Abstract
Hox transcription factors specify numerous cell fates along the anterior-posterior axis by regulating the expression of downstream target genes. While expression analysis has uncovered large numbers of de-regulated genes in cells with altered Hox activity, determining which are direct versus indirect targets has remained a significant challenge. Here, we characterize the DNA binding activity of Hox transcription factor complexes on eight experimentally verified cis-regulatory elements. Hox factors regulate the activity of each element by forming protein complexes with two cofactor proteins, Extradenticle (Exd) and Homothorax (Hth). Using comparative DNA binding assays, we found that a number of flexible arrangements of Hox, Exd, and Hth binding sites mediate cooperative transcription factor complexes. Moreover, analysis of a Distal-less regulatory element (DMXR) that is repressed by abdominal Hox factors revealed that suboptimal binding sites can be combined to form high affinity transcription complexes. Lastly, we determined that the anterior Hox factors are more dependent upon Exd and Hth for complex formation than posterior Hox factors. Based upon these findings, we suggest a general set of guidelines to serve as a basis for designing bioinformatics algorithms aimed at identifying Hox regulatory elements using the wealth of recently sequenced genomes.
Keywords: Hox, Extradenticle, Homothorax, transcription factor, cis-regulation
Introduction
Hox genes encode a widely conserved family of transcription factors that control diverse cell fates by directly regulating the expression of downstream target genes (Mann et al., 2009; Pearson et al., 2005). Species across numerous phyla contain anywhere from five to thirty-nine Hox genes that are often clustered on chromosomes (Carroll et al., 2001; Duboule, 2007; Garcia-Fernandez, 2005; Lemons and McGinnis, 2006). Hox genes are differentially expressed along the developing anterior-posterior axis of individuals to specify diverse cell fates of specific organs and morphological structures. Between species, the modification of Hox gene numbers (duplication and deletion), Hox gene expression patterns, and/or Hox protein functions are thought to underlie many of the morphological changes observed in divergent body plans (Carroll et al., 2001; Galant and Carroll, 2002; Garcia-Fernandez, 2005; Ronshaugen et al., 2002). In addition to their role in development, Hox genes are widely expressed in adult life and play key roles in vertebrate hematopoiesis with altered Hox activity being directly linked to human leukemia (Lawrence et al., 2005; Lawrence et al., 1997; Lawrence et al., 1999; McGonigle et al., 2008). Thus, determining how Hox factors identify the appropriate set of target genes is critical for better understanding development, evolution, and disease.
Hox transcription factors have two defining molecular features: a highly conserved homeodomain that binds DNA, and a short pentapeptide motif (usually containing YPWM residues) N-terminal to the homeodomain (Chang et al., 1996; Chang et al., 1995; Lu and Kamps, 1996; Mann, 1995; Phelan et al., 1995). The homeodomain folds into three α-helices with the third helix contacting nucleotides in the major groove of DNA (Qian et al., 1989; Qian et al., 1993). Since each Hox factor performs highly specific functions in vivo, it was initially predicted that each would bind a distinct set of nucleotides. However, numerous studies over the past twenty years have established that Hox factors bind short stretches of AT-rich DNA (TNAT) with relatively low specificity in vitro (Affolter et al., 2008; Berger et al., 2008; Ekker et al., 1994; Noyes et al., 2008). Thus, a fundamental question remains: how do Hox transcription factors direct region specific cell fates in vivo when they bind similar DNA sequences in vitro?
One answer to the question of target specificity is that Hox factors form transcription factor complexes on DNA. The two best-characterized Hox co-factor proteins are Extradenticle (Exd, Drosophila) and Homothorax (Hth, Drosophila) and their vertebrate homologues Pbx and Meis, respectively (Burglin, 1997; Mann and Affolter, 1998; Moens and Selleri, 2006). Exd and Hth have C-terminal TALE homeodomains that contain a three amino acid loop extension between their first and second α-helices. Biochemical and crystallographic studies revealed that the Exd TALE motif forms a hydrophobic pocket that mediates interactions with the Hox pentapeptide to cooperatively bind adjacent DNA sites (Chan et al., 1994; Joshi et al., 2007; LaRonde-LeBlanc and Wolberger, 2003; Passner et al., 1999; Piper et al., 1999). Although the exact domains of interaction between Hox and Hth are not as clear, Hth also interacts with Hox factors on adjacent DNA binding sites (Shen et al., 1997). Since Exd binds a core sequence of TGAT and Hth binds a consensus sequence of TGACAG, the interactions between Hox proteins and these cofactors expand their DNA binding footprint and enhance DNA binding selectivity (Chang et al., 1997). In addition, Exd and Hth are obligate heterodimers that require each other for their in vivo activities (Abu-Shaar et al., 1999; Rieckhof et al., 1997). Exd-Hth interactions through conserved N-terminal domains thereby add further flexibility to the formation of transcription factor complexes with Hox proteins as Hox/Exd/Hth, Hox/Hth/Exd, and even Hox/Exd/Hth/Hox complexes form on DNA (Chan et al., 1997; Ebner et al., 2005; Ferretti et al., 2005; Gebelein et al., 2002; Gebelein et al., 2004; Jacobs et al., 1999; Li-Kroeger et al., 2008; Manzanares et al., 2001; Popperl et al., 1995; Ryoo and Mann, 1999; Ryoo et al., 1999; Samad et al., 2004; Tumpel et al., 2007). Thus, three direct protein-protein interactions (Exd-Hox, Hth-Hox, and Exd-Hth) contribute to the cooperative formation of higher-order Hox transcription factor complexes with enhanced target selectivity and affinity.
The formation of large Hox transcription factor complexes in which each protein binds specific DNA sequences suggests it may be possible to predict cis-regulatory elements from primary DNA sequences. With the large amount of genomic sequence data available from a broad range of organisms, the successful application of a bioinformatics approach has the potential to illuminate the Hox target genes required for animal development as well as many of the Hox-mediated changes underlying morphological evolution between species. However, to accurately predict target sites, one needs to first understand the DNA sequence requirements for the formation of active Hox complexes. In this study, we begin to address this problem by performing a comparative study on eight confirmed Hox target sites identified in vertebrates and Drosophila melanogaster. Through a combination of protein and DNA site mutations, we find that a great deal of flexibility exists in forming Hox complexes with Exd and Hth. However, we have uncovered differences between the formation of anterior and posterior Hox complexes and suggest several general rules that should aid future efforts aimed at identifying new Hox target sequences.
Materials and Methods
Expression Vectors and Protein Purification
The following constructs were used to generate the Hox and Exd/Hth proteins for this study: His-Lab (Chan et al., 1996), His-Abd-A (Ryoo and Mann, 1999), His-Scr (Ryoo and Mann, 1999), His-Exd (Chan et al., 1997), His-Hth (Ryoo et al., 1999), Exd (untagged) (Gebelein et al., 2002), Hth (untagged) (Gebelein et al., 2002), His-Exd51A (Li-Kroeger et al., 2008), and His-Hth51A (Li-Kroeger et al., 2008). The Hox expression constructs were transformed in BL21 bacteria. Exd/Hth heterodimers were produced by co-transformation of a His-tagged Exd (or Hth) construct (pET14b, ampicillin) with an untagged Hth (or Exd) construct (pET9, kanamycin) placed under double antibiotic selection. Bacteria were grown in 100 mL of LB liquid culture to log phase, and induced using 0.2 mM IPTG for 1.5 hrs at 37°C. After centrifugation, bacteria were resuspended in 5 mLs lysis buffer (50 mM Tris, pH 7.5; 100 mM NaCl, 20 mM imidizole), and lysed on ice using sonication (4 × 20 seconds). Glycerol (final concentration of 10%) and Igepal (NP-40) detergent (final concentration of 0.5%) were added and samples incubated for five minutes with gentle rocking. Membranes and cellular debris were pelleted in eppendorf tubes (20 minutes at 13,000 rpm) at 4°C. Proteins were bound to 450 uLs of washed Ni-agarose beads with gentle rocking for 1.5 hrs at 4°C, washed three times with lysis buffer plus glycerol/Igepal, and eluted with the same buffer fortified with 250mM imidizole. Samples were dialyzed against 500 mL binding buffer (50 mM Tris, pH 7.5; 100 mM NaCl, 1 mM MgCl2, 10% glycerol) for 4 hrs at 4°C. Protein concentrations were measured by the Bradford assay and confirmed by SDS-PAGE and Coomassie blue analysis.
Oligonucleotides and Probe Preparation
The forward and reverse complement oligonucleotides of the DMXR, RhoA, Lab48/95, EVIII, Hoxb1 R3-PM2, Hoxb2-PP2, Hoxa2 PM-PH2, and Hoxa3 PHP1 probes used in EMSA analysis are shown in Figure 1. DMXR1 and DMXR2 sequences are shown in Figure 2A; DMXR-Hox1m, -Hox2m, -Hox12m sequences are shown in Figure 2B; DMXR2-G>T and RhoA T>G sequences are shown in Figure 3A; DMXR2-Exdm, -Hthm, and -Hox2m sequences are shown in Figure 3B; DMXR1-Con, -ΔA, and −HoxC sequences are shown in Figure 4B; RhoA-Exdm, -Hthm, and -Hoxm sequences were previously described (Li-Kroeger et al., 2008). In each case, reverse complement oligonucleotides were synthesized and double-stranded probes generated by annealing oligonucleotides to a final concentration of 1 uM in STE buffer (100 mM NaCl, 10 mM Tris pH 8.0, 1 mM EDTA pH 8.0). 1.75 uLs of each annealed oligonucleotide was end-labeled with T4 PNK and ATPγ-32P in a 5 uL reaction. The kinase reaction was terminated using 95 uLs of TE-stop solution (10 mM Tris pH 8.0, 7 mM EDTA pH 8.0).
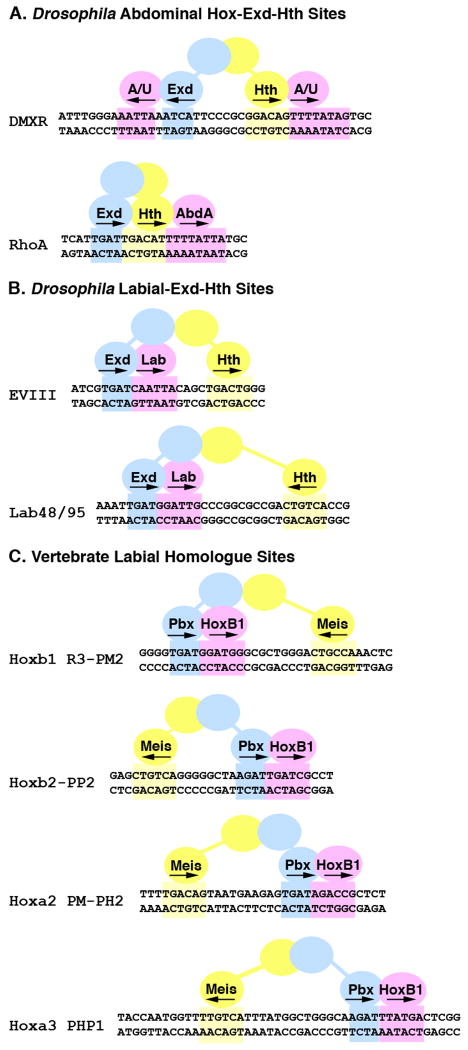
Figure 1. Configurations of Hox, Exd, and Hth binding sites in Drosophila and vertebrate cis-regulatory elements.
Schematics of eight Hox regulated enhancer elements with the Exd (blue), Hth (yellow) and Hox (pink) sites highlighted. A. Abdominal Hox target elements from Drosophila. The DMXR element contains both Exd/Hox and Hth/Hox sites that are bound by the Abd-A and Ubx (A/U) Hox factors to repress Dll expression in the abdomen (Gebelein et al., 2004). The RhoA element contains consecutive Exd/Hth/Hox binding sites that are bound by Abd-A to activate rhomboid expression in developing abdominal sensory cells (Li-Kroeger et al., 2008). B. The EVIII and Lab 48/95 elements are both activated by the Lab Hox factor in the developing gut endoderm and each contains a Hox, Exd, and Hth binding site (Ebner et al., 2005; Ryoo et al., 1999). C. Four mouse cis-regulatory elements containing Pbx/Hox and distant Meis binding sites. All are activated by the HoxB1 vertebrate homologue of Lab in collaboration with the Pbx and Meis proteins within the developing hindbrain (Ferretti et al., 2005; Ferretti et al., 2000; Manzanares et al., 2001; Popperl et al., 1995; Tumpel et al., 2007).
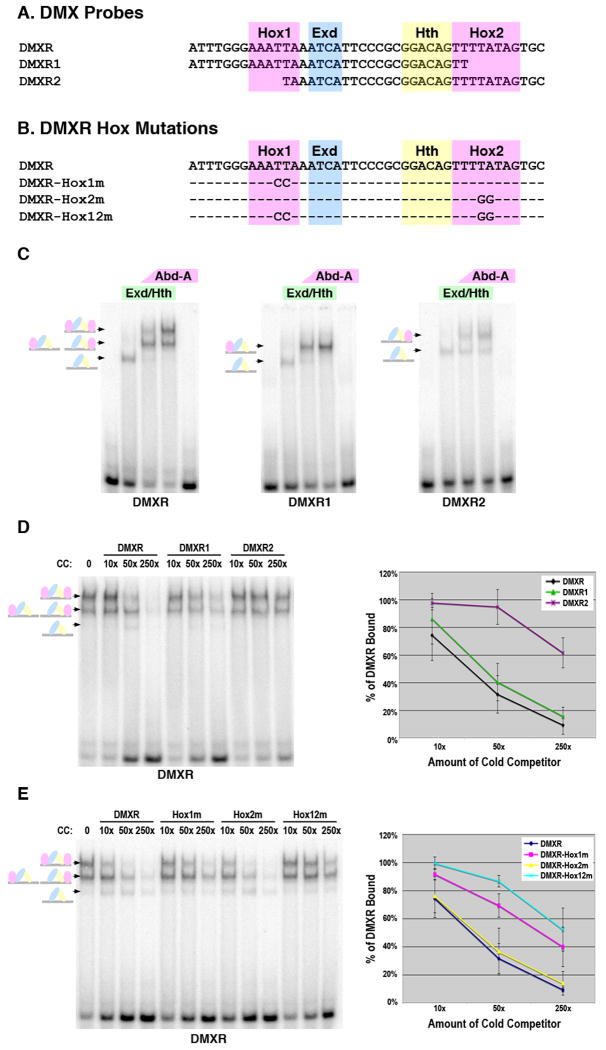
Figure 2. Dependence of Abd-A tetramer formation on the presence of two Hox sites within DMXR.
A. The DMXR probes used in EMSAs with the Hox1, Exd, Hth, and Hox2 sites highlighted. B. The DMXR Hox point mutations used in gel shift analysis. C. Comparative EMSAs of Hox complex formation on the DMXR, DMXR1, and DMXR2 probes. Conditions are as follows: First lane is probe alone, second lane is 75 × 10-9 M of Exd/Hth heterodimers, third and fourth lane contain the same amount of Exd/Hth with either 17 × 10-9 M (low amount) or 75 × 10-9 M (high amount) of Abd-A, respectively and the fifth lane contains 75 × 10-9 M of Abd-A alone. Schematics at left denote color-coded complexes formed (Exd, blue; Hth, yellow; Abd-A, pink). D and E. Competition DNA binding assays for Abd-A/Exd/Hth complexes on labeled DMXR. Each lane contains 75 × 10-9 M of Exd/Hth and Abd-A. The first lane contains no competitor DNA whereas subsequent lanes contain either 10×, 50×, or 250× of the indicated cold competitor. Schematics at left denote complexes (Exd, blue; Hth, yellow; Abd-A, pink). Graph depicts the average percent of DMXR probe bound from three different experiments in the presence of different amounts of each cold competitor probe. For this analysis, the amount of probe bound was determined using phosphor-imaging densitometry, and 100% binding was assigned to the amount of probe bound in the absence of competitor. Standard error is noted.
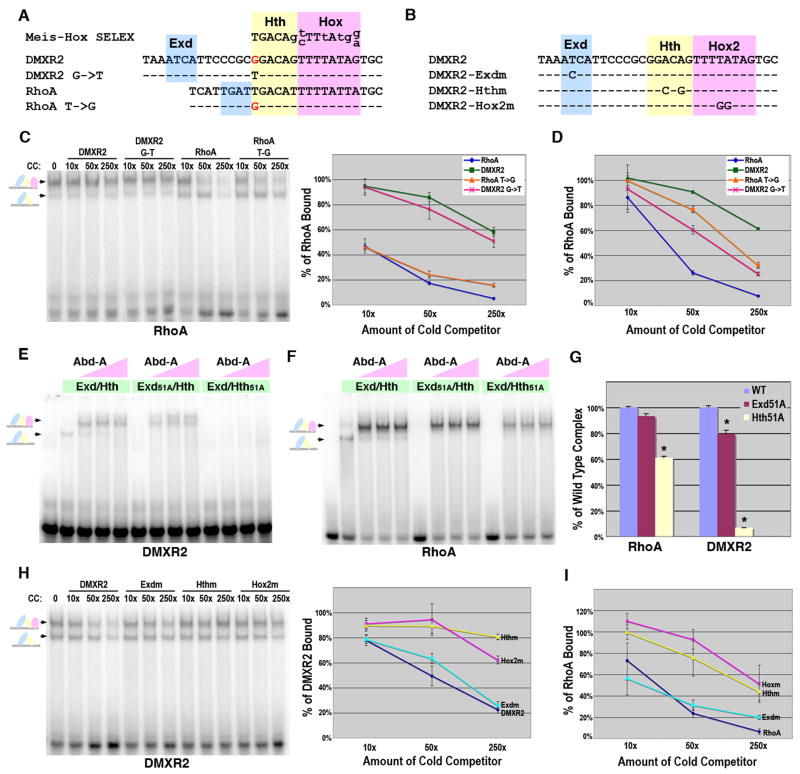
Figure 3. Dependence of Hox complex formation on the Exd, Hth, and Hox sites in DMXR2 and RhoA.
A. Sequence comparison of the DMXR2, RhoA, and Meis-Hox site previously identified using site selection assays (SELEX) (Shen et al., 1997). Lower case letters in the Meis-Hox SELEX denote nucleotides under less constraint. The Hth (yellow) and Hox (pink) sites are highlighted and mismatches from SELEX consensus are in red text. The Exd binding sites are highlighted in blue. B. The DMXR2 Exd, Hth, and Hox2 point mutations used in gel shift analysis. C. DNA binding competition assays for Abd-A, Exd and Hth complexes on RhoA, RhoA-T>G, DMXR2, and DMXR2-G>T. Labeled RhoA probe was bound with a constant amount (75 × 10-9 M) of Exd/Hth and Abd-A. Different amounts of competitor were added as indicated. Schematics at left denote color-coded complexes (Exd, blue; Hth, yellow; Abd-A, pink). The amount of probe bound in absence of competitor was assigned 100% binding and the graph (at right) depicts the average amount of probe bound in presence of competitor from three different experiments with standard error noted. D. Graph of DNA binding competition assays of Exd/Hth (75 × 10-9 M) binding on RhoA, RhoA-T>G, DMXR2, and DMXR2-G>T in the absence of Hox factors. Three experiments were performed and the average amount of probe bound in absence and presence of competitor was compared with standard error noted. E. Comparative EMSAs using wild type and mutant Exd/Hth heterodimers with Abd-A on DMXR2. Equimolar amounts of Exd/Hth, Exd51A/Hth, and Exd/Hth51A proteins (30 × 10-9 M) were used with three concentrations of Abd-A (7.5 × 10-9 M, 22.5 × 10-9 M, and 70 × 10-9 M). F. Comparative EMSAs using wild type and mutant Exd/Hth heterodimers with Abd-A on RhoA. Equimolar amounts of Exd/Hth, Exd51A/Hth, and Exd/Hth51A proteins (15 × 10-9 M) were used with three concentrations of Abd-A (7.5 × 10-9 M, 22.5 × 10-9 M, and 70 × 10-9 M). G. Assessing the dependence of Hox complex formation on Exd and Hth binding to DMXR2 and RhoA. Comparative DNA binding assays were performed in triplicate using equimolar amounts of Exd/Hth, Exd51A/Hth, and Exd/Hth51A proteins and Abd-A. The average amount of probe bound by Abd-A and the wild type Exd/Hth proteins was assigned to 100% for each probe tested (blue bar). The amount of probe bound by Exd51A/Hth (red bar) and Exd/Hth51A (yellow bar) compared to wild type was determined. Standard error bars are noted and * denotes significant difference from wild type binding (p-value < 0.001). H. DNA binding competition assays for Abd-A, Exd and Hth complexes on wild type, Exdm, Hthm, and Hox2m DMXR2 probes. Labeled DMXR2 probe was bound with a constant amount (75 × 10-9 M) of Exd/Hth and Abd-A. Different amounts of competitor were added as indicated. Schematics at left denote color-coded complexes (Exd, blue; Hth, yellow; Abd-A, pink). The amount of probe bound in absence of competitor was assigned 100% binding and the graph (at right) depicts the average amount of probe bound in presence of competitor in three different experiments. I. DNA binding competition assays for Abd-A, Exd and Hth complexes using wild type, Exdm, Hthm, and Hoxm RhoA probes. Labeled RhoA probe was bound with a constant amount (75 × 10-9 M) of Exd/Hth and Abd-A. The amount of probe bound in absence of competitor was assigned 100% binding and the graph depicts the average amount of probe bound in the presence of competitor in three different experiments.
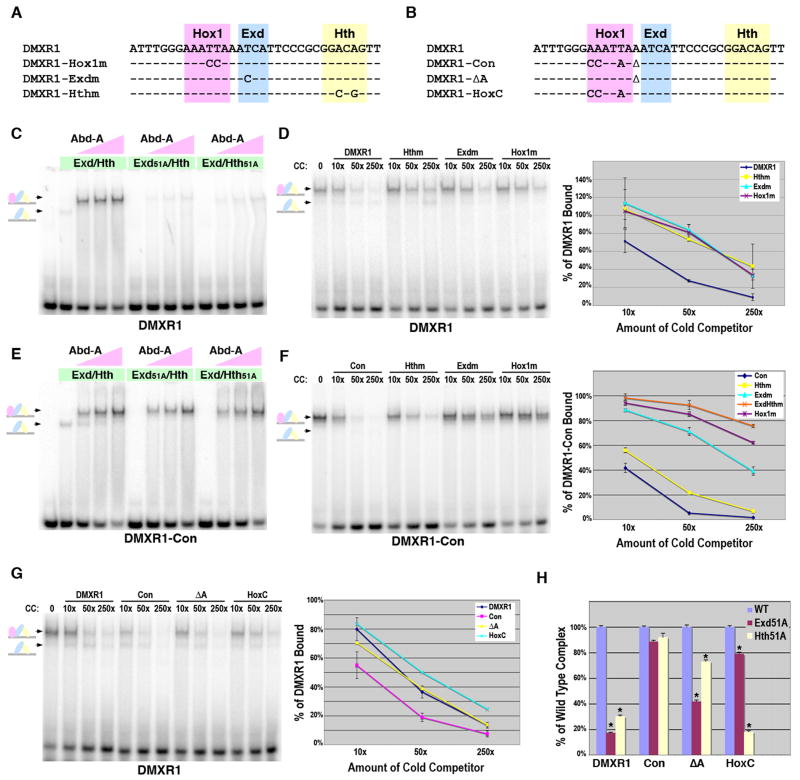
Figure 4. Role of Hth binding for Hox complex formation on Exd/Hox sites.
A. The DMXR1 Hox1, Exd, and Hth point mutations used in gel shift analysis. B. Sequence comparisons of the DMXR1, DMXR1-Con, DMXR1-ΔA, and DMXR1-HoxC probes used in gel shift assays. C. Comparative EMSAs using wild type and mutant Exd/Hth heterodimers with Abd-A on DMXR1. Equimolar amounts of Exd/Hth, Exd51A/Hth, and Exd/Hth51A proteins (30 × 10-9 M) were used with three concentrations of Abd-A (7.5 × 10-9 M, 22.5 × 10-9 M, and 70 × 10-9 M). D. Dependence of Abd-A/Exd/Hth binding to DMXR1 on the Hox1, Exd, and Hth sites. DNA competition assays were performed using labeled DMXR1 and DMXR1, Hox1m, Exdm, and Hthm probes as cold competitors. The amount of probe bound in absence of competitor was assigned 100% binding and the graph depicts the average amount of probe bound in presence of each competitor from three different experiments. E. Comparative EMSAs using wild type and mutant Exd/Hth heterodimers with Abd-A on DMXR1-Con. Equimolar amounts of Exd/Hth, Exd51A/Hth, and Exd/Hth51A proteins (30 × 10-9 M) were used with three amounts of Abd-A (7.5 × 10-9 M, 22.5 × 10-9 M, and 70 × 10-9 M). F. Dependence of Abd-A/Exd/Hth binding to DMXR1-Con on the Hox1, Exd, and Hth sites. DNA binding competition assays were performed using labeled DMXR1-Con and different amounts of DMXR1-Con wild type, Hox1m, Exdm, Hthm, and ExdmHthm probes as cold competitors. The amount of probe bound in absence of competitor was assigned 100% binding and the graph depicts the average amount of probe bound in presence of competitor from three different experiments. G. DNA binding competition assays of Abd-A, Exd, and Hth complex formation on the DMXR1, DMXR1-Con, DMXR1-ΔA, and DMXR1-HoxC probes. Labeled DMXR1 probe was bound with a constant amount of Exd/Hth and Abd-A. Different amounts of competitor were added as indicated. Schematics at left denote color-coded complexes (Exd, blue; Hth, yellow; Abd-A, pink). Data from three independent experiments is graphed at right. Note only the DMXR1-Con is significantly different from the wild type DMXR1 (p-value < 0.01) H. Assessing the dependence of Hox complex formation on Exd and Hth binding to DMXR1, DMXR1-Con, DMXR1-ΔA, and DMXR1-HoxC. Comparative DNA binding assays were performed in triplicate using equimolar amounts of Exd/Hth, Exd51A/Hth, or Exd/Hth51A proteins and Abd-A. The amount of probe bound by Abd-A and wild type Exd/Hth was assigned to 100% for each probe tested (blue bar). The amount of probe bound by Exd51A/Hth (red bar) and Exd/Hth51A (yellow bar) compared to wild type was then determined. * denotes a significant difference from wild type binding (p-value < 0.001).
Electromobility Shift Assays (EMSAs)
EMSAs were performed using native PAGE essentially as previously described (Gebelein and Urrutia, 2001). In brief, 1 uL of labeled 32P probe was used in a 20 uL binding reaction (bringing the final concentration of probe to 8.75 × 10-10 M). The binding reaction buffer consisted of a final concentration of 10 mM Tris, pH 7.5; 50 mM NaCl; 1 mM MgCl2; 4% glycerol; 0.5 mM DTT; 0.5 mM EDTA; 50 ug/mL poly(dI-dC); and 200 ug/mL of BSA. For non-competition EMSAs, protein samples (see figure legends for amount of each protein used) were gently mixed with labeled probes, incubated at room temperature for 15 min, and run on a 4% polyacrylamide gel for 75 min at 150V. For competition assays, the appropriate amount of cold competitor was added with the 32P-labeled probe prior to the 15 min incubation. All experiments were performed at least three times. The dried acrylamide gels were exposed to phosphor-screens and densitometry was performed using ImageQuant 5.1 software.
Results
To better understand the sequence requirements for Hox factors to bind DNA with Exd and Hth, we selected a set of Hox target sites from Drosophila and vertebrates for comparative analysis (Figure 1). The Drosophila target genes include: the Distal-less element (DMXR) that is repressed by the Abdominal-A (Abd-A) and Ultrabithorax (Ubx) Hox factors (A/U) in abdominal segments (Gebelein et al., 2002; Gebelein et al., 2004); a rhomboid (rho) activation element (RhoA) that is stimulated by Abd-A in the abdominal nervous system (Li-Kroeger et al., 2008); a labial (lab) auto-regulatory element (Lab48-95) and an element (EVIII) from CG11339 that are activated by Lab within the gut endoderm (Ryoo et al., 1999). The vertebrate target sites include a Hoxb1 auto-regulatory element (Hoxb1-R3-PM2) and elements within other anterior Hox genes (Hoxb2-PP2; Hoxa2-PM-PH2; and Hoxa3-PHP1), which are all regulated by HoxB1 (a lab homologue) and the Pbx and Meis co-factors within the hindbrain (Ferretti et al., 2005; Ferretti et al., 2000; Manzanares et al., 2001; Popperl et al., 1995; Tumpel et al., 2007). Importantly, each cis-element has been experimentally verified as a Hox target using a combination of genetic, DNA binding, and transgenic reporter assays.
Sequence comparisons between the eight cis-regulatory elements in Figure 1 reveals that while each contains an Exd(Pbx), Hth(Meis), and at least one Hox binding site, the order, orientation, and spacing among sites vary. For example, the two abdominal Hox targets contain neighboring Hth/Hox sites whereas the Lab and HoxB1 targets contain Exd(Pbx)/Hox sites with a distant Hth(Meis) site. Moreover, no specific order and/or orientation of the Hth(Meis) site relative the Exd(Pbx)/Hox site is favored as all four possibilities are represented in the six Lab/HoxB1 targets. These data indicate that a great deal of flexibility exists in forming Hox transcriptional complexes. However, as each site was studied independently, it is unclear how Hox complex formation compares between sites and if there are significant differences between the formation of anterior and posterior Hox complexes. In this study, we use a combination of protein and DNA binding site mutations to compare and contrast the ability of the Abd-A and the Lab Hox factors to form transcription factor complexes with Exd and Hth on their respective target sites.
Characterization of Hox tetramer formation on DMXR
Compared to other Hox-regulated elements, DMXR is unusual in that it contains Hox binding sites close to both an Exd site (Hox1) and a Hth site (Hox2). Previous studies revealed each independent Hox site is cooperatively bound by an Abd-A, Exd, and Hth complex, and that altogether, an abdominal Hox/Exd/Hth/Hox tetramer forms on DMXR to repress Dll and suppress leg development (Gebelein et al., 2004). However, it is unclear whether the two Hox sites synergize to further enhance binding cooperativity during Hox complex formation. To address this question, we compared the ability of Abd-A to form complexes with Exd and Hth on probes containing all four binding sites (DMXR), the Hox1/Exd/Hth sites (DMXR1), or the Exd/Hth/Hox2 sites (DMXR2, Figure 2A). Using a defined quantity of Exd and Hth with two concentrations of Abd-A, we found the amount of DMXR bound (68% and 81%) was approximately the sum of DMXR1 (47% and 62%) and DMXR2 (24% and 33%) (Figure 2C). Next, we analyzed the relative strength of Hox binding to each probe using unlabeled DMXR, DMXR1, and DMXR2 to compete with labeled DMXR. As shown in Figure 2D, the ability to compete for Hox complexes is in the following order: DMXR ≥ DMXR1 > DMXR2 with DMXR2 being significantly weaker than both DMXR and DMXR1. These findings suggest the Hox1 site mediates most of the cooperative binding to DMXR. Consistent with this idea, mutation of the Hox1 but not the Hox2 site significantly compromised binding to DMXR (Figure 2E). However, Hox1/Hox2 double mutations resulted in additional loss in competition, demonstrating both sites can mediate Hox complex formation (Figure 2E). Importantly, these DNA binding assays correlate well with transgenic reporter assays in Drosophila showing that Hox-mediated repression on DMXR is more sensitive to mutations in Hox1 than Hox2 but double mutations fully compromise gene repression (Gebelein et al., 2004). Nevertheless, these data indicate that the binding of either Abd-A/Exd/Hth on DMXR1 or Exd/Hth/Abd-A on DMXR2 is sufficient to mediate significant repression in vivo (Gebelein et al., 2004).
Comparing RhoA with DMXR2 – the role of Exd binding on Hth/Hox targets
While Abd-A directly regulates the expression of several other genes, only one has a confirmed set of Exd, Hth, and Hox sites. The RhoA element within rhomboid contains Exd/Hth/Hox sites that are bound by an Abd-A complex to stimulate gene expression in abdominal sensory cells (Figure 1A) (Gebelein et al., 2004; Li-Kroeger et al., 2008). Like DMXR2, RhoA has adjacent Hth/Hox sites that closely match a consensus sequence bound by the vertebrate Meis/HoxA9 factors (Figure 3A, (Shen et al., 1997)). However, the RhoA site more closely matches this consensus, as DMXR2 contains a single nucleotide difference at a constrained position within the Hth site. RhoA and DMXR2 also differ in regards to the location of their Exd binding sites. The Exd site in RhoA is directly adjacent to the Hth site, a configuration found in an optimal Pbx/Meis binding site defined using selection assays in the absence of Hox factors (TGATTGACAG, Pbx site is italicized; Meis site in bold (Chang et al., 1997)). In contrast, DMXR2 has the same Exd site (TGAT), but in the opposite orientation and separated from the Hth site by seven nucleotides.
To compare transcription factor binding to RhoA and DMXR2, we performed competition assays with Exd/Hth in the absence and presence of Abd-A. As shown in Figure 3, RhoA has much higher affinity than DMXR2 for both Exd/Hth and Exd/Hth/Abd-A complexes (Figure 3C-D). To determine if the single nucleotide change within the DMXR2 Hth site is sufficient to alter binding, we performed gel shift assays using probes containing reciprocal nucleotide changes: DMXR2 G->T and RhoA T->G. As shown in Figure 3D, DMXR2 can be made into a significantly better Exd/Hth site by changing the G to T, but not to wild type RhoA levels. Changing the T to G in RhoA also significantly compromises Exd/Hth binding, but again the RhoA T>G change competes better than DMXR2. Surprisingly, however, neither RhoA T>G nor DMXR2 G>T results in significant changes in competition for the Exd/Hth/Abd-A complex (Figure 3C). Thus, enhanced Exd/Hth binding to RhoA is not sufficient to explain high affinity Hox complex formation relative to DMXR2.
Prior biochemical studies showed that the vertebrate Meis/HoxA9 factors cooperatively bind DNA in the absence of Pbx (Shen et al., 1997). To determine if Abd-A complex formation on DMXR2 and RhoA depends on Exd binding, we generated DNA binding compromised Exd and Hth proteins by mutating the highly conserved asparagine 51 residue of each of their homeodomains to alanine (N51A) (Gehring et al., 1994). Using equimolar amounts of wild type Exd/Hth, Exd51A/Hth, or Exd/Hth51A with Abd-A, we found that Exd binding was largely dispensable for Abd-A complex formation on RhoA and DMXR2 (Figure 3E-F). Moreover, point mutations in the Exd binding sites (Exdm) of RhoA and DMXR2 also have a negligible effect in competing Abd-A complexes compared to wild type probes (Figure 3H-I). In sharp contrast, the Hth protein mutation (Hth51A) abolished Hox complexes on DMXR2 and greatly diminished Hox complexes on RhoA (Figure 3E-F). To more easily visualize these results, we assigned the amount of wild type Exd/Hth/Abd-A bound to each probe to 100% and graphed the relative amount bound by each mutant protein (Figure 3G). For example, the Exd/Hth51A heterodimer results in a 90% decrease in Abd-A complexes on DMXR2 versus a 40% decrease on RhoA. We also found that mutation of either the Hth (Hthm) or Hox (Hoxm) sites in DMXR2 and RhoA strongly compromised Hox complex formation in competition assays (Figure 3H-I). However, the Hth mutations did so to varying degrees as Hthm within DMXR2 nearly eliminated competition, whereas the same Hth mutation in RhoA still competes over 50% of the Hox complex. Altogether, these data demonstrate that while most cooperative Hox binding on DMXR2 and RhoA is mediated by Abd-A and Hth, the presence of a nearby Exd site greatly enhances complex formation on RhoA.
The role of Hth and the spacing between Exd and Hox binding sites
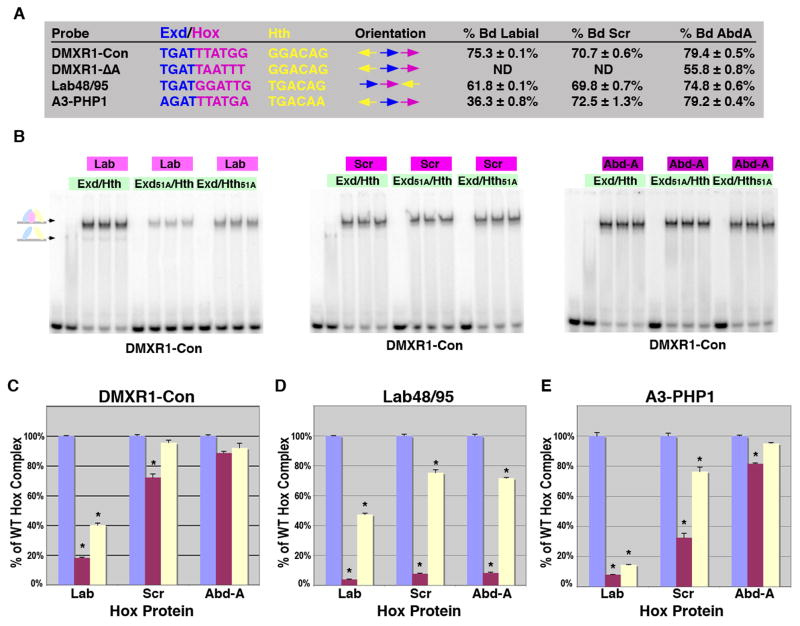
Currently, no other Abd-A targets contain Exd/Hox and Hth sites like DMXR1 for comparative purposes. However, over twenty-five cis-regulatory elements for other Hox factors contain adjacent Exd(Pbx)/Hox sites (Mann et al., 2009), and sequence comparisons reveal DMXR1 is the only element that contains a nucleotide (Adenine) inserted between its Hox and Exd sites (Figure 4A-B). In fact, a previous study using vertebrate proteins found that inserting an extra nucleotide within a Pbx/Hox binding site abolished DNA binding (Chang et al., 1996). However, that study was performed in the absence of a Hth(Meis) protein and binding site. Hence, to test the dependence of Hox complex formation on Hth, we compared binding to wild type DMXR1 and a modified DMXR1 probe (DMXR1-Con) that removes the extra nucleotide and changes the Hox site from TAATTT to TTATGG (reverse complement is listed as most studies use this orientation) (Figure 4B, (Gebelein et al., 2002)). Using Exd/Hth, Exd51A/Hth, and Exd/Hth51A with Abd-A, we found that both Exd and Hth binding are required for forming Abd-A/Exd/Hth complexes on DMXR1 (Figure 4C). In addition, DMXR1 probes containing Exd, Hth, or Hox1 site mutations all significantly decreased competition for Hox complexes compared to wild type DMXR1 (Figure 4D). In contrast, the same concentrations of Exd/Hth and Abd-A proteins on DMXR1-Con revealed that Hth and Exd binding are largely dispensable for complex binding (Figure 4E). These data indicate that either Exd or Hth is sufficient to mediate complex formation on DMXR1-Con. To better test this idea, competition assays using Exd, Hth, and Hox DMXR1-Con binding site mutations revealed that: 1) Hthm competed as well as wild type DMXR1-Con; 2) Exdm did not compete as well as wild type but did compete significantly better than Hoxm; and 3) ExdmHthm double mutations resulted in a significant decrease in competition when compared to either mutation alone (Figure 4F). These findings indicate that a Hth binding site is more critical for Abd-A complex formation on a suboptimal Exd/Hox site (DMXR1) than on an optimal binding site (DMXR1-Con). Surprisingly, we also found a significant amount of complex forms on DMXR1-Con but not DMXR1 in the absence of Exd binding, suggesting additional differences in Hox complex formation between these two probes.
DMXR1-Con alters both the spacing between Exd and Hox sites as well as the Hox binding sequence. To determine which of these changes permits Abd-A complex formation independent of Exd and/or Hth binding, we used two additional probes: DMXR1-ΔA deletes the extra Adenine between the Hox1 and Exd sites, and DMXR1-HoxC changes TAATTT to TTATGG but leaves the extra nucleotide (Figure 4B). We tested the relative strength of each probe in competition assays with DMXR1 and found each behaves similarly in competition assays except DMXR1-Con, which is a significantly better competitor (Figure 4G). We next used wild type versus 51A mutant Exd/Hth proteins to determine the relative dependence of Hox complex formation on Exd and Hth binding to each probe. For comparative purposes, the amount of wild type Abd-A/Exd/Hth bound to each probe was assigned to 100% and the relative amount bound by each mutant was determined (Figure 4H). On DMXR1, Exd51A/Hth results in an 80% decrease and Exd/Hth51A results in over a 70% decrease in Abd-A complex formation. In contrast, the same amount of Exd and Hth mutant proteins decrease binding to DMXR1-Con by only 15% and 10%, respectively. DMXR1-ΔA, on the other hand is highly dependent upon Exd (60% decrease) but not Hth (25% decrease), while DMXR1-HoxC is highly dependent upon Hth (80% decrease) and relatively independent of Exd (20% decrease). Altogether these findings indicate: 1) If the spacing between Hox and Exd sites is optimal, then Hox complexes form relatively independent of Hth (DMXR1-Con and DMXR1-ΔA). 2) If the Hox binding site is TTATGG as opposed to TAATTT, then a distant Hth site can mediate Hox complexes independent of Exd binding (DMXR1-Con and DMXR1-HoxC). Further studies revealed that merely changing the Hox site from TAATTT to TTATTT is sufficient to make DNA binding dependent upon distant Hth sites (data not shown).
Sequence preference for inserted nucleotide between the Exd and Hox sites
As mentioned above, previous studies using vertebrate proteins found that inserting an extra Cytosine nucleotide within a consensus Pbx/Hox binding site abolished DNA binding (Chang et al., 1996). To determine if the identity of the nucleotide inserted between the Hox1 and Exd sites makes a significant difference in DNA binding activity, we used three additional probes that change the Adenine to either Thymine (DMXR1-A>T), Cytosine (DMXR1-A>C), or Guanine (DMXR1-A>G) as cold competitors for binding Abd-A and Exd/Hth. We found that changing Adenine to any other nucleotide significantly decreased Hox complex formation revealing that Adenine is the preferred nucleotide, then Thymine, Cytosine, and lastly Guanine (Supplemental Figure 1).
Comparative analysis of Lab-Exd-Hth cis-regulatory elements
We next tested if the behavior of the abdominal Hox complex on DMXR1 and DMXR1-Con can be extrapolated to other Hox factors and their binding sites. For this analysis, we selected a set of six cis-elements regulated by lab Hox genes. As opposed to the posterior Abd-A Hox factor, lab and its homologues are the most anterior Hox genes and regulate head structures in both vertebrates and invertebrates (Carpenter et al., 1993; Dolle et al., 1993; Mark et al., 1993; Merrill et al., 1989). Functional conservation between lab and vertebrate genes has been demonstrated by showing Hoxb1 rescues a lab null allele in Drosophila, and a vertebrate cis-regulatory element (Hoxb1 R3-PM2) drives gene expression in a lab-dependent pattern in transgenic Drosophila (Lutz et al., 1996; Popperl et al., 1995). Thus, we used purified Drosophila Exd, Hth, and Lab proteins to analyze complex formation on six Drosophila and vertebrate cis-regulatory elements that contain adjacent Exd(Pbx)/Hox sites with variably spaced/oriented Hth(Meis) sites (Figure 1B-C).
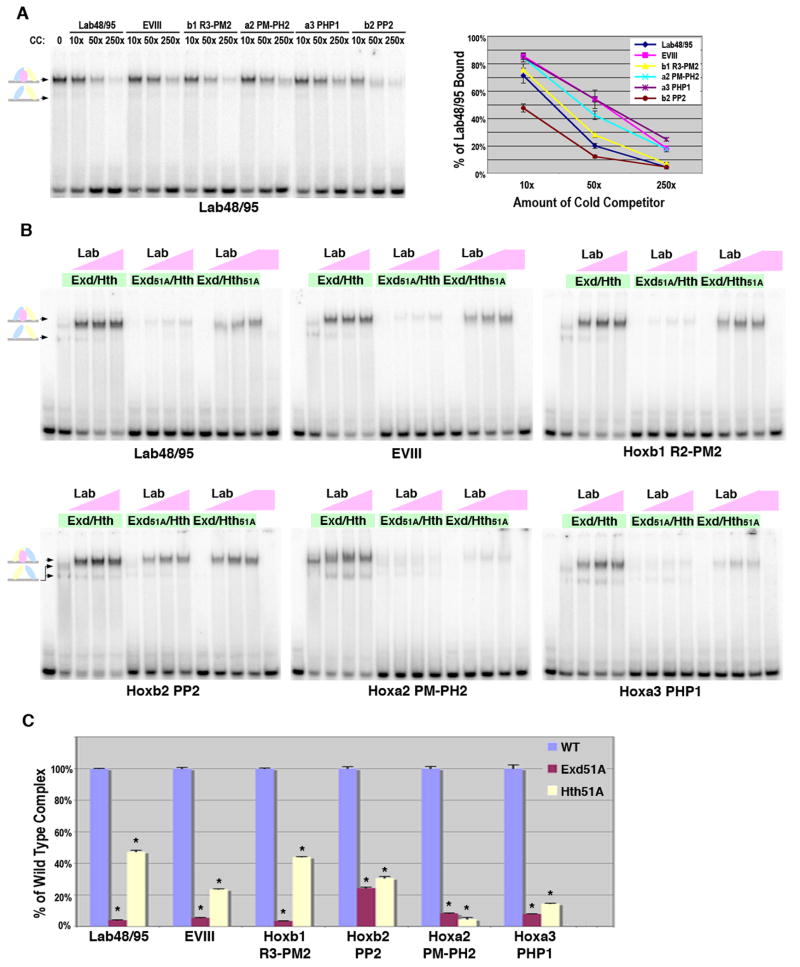
Lab complex formation on all six probes was first analyzed using direct competition analysis. As shown in Figure 5A, a constant amount of Lab and Exd/Hth was bound to the labeled Lab48/95 probe in the absence and presence of different concentrations of each probe as cold competitors. Importantly, while each probe significantly competes for Lab/Exd/Hth complexes, they did so to differing degrees and in the following order: Hoxb2-PP2 > Lab48/95 ≥ Hoxb1 R3-PM2 > Hoxa2 PM-PH2 ≥ EVIII = Hoxa3 PHP1. Similar results were found using comparative gel shift analysis and calculating the percent probe bound under identical Exd/Hth and Lab protein conditions (data not shown). In Table 1, we list these cis-elements from strongest to weakest in terms of Lab complex formation and compare strength of binding with cis-element architecture and sequence. Importantly, we find that no one orientation of binding sites is favored as both the strongest and weakest sites share the same configuration. We also compared the Exd/Hox and Hth binding site sequences using position weight based matrices (PWMs).
Figure 5. Comparisons between Lab-Exd-Hth binding sites.
A. DNA binding competition assays for Lab, Exd and Hth complexes on Lab48/95, EVIII, Hoxb1 R3-PM2, Hoxa2 PM-PH2, Hoxa3-PHP1, and Hoxb2-PP2 probes (see Figure 1 for sequences). Labeled Lab48/95 probe was bound with a constant amount of Exd/Hth (75 × 10-9 M) and Lab (110 × 10-9 M). Different amounts of competitor were added as indicated. Schematics at left denote color-coded complexes (Exd, blue; Hth, yellow; Lab, pink). The amount of probe bound in absence of competitor was assigned 100% binding and the graph (at right) depicts the average amount of probe bound in presence of competitor from three different experiments with standard error noted. B. Comparative EMSAs using wild type and mutant Exd/Hth heterodimers with Lab on Lab48/95, EVIII, Hoxb1 R3-PM2, Hoxa2 PM-PH2, Hoxa3-PHP1, and Hoxb2-PP2 probes. Equimolar amounts of Exd/Hth, Exd51A/Hth, and Exd/Hth51A proteins (30 × 10-9 M) were used with three different amounts of Lab (12 × 10-9 M, 36 × 10-9 M, or 110 × 10-9 M). C. Assessing the dependence of Lab complex formation on Exd and Hth binding to Lab48/95, EVIII, Hoxb1 R3-PM2, Hoxa2 PM-PH2, Hoxa3-PHP1, and Hoxb2-PP2. Comparative DNA binding assays were performed in triplicate using equimolar amounts of Exd/Hth, Exd51A/Hth, and Exd/Hth51A proteins (30 × 10-9 M) and Lab (110 × 10-9 M). The amount of probe bound by Abd-A and wild type Exd/Hth was assigned to 100% for each probe tested (blue bar). The amount of probe bound by Exd51A/Hth (red bar) and Exd/Hth51A (yellow bar) compared to wild type was then determined. * denotes a significant difference from wild type binding (p-value < 0.001).
Table 1.
Comparisons of Hox cis-regulatory elements and Lab complex formation. Probes are listed from strongest (Top) to weakest (Bottom) in terms of Lab complex formation. The Exd (blue), Lab (red), and Hth (yellow) sites are listed in the same direction for each probe. The orientation of the Hth site relative to the Exd/Lab sites are denoted by colored arrows (same colors as sites). The PWM score of each sample is listed for the Bacterial One-hybrid and SELEX approaches. Totals are calculated by summating each PWM score.
| Probe | Hth | Orientation | Bacterial One-hybrid PWM Score | SELEX PWM Score | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Exd | Lab | Hth | Total | Exd/Lab | Hth | Total | |||||
| Strongest | Hoxb2-PP2 | TGACAG | -3.06 | -0.57 | 5.84 | 2.21 | 7.00 | 8.05 | 15.05 | ||
| Lab48/95 | TGACAG | 3.89 | -4.91 | 5.84 | 4.82 | 2.94 | 8.05 | 10.99 | |||
| Hoxb1 R3-PM2 | TGGCAG | 0.77 | -4.88 | 1.63 | -2.48 | 4.74 | 4.05 | 8.79 | |||
| Hoxa2 PM-PH2 | TGACAG | -0.28 | -5.71 | 5.84 | -0.15 | 0.57 | 8.05 | 8.62 | |||
| EVIII | TGACTG | -3.06 | 4.27 | 1.98 | 3.19 | 4.34* | 3.88 | 8.22* | |||
| Weakest | Hoxa3-PHP1 | TGACAA | -4.92 | 2.87 | 5.84 | 3.79 | 2.48 | 3.48 | 5.96 | ||
denotes that the PWM calculated the Exd/Lab value using the reverse complement of the listed site such that the Exd site is TAAT and the Lab site is TGATCA. Note that if the reverse complement site is used, the relative orientation of the Exd/Lab site to the Hth site would change.
For this purpose, we generated PWMs from two data sets. First, we used the recently published binding sequences for the individual Exd, Hth, and Lab proteins identified using a bacterial one-hybrid assay (Noyes et al., 2008). Second, we used the published sequences identified using purified Meis1 and Pbx1/HoxB1 heterodimers bound to a random oligonucleotide library followed by reiterative purification/amplification (SELEX) (Chang et al., 1996; Shen et al., 1997). Sequences were imported into Target Explorer (http://luna.bioc.columbia.edu/Target_Explorer/) and the program assigned the best matrix for each binding site (Supplemental Data) (Sosinsky et al., 2003). Analysis of the six Lab/HoxB1 regulatory elements using each PWM revealed the following: 1) scores for the six Hth sites are uniformly positive using the bacterial one-hybrid PWMs, but the Exd and Lab scores vary greatly and when summated the total scores do not correlate well with strength of Lab complex formation. 2) In contrast, all six probes scored positively using the in vitro SELEX sites for Meis1 and Pbx1/HoxB1 and there is a strong correlation between strength of Lab complex formation and total PWM score. An additional interesting result that came from this analysis is that the reverse complement of the Exd/Lab site (TAATTGATCA; Exd in italics, Hox in bold) in the EVIII probe scored significantly higher than the suggested sequence (TGATCAATTA). This finding indicates that the orientation of these sites may differ from the original published report (Ebner et al., 2005). However, since our data cannot discriminate between these two possibilities and structural studies would be required to determine the correct orientation, we left the orientation of binding sites as previously published.
We next determined the dependence of Lab complex formation on Exd and Hth binding using the wild type and mutant Exd/Hth proteins. As shown in Figure 5B, all six Lab complexes are heavily dependent upon Exd binding as Exd51A abolishes nearly all complex formation to each probe. The one exception is that some Lab/Exd51A/Hth complex forms on HoxB2-PP2, but when normalized, even this binding is 75% less than wild type (Figure 5C). The Hth51A protein also significantly disrupted Lab complexes but to a variable degree. For example, Lab/Exd/Hth51A disrupted over 90% of binding to Hoxa2 PM-PH2 but only 55% of binding to Lab48/95 and Hoxb1 R3-PM2. Interestingly, the Hoxa2 element has a poor Hox binding site (AGACCG) compared to Lab48/95 (GGATTG) and Hoxb1 (GGATGG), suggesting that like abdominal Hox complexes on DMXR1, Lab complex formation is more highly dependent upon Hth binding if the Exd/Hox site is suboptimal.
Anterior and posterior Hox factors differ in their Exd and Hth binding requirements for Hox complex formation
Unlike our findings using Abd-A on DMXR1-Con, all six of the Lab complexes required wild type Exd and Hth proteins for strong binding. This finding could be due to inherent differences between the Abd-A and Lab Hox factors or due to differences between the cis-regulatory elements tested. To distinguish between these possibilities, we performed gel shift analysis using Lab with wild type Exd/Hth on DMXR1-Con and found Lab readily forms complexes on this probe (Figure 6A-B, note: slightly more Lab protein was used than Abd-A to achieve a similar level of Hox complex formation, 75.3% vs 79.4% respectively). To determine if Lab can bind DMXR1-Con independent of either Exd or Hth binding, we performed comparative gel shifts using wild type and Exd/Hth mutant proteins and found that both Exd51A and Hth51A formed significantly less complex with Lab than wild type proteins (an 80% decrease with Exd51A and a 60% decrease with Hth51A, Figure 6C). In contrast, the same amount of mutant Exd and Hth proteins disrupted less than 15% of binding with Abd-A (Figure 6B-C). As Lab is the most anterior Hox factor and Abd-A is one of the most posterior Hox factors, we also performed the same set of assays using the central Hox factor Sex Combs Reduced (Scr). As shown in Figure 6, Scr behaved much like Abd-A in that it is able to form a significant amount of Hox complex on DMXR1-Con with either Exd51A or Hth51A.
Figure 6. Differences in complex formation between anterior and posterior Hox factors.
A. The DNA probes tested for Hox complex formation in gel shift assays using Lab, Scr and Abd-A. Comparisons between the Exd/Hox sites, Hth sites and orientations between sites are highlighted. The percent of probe bound by Lab (110 × 10-9 M), Scr (100 × 10-9 M) and Abd-A (75 × 10-9 M) with a constant amount of Exd/Hth (75 × 10-9 M) in triplicate is noted. B. Comparative EMSAs on the DMXR1-Con probe using wild type and mutant Exd/Hth heterodimers (30 × 10-9 M) with Lab (110 × 10-9 M), Scr (100 × 10-9 M) or Abd-A (75 × 10-9 M) as indicated. C-E. Assessing the dependence of Hox complex formation on Exd and Hth binding to DMXR1-Con (C), Lab48/95 (D), and, Hoxa3-PHP1 (E). Comparative DNA binding assays were performed in triplicate using equimolar amounts of Exd/Hth, Exd51A/Hth, and Exd/Hth51A proteins (30 × 10-9 M) and Lab (110 × 10-9 M), Scr (100 × 10-9 M) or Abd-A (75 × 10-9 M). The amount of probe bound by each Hox factor with wild type Exd/Hth was assigned to 100% (blue bar). The amount of probe bound by Exd51A/Hth (red bar) and Exd/Hth51A (yellow bar) compared to wild type was then determined. * denotes a significant difference from wild type binding (p-value < 0.001).
We next determined if Abd-A or Scr could form Hox complexes with Exd51A and Hth51A on two of the Lab-regulated cis-elements. For this purpose, we first selected the Hoxa3-PHP1 site because it closely resembles the Exd/Hox site of DMXR1-Con (Figure 6A). Using the same amount of each respective Hox factor as for the DMXR1-Con gel shifts, we found that Scr and Abd-A bind significantly more Hoxa3-PHP1 than Lab (Figure 6A). Moreover, when tested with equal amounts of Exd51A/Hth and Exd/Hth51A, the Abd-A Hox factor was able to significantly bind Hoxa3-PHP1 in the presence of either protein mutant (Figure 6E). Scr, on the other hand, was intermediate between Lab and Abd-A in terms of its ability to form complexes on Hoxa3-PHP1 in the absence of Exd binding (Exd51A). Lastly, we selected the Lab48/95 probe, which contains an Exd/Hox site that is significantly different from DMXR1-Con. This probe was similarly bound by all three Hox factors with wild type Exd/Hth (Figure 6A). However, unlike on DMXR1-Con and Hoxa3-PHP1, Lab, Scr and Abd-A mediated Hox complex formation is heavily dependent upon Exd binding (Exd51A). Thus, Abd-A and to a lesser extent Scr forms complexes on Exd/Hox and Hth sites in the absence of Exd binding, but only on specific Hox sites (TTAT). Moreover, the anterior and posterior Hox factors significantly differ in their ability to form complexes independent of Exd binding. As discussed below, these data have implications for the mechanisms underlying the general ability of posterior Hox factors to phenotypically suppress anterior Hox factors during development.
Discussion
The Hox genes encode a family of conserved transcription factors that regulate numerous fundamental cell behaviors throughout the development and life of an organism (Carroll et al., 2001; Mann et al., 2009; Mann and Morata, 2000; McGinnis and Krumlauf, 1992). Expression experiments (mainly microarrays) from several organisms and/or tissues revealed that hundreds if not thousands of genes are affected by alterations in Hox gene activity (Chung et al., 2006; Ghannam et al., 2004; Hueber et al., 2007; Lei et al., 2006; Lei et al., 2005; Lu et al., 2008; Salsi et al., 2008; Takeda et al., 2006; Williams et al., 2005). Identifying which of these genes are direct targets is of primary importance to understand how Hox factors regulate cell growth and differentiation. Moreover, large-scale genomics have provided a wealth of sequence data that can be mined for the presence of potential Hox binding sites (as well as their co-factor proteins) near candidate target genes. However, our ability to use bioinformatics to predict Hox target sites is limited by our understanding of what Hox regulatory elements look like and the characteristics they share. In this study, we used protein and DNA site mutations to explore the binding characteristics of both anterior (Labial) and posterior (Abd-A) Hox complexes with the Exd and Hth transcription factors on cis-regulatory elements. Through this analysis, we determined that: 1) Hox factors interact with Exd/Hth on a variety of binding site combinations, and that suboptimal sites can be combined to yield functional Hox complexes; 2) in general, posterior Hox factors are less dependent on Exd/Hth binding than anterior Hox factors on the same DNA probes. Here, we discuss the implications of these findings and assess the relative accuracy of using position weight matrices (PWMs) to predict Hox complex binding sites.
The DMXR element: Integration of multiple suboptimal Hox binding sites
The DMXR element provides a good example of the flexible nature of Hox complex formation on DNA. Unlike other previously characterized Hox regulatory elements, DMXR contains both Exd/Hox and Hth/Hox sites that mediate abdominal Hox tetramer formation. Importantly, each on its own is a relatively low-affinity binding site due to suboptimal spacing between sites. Together, however, they constitute a strong binding site that mediates abdominal Hox complex formation to repress Dll expression (Gebelein et al., 2004). Moreover, this combination of binding sites results in DMXR being a robust repression element. For example, mutating either the Exd or Hth sites in the context of the full DMXR element results in only a modest loss of both DNA binding (Supplemental Figure 2) and repression activity in vivo (Gebelein et al., 2004). In contrast, when the same mutations are introduced into the DMXR1 probe that lacks the Hox2 site, abdominal Hox complex formation is severely disrupted (Figure 4) and abdominal Hox-mediated repression is significantly more compromised than on the full DMXR element (Gebelein et al., 2002). Thus, suboptimal Hox binding sites can be combined to form robust regulatory elements, and as relatively few Hox cis-elements have been thoroughly characterized, we propose additional configurations of Exd, Hth, and Hox sites are likely to result in cooperative complexes on cis-regulatory elements.
Differences in Hox complex formation between Anterior and Posterior Hox factors
By characterizing the dependence of Hox complex formation upon Exd and Hth DNA binding on several cis-regulatory elements, we made the unanticipated finding that in general posterior Hox factors (Abd-A) form more robust complexes than anterior Hox factors (Lab). For example, mutations within the Hth protein that disrupt DNA binding (Hth51A) consistently had a greater affect on Lab complexes than Abd-A complexes on the same DNA probes (Figure 6). More surprisingly, we found that a distant Hth site can mediate cooperative Hox complexes with posterior but not anterior Hox factors in the absence of Exd binding (Figure 6). On both Hoxa3-PHP1 and DMXR1-Con, for instance, Abd-A but not Lab, readily formed complexes with a DNA binding compromised Exd protein (Exd51A). Intriguingly, the ability of Abd-A to form these complexes was Hox site dependent, as DMXR1 and Lab48/95 were unable to mediate similar complexes in the presence of Exd51A. Further analysis revealed a single nucleotide change in DMXR1 (TAAT to TTAT) makes Hox complex formation relatively independent of Exd binding. Moreover, we found that a central Hox factor (Scr, Hox5 homologue), displays an intermediate ability to form Hox complexes independent of Exd binding on the same probes. Thus, at least for these three Hox proteins, the more posterior Hox factors are better able than anterior Hox factors to tolerate loss of either Exd or Hth binding and retain Hox complex formation.
The differential ability of anterior and posterior Hox factors to form complexes with Exd and Hth has two important implications. First, if anterior Hox complex formation is more dependent upon Exd and Hth binding, then anterior Hox cis-regulatory elements should be more likely to have binding sites for all three factors than posterior Hox regulatory elements. While the total number of characterized regulatory elements is still relatively small, it is interesting to note that of the eleven cis-elements regulated by Lab (or Lab homologues) seven contain characterized Hth/Meis binding sites (Mann et al., 2009). In contrast of the thirty-three cis-elements regulated by the central/posterior Hox factors, only two contain characterized Hth binding sites (Mann et al., 2009). This model also predicts that anterior Hox factors should be more sensitive to hypomorphic exd and hth gene mutations than posterior Hox factors. At least for cuticle formation in Drosophila, this prediction holds, as posterior segments have relatively normal cuticles in both exd and hth mutant embryos whereas anterior segments are transformed into posterior fates (Peifer and Wieschaus, 1990; Rieckhof et al., 1997).
A second implication of our work relates to the phenomenon of posterior dominance (also known as prevalence) of the Hox factors. Studies in Drosophila have shown that when an anterior and posterior Hox factor are co-expressed in the same segment, the fate of the segment is predominantly determined by the posterior Hox factor. Several explanations have been proposed regarding this phenomenon, including that posterior Hox factors make additional contacts with both DNA and Exd to raise their DNA binding affinity (LaRonde-LeBlanc and Wolberger, 2003; Merabet et al., 2007). Our studies are consistent with and add to this model by showing that posterior Hox factors form more robust complexes than anterior Hox factors. In particular, the ability of posterior Hox factors to bind DNA sequences relatively independent of Exd or Hth suggests the posterior Hox factors would bind additional target genes than anterior Hox factors. Congruent with this possibility, Hueber et al used microarray studies to show that Abd-A regulated many more target genes than the anterior Hox factors and that most of these targets were unique to Abd-A (i.e. not regulated by other Hox factors) (Hueber et al., 2007). Future studies focused on the identification and characterization of additional cis-regulatory elements will be required to determine how the Abd-A factor can specifically affect the regulation of so many downstream target genes.
How good are we at predicting Hox cis-regulatory elements using PWMs?
Advances in DNA sequencing technologies have provided a wealth of genomic sequence data that can be searched for potential cis-regulatory elements. To successfully do so, we first need to accurately predict transcription factor binding sites. For the Hox factors, several biochemical and high throughput approaches have been used to identify sequences bound by individual Hox factors or in combination with Exd(Pbx) and/or Hth(Meis) proteins (Chang et al., 1996; Noyes et al., 2008; Shen et al., 1997). Here, we applied the results from these searches to generate PWMs and correlated their scores with the relative DNA binding activity of Lab Hox complexes. We found that using the bacterial one-hybrid PWMs did not result in a strong correlation with our DNA binding analysis. This result may be due to the Lab and Exd proteins being tested in isolation in the one-hybrid assays, whereas the cooperative binding of Exd/Lab heterodimers may alter/restrict the sequences that can be bound relative to monomer proteins. In contrast, the in vitro SELEX sites were identified using Pbx1/HoxB1 proteins together and the Meis1 protein alone, and the PWMs from these sites strongly correlated with Lab complex formation. However, there is an important caveat with this analysis. Since the SELEX sites were published prior to the identification of the cis-regulatory elements, the SELEX sequences are likely to have been used as a guide to identify the cis-elements in the first place. Nevertheless, the direct correlation between strength of Lab complex and total PWM score suggests using the SELEX matrices in a bioinformatics approach is likely a good strategy to identify additional Lab cis-regulatory elements.
What about using PWMs for the posterior Hox factors? Our biochemical data indicate that using a similarly strict search for both Exd/Hox and Hth sites for the posterior Hox cis-regulatory elements may not work as well due to posterior Hox complex formation being less dependent upon both Exd and Hth binding. However, we do believe some general guidelines can be useful in searching for Hox cis-regulatory elements. 1) As mentioned above, anterior Hox factors are likely to be more highly dependent upon Hth binding than posterior Hox factors. 2) Optimal Hth/Hox sites can mediate posterior Hox complex formation relatively independent of Exd binding. 3) A distant Hth binding site can permit posterior Hox complex formation on suboptimal Exd/Hox binding sites or on specific Hox binding sites independent of Exd binding. 4) Nearby low affinity (suboptimal) binding sites can be combined to mediate functional Hox regulatory complexes.
Supplementary Material
Acknowledgments
We would like to acknowledge the Gebelein and Cook laboratories for their helpful discussions of this project. This work was supported by the National Institutes of Health (GM079428 to BG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Shaar M, Ryoo HD, Mann RS. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 1999;13:935–945. doi: 10.1101/gad.13.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affolter M, Slattery M, Mann RS. A lexicon for homeodomain-DNA recognition. Cell. 2008;133:1133–1135. doi: 10.1016/j.cell.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, Khalid F, Zhang W, Newburger D, Jaeger SA, Morris QD, Bulyk ML, Hughes TR. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter EM, Goddard JM, Chisaka O, Manley NR, Capecchi MR. Loss of Hox-A1 (Hox-1.6) function results in the reorganization of the murine hindbrain. Development. 1993;118:1063–1075. doi: 10.1242/dev.118.4.1063. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity. Blackwell Science; Malden, MA: 2001. [Google Scholar]
- Chan SK, Jaffe L, Capovilla M, Botas J, Mann RS. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- Chan SK, Popperl H, Krumlauf R, Mann RS. An extradenticle-induced conformational change in a HOX protein overcomes an inhibitory function of the conserved hexapeptide motif. Embo J. 1996;15:2476–2487. [PMC free article] [PubMed] [Google Scholar]
- Chan SK, Ryoo HD, Gould A, Krumlauf R, Mann RS. Switching the in vivo specificity of a minimal Hox-responsive element. Development. 1997;124:2007–2014. doi: 10.1242/dev.124.10.2007. [DOI] [PubMed] [Google Scholar]
- Chang CP, Brocchieri L, Shen WF, Largman C, Cleary ML. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol Cell Biol. 1996;16:1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Jacobs Y, Nakamura T, Jenkins NA, Copeland NG, Cleary ML. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- Chung KY, Morrone G, Schuringa JJ, Plasilova M, Shieh JH, Zhang Y, Zhou P, Moore MA. Enforced expression of NUP98-HOXA9 in human CD34(+) cells enhances stem cell proliferation. Cancer Res. 2006;66:11781–11791. doi: 10.1158/0008-5472.CAN-06-0706. [DOI] [PubMed] [Google Scholar]
- Dolle P, Lufkin T, Krumlauf R, Mark M, Duboule D, Chambon P. Local alterations of Krox-20 and Hox gene expression in the hindbrain suggest lack of rhombomeres 4 and 5 in homozygote null Hoxa-1 (Hox-1.6) mutant embryos. Proc Natl Acad Sci U S A. 1993;90:7666–7670. doi: 10.1073/pnas.90.16.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- Ebner A, Cabernard C, Affolter M, Merabet S. Recognition of distinct target sites by a unique Labial/Extradenticle/Homothorax complex. Development. 2005;132:1591–1600. doi: 10.1242/dev.01721. [DOI] [PubMed] [Google Scholar]
- Ekker SC, Jackson DG, von Kessler DP, Sun BI, Young KE, Beachy PA. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. Embo J. 1994;13:3551–3560. doi: 10.1002/j.1460-2075.1994.tb06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Cambronero F, Tumpel S, Longobardi E, Wiedemann LM, Blasi F, Krumlauf R. Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol Cell Biol. 2005;25:8541–8552. doi: 10.1128/MCB.25.19.8541-8552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Marshall H, Popperl H, Maconochie M, Krumlauf R, Blasi F. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development. 2000;127:155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- Galant R, Carroll SB. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415:910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6:881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- Gebelein B, Culi J, Ryoo HD, Zhang W, Mann RS. Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev Cell. 2002;3:487–498. doi: 10.1016/s1534-5807(02)00257-5. [DOI] [PubMed] [Google Scholar]
- Gebelein B, McKay DJ, Mann RS. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature. 2004;431:653–659. doi: 10.1038/nature02946. [DOI] [PubMed] [Google Scholar]
- Gebelein B, Urrutia R. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Kruppel-associated box protein. Mol Cell Biol. 2001;21:928–939. doi: 10.1128/MCB.21.3.928-939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- Ghannam G, Takeda A, Camarata T, Moore MA, Viale A, Yaseen NR. The oncogene Nup98-HOXA9 induces gene transcription in myeloid cells. J Biol Chem. 2004;279:866–875. doi: 10.1074/jbc.M307280200. [DOI] [PubMed] [Google Scholar]
- Hueber SD, Bezdan D, Henz SR, Blank M, Wu H, Lohmann I. Comparative analysis of Hox downstream genes in Drosophila. Development. 2007;134:381–392. doi: 10.1242/dev.02746. [DOI] [PubMed] [Google Scholar]
- Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRonde-LeBlanc NA, Wolberger C. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 2003;17:2060–2072. doi: 10.1101/gad.1103303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89:1922–1930. [PubMed] [Google Scholar]
- Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Komuves L, Buchberg AM, Largman C. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13:1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- Lei H, Juan AH, Kim MS, Ruddle FH. Identification of a Hoxc8-regulated transcriptional network in mouse embryo fibroblast cells. Proc Natl Acad Sci U S A. 2006;103:10305–10309. doi: 10.1073/pnas.0603552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Wang H, Juan AH, Ruddle FH. The identification of Hoxc8 target genes. Proc Natl Acad Sci U S A. 2005;102:2420–2424. doi: 10.1073/pnas.0409700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons D, McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006;313:1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- Li-Kroeger D, Witt LM, Grimes HL, Cook TA, Gebelein B. Hox and senseless antagonism functions as a molecular switch to regulate EGF secretion in the Drosophila PNS. Dev Cell. 2008;15:298–308. doi: 10.1016/j.devcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Kamps MP. Structural determinants within Pbx1 that mediate cooperative DNA binding with pentapeptide-containing Hox proteins: proposal for a model of a Pbx1-Hox-DNA complex. Mol Cell Biol. 1996;16:1632–1640. doi: 10.1128/mcb.16.4.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hardt J, Kim JJ. Global analysis of genes regulated by HOXA10 in decidualization reveals a role in cell proliferation. Mol Hum Reprod. 2008;14:357–366. doi: 10.1093/molehr/gan023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz B, Lu HC, Eichele G, Miller D, Kaufman TC. Rescue of Drosophila labial null mutant by the chicken ortholog Hoxb-1 demonstrates that the function of Hox genes is phylogenetically conserved. Genes Dev. 1996;10:176–184. doi: 10.1101/gad.10.2.176. [DOI] [PubMed] [Google Scholar]
- Mann RS. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Morata G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol. 2000;16:243–271. doi: 10.1146/annurev.cellbio.16.1.243. [DOI] [PubMed] [Google Scholar]
- Manzanares M, Bel-Vialar S, Ariza-McNaughton L, Ferretti E, Marshall H, Maconochie MM, Blasi F, Krumlauf R. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involve auto- and cross-regulatory mechanisms. Development. 2001;128:3595–3607. doi: 10.1242/dev.128.18.3595. [DOI] [PubMed] [Google Scholar]
- Mark M, Lufkin T, Dolle P, Dierich A, LeMeur M, Chambon P. Roles of Hox genes: what we have learnt from gain of function and loss of function mutations in the mouse. C R Acad Sci III. 1993;316:995–1008. [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- McGonigle GJ, Lappin TR, Thompson A. Grappling with the HOX network in hematopoiesis and leukemia. Front Biosci. 2008;13:4297–4308. doi: 10.2741/3006. [DOI] [PubMed] [Google Scholar]
- Merabet S, Saadaoui M, Sambrani N, Hudry B, Pradel J, Affolter M, Graba Y. A unique Extradenticle recruitment mode in the Drosophila Hox protein Ultrabithorax. Proc Natl Acad Sci U S A. 2007;104:16946–16951. doi: 10.1073/pnas.0705832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill VK, Diederich RJ, Turner FR, Kaufman TC. A genetic and developmental analysis of mutations in labial, a gene necessary for proper head formation in Drosophila melanogaster. Dev Biol. 1989;135:376–391. doi: 10.1016/0012-1606(89)90187-5. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature. 1999;397:714–719. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Peifer M, Wieschaus E. Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev. 1990;4:1209–1223. doi: 10.1101/gad.4.7.1209. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Rambaldi I, Featherstone MS. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–597. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Qian YQ, Billeter M, Otting G, Muller M, Gehring WJ, Wuthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989;59:573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- Qian YQ, Otting G, Billeter M, Muller M, Gehring W, Wuthrich K. Nuclear magnetic resonance spectroscopy of a DNA complex with the uniformly 13C-labeled Antennapedia homeodomain and structure determination of the DNA-bound homeodomain. J Mol Biol. 1993;234:1070–1083. doi: 10.1006/jmbi.1993.1660. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Ronshaugen M, McGinnis N, McGinnis W. Hox protein mutation and macroevolution of the insect body plan. Nature. 2002;415:914–917. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Mann RS. The control of trunk Hox specificity and activity by Extradenticle. Genes Dev. 1999;13:1704–1716. doi: 10.1101/gad.13.13.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Marty T, Casares F, Affolter M, Mann RS. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development. 1999;126:5137–5148. doi: 10.1242/dev.126.22.5137. [DOI] [PubMed] [Google Scholar]
- Salsi V, Vigano MA, Cocchiarella F, Mantovani R, Zappavigna V. Hoxd13 binds in vivo and regulates the expression of genes acting in key pathways for early limb and skeletal patterning. Dev Biol. 2008;317:497–507. doi: 10.1016/j.ydbio.2008.02.048. [DOI] [PubMed] [Google Scholar]
- Samad OA, Geisen MJ, Caronia G, Varlet I, Zappavigna V, Ericson J, Goridis C, Rijli FM. Integration of anteroposterior and dorsoventral regulation of Phox2b transcription in cranial motoneuron progenitors by homeodomain proteins. Development. 2004;131:4071–4083. doi: 10.1242/dev.01282. [DOI] [PubMed] [Google Scholar]
- Shen WF, Montgomery JC, Rozenfeld S, Moskow JJ, Lawrence HJ, Buchberg AM, Largman C. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky A, Bonin CP, Mann RS, Honig B. Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 2003;31:3589–3592. doi: 10.1093/nar/gkg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Goolsby C, Yaseen NR. NUP98-HOXA9 induces long-term proliferation and blocks differentiation of primary human CD34+ hematopoietic cells. Cancer Res. 2006;66:6628–6637. doi: 10.1158/0008-5472.CAN-06-0458. [DOI] [PubMed] [Google Scholar]
- Tumpel S, Cambronero F, Ferretti E, Blasi F, Wiedemann LM, Krumlauf R. Expression of Hoxa2 in rhombomere 4 is regulated by a conserved cross-regulatory mechanism dependent upon Hoxb1. Dev Biol. 2007;302:646–660. doi: 10.1016/j.ydbio.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Williams TM, Williams ME, Kuick R, Misek D, McDonagh K, Hanash S, Innis JW. Candidate downstream regulated genes of HOX group 13 transcription factors with and without monomeric DNA binding capability. Dev Biol. 2005;279:462–480. doi: 10.1016/j.ydbio.2004.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.