Abstract
Antimicrobial peptides (AMPs) are small, usually cationic peptides, which permeabilize biological membranes. Their mechanism of action is still not well understood. Here we investigate the preference of alamethicin and melittin for pores of different shapes, using molecular dynamics (MD) simulations of the peptides in pre-formed toroidal and cylindrical pores. When an alamethicin hexamer is initially embedded in a cylindrical pore, at the end of the simulation the pore remains cylindrical or closes if glutamines in the N-termini are not located within the pore. On the other hand, when a melittin tetramer is embedded in toroidal pore or in a cylindrical pore, at the end of the simulation the pore is lined both with peptides and lipid headgroups, and, thus, can be classified as a toroidal pore. These observations agree with the prevailing views that alamethicin forms barrel-stave pores whereas melittin forms toroidal pores. Both alamethicin and melittin form amphiphilic helices in the presence of membranes, but their net charge differs; at pH ~7, the net charge of alamethicin is −1 whereas that of melittin is +5. This gives rise to stronger electrostatic interactions of melittin with membranes than those of alamethicin. The melittin tetramer interacts more strongly with lipids in the toroidal pore than in the cylindrical one, due to more favorable electrostatic interactions.
Keywords: molecular dynamics simulations, antimicrobial peptides, toroidal pore, barrel-stave pore, melittin, alamethicin
Introduction
Antimicrobial peptides (AMPs) are produced by virtually all organisms, from invertebrates and plants to animals and humans, and play an important role in their defense system [1–3]. AMPs are active against bacteria, fungi, parasites, enveloped viruses such as HIV, and even cancer cells [1, 2, 4–8]. In recent years, antimicrobial peptides have attracted worldwide attention due to their potential as a replacement for conventional antibiotics [9]. A potential advantage over conventional antibiotics is that, since they are thought to target cell membranes directly, they are less likely to be affected by antibiotic resistance. The origin of their cell selectivity and their mechanism of action, however, are still not fully understood. The cell selectivity seems to be related to different properties of prokaryotic and eukaryotic membranes [10–12].
Most AMPs are small (consisting of 12–50 amino acids), usually cationic peptides, which, in the presence of membranes, fold into amphiphilic α-helices (or, less often, β-sheets, or are unfolded). Generally, AMPs are believed to interact directly with membranes. Cationic peptides normally bind more strongly to anionic bacterial membranes than to zwitterionic mammalian membranes [10, 11]. The configuration of AMPs in membranes depends on peptide-to-lipid ratio [13]. At low ratio, peptides are adsorbed on the target membrane in an interfacial orientation. Above a threshold peptide-to-lipid ratio, which depends on peptide and type of lipids, the peptides reorient to a transmembrane orientation. The inserted peptides may aggregate and form pores that depolarize the membrane, induce leakage of cell components and finally kill a cell [13, 14], or they may disintegrate and/or micellize the membrane (the carpet mechanism) [15]. Two types of pores have been proposed: barrel-stave and toroidal pores. The classical picture of the barrel-stave pore is a highly ordered cylindrical water pore surrounded by peptides in the transmembrane orientation; the peptides are in direct contact with each other [16, 17]. Peptides acting via the toroidal pore mechanism first adsorb on a membrane surface, aggregate and impose thinning of the membrane and the expansion of the headgroup region; this induces a bending of the bilayer so that the upper and lower leaflets meet, giving a toroidal appearance to the pore formed. The pore is usually thought of as a well ordered structure, with a mixture of peptides and lipid headgroups lining its interior; it is smallest in the middle and largest at the ends [6, 18–20].
Recent atomistic MD simulations of pore formation induced by magainin (MG-H2) revealed a different picture of toroidal pores [21]. The pores are significantly irregular with only one or two monomers located in the center of the pore and other peptides located at the pore rim; the peptides adopt a variety of orientations. The authors termed this pore “disordered toroidal pore”. A similar picture emerged from simulations of melittin in a DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine) bilayer [22]. Protegrin-1 pores also appear to be disordered [23]. Combined coarse-grained and atomistic MD simulations of alamethicin in a DMPC (1,2-dimyristoyl-sn-glycero-3-phophocholine) bilayer suggest that barrel-stave pores are also more disordered than in the classical view [24]. It is surprising that only one or two peptides are found in the center of pores produced by MG-H2 and melittin; previously, it was estimated that 4–7 monomers of magainin 2 (which is similar to MG-H2) are embedded in a pore [19]. The simulations were fairly long (~250 ns) but, given the long relaxation times in atomistic MD simulations, they might not be long enough to observe aggregation of peptides in pores or pores that resemble more closely the classical view.
In this paper we report a computational study of two peptides, alamethicin and melittin. Alamethicin is a 20-residue peptide produced by the fungus Trichoderma viride [25]; it belongs to the family of peptaibols, characteristic for a high content of non-standard α-methylalanine (Aib) residues. Alamethicin forms an amphiphilic α-helix, with a kink at Pro 14; the C-terminus is more polar than the N-terminus [26]. A previous study from our group using an implicit membrane model showed that, in the absence of voltage, alamethicin monomer preferentially binds to a membrane in a slightly tilted, interfacial orientation, while voltage favors a more deeply inserted, almost transmembrane orientation [27].
Melittin is a 26-residue peptide isolated from bee venom [28]. It is unstructured in solution [29] but folds into an α-helix in the presence of lipids [30]. Both peptides have been extensively studied experimentally and computationally [17, 18, 20, 24, 31–49]. Common for alamethicin and melittin is that they bind to membrane surfaces in an interfacial orientation at low peptide-to lipid ratios whereas they insert into the membrane at higher concentrations [20, 50–52]. The mechanism of action of alamethicin and melittin, however, differs. Pores of alamethicin are generally described as barrel-stave [16, 17, 24], with a central water pore surrounded by 4–11 peptides [53]. Melittin is commonly believed to form toroidal pores [18, 20, 22, 49], although others reported that it acts via a barrel-stave mechanism [54] or, in anionic lipids, as a detergent [48].
The goal of this study is to investigate not the formation of pores starting from a flat lipid bilayer (as has been done in the aforementioned computational studies [22, 24]) but the reported preference of the peptides for different pore shapes. Thus, we conduct atomistic MD simulations of alamethicin and melittin monomers and oligomers inserted into pre-formed toroidal and cylindrical pores. This strategy allows us to circumvent the potential problem of long time scales and to explore alternative scenarios for pore formation, closer to the classical view established experimentally. During the simulations we observed a melittin tetramer transforming a cylindrical pore into a toroidal pore and an alamethicin hexamer preserving a cylindrical pore, if its glutamines (Q7) are located within the pore region. The melittin tetramer interacts more strongly with lipids in a toroidal pore than in a cylindrical pore, which, we argue, might be key to its mechanism of action. In a companion paper reporting MD simulations of antimicrobial peptides in implicit cylindrical and toroidal pores, we show that melittin is better solvated in curved pores than in cylindrical ones [55].
Methods
Initial structures
The coordinates for alamethicin and melittin were obtained from the Protein Data Bank, entries 1AMT [26] and 2MLT [56, 57], respectively (chain A from the PDB files was used for both peptides). Oligomers of alamethicin and melittin were formed by replicating the monomers. The sequence of alamethicin is: Ace-Aib-Pro-Aib-Ala-Aib-Ala-Gln-Aib-Val-Aib-Gly-Leu-Aib-Pro-Val-Aib-Aib-Glu-Gln-Phl, where Ace is acetylated N-terminus, Aib is α-methylalanine and Phl is phenylalaninol; the sequence of melittin is: Gly-Ile-Gly-Ala-Val-Leu-Lys-Val-Leu-Thr-Thr-Gly-Leu-Pro-Ala-Leu-Ile-Ser-Trp-Ile-Lys-Arg-Lys-Arg-Gln-Gln. All charged residues were in the standard ionization state corresponding to pH ~7. In a membrane with a pore, charged residues are located either at the membrane-water interface or within the water pore, which reduces the desolvation penalty for transferring charges into the hydrophobic region of the membrane. Charged residues might be important for the preferred pore shape (i.e. barrel-stave versus toroidal), as has been discussed in the mean-field study by Zemel et al. [58]. Ideally, one should do constant-pH MD simulations that allow the ionization state to change, but these are quite expensive.
Simulation setup
Membrane with a pre-formed cylindrical pore
Membrane Builder in the CHARMM-GUI website (http://www.charmm-gui.org) [59] was used to build the initial setup of the membrane with a cylindrical pore. It consisted of 71 DMPC lipids, 4096 water molecules (modeled as TIP3P), 7 K+ and 7 Cl− ions; the radius of the pore was 15 Å. A MD simulation was performed using the CHARMM software [60]. The miscellaneous mean field potential (MMFP) was applied to keep the headgroups close to ±17Å (planar restraints) and to keep the lipid tails away from the pore (cylindrical restraints, the cylinder having radius of 15 Å and its long axis along the z axis). The energy of the system was minimized before the onset of a 375-ps equilibration and a 4-ns of constant pressure (P=1atm) and temperature (T=303.15K) (CPT) MD simulation. Particle Mesh Ewald was used for the calculation of electrostatics and periodic boundary conditions were applied in all three dimensions; the initial size of the primary box was 56.4 Å × 56.4 Å × 64 Å.
Membrane with a pre-formed toroidal pore
The initial simulation setup was the same as for the membrane with the cylindrical pore. However, after the energy minimization, MMFP restraints were gradually released during a 375-ps equilibration, followed by a 4-ns unrestrained CPT MD simulation; this resulted in the formation of a toroidal pore (i.e., a pore with headgroups located within the pore region; see below for more details). The last membrane structure from the simulations (see the snapshots at 0 ns in Figures 1, 2 and 4) was used in the subsequent simulations of peptides in pores.
Figure 1.

Snapshots from MD simulations of a melittin monomer started from a toroidal pore (A), a melittin monomer started from a cylindrical pore (B), an alamethicin monomer started from a toroidal pore (C), and an alamethicin monomer started from a cylindrical pore (D). The green balls represent the phosphocholines; the red balls represent ions; the blue balls represent water; the nonpolar residues are shown in black, the polar residues are yellow and the charged residues are magenta; the lipid tails have been removed for clarity.
Figure 2.
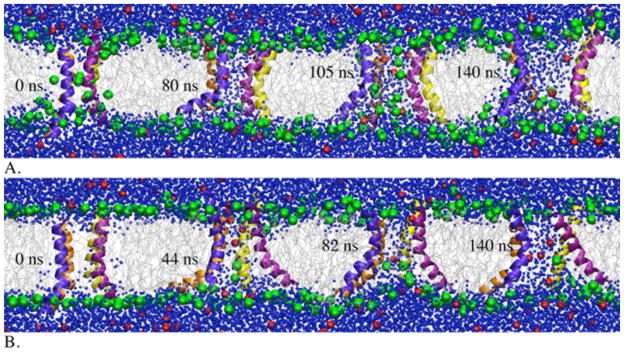
Snapshots from MD simulations of a melittin tetramer started from a toroidal pore (A) and started from a cylindrical pore (B). The green balls represent the phosphocholines; the red balls represent Cl− ions; the gray lines represent lipid tails; the blue balls represent water.
Figure 4.
Snapshots from MD simulations of an alamethicin tetramer started from a toroidal pore (A) and an alamethicin hexamer started from a cylindrical pore: closed pore (B) and wet pore (C). The green balls represent the phosphocholines; the red balls represent K+ ions; the gray lines represent lipid tails; the blue balls represent water; in (C), the peptide are shown as ribbons and the lipid tails are removed for clarity.
Peptides inserted into a pore
The number of monomers in alamethicin pores is estimated to be 4–11, depending on type of lipids (6 in a DMPC bilayer) [53]. Here, we have performed simulations of an alamethicin tetramer and hexamer. Yang et al. reported that at peptide-to-lipid ratio (P/L) of 1/30 melittin is always oriented perpendicular to the membrane surface of a DMPC bilayer [20]. We thus chose a melittin tetramer for our simulations to ensure that peptide-to-lipid ratio is in the range at which pores were observed (in the simulations, P/L = 4/71).
Alamethicin or melittin monomers or oligomers were placed in a cylindrical or toroidal pore, with their apolar face facing the lipids or in a random orientation (see below), as shown in the snapshots at 0 ns in Figures 1, 2 and 4, and the system was solvated in water. In one set of simulations of peptides in the cylindrical pore, geometric restraints were applied to the phosphocholines to keep them above ±13Å and away from the pore and to methyls to prevent lipid tails from entering the pore (we refer to this set of simulations as “constrained cylinder”); in the other set of simulations, the constraints were removed. Details of the systems simulated are given in Table 1. Counterions (Cl− or K+) were added to neutralize the charges of the peptides; no additional ions were present. All simulations were performed using the NAMD software [61]. Each system was energy-minimized, equilibrated for 575 ps, and subjected to a CPT MD simulation (P=1atm; T=303.15K), with periodic boundary conditions in all three dimensions and particle mesh Ewald used for the calculation of electrostatics; the initial size of the primary box was 56.4 Å × 56.4 Å × 64 Å.
Table 1.
Summary of the MD simulations performed.1
| peptide/pore2 | number of monomers | number of water molecules | number of K+ ions | number of Cl− ions | Time (ns) |
|---|---|---|---|---|---|
| melittin/torus | 1 | 3763 | 0 | 5 | 13 |
| 4 | 3278 | 0 | 20 | 140 | |
| melittin/cylinder | 1 | 3767 | 0 | 5 | 13 |
| 4 | 3100 | 0 | 20 | 140 | |
| melittin/constrained cylinder | 1 | 3767 | 0 | 5 | 13 |
| 4 | 3100 | 0 | 20 | 13 | |
| alamethicin/torus | 1 | 3869 | 1 | 0 | 13 |
| 4 | 3525 | 4 | 0 | 130 | |
| alamethicin/cylinder | 1 | 3903 | 1 | 0 | 13 |
| 6 | 3140 | 6 | 0 | 100 | |
| 63 | 3113 | 6 | 0 | 160 | |
| alamethicin/constrained cylinder | 1 | 3903 | 1 | 0 | 13 |
Each system contains 71 DMPC lipids.
The pore configuration at the beginning of the simulation.
The simulation is started with glutamines (Q7) oriented towards the pore interior.
The CHARMM27 force field [62, 63] was used in all simulations. Since alamethicin has non-standard residues (α-methylalanine, Aib, and phenylalaninol, Phl), new entries have been added to the topology and parameter files. The topology and parameters for Aib were developed based on similarities with alanine and those for Phl based on similarities with phenylalanine and serine side chain.
For data analysis, interaction energies between lipids and peptides were calculated from simulation trajectories saved every 200 ps; the average energies were calculated from 1 ns until the end of the simulations. The pore radius was calculated using the COOR SEARCH command in the CHARMM software [60]. This command searches for a hole within a protein, big enough to hold a water molecule. The tilt angles were calculated as the angle between a helix axis and the bilayer normal. The COOR HELIX command in CHARMM was used to calculate the helix axis of peptides from simulation trajectories. The helix axis of alamethicin was defined by the Cα atoms of residues 5 to 15; the helix axis of melittin was defined by the Cα atoms of residues 5 to 22 whereas the helix axes of the N-terminal and C-terminal helices of melittin were defined by the Cα atoms of residues 1 to 10 and 13 to 26, respectively.
In the classical view of a toroidal pore, the two leaflets of the bilayer are thought to bend toward each other so that their headgroups meet. The pores formed in the MD simulations are, however, more inhomogeneous. Here, we consider that a pore is toroidal if several headgroups enter the pore region (as opposed to a barrel-stave pore in which headgroups are located exclusively at the membrane surface). In some of the simulations, headgroups from the two leaflets completely meet at one or both faces of the pore; in others, a few headgroups transiently enter and exit the pore region but the leaflets are not in contact with each other.
Results
1. Monomers inserted into pores
A monomer of melittin or of alamethicin was simulated starting from toroidal and cylindrical pores. As Figure 1 shows, during the 13-ns MD simulations in the pores, the apolar face of the helical peptides interacts with lipid tails whereas the polar face is in contact with water and headgroups. Soon after the onset of the simulations in the cylindrical pores, headgroups enter the pore region, shielding lipid tails from water, and the pore evolves into a disordered toroidal pore. Disordered toroidal pores have previously been observed in atomistic MD simulations of magainin and melittin, in DPPC bilayers [21, 22]. These studies have reported that a monomer is unable to induce pore formation, but after the pore is formed, only one or two monomers are inserted in the pore, in a variety of orientations and rarely in a perfect transmembrane orientation. Although we did not simulate pore formation per se and, thus, cannot draw conclusions about the ability of monomers to form pores, our simulations suggest that, regardless of which monomer is inserted and what is the starting shape of the pore, the ending pore configuration is toroidal. The same result is obtained for pure bilayers (see Methods). Toroidal pores are preferred in pure bilayers because they allow lipid tails to be shielded from water by headgroups located within the pore. Obviously, a single peptide does not suffice to counteract this preference.
2. Melittin tetramer inserted into pores
Since oligomerization of peptides is thought to be a prerequisite for pore formation, we next performed MD simulations of melittin and alamethicin oligomers inserted into pores. Snapshots of a melittin tetramer in toroidal and cylindrical pores are shown in Figures 2A and 2B. The more hydrophilic C-termini of peptides are anchored to the headgroup region of the upper leaflet whereas the N-termini, which appear to be more mobile, are in contact with the lower leaflet of the membrane. The apolar face of the helices interacts with lipids whereas the polar face is oriented toward the interior of the pore.
The melittin tetramer embedded into a toroidal pore (Figure 2A) preserves the toroidal pore shape (with the headgroups located within the pore region). At the end of the 140-ns MD simulation, the inner radius of the pore is ~10 Å and the outer radius ~21 Å. These radii are significantly smaller than the radii of melittin pores determined using neutron scattering (~22 Å and ~38 Å) [20], but comparable to the values determined in leakage measurements (inner radius of 12.5 – 15 Å) [47], as well as to the radii obtained from MD simulations of melittin in DPPC (inner radius of ~15 Å; outer radius of ~20–25 Å) [22].
About ~10 ns after insertion of a melittin tetramer into a cylindrical pore, a few headgroups from the lower leaflet start to move into the pore interior, giving the pore a semitoroidal appearance (i.e., the headgroups within the pore region do not connect the two leaflets) (Figure 2B). The pore obtained at the end of the MD simulation is of a similar size as the pore shown in Figure 2A, with inner radius ~7 Å and outer radius ~20 Å.
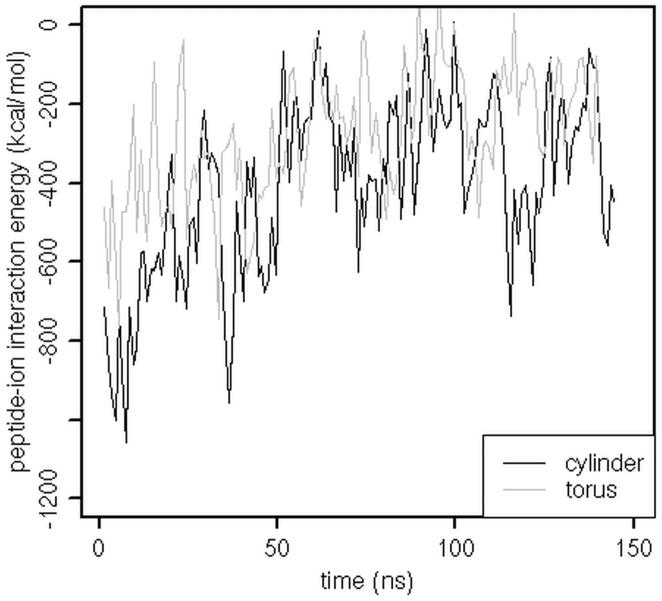
The pores shown in Figures 2A and 2B are filled with water and some Cl− ions (shown as red balls). The ions transiently neutralize some of the charged residues within the pore, but not all the charges. The ions are more abundant in the pore in Figure 2B, which gives rise to stronger ion-peptide interaction energy (see Figure 3). In the pores of Figures 2A and 2B, the calculated interactions of Cl− ions with K7 are −114.51 ± 86.25 kcal/mol and −148.32 ± 97.23 kcal/mol, respectively; those with K21 are −103.40 ± 92.15 kcal/mol and −220.11 ± 147.54 kcal/mol, respectively; those with R22, K23 or R24 are the same in both pores. The large error bars point toward the transient nature of the interactions. Sengupta et al. observed that the presence of counterions delayed formation of pores during atomistic MD simulations of melittin in DPPC [22]. It is possible that in the simulation of Figure 2B the pore evolves into only a semitoroidal pore due to the ions within the pore. Interactions between Cl− ions and K7 might interfere with K7-lower leaflet headgroup interactions while those between the ions and K21 might preclude headgroups from the upper leaflet to enter the pore region.
Figure 3.
The interaction energy between a melittin tetramer and Cl− ions versus time. ‘Cylinder’ refers to the MD simulation of melittin started from a cylindrical pore; ‘torus’ refers to the simulation started from a toroidal pore. The energies are averaged over 1 ns.
The orientation of melittin monomers in the pores can be described as a tilted transmembrane orientation. The calculated tilt angles are similar in both pores and range from ~10° to ~33°, relative to the bilayer normal. Previous solid-state NMR studies reported that melittin adopts a bent transmembrane orientation in bilayers [64, 65], with tilt angles of 30° for the N-terminal helix (residues 1–10) and 10° for the C-terminal helix (residues 13–26) in a DMPC bilayer [64]. Our simulations agree that melittin peptides are in a bent transmembrane orientation although the calculated tilt angles of the terminal helices are somewhat larger (~26° – 60° for the N-terminal helix and ~20° – 35° for the C-terminal helix) than the corresponding values from the experiment. Although Naito et al. reported that they observed pore formation in their experiments [64], it is not clear whether the reported bent orientation of melittin monomers pertains to the pore state. The calculated kink angle (i.e., the angle between the N-terminal and the C-terminal helices) is ~124° – 144°, which is comparable to the values reported by Toraya et al. for melittin in a DLPC (1,2-dilauroyl-sn-glycero-3-phosphocholine) bilayer (~126°) and in a DPPC bilayer (~119°) [65].
3. Alamethicin oligomers inserted into pores
Figures 4A and 4B show snapshots from MD simulations of an alamethicin tetramer embedded into a toroidal pore and of an alamethicin hexamer embedded into a cylindrical pore, respectively. In both simulations, the C-termini of peptides interact with the headgroup region of the upper leaflet and the N-temini are inserted into the lower leaflet and interact loosely with the headgroups. In the simulation started from a toroidal pore (Figure 4A), the alamethicin tetramer moves away from the pore region, which remains filled with water and headgroups. As the simulation advances, the number of headgroups and water molecules in the pore gradually decreases and at ~100 ns the water-filled pore disappears completely and all headgroups are at the membrane surface. In the simulation of an alamethicin hexamer inserted into a cylindrical pore (Figure 4B), the pore shape does not change significantly, that is, headgroups do not enter the pore region. However, as in the tetramer simulation, water molecules initially present within the hexamer interior move away.
A possible explanation for the drying of the pores in the above simulations is the orientation of Q7. In these simulations Q7 was initially oriented randomly and during the simulations the peptides did not reorient so that Q7 points towards the pore region. We thus performed a 160-ns MD simulation of the alamethicin hexamer in a cylindrical pore in which Q7 was initially located within the pore. As snapshots from the simulation show (Figure 4C), the pore remains filled with water. The radius of the pore formed by the hexamer is ~18 Å at the ends but the pore is constricted in the vicinity of Q7. Huang and coworkers [17], using neutron scattering, found that in DLPC the inner radius (i.e., the radius of the water pore) of alamethicin pores is ~9 Å and the outer radius (i.e., the radius of the pore including peptides) of ~20 Å.
The water pore is surrounded by five alamethicin monomers tilted by ~10–21° and one monomer tilted by ~33° relative to the membrane normal. The calculated tilt angles are in good agreement with the tilt angles of alamethicin in a DMPC bilayer obtained from solid-state NMR at peptide-to-lipid ratios at which it is expected that alamethicin forms pores [66, 67] and from coarse-grained MD simulations in which pores were formed [24]. In contrast to what was observed with melittin, the pore remains cylindrical, that is no headgroups move into the pore. This is likely due to differences in polarity of the N-termini; melittin carries one lysine (K7) and two threonines (T10 and T11) in its N-terminus whereas the N-terminus of alamethicin is more hydrophobic (it has only one polar residue, Q7).
The secondary structure of the peptides is more stable in the hexamer than in the tetramer. In simulations of an alamethicin hexamer reported by Thøgersen et al. unfolding of helices was especially pronounced for the peptides interacting strongly with water within a pore and for the surface-bound peptides [24]. In the case of the tetramer, two peptides (shown in yellow and violet in Figure 2C) stay helical until the end of the simulation. The orange peptide partially unfolds but, after the pore dries out, it refolds. The N-terminus of the purple peptide is helical and inserted into the membrane, its middle section is unfolded, and the C-terminus is helical and surface-bound.
4. Peptide-lipid interaction energy
Simulations of monomers and oligomers in constrained cylindrical pores (i.e., cylindrical pores whose shape was not allowed to change during the MD simulation) were also performed in order to compare the peptide-lipid interaction energy to that in the pores generated spontaneously during MD simulations.
The calculated interaction energy [electrostatic interaction energy] between a monomer and lipids in the constrained cylindrical pores is within statistical uncertainty to that in the toroidal pores (for melittin: −458.51 ± 100.08 [−364.34 ± 98.61] kcal/mol in the toroidal pore, −469.14 ± 94.33 [−366.31 ± 83.93] kcal/mol in the initially cylindrical pore and −472.35 ± 137.93 [−378.67 ± 130.31] kcal/mol in the constrained cylindrical pore; for alamethicin: −128.70 ± 41.00 [−52.79 ± 33.57] kcal/mol in the toroidal pore, −145.81 ± 48.29 [−56.61 ± 43.77] kcal/mol in the initially cylindrical pore and −139.99 ± 27.38 [−58.31 ± 22.07] kcal/mol in the constrained cylindrical pore).
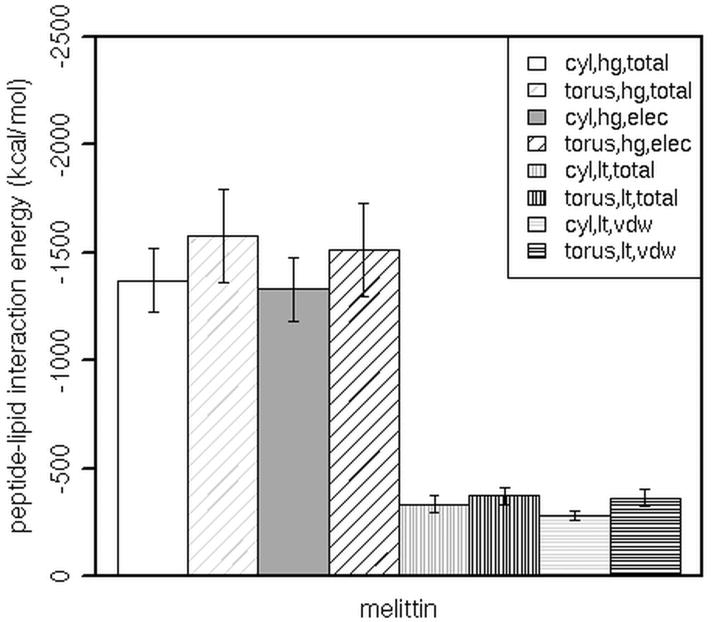
Better insight is obtained from the results on the oligomers. Figure 5 displays the average interaction energy of the melittin tetramer with lipid headgroups (‘hg’) and lipid tails (‘lt’). The trends in tetramer-lipid interaction energies are the same as those in tetramer-headgroup interaction energies. The energies calculated from simulations of the tetramer in the toroidal and in the initially cylindrical pores are close (the total tetramer-lipid interaction energy is −1949.10 ± 224.70 kcal/mol in the toroidal pore and −2015.59 ± 204.93 kcal/mol in the cylindrical pore, which becomes semitoroidal by the end of the simulation). The interactions in the constrained cylindrical pore are less favorable (−1703.15 ± 155.98 kcal/mol). Melittin interacts more strongly with lipid headgroups in the toroidal pore than with those in the constrained cylindrical pore. The interactions of the tetramer with lipid tails are also stronger in the toroidal pore than in the constrained cylindrical pore, but to a lesser extent. The calculated interaction energies are similar in magnitude to melittin-lipid interaction energies from a recent MD study by Manna and Mukhopadhyay (~ −350 kcal/mol per monomer) [68], although the direct comparison of our and their results is not possible due to different lipids (DMPC vs POPC), simulation times (140 ns vs 15 ns) and pore configurations (in their simulations, a toroidal pore spans only the lower leaflet of a bilayer).
Figure 5.
The total, electrostatic (elec) or van der Waals (vdw) interaction energy between a melittin tetramer and the lipid headgroups (hg) or tails (lt) in toroidal pores (torus) or in constrained cylindrical pores (cyl). Error bars are the standard deviation. The Welch two sample t-test was used to determine significant differences between the peptide-lipid interaction energies in the constrained cylindrical pore and in the toroidal pore; in all the cases p value is less than 0.05.
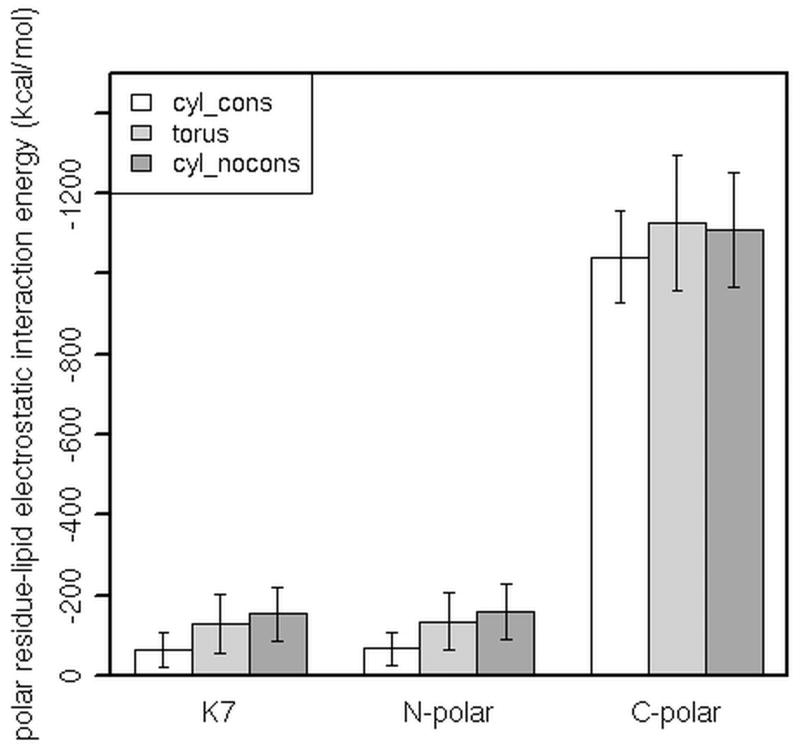
Given that the tetramer-lipid interactions are dominated by the electrostatic interactions with headgroups, we further looked at the electrostatic interaction energy between polar residues of the tetramer and headgroups in different pores (here, ‘polar’ refers to both polar and charged residues). The results are shown in Figure 6. In general, the interactions are stronger in the toroidal (‘torus’) and in the initially cylindrical (‘cyl_nocons’) pores than in the constrained cylindrical (‘cyl_cons’) pore. The N-terminus of melittin contains three polar residues: K7, T10 and T11 (denoted as ‘N-polar’ in Figure 6). Here, the interactions with headgroups are dominated by the K7-headgroup electrostatic interactions and they are more favorable when headgroups from the lower leaflet are allowed to move up into the pore and approach K7 at shorter distances (i.e., in toroidal pores). The contribution of T10 and T11 to the interaction energy is minor. The electrostatic interactions between polar residues in the C-terminus (S18, W19, K21, R22, K23, R24, Q25 and Q26) are also stronger in toroidal pores than in the constrained cylindrical pore. Figure 7 shows the position of charged (shown in blue) and polar residues (shown in green) in pores, at the end of simulations. The snapshots illustrate how charged and polar residues attract headgroups (green balls), as well as Cl− ions (red balls), and, in toroidal pores, facilitate their entry into the pore region.
Figure 6.
The electrostatic interaction energy between polar residues of a melittin tetramer and lipid headgroups. ‘N-polar’ refers to K7, T10 and T11; ‘C-polar’ refers to S18, W19, K21, R22, K23, R24, Q25 and Q26; ‘cyl_cons’ refers to the MD simulation in a constrained cylindrical pore; ‘torus’ refers to the MD simulation started from a toroidal pore; ‘cyl_nocons’ refers to the MD simulation started from a cylindrical pore. The Welch two sample t-test was used to determine significant differences between the polar residue-lipid headgroup interaction energies in the constrained cylindrical pore and in the toroidal or the initially cylindrical pore; in all the cases p value is less than 0.05.
Figure 7.
Polar and charged residues of a melittin tetramer interacting with lipid headgroups and Cl− ions in a constrained cylindrical pore (A) and toroidal pores (B and C). Snapshots (B) and (C) are from the MD simulations started from a cylindrical pore and a toroidal pore, respectively. Water molecules are removed for clarity. Polar residues are shown as yellow licorice, charged residues as blue licorice, W19 as black licorice, phosphocholines as green balls, ions as red balls, lipid tails as gray lines, and peptides as blue, orange, yellow and purple ribbons.
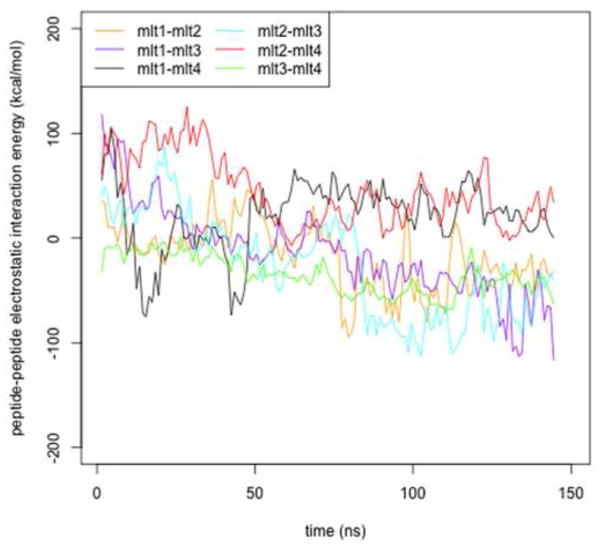
Since melittin carries five charged residues (K7, K21, R22, K23 and R24) one can expect significant peptide-peptide repulsions that might cause separation of monomers comprising a tetramer. Indeed, in our simulation of the tetramer embedded into the cylindrical pore, we observed predominantly repulsive interactions during the first ~80 ns (see Figure 8). However, as the radius of the pore enlarged, the interactions between most of the peptides became favorable. This finding suggests that the size of the pore might be determined by electrostatic interactions, which is in agreement with a theoretical study by Zemel et al [58].
Figure 8.
The peptide-peptide electrostatic interactions versus time. The interaction energies are calculated for melittin tetramer embedded into a cylindrical pore and averaged over 1ns; mlt1, mlt2, mlt3 and mlt4 denote monomers 1 through 4.
As mentioned above, we observed that the alamethicin pore closes if Q7 is not oriented towards the pore interior. Thus, we calculated the peptide-lipid interaction energies and the peptide-peptide interaction energies in the dry and in the wet pore and compare them here in order to gain insight into the interactions that hold the pore together. The calculated alamethicin hexamer-lipid interaction energies in the dry and in the wet pore are −750.36 ± 91.33 [−346.31 ± 83.88; −404.05 ± 21.49] kcal/mol and −833.37 ± 108.78 [−431.16 ± 94.70; −402.21 ± 25.44], respectively, where the first number in the brackets is the contribution from electrostatic interactions and the second number is the contribution from van der Waals interactions. The lipid-peptide interactions appear to be more favorable in the wet pore, although the large statistical uncertainty precludes a definite conclusion. The total peptide-peptide interaction energy, calculated as the sum of the interaction energy of pairs of the peptides, in the dry and in the wet pore are 99.14 ± 30.26 [262.87 ± 30.83; −163.73 ± 8.00] kcal/mol and 190.74 ± 51.66 [351.84 ± 52.05; −161.10 ± 9.06] kcal/mol, respectively. Thus, peptide-peptide interactions are unfavorable, but they are apparently offset by the lipid-peptide interactions.
Discussion
This paper reports atomistic MD simulations of melittin and alamethicin starting from toroidal and cylindrical pores aiming to investigate their preference for a particular pore type. During the simulations, we observed that a melittin tetramer reshaped a cylindrical pore to a toroidal pore whereas an alamethicin hexamer preserved the cylindrical pore. This finding is in agreement with the prevailing view that melittin forms toroidal pores whereas alamethicin forms barrel-stave pores. However, the pores formed in our simulations deviate from the classical pictures of highly ordered pores [16–18, 20]; they are more similar to disordered, irregular pores observed in other computational studies [22, 24]. The peptides rarely adopt the perfect transmembrane orientation.
Fox and Richards [26] postulated that glutamine (Q7) is a key residue in an alamethicin channel since it may form hydrogen bonds with Q7 of the neighboring peptide in the bundle and with water inside the pore, thus forming a hydrophilic patch within the pore. Later studies reported that mutations of Q7 perturbed the channel stability [32, 34]. In the simulations of the alamethicin tetramer in a toroidal pore and of the alamethicin hexamer in a cylindrical pore, we observed that the pores dried up. This seems to be related to the position of Q7, which in these simulations are located at the peptide-peptide interface or are hydrogen bonded with the backbone atoms or with water outside the pore region, making the pore essentially hydrophobic. In the simulations of a monomer, the monomer quickly reoriented so that Q7 was exposed to the pore water, but this did not happen in the case of oligomers. The simulations of oligomers may not be long enough for the helices to reorient and position the glutamines inside the pore. The MD simulation of the alamethicin hexamer in a cylindrical pore in which the helices were initially oriented with Q7 pointing toward the pore interior supports the importance of Q7 for stabilizing a wet pore since water remained within the pore. It would be interesting in future work to determine the free energy difference between the wet and the dry pore, information that would shed light into the mechanism of alamethicin ion channel formation.
A notable difference between alamethicin and melittin is the polarity of their N-termini; alamethicin has only one polar residue there (Q7) whereas melittin has one charged residue (K7) and two polar residues (T10, and T11). We focus here on the N-termini since they appear to initialize formation of a toroidal pore by perturbing headgroups in the lower leaflet and “pulling” some of them up into the pore (see Figures 2B and 7). From the calculated interaction energies, one can see that melittin interacts more strongly with headgroups in toroidal pores than in a constrained cylindrical pore (see Figure 5). This is true not only for the highly polar and charged C-terminus but also for the N-terminus (see Figure 6). The importance of the lysine 7 has also been reported in literature previously; e.g., Bachar and Becker observed in MD simulations that K7 facilitates pore formation by allowing water entry from both sides of a lipid bilayer [43] whereas Blondelle and Houghten measured significantly decreased hemolytic activity of a melittin mutant lacking K7 [69].
In this paper, interaction energies between peptides and lipids were compared in order to gain insights into the physical origin of the observed preferences for a given pore type. A melittin tetramer appears to interact more strongly with lipids in toroidal pores than in a cylindrical pore, which might be related to its preference for toroidal pores. Of course, one should ideally compute free energies of peptide solvation and also take into account the free energy of membrane deformation, but such calculations are methodologically and computationally demanding.
Melittin and alamethicin peptides in pores are in a tilted transmembrane orientation. This is in disagreement with the picture derived from oriented circular dichroism (OCD), in which the peptides are in a perfect transmembrane orientation [20, 50]. This discrepancy might be related to an assumption used in analyzing the OCD data that only parallel or perpendicular orientation of a helix is possible. The calculated tilted angles are more comparable to the tilt angles obtained from solid-state NMR [64–67]. However, although the experiments were conducted at high peptide-to-lipid ratios, peptide pores have not been characterized and it is not clear that the reported orientations correspond to the pore state.
The simulations reported herein range in length from 100 to 160 ns. We clearly make no claim that equilibrium has been reached within this time scale, nor was this the aim of the current investigation. Pores may well be transient or metastable structures. However, relaxation between pore types seems to take place on a time scale of ~10 ns (see Figure 1), therefore the length of the present simulations should be sufficient for investigating the preference for a given pore type.
Acknowledgments
This work was supported by the National Science Foundation (MCB-0615552) and the NIH (SC1GM087190). Infrastructure support was provided in part by RCMI grant RR03060 from NIH. A grant of computer time from the City University of New York’s High Performance Computing Research Center is also acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Maja Mihajlovic, Email: maja@sci.ccny.cuny.edu.
Themis Lazaridis, Email: tlazaridis@ccny.cuny.edu.
References
- 1.Hancock REW. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis. 2001;1:156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 2.Brown KL, Hancock REW. Cationic host defense (antimicrobial) peptides. Curr Opin Immunol. 2006;18:24–30. doi: 10.1016/j.coi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Brogden K. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 4.Bechinger B. Structure and function of membrane-lytic peptides. Crit Rev Plant Sci. 2004;23:271–292. [Google Scholar]
- 5.Cole AM. Antimicrobial peptide microbicides targeting HIV. Protein Pept Lett. 2005;12:41–47. doi: 10.2174/0929866053406101. [DOI] [PubMed] [Google Scholar]
- 6.Matsuzaki K, Murase O, Fujii N, Miyajima K. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry. 1996;35:11361–11368. doi: 10.1021/bi960016v. [DOI] [PubMed] [Google Scholar]
- 7.Papo N, Shai Y. Host defense peptides as new weapons in cancer treatment. Cell Mol Life Sci. 2005;62:784–790. doi: 10.1007/s00018-005-4560-2. [DOI] [PubMed] [Google Scholar]
- 8.Silphaduang U, Noga EJ. Peptide antibiotics in mast cells of fish. Nature. 2001;414:268–269. doi: 10.1038/35104690. [DOI] [PubMed] [Google Scholar]
- 9.Bradshow JP. Cationic antimicrobial peptides. Issues for potential clinical use. Biodrugs. 2003;17:233–240. doi: 10.2165/00063030-200317040-00002. [DOI] [PubMed] [Google Scholar]
- 10.Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim Biophys Acta. 1999;1462:1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 11.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim Biophys Acta. 2009;1788:1687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Matsuzaki K, Sugishita K, Fujii N, Miyajima K. Molecular basis for membrane selectivity of an antimicrobial peptide, magainin 2. Biochemistry. 1995;34:3423–3429. doi: 10.1021/bi00010a034. [DOI] [PubMed] [Google Scholar]
- 13.Huang HW. Action of antimicrobial peptides: Two-state model. Biochemistry. 2000;39:8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- 14.Huang HW. Molecular mechanism of antimicrobial peptides: The origin of cooperativity. Biochim Biophys Acta. 2006;1758:1292–1302. doi: 10.1016/j.bbamem.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Oren Z, Shai Y. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopoly. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 16.He K, Ludtke SJ, Huang HW. Antimicrobial peptide pores in membranes detected by neutron in-plane scattering. Biochemistry. 1995;34:15614–15618. doi: 10.1021/bi00048a002. [DOI] [PubMed] [Google Scholar]
- 17.He K, Ludtke SJ, Worcester DL, Huang HW. Neutron scattering in the plane of membranes: Structure of alamethicin pores. Biophys J. 1996;70:2659–2666. doi: 10.1016/S0006-3495(96)79835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allende D, Simon SA, McIntosh TJ. Melittin-induced bilayer leakage depends on lipid material properties: Evidence for toroidal pores. Biophys J. 2005;88:1828–1837. doi: 10.1529/biophysj.104.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. Membrane pores induced by magainin. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Harroun TA, Weiss TM, Ding L, Huang HW. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys J. 2001;81:1475–1485. doi: 10.1016/S0006-3495(01)75802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leontiadou H, Mark AE, Marrink SJ. Antimicrobial peptides in action. J Am Chem Soc. 2006;128:12156–12161. doi: 10.1021/ja062927q. [DOI] [PubMed] [Google Scholar]
- 22.Sengupta D, Leontiadou H, Mark AE, Marrink SJ. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim Biophys Acta. 2008;1778:2308–2317. doi: 10.1016/j.bbamem.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Jang H, Ma B, Lal R, Nussinov R. Models of toxic beta-sheet channels of protegrin-1 suggest a common subunit organization motif shared with toxic Alzheimer beta-amyloid ion channels. Biophys J. 2008;95:4631–4642. doi: 10.1529/biophysj.108.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thøgersen L, Schiøtt B, Vosegaard T, Nielsen NC, Tajkhorshid E. Peptide aggregation and pore formation in a lipid bilayer: A combined coarse-grained and all atom molecular dynamics study. Biophys J. 2008;95:4337–4347. doi: 10.1529/biophysj.108.133330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer CE, Reusser F. A polypeptide antibacterial agent isolated from trichoderma viride. Experentia. 1967;23:85–86. doi: 10.1007/BF02135929. [DOI] [PubMed] [Google Scholar]
- 26.Fox RO, Jr, Richards FM. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-Å resolution. Nature. 1982;300:325–330. doi: 10.1038/300325a0. [DOI] [PubMed] [Google Scholar]
- 27.Mottamal M, Lazaridis T. Voltage-dependent energetics of alamethicin monomers in the membrane. Biophys Chem. 2006;122:50–57. doi: 10.1016/j.bpc.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Gauldie J, Hanson JM, Rumjanek FD, Shipolini RA, Vernon CA. The peptide components of bee venum. Eur J Biochem. 1976;61:369–376. doi: 10.1111/j.1432-1033.1976.tb10030.x. [DOI] [PubMed] [Google Scholar]
- 29.Lauterwein LR, Brown K, Wuethrich K. High-resolution 1H-NMR studies of monomeric melittin in aqueous solution. Biochim Biophys Acta. 1980;622:219–230. doi: 10.1016/0005-2795(80)90033-1. [DOI] [PubMed] [Google Scholar]
- 30.Vogel H, Jähnig F. The structure of melittin in membranes. Biophys J. 1986;50:573–582. doi: 10.1016/S0006-3495(86)83497-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barranger-Mathys M, Cafiso DS. Membrane structure of voltage-gated channel forming peptides by site-directed spin-labeling. Biochemistry. 1996;35:498–505. doi: 10.1021/bi951985d. [DOI] [PubMed] [Google Scholar]
- 32.Breed J, Kerr ID, Molle G, Duclohier H, Sansom MSP. Ion channel stability and hydrogen bonding. Molecular modelling of channels formed by synthetic alamethicin analogues. Biochim Biophys Acta. 1997;1330:103–109. doi: 10.1016/s0005-2736(97)00163-6. [DOI] [PubMed] [Google Scholar]
- 33.He K, Ludtke SJ, Heller WT, Huang HW. Mechanism of alamethicin insertion into lipid bilayers. Biophys J. 1996;71:2669–2679. doi: 10.1016/S0006-3495(96)79458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molle G, Dugast JY, Spach G, Duclohier H. Ion channel stabilization of synthetic alamethicin analogs by rings of inter-helix H-bonds. Biophys J. 1996;70:1669–1675. doi: 10.1016/S0006-3495(96)79729-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tieleman DP, Berendsen HJC, Sansom MSP. An alamethicin channel in a lipid bilayer: Molecular dynamics simulations. Biophys J. 1999;76:1757–1769. doi: 10.1016/s0006-3495(99)77337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, Weiss TM, Harroun TA, Heller WT, Huang HW. Supramolecular structures of peptide assemblies in membranes by neutron off-plane scattering: Method of analysis. Biophys J. 1999;77:2648–2656. doi: 10.1016/S0006-3495(99)77099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dittmer J, Thøgersen L, Underhaug J, Bertelsen K, Vosegaard T, Pedersen JM, Schiøtt B, Tajkhorshid E, Skrydstrup T, Nielsen NC. Incorporation of antimicrobial peptides into membranes: A combined liquid-state NMR and molecular dynamics study of alamethicin in DMPC/DHPC bicelles. J Phys Chem B. 2009;113:6928–6937. doi: 10.1021/jp811494p. [DOI] [PubMed] [Google Scholar]
- 38.Pan J, Tieleman DP, Nagle JF, Kucerka N, Tristram-Nagle S. Alamethicin in lipid bilayer: Combined use of X-ray scattering and MD simulations. Biochim Biophys Acta. 2009;1788:1387–1397. doi: 10.1016/j.bbamem.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sansom MSP, Kerr ID, Mellor IR. Ion channels formed by amphipathic helical peptides. A molecular modelling study. Eur Biophys J. 1991;20:229–240. doi: 10.1007/BF00183460. [DOI] [PubMed] [Google Scholar]
- 40.Breed J, Biggin PC, Kerr ID, Smart OS, Sansom MSP. Alamethicin channel - modelling via restrained molecular dynamics simulations. Biochim Biophys Acta. 1997;1325:235–249. doi: 10.1016/s0005-2736(96)00262-3. [DOI] [PubMed] [Google Scholar]
- 41.Tieleman DP, Berendsen HJC, Sansom MSP. Voltage-dependent insertion of alamethicin at phospholipid/water and octane/water interfaces. Biophys J. 2001;80:331–346. doi: 10.1016/S0006-3495(01)76018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tieleman DP, Hess B, Sansom MSP. Analysis and evaluation of channel models: Simultions of alamethicin. Biophys J. 2002;83:2393–2407. doi: 10.1016/s0006-3495(02)75253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachar M, Becker OM. Melittin at a membrane/water interface: Effects on water orientation and water penetration. J Chem Phys. 1999;111:8672–8685. [Google Scholar]
- 44.Bernèche S, Nina M, Roux B. Molecular dynamics simulations of melittin in dimyristoylphosphatidylcholine bilayer membrane. Biophys J. 1998;75:1603–1618. doi: 10.1016/S0006-3495(98)77604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin JH, Baumgaertner A. Stability of a melittin pore in a lipid bilayer: A molecular dynamics study. Biophys J. 2000;78:1714–1724. doi: 10.1016/S0006-3495(00)76723-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee MT, Chen FY, Huang HW. Energetics of pore formation induced by membrane active peptides. Biochemistry. 2004;43:3590–3599. doi: 10.1021/bi036153r. [DOI] [PubMed] [Google Scholar]
- 47.Ladokhin AS, Selsted ME, White SH. Sizing membrane pores in lipid vesicles by leakage of co-encapsulated markers: pore formation by melittin. Biophys J. 1997;72:1762–1766. doi: 10.1016/S0006-3495(97)78822-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ladokhin AS, White SH. ‘Detergent-like’ permeabilization of anionic lipid vesicles by melittin. Biochim Biophys Acta. 2001;1514:253–260. doi: 10.1016/s0005-2736(01)00382-0. [DOI] [PubMed] [Google Scholar]
- 49.Matsuzaki K, Yoneyama S, Miyajima K. Pore formation and translocation of melittin. Biochemistry. 1997;73:831–838. doi: 10.1016/S0006-3495(97)78115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang HW, Wu Y. Lipid-alamethicin interactions influence alamethicin orientation. Biophys J. 1991;60:1079–1087. doi: 10.1016/S0006-3495(91)82144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen FY, Lee MT, Huang HW. Sigmoidal concentration dependence of antimicrobial peptide activities: a case of alamethicin. Biophys J. 2002;82:908–914. doi: 10.1016/S0006-3495(02)75452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hristova K, Dempsey CE, White SH. Structure, location, and lipid perturbations of melittin at the membrane interface. Biophys J. 2001;80:801–811. doi: 10.1016/S0006-3495(01)76059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hall JE, Vodyanoy I, Balasubramanian TM, Marshall R. Alamethicin. A rich model for channel behavior. Biophys J. 1984;45:233–247. doi: 10.1016/S0006-3495(84)84151-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rex S, Schwarz G. Quantitative studies on the melittin-induced leakage mechanism of lipid vesicles. Biochemistry. 1998;37:2336–2345. doi: 10.1021/bi971009p. [DOI] [PubMed] [Google Scholar]
- 55.Mihajlovic M, Lazaridis T. Antimicrobial peptides bind more strongly to membrane pores. Biochim Biophys Acta. doi: 10.1016/j.bbamem.2010.02.023. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terwilliger TC, Eisenberg D. The structure of melittin. I. Structure determination and partial refinement. J Biol Chem. 1982;257:6010–6015. doi: 10.2210/pdb1mlt/pdb. [DOI] [PubMed] [Google Scholar]
- 57.Terwilliger TC, Eisenberg D. The structure of melittin. II. Interpretation of the structure. J Biol Chem. 1982;257:6016–6022. [PubMed] [Google Scholar]
- 58.Zemel A, Fattal DR, Ben-Shaul A. Energetics and self-assembly of amphiphatic peptide pores in lipid membranes. Biophys J. 2003;84:2242–2255. doi: 10.1016/S0006-3495(03)75030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brooks BR, Brooks CL, III, MAD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch A, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacKerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 63.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 64.Naito A, Nagao T, Norisada K, Mizuno T, Tuzi S, Saitô H. Conformation and dynamics of melittin bound to magnetically oriented lipid bilayers by solid-state 31P and 13C NMR spectroscopy. Biophys J. 2000;78:2405–2417. doi: 10.1016/S0006-3495(00)76784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toraya S, Nishimura K, Naito A. Dynamic structure of vesicle-bound melittin in a variety of lipid chain legths by solid-state NMR. Biophys J. 2004;87:3323–3335. doi: 10.1529/biophysj.104.046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bak M, Bywater RP, Hohwy M, Thomsen JK, Adelhorst K, Jakobsen HJ, Sørensen OW, Nielsen NC. Conformation of alamethicin in oriented phospholipid bilayers determined by 15N solid-state nuclear magnetic resonance. Biophys J. 2001;81:1684–1698. doi: 10.1016/S0006-3495(01)75822-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertelsen K, Paaske B, Thøgersen L, Tajkhorshid E, Schiøtt B, Skrydstrup T, Nielsen NC, Vosegaard T. Residue-specific information about the dynamics of antimicrobial peptides from 1H-15N and 2H solid-state NMR spectroscopy. J Am Chem Soc. 2009;131:18335–18342. doi: 10.1021/ja908604u. [DOI] [PubMed] [Google Scholar]
- 68.Manna M, Mukhopadhyay C. Cause and effect of melittin-induced pore formation: A computational approach. Langmuir. 2009;25:12235–12242. doi: 10.1021/la902660q. [DOI] [PubMed] [Google Scholar]
- 69.Blondelle SE, Houghten RA. Hemolytic and antimicrobial activities of the twenty-four individual omission analogues of melittin. Biochemistry. 1991;30:4671–4678. doi: 10.1021/bi00233a006. [DOI] [PubMed] [Google Scholar]