike all previously characterized TrpRS, the Giardia protein as seen in the crystal forms an (α2)2 homotetramer. An unexpected monomer:monomer interface is formed by reciprocal interdigitation of the N-terminal α-helix containing residues 6–22. The unanticipated N-terminal helix structurally replaces a β-hairpin that has been assumed to be a feature common to eukaryotic TrpRS homologs and has been implicated as being crucial for ATP binding in human TrpRS (Yang et al., 2007). This structural difference between the G. lamblia TrpRS and previously studied eukaryotic TrpRS corresponds to a recognizable sequence difference. The Giardia sequence lacks a set of hydrophobic residues that stabilize previously observed interactions between the β-hairpin element and the ATP binding site (Figure 1).
2. Target choice, protein expression and crystallization strategy
The tryptophanyl-tRNA synthetase from Giardia was chosen for study as part of a broader effort by the Medical Structural Genomics of Pathogenic Protozoa (MSGPP) collaboration to investigate aminoacyl-tRNA synthetases from parasitic protozoa as possible targets for drug discovery (Fan et al., 2008). The structure reported here corresponds to the sole TrpRS found in the G. lamblia genome. The gene encoding the full-length 429-residue G. lamblia tryptophanyl tRNA synthetase (GiardiaDB GL50803 3032) was cloned from strain WB genomic DNA into vector AVA421 (Alexandrov et al., 2004) containing a cleavable N-terminal His6-tag, and expressed in E. coli (Mehlin et al., 2006; Arakaki et al., 2006). The protein was purified using a Ni-NTA column and cleaved on the column by protease 3C overnight at 4° C. The released protein was further purified by gel filtration on a HiLoad Superdex 75 26/60 at a flow rate of 0.5 ml/min and eluted in a single peak corresponding to a dimer. DTT was added to a final concentration of 2mM prior to loading on the gel filtration column. The dimeric state in solution was later confirmed by HPLC (Shimadzu Prominence), which showed an apparent molecular weight of 105.8 kDa.
Purified protein was screened at the high-throughput crystallization facility at Hauptman-Woodward Institute as previously described (Luft et al., 2003; Arakaki et al., 2008). Initial crystallization condition hits were further optimized to produce crystals used for data collection. Crystals were grown at room temperature by vapor equilibration from sitting drops containing 2µL protein solution (11 mg/ml protein, 500 mM NaCl, 20 mM HEPES, 2mM BME, 5% glycerol, 0.025% NaAzide, pH 7.5) mixed with 1µL crystallization buffer (2.7M ammonium sulfate, 5 mM DTT, 0.1 M citric acid, pH 5.2).
3. X-ray diffraction and structure determination
Crystals of G. lamblia TrpRS were briefly soaked in paraffin oil and then frozen directly in liquid nitrogen. Data were collected using SSRL beamline 9.1 at an X-ray energy of 12.658 keV. Data were processed and scaled using HKL2000 (Otwinowski and Minor, 1997). The program Molrep (Vagin and Teplyakov, 1997) found an initial molecular replacement solution using a model based on the structure of yeast TrpRS (Malkowski et al., 2007) (PDB accession code 2ip1, 38% overall sequence identity). Initial structure determination and refinement used data to 2.8 Å resolution from a single crystal. Iterative manual model building using Coot (Emsley and Cowtan, 2004) and in Refmac (Murshudov et al., 1997) continued until R and Rfree reached 0.243 and 0.304, respectively. This initial model was later refined against a higher resolution data set obtained from a second crystal at 2.1 Å resolution to final values R = 0.199, Rfree= 0.234. Model quality was validated using Coot and MolProbity (Lovell et al., 2003). Crystallographic data statistics are given in Table 1. The refined model has been deposited in the Protein Data Bank with accession code 3foc.
Table 1.
Data collection and refinement statistics
| Crystal 1 | Crystal 2 | |
|---|---|---|
| Data collection | ||
| Space Group | P212121 | P212121 |
| Unit cell (Å) | a = 107.8 b = 140.8 c = 91.0 | a = 106.7 b = 140.1 c = 90.3 |
| Wavelength (Å) | 0.9795 | 0.9795 |
| Resolution (Å) | 40-2.8 (2.9-2.8) | 40-2.1 (2.18-2.10) |
| Total reflections | 200381 | 535273 |
| Unique reflections | 32421 | 77780 |
| Rmerge | 0.127 (0.480) | 0.089 (0.610) |
| Completeness (%) | 94(57) | 97(82) |
| Redundancy | 6.2 (2.4) | 6.9 (5.1) |
| I/σ(I) | 12.7 (1.5) | 21 (2.2) |
| Wilson B (Å2) | 66 | 49 |
| Refinement | ||
| Resolution (Å) | 40-2.09 (2.15-2.09) | |
| R | 0.199 (0.284) | |
| Rfree | 0.234 (0.274) | |
| RMSD non-ideality of bond lengths (Å) | 0.007 | |
| RMSD non-ideality of bond angles (°) | 1.003 | |
| Mean BTLS + Biso for 6504 protein atoms (Å2) | 49.4 | |
| Mean Biso for 365 waters (Å2) | 32.9 | |
| Mean Biso for 16 other atoms (Å2) | 77.5 | |
| Residues with ϕ/ψ in favored regions | 798 | |
| Residues with ϕ/ψ in allowed regions | 16 | |
| Residues with ϕ/ψ in disallowed regions | 2 | |
| TLS groups: | A: 1–35, 36–303, 304–356, 357–429 | |
| B: 3–39, 40–103, 104–300, 301–429 |
4. Overall structure
Monomer
The overall structure of the Giardia TrpRS monomer is that of a typical class I aminoacyl-tRNA synthetase. It contains an N-terminal extension (residues 1–64), a Rossman fold catalytic domain (residues 65–324, 415–429), and an anticodon recognition domain (residues 325–414). The crystallographic asymmetric unit contains two monomers, related by a rotation of 177°. The two monomers were treated independently during crystallographic refinement; their respective Cα atoms superimpose with an RMSD of 0.6 Å aligned over 395 residues. Residues 1–219 and 244–420 are well-defined by electron density in monomer A and residues 3–220, 244–344, and 351–429 are well-defined in monomer B. The electron density corresponding to residues of 220–243 in A and 221–243 in B was very weak and these residues, part of a Giardia-specific insertion in the catalytic domain, were not modeled. One sulfate was found at the surface of each monomer between the side chains of residues Arg280 and Arg283.
N-terminal dimer interface
The two monomers in the asymmetric unit form an unusual dimer in which helix α1 in each monomer’s N-terminal extension intertwines with helix α1 of the other monomer (Figure 2). The α1 helices from monomer A monomer B cross at an angle of 70°. Each monomer’s α1 helix is surrounded on all sides by residues from the other monomer, burying a surface area of 1380 Å2 from each monomer (Figure 2). An additional 160 Å2 of surface from each monomer is buried by reciprocal interaction of residues 175–185 from α11, for a total buried surface area of 1540 Å2 on each monomer due to formation of the A:B dimer. While the buried surface is 44% hydrophobic, the interface is further stabilized by three salt-bridges (Glu15B-Arg363A, Arg20B-Asp274A, Asp274B-Arg20A). When monomers A and B of the asymmetric unit are superimposed overall, the Cα atoms of the respective α1-helices differ by 1.5 – 1.7 Å. Thus the helix-helix interaction is not perfectly symmetric.
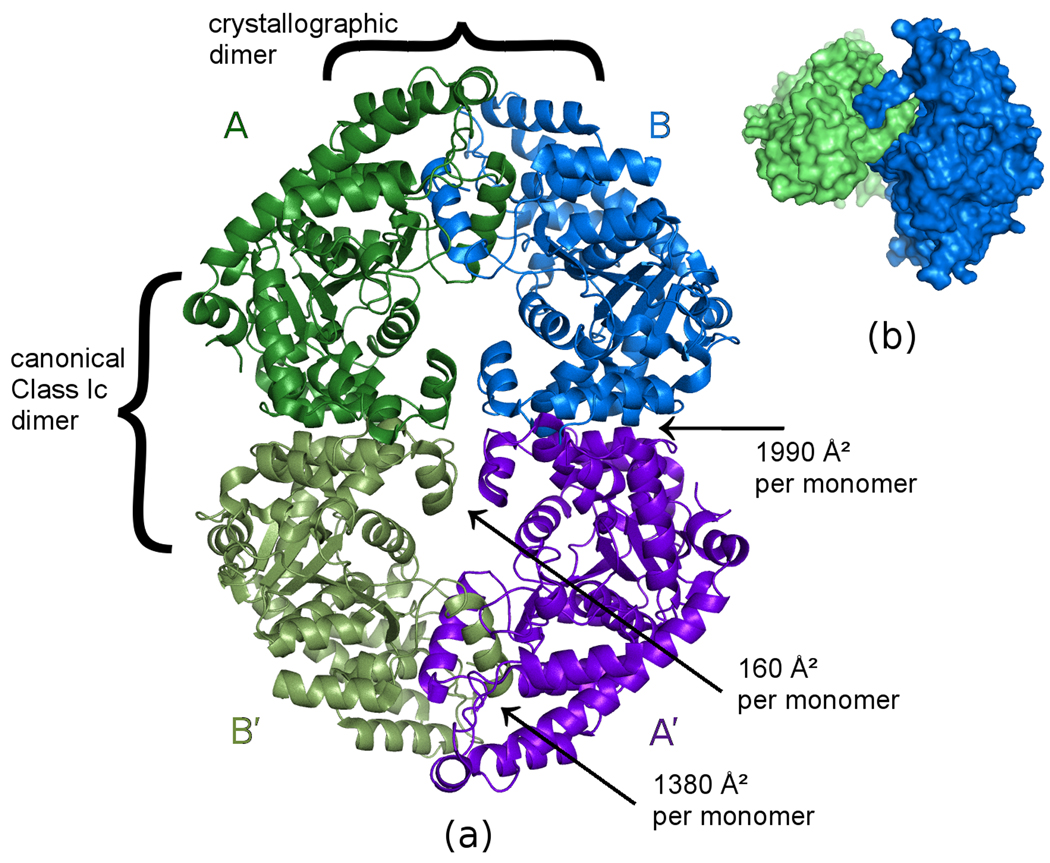
Figure 2. Crystallographically observed tetramer of G. lamblia TrpRS.
(a) The two monomers in the upper half of the figure are crystallographically independent. The view is along the crystallographic 2-fold axis relating the monomers in the upper half of the figure to those in the lower half. The pair of monomers on the left (dark green, light green) and the pair monomers on the right (light blue, dark blue) each form the canonical dimer previously observed for all class Ic aminoacyl-tRNA synthetases. The dimer formed by the two crystallographically independent monomers (dark green, light blue) buries 1380 Å2 of accessible surface on the N-terminus of each monomer due to the interlocked α1 helices, plus an additional 160 Å2 per monomer due to reciprocal interaction of the α11 helices. Buried surface areas were calculated using PISA (Krissinel and Henrick, 2007). (b) Surface representation of the two crystallographically independent monomers, showing how they reciprocally interdigitate to form a dimer. The view is rotated roughly 90° from the view in (a).
Canonical dimer interface
A second dimer interface A:B′ is formed by crystallographic 2-fold symmetry, generating the expected canonical dimer previously observed for all class Ic aminoacyl-tRNA synthetases. The canonical dimer interface buries 1990 Å2 of surface on each monomer, primarily from helices α10, α11, α12 and α16. This interaction surface is larger than that seen in previous structures of eukaryotic class Ic structures, typically 1400–1500 Å2 for TrpRS and 1300–1400 Å2 for TyrRS. The additional interaction surface in the Giardia interface is contributed by residues 246–255, including helix α16, belonging to the Giardia-specific 49-residue insertion in the catalytic domain.
Tetramer
Together the N-terminal and the canonical dimer interactions form a crystallographic (α2)2 homotetramer. The tetrameric assembly has the form of a symmetric figure-eight, with a narrow hole through center of both the upper and lower lobes (Figure 2). We have no evidence that the biologically active form is tetrameric, and indeed the protein appeared to be dimeric on sizing columns (not shown). Nevertheless, the interdigitation of structural elements and the extensive buried surface area of the N-terminal dimer interface, which is actually larger than that of a typical class Ic aaRS dimer interface, both argue against dismissing the observed tetramer as a packing artifact. Furthermore, analysis of the estimated solvation free energy gain ΔiG by PISA (Krissinel and Henrick, 2007) predicts the tetramer to be a stable assembly in solution. The estimated ΔiG due to monomer:monomer association at each N-terminal dimer interface is −25.6 kcal/mol, while the estimated ΔiG due to monomer:monomer association at each canonical dimer interface is −17.5 kcal/mol. This would not be the first time that crystallization stabilized formation of a biologically relevant tetramer that is present only transiently in solution (Yuan et al., 2008).
5. Active site
TrpRS carries out two successive reactions at a single site:
The active site is located in a deep, long pocket within the catalytic domain, and is surrounded by several conserved features. One is the loop formed by the residues 311–315 of the KMSAS motif, characteristic of all class I aminoacyl-tRNA synthetases. The ATP binding site is sandwiched by the characteristic class I HIGH motif (residues 84–87) and by residues 272–275 of a GIEQ motif that is weakly conserved among TrpRS sequences. The Giardia structure exhibits only small differences in these features compared to previously reported eukaryotic TrpRS structures. Helix α5, containing the HIGH motif residues, is shifted along the helix axis by slightly more than 1 Å relative to the previous structures. The GIEQ loop in both monomers is intermediate in position between that previously seen for apo structures (e.g. PDB entries 1o5t, 2ip1, 2quk) and that consistently seen in homologous eukaryotic structures with substrates bound (1r6u, 1r6t, 2azx, 2dr2, 2qui). This suggests that the Giardia active site may require less conformational adjustment on substrate binding than the active site of the human or yeast enzymes.
6. Structural divergence of the N-terminal extension in eukaryotes
Archaeal and eukaryotic cytosolic TrpRS homologs contain additional residues N-terminal to the conserved catalytic core domain. The length of this additional region varies with the organism. The sequence immediately preceding the catalytic core is itself weakly conserved across eukaryotes. This “eukaryote-specific extension” has been characterized as corresponding to residues 82–154 of the human TrpRS sequence (Yang et al., 2007). By this definition it is a 73-residue element that would subsume the first 68 residues of the G. lamblia TrpRS sequence. Residues 23–68 of the present Giardia structure, containing the structural elements α2, α3, β1, and α4, indeed superimpose very closely onto the human and yeast TrpRS structures. The root mean square difference for the 46 Cα atoms after superposition is 0.7 Å and 1.0 Å, respectively. This suggests that the structural homology, and plausibly the functional significance, of these 46 residues does in fact span eukaryotes from protozoans to plants to mammals. However, the present structure provides evidence for both structural and functional divergence among eukaryote sequences N-terminal to α2 of the Giardia TrpRS.
The functional role of the entire 150-residue N-terminal extension in human TrpRS has been extensively characterized. A short mini-TrpRS splicing variant lacking residues 1–47 retains normal activity as an aminoacyl-tRNA synthetase (Wakasugi et al., 2002; Kise et al., 2004), as does the proteolytic cleavage product T1-TrpRS lacking residues 1–70. A shorter naturally occurring proteolytic cleavage product T2-TrpRS lacking residues 1–93 shows 25-fold less affinity for ATP (Yang et al., 2007) and is inactive as a synthetase (Otani et al., 2002; Yu et al., 2004). The interesting point here is the implication that residues 71–93 of human TrpRS are required for normal synthetase activity even though they lie outside of the conserved canonical class I catalytic domain. Structures of human TrpRS in complex with relevant substrate molecules (Yang et al., 2003; Yu et al., 2004; Shen et al., 2008) have shown that human TrpRS residues 82–93 form a two strand β-hairpin adjacent to the active site, but only do so in the presence of bound Trp-AMP or ATP + Trp-amide. This hairpin, and indeed the entire region N-terminal to residue 93, is disordered in structures of human TrpRS with an empty active site, with bound tryptophan alone, or in complex with the cognate tRNATrp(Shen et al., 2006). In the pre-transition and product states of the tryptophan activation reaction, the β1/β2−hairpin is well-ordered and caps the ATP binding site, although it does not interact directly with with the substrates (Shen et al., 2008). The position of the hairpin relative to the core domain is stabilized by hydrophobic interactions; hairpin residue Trp88 stacks with Trp203, while Val90 lies against Phe339. This phenylalanine sidechain forms part of the binding site for the purine ring of the ATP substrate, and is conserved across eukaryotic TrpRS homologs including that from Giardia. Residues 92–94 at the base of the hairpin are adjacent to the residues forming the active site KMSAS motif. Mutational studies showing that Val85 is important to tryptophan activation by the human TrpRS (Guo et al., 2007) further support the conclusion that this β-hairpin is essential for binding the ATP substrate and guiding the activation of tryptophan into Trp-AMP.
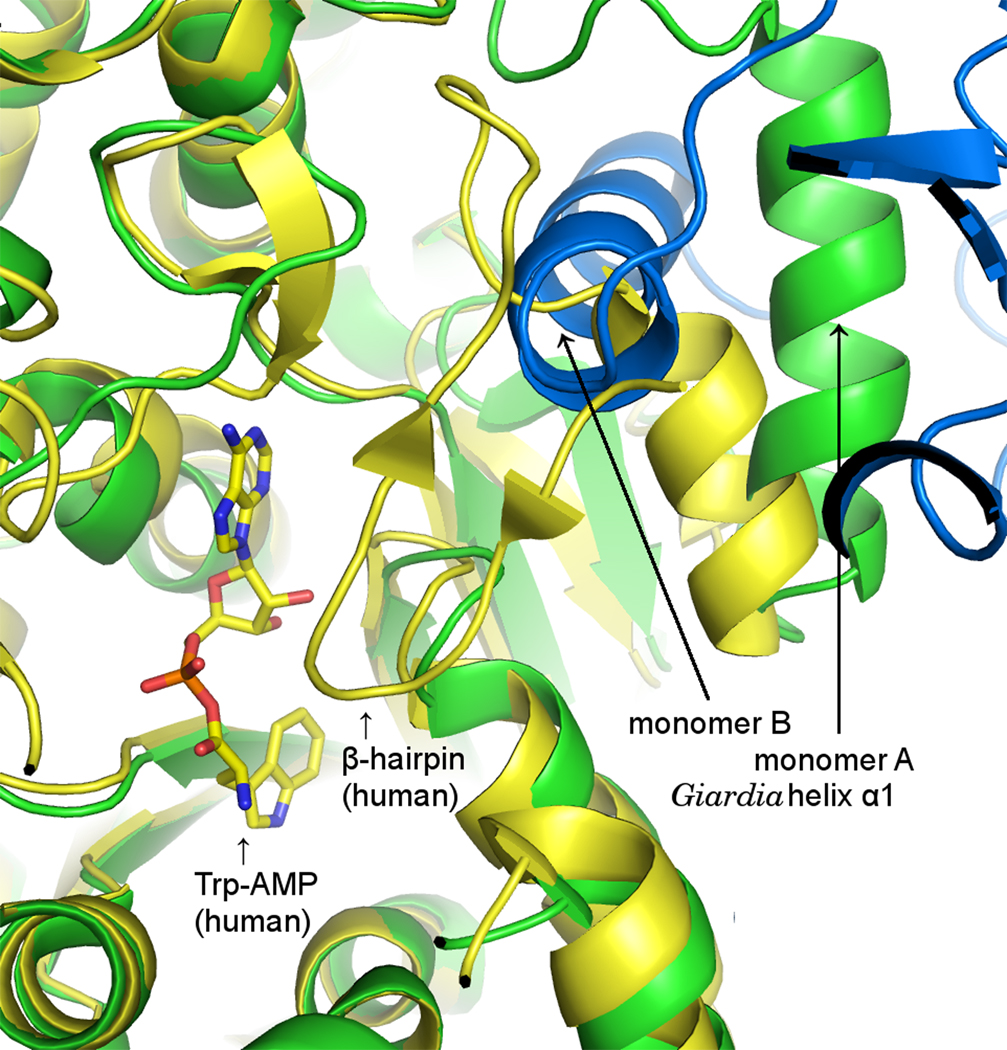
Given that hydrophobic residues in the β1/β2 hairpin are instrumental for tryptophan activation by human TrpRS, it is somewhat surprising that the G. lamblia TrpRS sequence instead contains hydrophilic residues (Thr, Glu) at the positions equivalent to hairpin residues Val85 and Val90 of the human sequence. It also lacks homologs for the interacting pair of hairpin/active-site tryptophans, Trp88/Trp203 in the human sequence. These sequence differences are explained by the present crystal structure, in which the β1/β2−hairpin is replaced structurally by a well-ordered α1 helix even in the absence of bound ligands. The α1 helix does not cap the ATP-binding site of its own monomer; instead it extends into a cavity of another monomer to form the reciprocally interdigitated tetramer interface. This reciprocal swap places the α1 helix of the neighboring monomer adjacent to the active site residues forming the the KMSAS motif (Figure 3), although it does not limit access to the ATP-binding site as severely as the tip of the hairpin does in the human TrpRS structure. While tRNA-binding to human TrpRS requires displacement of the β-hairpin, our initial modeling suggests that tRNA could bind to the full tetrameric assembly of Giardia TrpRS without requiring substantial displacement of the α1 helices.
Figure 3. Superposition of human and Giardia TrpRS active sites.
The complex of human TrpRS with Trp-AMP (PDB 1r6t; Yang et al., 2003) is shown in yellow. The β1β2 hairpin in the center of the figure is necessary for ATP binding and subsequent tRNA-independent activation of tryptophan to yield Trp-AMP (Otani et al., 2002; Guo et al., 2007). The hairpin is necessarily displaced to allow access by the acceptor arm of tRNA so that the activated tryptophan may be transferred to the tRNA. One monomer of the Giardia TrpRS (green) is superimposed onto the human structure (yellow). The short helix + β1β2 hairpin of the human structure is replaced by a single, longer helix α1 at the N-terminus of the Giardia TrpRS monomer (green). The Giardia helix α1 interlocks with the equivalent helix α1 from a second monomer (blue) in the tetramer, so that the active site is bounded by residues from two contributing monomers of the tetramer.
Whether or not the (α2)2 homotetramer observed in the present Giardia TrpRS structure is the biologically active form, we may nevertheless infer from the absence of residues homologous to human TrpRS residues Val85, Trp88, and Val90, and the concomitant replacement of the β-hairpin by α1, that the hairpin-mediated control of tryptophan activation is not shared by all eukaryotic TrpRS homologs. The N-terminal extension in TrpRS homologs from the parabasalid protozoa Trichomonas vaginalis is essentially the same length as the Giardia homolog and likewise does not exhibit conservation of the triplet of hydrophobic hairpin residues or the hydrophobic interaction partner (Trp203 in human) at the active site. The N-terminal TrpRS extension from the more distantly related apicoplexan protozoan Toxoplasma gondii also lacks homologous residues for the signature Val/Trp/Val hairpin residues, although they are present in other apicomplexan species. This appears to indicate that in some protozoa tryptophan activation proceeds without mediation by a β-hairpin.
7. Speculative model for tRNA binding
The genetic code contains only one Trp codon, and tRNATrp is recognized via its unique anticodon sequence CCA. The Giardia TrpRS anticodon binding site is formed by residues 335–345 and 388–393. It is structurally similar to the anticodon binding site in other TrpRS structures. All sidechains previously observed to make direct contact with the anticodon bases in the human TrpRS:tRNA complex are conserved in the Giardia TrpRS sequence (Ser 340, Asp 344, Thr 389, Lys 393).
In order to determine if the observed tetrameric assembly of Giardia TrpRS is consistent with tRNA binding, we superimposed the structure of the human TrpRS in complex with tRNATrp onto the Giardia TrpRS tetramer (Figure 4). The biological dimer of the human structure (PDB 2dr2) was first generated by application of crystallographic symmetry to yield an A:A′ dimer in complex with two molecules of tRNATrp. Only one tRNA molecule was then retained. Superposition of monomer A from the human structure, associated with the acceptor stem of the tRNA, onto monomer A of the Giardia structure using the algorithm SSM (Krissinel and Henrick, 2004) yielded an RMSD fit of 1.42 Å for 351 Cα atoms. This resulted in an imperfect superposition of monomer A′ from the human structure, to which the anticodon loop of the tRNA was bound, onto monomer B′ of the Giardia tetramer, yielding a displacement of 9 Å between their respective anticodon binding sites. In constructing a model for tRNA binding to the Giardia tetramer, we assumed that the monomer:monomer orientation observed in the crystal structure would remain constant. The displacement of the anticodon binding site can be accommodated by pivoting the tRNA by approximately 7° about a point near the active site, but we did not attempt to model this precisely.
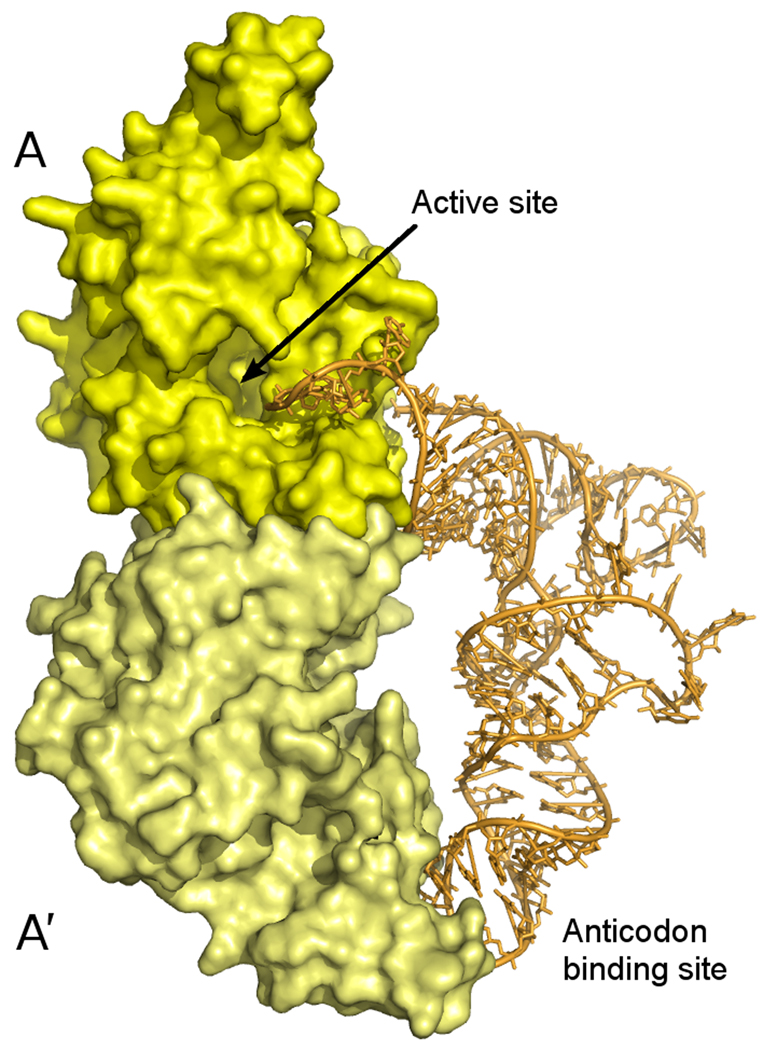
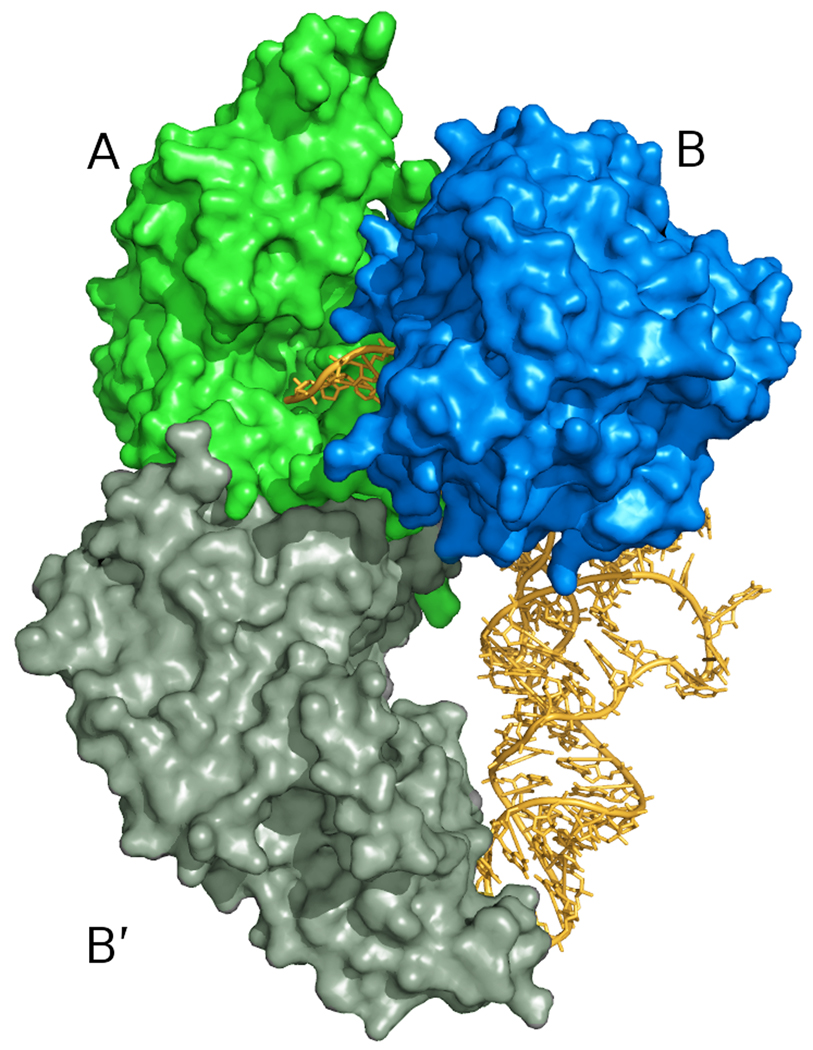
Figure 4. Model for tRNA binding.
(a) tRNA bound to the dimeric human TrpRS (PDB 2dr2). Although two tRNA molecules are bound to each dimer in the crystal structure, for clarity only one is shown here. (b) A crude model for tRNA binding to the Giardia TrpRS tetramer. The human TrpRS:tRNA complex was positioned onto the full Giardia tetramer by superposition of the monomer containing the active site (upper monomer in (a), green monomer in (b)). The fourth monomer of the Giardia structure is omitted for clarity. The acceptor arm of the tRNA molecule extends through the central hole in the upper lobe of the tetramer to enter the active site of the monomer at the upper left.
In the context of the tetrameric assembly, this simple superposition results in a crude model for tRNA binding in which the anticodon loop of the tRNA binds to monomer B′ in the lower lobe of the figure-eight shaped tetramer, the acceptor arm of the tRNA passes through the central hole of the A:B dimer constituting the upper lobe of the figure-eight, and the terminal CCA of the acceptor arm extends into the active site of monomer A (Figure 4). Two clashes between the tRNA and the protein result from this very approximate superposition. A clash between residues from helix α11 and the pyrimidine ring of C74 at the base of the tRNA acceptor arm can plausibly be resolved by a conformational shift within this one cytidine residue. A second clash occurs between the acceptor stem proper and the loop consisting of residues 183–186 in monomer B. This loop has no regular secondary structure, and is the point of maximal conformational difference between the two crystallographically independent monomers in the current Giardia TrpRS structure. Again it seems plausible that the clash can be resolved, in this case by a relatively minor conformational shift in this flexible loop.
The inherent symmetry of the tetramer makes it possible to model simultaneous binding of a second tRNA molecule to the same tetramer. Each tRNA would thus fill one of the two holes through the figure-eight shape of the tetramer (Figure 2). Because the tetramer has only two holes, no additional tRNA molecules can be accommodated. As a consequence, this model predicts a 1:2 stoichiometry of tRNA binding to the tetrameric TrpRS rather than the 1:1 stoichiometry observed for dimeric TrpRS, where two tRNA molecules can bind per dimer.
Acknowledgments
This work was funded by NIAID award P01AI067921 (Medical Structural Genomics of Pathogenic Protozoa). Genomic DNA was kindly provided by Dr. Rod Adam, University of Arizona Department of Immunobiology. We thank Eric Larson for aid in data collection. We are also reliant on the efforts of other members of the MSGPP consortium, including Christine Stewart, Lisa Castaneda, and Angela Kelley. We would especially like to thank the Hauptman-Woodward Institute for providing high-throughput screening for crystallization conditions. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrov A, Vignali M, LaCount DJ, Quartley E, de Vries C, Rosa DD, Babulski J, Mitchell SF, Schoenfeld LW, Fields S, Hol WG, Dumont ME, Phizicky EM, Grayhack EJ. A facile method for high-throughput co-expression of protein pairs. Mol. Cell. Proteomics. 2004 Sep;3:934–938. doi: 10.1074/mcp.T400008-MCP200. [DOI] [PubMed] [Google Scholar]
- Arakaki T, Trong IL, Phizicky E, Quartley E, DeTitta G, Luft J, Lauricella A, Anderson L, Kalyuzhniy O, Worthey E, Myler PJ, Kim D, Baker D, Hol WGJ, Merritt EA. Structure of Lmaj006129AAA, a hypothetical protein from Leishmania major. Acta Cryst. 2006;F62:175–179. doi: 10.1107/S1744309106005902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki TL, Buckner FS, Gillespie JR, Malmquist NA, Phillips MA, Kalyuzhniy O, Luft JR, DeTitta GT, Verlinde CLMJ, Van Voorhis WC, Hol WGJ, Merritt EA. Characterization of Trypanosoma brucei dihydroorotate dehydrogenase as a possible drug target; structural, kinetic and RNAi studies. Molecular Microbiology. 2008;68:37–50. doi: 10.1111/j.1365-2958.2008.06131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fan E, Baker D, Gelb MH, Buckner FS, Van Voorhis WC, Phizicky E, Dumont M, Mehlin C, Grayhack EJ, Sullivan M, Verlinde CL, DeTitta G, Meldrum D, Merritt EA, Earnest TN, Soltis M, Zucker F, Myler P, Schoenfeld L, Kim D, Worthey EA, LaCount D, Vignali M, Li J, Mondal S, Massey A, Carroll B, Gulde S, Luft JR, DeSoto L, Holl M, Caruthers JM, Bosch J, Robien MA, Arakaki T, Holmes MA, LeTrong I, Hol WG. Structural genomics of pathogenic protozoa: An overview. Methods in Molecular Biology. 2008;426:497–513. doi: 10.1007/978-1-60327-058-8_33. [DOI] [PubMed] [Google Scholar]
- Guo L-T, Chen X-L, Zhao B-T, Shi Y, Li W, Xue H, Jin Y-X. Human tryptophanyl-tRNA synthetase is switched to a tRNA-dependent mode for tryptophan activation by mutations at V85 and I311. Nucleic Acids Res. 2007;35:5934–5943. doi: 10.1093/nar/gkm633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kise Y, Lee SW, Park SG, Fukai S, Sengoku T, Ishii R, Yokoyama S, Kim S, Nureki O. A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat Struct Mol Biol. 2004;11:149–156. doi: 10.1038/nsmb722. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallographica Section D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. URL http://dx.doi.org/10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Molec. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lovell S, Davis I, Arendall WB, III, de Bakker P, Word J, Prisant M, Richardson J, Richardson D. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins: Structure, Function, and Genetics. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J. Struct. Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- Malkowski M, Quartley E, Friedman A, Babulski J, Kon Y, Wolfley J, Said M, Luft J, Phizicky E, DeTitta G, Grayhack E. Blocking S-adenosylmethionine synthesis in yeast allows selenomethionine incorporation and multiwavelength anomalous dispersion phasing. Proc Natl Acad Sci U S A. 2007;104:6678–6683. doi: 10.1073/pnas.0610337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlin C, Boni E, Buckner FS, Engel L, Feist T, Gelb M, Haji L, Kim D, Liu C, Mueller N, Myler PJ, Reddy JT, Sampson JN, Subramanian E, Van Voorhis WC, Worthey E, Zucker F, Hol WGJ. Heterologous expression of proteins from plasmodium falciparum: results from 1000 genes. Molecular and Biochemical Parasitology. 2006;148:144–160. doi: 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, Davids BJ, Dawson SC, Elmendorf HG, Hehl AB, Holder ME, Huse SM, Kim UU, Lasek-Nesselquist E, Manning G, Nigam A, Nixon JEJ, Palm D, Passamaneck NE, Prabhu A, Reich CI, Reiner DS, Samuelson J, Svard SG, Sogin ML. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otani A, Slike BM, Dorrell MI, Hood J, Kinder K, Ewalt KL, Cheresh D, Schimmel P, Friedlander M. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc Natl Acad Sci U S A. 2002;99:178–183. doi: 10.1073/pnas.012601899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Shen N, Guo L, Yang B, Jin Y, Ding J. Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res. 2006;34:3246–3258. doi: 10.1093/nar/gkl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Zhou M, Yang B, Yu Y, Dong X, Ding J. Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states. Nucleic Acids Res. 2008;36:1288–1299. doi: 10.1093/nar/gkm1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda M, Kusakabe Y, Tanaka N, Ohno S, Nakamura M, Senda T, Moriguchi T, Asai N, Sekine M, Yokogawa T, Nishikawa K, Nakamura KT. Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms. Nucleic Acids Res. 2007;35:4289–4300. doi: 10.1093/nar/gkm417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molrep: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- Wakasugi K, Slike BM, Hood J, Friedlander KLEM, Cheresh DA, Schimmel P. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci U S A. 2002;99:173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Otero F, Skene R, McRee D, Schimmel P, de Pouplana L. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc Natl Acad Sci U S A. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-L, Guo M, Kapoor M, Ewalt KL, Otero FJ, Skene RJ, McRee DE, Schimmel P. Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase. Structure. 2007;15:793–805. doi: 10.1016/j.str.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-L, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Kohrer C, RajBhandary UL, Skene RJ, McRee DE, Schimmel P. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaremchuk A, Kriklivyi I, Tukalo M, Cusack S. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 2002;21:3829–3840. doi: 10.1093/emboj/cdf373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Liu Y, Shen N, Xu X, Xu F, Jia J, Jin Y, Arnold E, Ding J. Crystal structure of human tryptophanyl-tRNA synthetase catalytic fragment - Insights into substrate recognition, tRNA binding, and angiogenesis activity. J Biol Chem. 2004;279:8378–8388. doi: 10.1074/jbc.M311284200. [DOI] [PubMed] [Google Scholar]
- Yuan P, Gupta K, Duyne GDV. Tetrameric structure of a serine integrase catalytic domain. Structure. 2008;16:1275–1286. doi: 10.1016/j.str.2008.04.018. [DOI] [PubMed] [Google Scholar]