Abstract
Protein tyrosine kinase 6 (PTK6), also referred to as breast tumor kinase BRK, is a member of a distinct family of kinases that is evolutionarily related to the SRC family of tyrosine kinases. While not expressed in the normal mammary gland, PTK6 expression is detected in a large proportion of human mammary gland tumors. In breast tumor cells, PTK6 promotes growth factor signaling and cell migration. PTK6 expression is also increased in a number of other epithelial tumors, including ovarian and colon cancer. In contrast, PTK6 is expressed in diverse normal epithelia, including the linings of the gastrointestinal tract, skin and prostate, where its expression correlates with cell cycle exit and differentiation. Disruption of the mouse Ptk6 gene leads to increased growth and impaired differentiation in the small intestine that is accompanied by increased AKT and Wnt signaling. Following total body irradiation, PTK6 expression is induced in proliferating progenitor cells of the intestine, where it plays an essential role in DNA-damage induced apoptosis. A distinguishing feature of PTK6 is its flexibility in intracellular localization, due to a lack of amino-terminal myristoylation/palmitoylation. Recently a number of substrates of PTK6 have been identified, including nuclear RNA-binding proteins and transcription factors. We discuss PTK6 signaling, its apparent conflicting roles in cancer and normal epithelia, and its potential as a therapeutic target in epithelial cancers.
Keywords: PTK6, BRK, Sik, Tyrosine kinase, Epithelia, Breast cancer, Colon cancer
1. Introduction
The intracellular protein tyrosine kinase 6 (PTK6) has been implicated in the regulation of a variety of signaling pathways that control the differentiation and maintenance of normal epithelia, as well as tumor growth. A rapidly growing number of publications have identified new PTK6 substrates and binding partners, and expanded the range of tissues and cancers in which PTK6 is expressed. However, some questions still exist about the involvement of PTK6 in cancer. Is PTK6 a useful tumor marker that is induced by oncogenic signaling? Does its expression promote tumorigenesis and/or metastases in vivo? Does PTK6 expression/localization correlate with tumor cell differentiation? Recent findings suggest that functions of PTK6 are context dependent and differ depending on cell type, as well as its intracellular localization.
PTK6 was first identified in a survey of protein tyrosine kinase mRNAs expressed in normal human melanocytes [1], and shortly thereafter cloned from human breast cancer cells as BRK (breast tumor kinase) [2], as well as from the mouse gastrointestinal tract as Sik (Src-related intestinal kinase) [3]. Other members of the PTK6 family include FRK (Fyn-related kinase; also referred to as RAK and in rodent BSK, GTK, IYK) (reviewed in [4]), and SRMS (SRC-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristoylation sites; also known as SRM). Like PTK6, FRK was identified in breast cancer cells and the normal intestinal epithelium (reviewed in [4]). SRMS was cloned from mouse embryonic neuroepithelial cells [5] and the mouse skin [6], and its functions remain poorly characterized.
2. PTK6 structure
2.1. PTK6 gene structure
The PTK6 gene contains eight exons, with intron/exon boundaries distinct from the SRC family of tyrosine kinases [7–9]. A common ancestral gene in metazoans is thought to have given rise to the two related, yet distinct PTK6 and SRC families [10]. A series of gene duplication events gave rise to the three PTK6 family members and the PTK6 and SRMS genes have remained tightly linked.
Human PTK6 maps to chromosome 20q13.3 [11] (chromosome 2 in the mouse [12]), and the 813 base pair (bp) region upstream of the translation initiation site has 60% of the activity of the SV40 promoter [13]. The region between −93 to −76 bp has been identified as the minimal promoter region, and −702 to −655 bp contains two cis-acting elements with binding sites for NFκB (−706 to −688 bp) and SP1 (−688 to −669 bp) [14]. The PTK6 transcript was shown to start at approximately −104 bp upstream of the translation start codon [14]. It was also demonstrated that Krüppel-Like Factor 9 has transcriptional activity at the PTK6 promoter in the colon, and indirectly affects PTK6 expression in the jejunum [15]. Analysis of the PTK6 regulatory region is complicated due to the tight linkage with the SRMS gene, which maps to chromosome 20q13.33, only 1.5 kbp upstream of the PTK6 gene.
The human PTK6 gene encodes at least one alternatively spliced transcript that lacks exon 2, encoding a truncated, catalytically inactive protein that shares its amino terminus and SH3 domain with full length PTK6 and has a novel carboxy-terminus (Fig. 1). This alternative PTK6 transcript (ALT-PTK6), originally referred to as λm5, was shown to be expressed in breast cancer cells but its function has not been explored [8]. Recently we identified ALT-PTK6 in a number of prostate and intestinal cancer cell lines (Brauer and Tyner, unpublished data) suggesting that the alternative splicing is not breast cancer specific. In most published studies, siRNA-mediated knockdown of PTK6 should target both full length PTK6 as well as its alternative spliced isoform ALT-PTK6, although consequences of ALT-PTK6 knockdown have not been assessed.
Fig. 1.
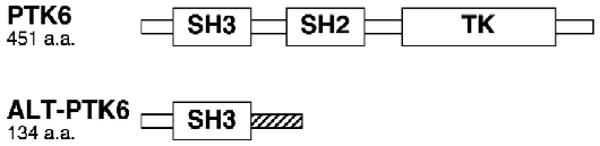
Proteins encoded by the PTK6 gene. PTK6 is a 48 kDa protein that consists of Src-homology (SH) 2, SH3, and tyrosine kinase (TK) domains. PTK6 lacks an amino-terminal SH4 domain required for myristoylation/palmitoylation found in the SRC family of tyrosine kinases. The alternatively spliced isoform of PTK6, ALT-PTK6, has a predicted molecular weight of 15 kDa and contains the SH3 domain and a novel proline-rich carboxy terminal sequence (striped area).
2.2. PTK6 protein structure
PTK6 is a 451 amino acid protein that consists of a tyrosine kinase domain, as well as SH2 and SH3 (SRC-homology) domains that are involved in protein interactions and autoregulation (Fig. 1). PTK6 autophosphorylates itself at tyrosine residues 13, 61, 66, 114, 351, as well as at tyrosine 342 within the kinase activation loop, which increases its catalytic activity [2,16]. In contrast to SRC, the interaction between the tryptophan 184 residue within the proline-rich SH2-Kinase linker region and the catalytic domain appears essential for kinase activity [17]. Like SRC, the C-terminal tyrosine 447 residue binds to the SH2 domain when phosphorylated and negatively regulates kinase activity, which can be prevented by mutating this residue to phenylalanine [16,18]. The kinase that phosphorylates the PTK6 C-terminal tyrosine has not been identified, although CSK (c-Src tyrosine Kinase) that phosphorylates the SRC C-terminal tyrosine probably does not phosphorylate PTK6 [16]. Interactions between the SH3 domain and the proline-rich linker region are also involved in autoinhibition [16,19,20] and are thought to stabilize the inactive state of the enzyme similar to SRC. The SH3 domain consists mainly of β-sheets, which exhibit a unique and folded structure at neutral pH that is sensitive to pH changes [21]. Aside from intramolecular interactions [19,20], the SH3 domain of PTK6 plays a major role in substrate interactions [18,22–24].
The SH2 domain of PTK6 consists of an α/β fold with a phosphotyrosine binding surface and two α-helices that are opposite to a central β-sheet made up of four anti-parallel strands [25,26]. Although phosphotyrosine binding is weaker than that of SRC-family members [26], the SH2 domain of PTK6 plays a role in protein–protein interactions [18,23,27,28], but is likely more important for the regulation of catalytic activity [16]. PTK6 family members lack myristoylation and palmitoylation signals, which set them apart from SRC-family members, allowing them a greater flexibility in their subcellular localization, and also giving them a different range of binding partners and substrates.
3. PTK6 substrates and binding partners
A growing number of PTK6 substrates and interacting proteins are being discovered (summarized in Table 1). To date, substrates include AKT [29], β-catenin [30], KAP3A [31], p190RhoGAP [32], Paxillin [23], PSF [24], Sam68 [18,22,33–35], SLM1 and SLM2 [36], BKS/STAP2 [28,37], STAT3 [38], and STAT5b [39]. Other potential substrates that have not been fully validated at this time include β-Tubulin [31], FLJ39441/SPTY2D1 [31], GNAS [31], and other unidentified phosphotyrosine bands such as the 100 kDa STAP2 associated protein [28].
Table 1.
PTK6 substrates and binding partners.
| Protein | Association | Substrate target | Proposed function/role | Reference |
|---|---|---|---|---|
| ADAM-15A and E | Yes | N/D | Not validated | [41] |
| AKT | Yes | Yes | Growth regulation | [29], Zheng and Tyner, unpub. |
| beta-catenin | Yes | Y64, Y331/333, Y142 | Growth regulation | [30] |
| beta-tubulin | No | Yes | Not validated | [31] |
| ErbB1 | Yes | N/D | Growth regulation | [42] |
| ErbB2 | Yes | N/D | Growth regulation | [35,43] |
| ErbB3 | Yes | N/D | Growth regulation | [35,46] |
| ErbB4 | Yes | N/D | Not validated | [35] |
| FLJ39441 | No | Yes | Not validated | [31] |
| GapA (p65) | PTK6 SH2 | Not likely | Differentiation | [27] |
| GNAS | No | Yes | Not validated | [31] |
| IRS-4 | PTK6 SH2 and SH3 | N/D | Growth and migration | [23] |
| KAP3A | Yes | C-terminal tyrosines | Migration | [31] |
| MAPK | Yes | N/D | Not validated | [35] |
| p190RhoGAP | Yes | Y1105 | Growth and migration | [32] |
| Paxilin | Yes | Y31, Y118 | Migration and invasion | [23] |
| PSF | PTK6 SH3 | C-terminal tyrosines | Growth regulation | [24] |
| PTEN | YES | N/D | Not validated | [35] |
| PTK6 | Intramolecular | Y13, Y61, Y66, Y114, Y342, Y351 | Regulation of kinase activity | [2,16,17,19,20] |
| Sam68 | PTK6 SH2 and SH3 | Y435; Y440; Y443 | Inhibition of Sam68 | [18,22,33–35] |
| SLM1; SLM2 | N/D | Yes | Inhibition of SLM1 and SLM2 | [36] |
| STAP2 | STAP2 SH2 | Y250 | STAT3 activation | [28,37] |
| STAT3 | N/D | Yes | Growth regulation | [38] |
| STAT5b | N/D | Y699 | Growth regulation | [39] |
| 23 kDa protein | Yes | N/D | Protein not identified | [56] |
| 100 kDa protein | (STAP2-mediated) | Likely | Protein not identified | [28] |
NLS = Nuclear localization signal; SH = Src-homology domain; N/D = Not determined.
In a study that focused on screening for PTK6 substrate specificity, the preferred target sequence was X-(E/I/L/N)-Y-(D/E)-(D/E), where X can be any amino acid [40]. Within the substrates for which the targeted tyrosine residue is known, including β-catenin (Tyr 64, Tyr 142, Tyr 333), p190RhoGAP (Tyr 1105), PTK6 (Tyr 66), Sam68 (Tyr 435), and STAP2 (Tyr 250), at least one critical residue conforms to the consensus sequence (summarized in Table 2). The sequence pY-(D/E)-(D/E)-Y was identified as a binding site for the SH2 domain of PTK6 [18].
Table 2.
PTK6 target sequences.
| Protein | Residue | Sequence | Reference |
|---|---|---|---|
| β-Catenin | Y64 | VLYEWE | [30] |
| Y142 | INYQDD | ||
| Y331* | RTYTYE | ||
| Y333* | YTYEKL | ||
| p190RhoGAP | Y1105 | NIYSVP | [32] |
| Paxillin | Y31 | TPYSYP | [23] |
| Y118 | HVYSFP | ||
| PTK6 | Y13 | PKYVGL | [16] |
| Y61 | QGYVPH | ||
| Y66 | HNYLAE | ||
| Y114 | ADYVLS | ||
| Y342 | DVYLSH | ||
| Y351 | IPYKWT | ||
| Sam68 | Y435 | GAYREH | [31] |
| Y440 | HPYGRY | ||
| Y443 | GRY. | ||
| STAP2 | Y250 | EDYEKV | [37] |
| STAT5b | Y699 | DGYVKP | [39] |
The tyrosine targeted for phosphorylation is in bold.
Amino acids conforming to the consensus target sequence X-(E/I/L/N)-Y-(D/E)-(D/E) are underlined. Tyrosines marked with * were too close in proximity with each other to distinguish between them by mass spec analysis.
PTK6 has been shown to associate with a variety of proteins that are likely upstream of PTK6 in various signaling pathways, or for which PTK6 may play an adaptor-like role. These proteins include ADAM-15A and ADAM-15B [41], ErbB1 [42], ErbB2 [35,43], GapA-p65 [27], and IRS-4 [23]. ADAM-15 (A Disintegrin And Metalloproteinase) has been implicated in inflammation, differentiation, cell–cell interactions, and angiogenesis ([44] and reviewed in [45]). Association with ErbB1, ErbB2, and ErbB3 may contribute to mammary tumor development and growth through enhancement of EGF and heregulin induced signaling [42,43,46,47]. Interestingly, ADAM-15 targets the extracellular domain of E-cadherin resulting in ErbB2/ErbB3 heterodimer activation, and subsequent proliferation and migration through increased Erk signaling [48]. GapA-p65 binds to the Ras-GTPase activating protein (GAP) [27], which can associate with p190 and the p62/DOK adaptor family [49–51]. The Insulin Receptor Substrate (IRS) 4 localizes to the plasma membrane, and not to intracellular structures like IRS-1 and IRS-2, which may confer functional differences in insulin and insulin-like growth factor signaling ([52] and reviewed in [53–55]). Some data also exist that other potential binding partners include ErbB4, MAPK, PTEN [35], and an unidentified 23 kDa phosphotyrosine band that also associates with SRC [56].
4. PTK6 expression
4.1. PTK6 in normal tissues
PTK6 expression is detected in a variety of epithelial linings, where it is generally localized to differentiated cells outside of a zone of proliferating progenitor cells (summarized in Table 3). Its expression is developmentally regulated and correlates with the differentiation of epithelial linings [57]. In mature tissues, expression is highest in the gut, where PTK6 is expressed in the nondividing villus epithelium of the small intestine and the surface lining of the colon [57–59]. PTK6 is expressed in the prostate [13], where it is localized to nuclei of epithelial cells [60], the skin [1,57,61], and oral epithelium [62]. PTK6 expression has not been detected in the normal mammary gland of the mouse [12] or human [2]. Expression of PTK6 was also detected in normal T-cells upon activation, cutaneous T-cell lymphomas, and transformed B- and T-cell lymphomas [63]. Microarray studies suggest that the expression of the PTK6 gene may be affected by neuregulin 3 [64], estradiol [65], and cell growth in Whitten's medium [66], but these findings need to be confirmed.
Table 3.
Intracellular localization of PTK6 in normal tissues and cancer cells.
| Group | Tissue/cell | Sub-category | Localization (method) | Reference |
|---|---|---|---|---|
| Breast | Breast cancer | N/D | [2,69,83] | |
| Grade I; Grade III | C; PN (IHC) | [93] | ||
| Grade II; low-mod. PTK6 | C+N (IHC; IF) | [34,47,85] | ||
| Grade III; high PTK6 | C+N; N (IHC) | [47,93,106] | ||
| Low PTK6; T47D | C (IHC; IF) | [56,106] | ||
| Grade III; high PTK6 | C+N; C (IHC) | [85] | ||
| MCF7 cells | C+SNB (IF) | [18] | ||
| G.I. tract | Esophagus | N/D | [12] | |
| Stomach | N/D | [12] | ||
| Small intestine | N/D | [3,13,57] | ||
| villus; crypt 72 h after irr. | C+N; isolated N (IHC) | [59,68] | ||
| crypt 6 h after irradiation | C+N (IHC) | [68] | ||
| Colon mucosa | N/D | [3,13,57,72] | ||
| C; C+N (IHC) | [12] | |||
| Colon cancer; HT29 cells | C; C+N; C+SNB (IHC; IF) | [12,18] | ||
| Head and neck | Normal oral epithelium | high PTK6 | C; C+N; N (IHC) | [62] |
| Oral SCC; OSCC25 cells | moderate PTK6 | C+N; N (IHC) | [62] | |
| OSCC3 cells | low PTK6 | C; PN (IF; Frac) | [62] | |
| Laryngeal cancer | N; C+N (IHC) | [73] | ||
| Lymphocytes | T-cells | N/D | [63] | |
| T and B-cell lymphomas | N (Frac) | [63] | ||
| Ovary | Ovarian cancer | C; C+N; C+N (IHC) | [71] | |
| Prostate | Normal | N/D | [13] | |
| prostate | N (IHC) | [60] | ||
| Prostatic | BPH | C+N (IHC) | [60] | |
| disease | PIN | C+PN (IHC) | [60] | |
| Prostate cancer | Gleason 2 | C+N (IHC) | [60] | |
| Gleason 3 | C+N (IHC) | [60] | ||
| Gleason 3–4; LNCaP cells | C+PN+SNB+N (IHC; IF) | [60] | ||
| Gleason 4; PC3 cells | C (IHC; IF) | [60] | ||
| Epidermis | Skin | N/D | [1,57] | |
| epithelium | C; C+N (IHC) | [61] | ||
| SCC | C; C+N (IHC) | [61] | ||
| HaCaT cells | Low density; low PTK6 | C+N (IF) | [61] | |
| High density; high PTK6 | C+N (IF) | [61] | ||
| Epid. cancer (A431 cells) | High density | C+N (IF) | [84] | |
| 2 h scratch; +EGF | C+N+MR (IF) | [84] | ||
| 6 h scratch | C+N (IF) | [84] |
SCC = Squamous cell carcinoma; BPH = Benign prostatic hyperplasia; PIN = Preneoplatic intraepithelial neoplasia; C = Cytoplasm; N = Nucleus; PN = Perinuclear; SNB = SAM68 nuclear bodies; MR = Membrane ruffles; IHC = Immunohistochemistry; IF = Immunofluorescence; Frac = Fractionation; N/D = Not determined; Epid. = Epidermoid. Predominant localization is in bold.
A variety of in vitro and in vivo studies indicate that PTK6 may promote differentiation of epithelial cells. PTK6 plays a role in calcium-induced differentiation of cultured mouse and human keratinocytes and it promotes expression of the epidermal differentiation markers Filaggrin [27] and Keratin-10 [61]. PTK6 expression increases during differentiation of the Caco-2 colon carcinoma cell line, which spontaneously polarizes and expresses enzymes characteristic of differentiated small intestinal absorptive cells [12]. Disruption of the Ptk6 gene in the mouse demonstrated that PTK6 positively regulates intestinal epithelial cell differentiation in vivo. Ptk6 null mice were characterized as having increased cell turnover in the small intestine, which was accompanied by increased villus length and crypt depth, and delayed enterocyte differentiation [59]. A cell line was generated from the colonic mucosa of the Ptk6 null mice [67], which will be useful to explore the role of PTK6 in differentiation. The ability of PTK6 to inhibit both AKT signaling [29,59] and the Wnt pathway [30,59] could contribute to its growth suppressive activities in vivo.
In addition to regulating epithelial cell differentiation, PTK6 modulates survival of normal cells. Ectopic overexpression of PTK6 in the immortalized nontransformed Rat1A rat embryo fibroblast cell line sensitized these cells to apoptotic stimuli [58]. Subjecting mice to total body gamma-irradiation resulted in induction of PTK6 in progenitor cells of the small intestine, where PTK6 promoted DNA-damage induced apoptosis [68]. PTK6-mediated inhibition of AKT and MAPK prosurvival signaling pathways appears important, as disruption of Ptk6 led to increased activation of AKT and ERK1/2 and impaired apoptosis in irradiated mice [68].
4.2. PTK6 in cancer
PTK6 was detected in a screen for protein tyrosine kinases expressed in human breast cancer, and was found in greater than 60% of breast tumors and breast cancer derived cell lines, but was absent in normal mammary tissue and benign lesions [2,69]. A recent examination of PTK6 expression in a human breast tissue microarray of 250 samples revealed PTK6 protein expression in 86% of invasive ductal breast tumors [47], and PTK6 mRNA was overexpressed in 85% of 44 breast carcinoma samples [70]. PTK6 expression has also been detected in human ovarian tumor cells but not the normal ovary [71], in primary and metastasized colon tumors [12,72], head and neck squamous cell carcinoma (SCC) [73], prostate tumors [60], B- and T-cell lymphomas [63], and the HeLa cervical cancer cell line [32]. PTK6 gene expression has been detected in the cancers of the lung [74–78], bladder [78], pancreas [79], and gastric cancer [80], but protein expression or functions have not been validated.
Comparison of the PTK6 cDNA sequence from normal and tumor tissues did not reveal the presence of any specific mutations within the PTK6 gene in breast cancer [8]. However, PTK6 somatic mutations were identified in 2.5% of human melanomas that were recently examined [81]. Mutations of PTK6 were also detected in the RT-4 bladder and SW900 lung cancer cell lines [78], suggesting that genetic alterations in PTK6 might in some cases contribute to cancer. Amplification of the PTK6 gene correlated with amplification of the ErbB2 locus in a panel of 202 human breast cancer samples [43]. In another study it was found that an increased PTK6 copy number in a cohort of 426 invasive breast carcinomas was largely due to polysomy of chromosome 20, and that PTK6 was amplified only at a low level relative to ErbB2 [82], suggesting that additional mechanisms may also lead to an increase in PTK6. Evaluation of a variety of breast cancer cell lines showed a wide range of ErbB2 and PTK6 expression [83]. Low level PTK6 gene amplification was observed in 6/7 primary ovarian tumors [71].
5. PTK6 signaling pathways
5.1. Growth-promoting signaling pathways activated by PTK6 in cancer
A variety of studies indicate that PTK6 stimulates ErbB receptor tyrosine kinase signaling in cancer cells. Overexpression of PTK6 alone sensitizes mammary epithelial cells to mitogenic effects of EGF [42] and its coexpression with ErbB3 markedly enhances EGF signaling via AKT and PI-3 kinase [46]. Different ErbB receptor ligands, including EGF and heregulin, stimulate PTK6 activity [42,46,47,84]. Expression of ErbB2 that lacks a known ligand also enhances PTK6 activity [43]. Other factors that activate PTK6 include calcium and ionomycin [27,61], fetal bovine serum [29], ErbB2 [43], IGF-1 [23], and osteopontin [85]. The finding that osteopontin is an activator of PTK6 is particularly striking since osteopontin is secreted from the bone and brain, a frequent site of metastasis in aggressive breast and prostate cancer ([86,87] and reviewed in [88,89]). Proteins and molecules reported to be upstream or downstream of PTK6 are shown in Fig. 2.
Fig. 2.
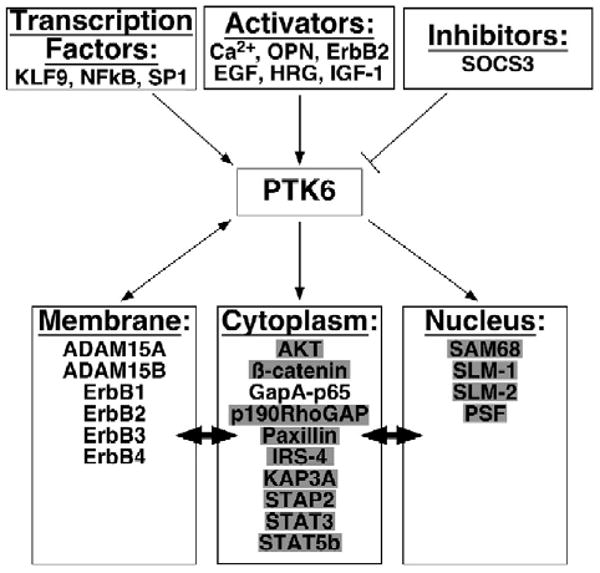
PTK6 activation and downstream signaling. PTK6 is regulated by a variety of factors. Substrates (grey boxes) and interacting proteins may be localized to different cellular compartments (as indicated by double headed arrows). For example, β-catenin has well-established functions at the membrane and in the nucleus, and PTK6 influences β-catenin in both compartments. In most cases the effects that PTK6 has on substrates and binding partners in different cellular compartments merit further exploration. (OPN: osteopontin; HRG: heregulin).
In a variety of breast cancer model systems, using both knockdown and overexpression systems, PTK6 was shown to increase proliferation, anchorage-independent growth, cell migration, and tumor growth [32,38,39,42,43,47,63,70,84,90,91]. While knockdown of PTK6 expression in breast tumor cells resulted in decreased proliferation, expression of catalytically inactive PTK6 increased cell proliferation [90], suggesting that kinase-independent PTK6 adaptor-like functions could promote breast cancer cell growth. PTK6 was shown to contribute to migration and proliferation by contributing to EGF-mediated phosphorylation of p190RhoGAP, which promotes association with p120RasGAP, inactivating RhoA while activating Ras [32]. EGF stimulation resulted in phosphorylation of Paxillin by PTK6 and activation of Rac1 via CrkII, thereby promoting migration and invasion [84]. However, it has also been suggested that a significant correlation between PTK6 and the estrogen receptor [92], as well as co-overexpression of PTK6 with ErbB3 and ErbB4, provides evidence that PTK6 may promote differentiation in tumors [93].
PTK6 is overexpressed in ER/PR positive and negative cell lines, in cell lines with high and low levels of ErbB2 expression, and in triple negative tumor cell lines [69]. Knockdown of PTK6 using siRNA led to impaired growth and migration in the ER positive T47D cell line [47,90]. Recently, PTK6 was shown to promote survival of nonadherent breast cancer cells by negatively regulating the autophagy protein Beclin [70]. Involvement of PTK6 in Mitogen-Activated Protein Kinase (MAPK) cascades is evident, and these pathways are typically activated by stimuli including growth factors, cytokine signals or stress. Stable expression of ErbB2 and PTK6 in the non-tumorigenic MCF10A cell line resulted in activated MEK1/2 and ERK1/2 [43]. EGF and heregulin stimulation of the SK-BR-3 and T47D breast cancer cell lines resulted in activation of p38, Erk5 and MEF2, which was abrogated by PTK6 shRNA [47].
The signal transducer and activator of transcription (STAT) family of proteins regulate a variety of biological processes including inflammation, differentiation, apoptosis, and transformation (reviewed in [94–96]). PTK6 activates STAT3 [38] and STAT5b [39] to promote proliferation, and this may be facilitated by the signal transducing adaptor protein-2 (STAP2) [28]. A recent study reported that concurrent activation of both STAT3 and STAT5b leads to decreased proliferation, increased differentiation and a more favorable prognosis compared to solely STAT3 activation [97]. The suppressor of cytokine signaling 3 (SOCS3), which acts as a negative regulator of JAK–STAT signaling, inhibits PTK6 [38]. PTK6 could play a role in activating STAT-mediated responses to inflammation. ErbB receptors may regulate inflammation [98], and PTK6 potentiates ErbB signaling. Additional evidence linking PTK6 to inflammation include identification of a binding site for the transcription factor NFκB in the PTK6 promoter [14].
AKT is frequently activated in cancer where it promotes cell growth, proliferation, and survival. Evidence in breast cancer suggests that PTK6 stimulates AKT signaling by increasing ErbB receptor signaling [42,43,46]. The IGF-1 and Insulin receptors also regulate AKT through IRS and PI3K, and IGF-1 is an activator of PTK6 that can bind to either receptor [23]. Although PTK6 associated with and promoted tyrosine phosphorylation of AKT in unstimulated cells, where AKT activity was inhibited, other results suggest that PTK6 may promote AKT activation through the formation of different complexes in breast cancer cells [29]. Our recent findings suggest that AKT is a direct PTK6 substrate, and that PTK6 mediated tyrosine phosphorylation of AKT can lead to an increase in its activity (Y. Zheng and A. L. Tyner, unpublished). Currently PTK6 regulation of AKT signaling appears complex and might be activating or inhibitory and dependent on cellular context; in immortalized nontransformed Rat1a cells, serum stimulation and expression of PTK6 did not lead to activation of Akt, p38, or p44/42 [58].
5.2. Growth-repressing signaling pathways regulated by PTK6
Only a handful of studies have explored PTK6 regulated signaling pathways in a normal physiological context. Disruption of the Ptk6 gene in the mouse led to increased growth and impaired enterocyte differentiation that was accompanied by an increase in AKT activation and decreased nuclear localization of the AKT substrate FoxO1 [59,68], supporting the notion that PTK6 inhibits AKT activity in normal tissue. In the mouse small intestine, PTK6 is induced in epithelial cells of the crypt following gamma-irradiation, where it plays a role in inhibiting AKT and ERK1/2 prosurvival functions and promotes apoptosis [58,68]. It was recently reported that the PTK6 family member FRK also negatively regulates AKT activity by directly phosphorylating and stabilizing PTEN (phosphatase and tensin homologue on chromosome ten), affecting AKT function [99].
Enhanced nuclear localization of β-catenin was detected in intestinal crypts of Ptk6 null mice, suggesting that PTK6 negatively regulates β-catenin in the normal intestine [59]. While PTK6 directly phosphorylates β-catenin at several tyrosine residues, the tyrosine kinase activity of PTK6 was not essential for inhibition of β-catenin transcription and did not alter levels of β-catenin protein, but increased nuclear levels of TCF and the transcriptional corepressor TLE/Groucho. BAT-GAL mice that contain a β-catenin-activated lacZ reporter transgene were crossed with Ptk6 null mice, and increased β-catenin transcriptional activity was indeed observed in intestines of Ptk6 null mice compared with wild-type mice. These data provide further evidence that PTK6 inhibits Wnt signaling by regulation of β-catenin in vivo, and may thus promote the transition of cells from a proliferative to a differentiated phenotype. [30]
In addition to regulating β-catenin nuclear activities, PTK6 targets a number of nuclear RNA-binding proteins including Sam68 (Src-associated in mitosis 68), which was originally identified as a target of SRC in mitosis when the nuclear membrane breaks down [18], and the Sam68-like mammalian proteins SLM1 and SLM2 [36]. Phosphorylation of these proteins on tyrosine by PTK6 leads to inhibition of their RNA-binding activities, including RNA transport [18] and expression [33]. In response to EGF stimulation, PTK6 also regulates Sam68 tyrosine phosphorylation and nuclear localization [34]. Sam68 haploinsufficiency delayed mammary tumor onset in vivo, suggesting a possible oncogenic role for Sam68 in breast cancer [100] and it will be interesting to examine crosstalk between PTK6 and Sam68 in mammary gland tumorigenesis. PTK6 also associates with and phosphorylates the nuclear protein PSF (Polypyrimidine tract-binding protein-associated Splicing Factor), leading to its cytoplasmic relocalization and causing growth arrest [24].
5.3. Context dependent functions of PTK6
While PTK6 expression can be detected in normal regenerating epithelial cell linings, it is also expressed in most epithelial cancers, and a variety of data indicate that PTK6 has opposing functions in normal tissues and cancer cells. Functions of PTK6 appear highly context and cell type specific, and may be dependent on overall expression levels and the availability of substrates and interacting proteins. Intracellular localization of PTK6 appears to play a key role in determining the outcomes of PTK6 signaling. A summary of PTK6 expression and localization in normal tissues and cancers is shown in Table 3. PTK6 has been observed in both the cytoplasm and nucleus in the normal intestine, skin, oral epithelium, breast and colon tumors, but further studies are required to assess the significance of PTK6 subcellular localization as well as the flexibility of PTK6 localization in each of these cell types. Differences in PTK6 intracellular localization have been reported in the epithelial cells of the head and neck [62] and HaCaT human keratinocyte cell line [61]. PTK6 is observed in the nucleus of the normal differentiated prostate epithelium and in well-differentiated prostate tumors, but it is cytoplasmic in poorly differentiated tumors, suggesting a role in differentiation of the prostate [60]. It has been proposed that nuclear PTK6 may be important for regulating growth in normal epithelia, while cytoplasmic PTK6 might activate oncogenic signaling pathways, since its access to substrates would be subsequently altered [36,60]. This idea was supported by the finding that targeting PTK6 to the membrane of HEK293 cells, which do not normally express PTK6, resulted in an increase in proliferation, survival, migration, and anchorage-independent growth [91]. In the A431 epidermoid cancer cell line, PTK6 localizes to membrane ruffles during wound healing as well as after EGF treatment [84], suggesting that PTK6 is involved in cell migration. Targeting PTK6 to the membrane or nucleus in the SW620 colon cancer cell line resulted in either activation or inhibition of β-catenin regulated transcription respectively, providing further evidence that PTK6 intracellular localization influences the consequences of PTK6 expression or activation [30].
Currently it is not understood how PTK6 intracellular localization is regulated. PTK6 does not contain any typically identifiable nuclear localization or export sequences. It may passively diffuse into the nucleus as it has a molecular weight of 48 kDa, and the threshold range for passive diffusion through the nuclear pore complex is 40–60 kDa (reviewed in [101]). It is also possible that PTK6 is transported to different cellular compartments through association with its targeted substrates or other binding proteins. Work in our laboratory suggests that PTK6 may be retained in the cytoplasm of prostate cancer by an unidentified protein (Brauer and Tyner, unpublished data).
6. PTK6 as a target for anticancer therapies
The recent trend toward targeted therapies has seen the development of drugs to block the function of proteins associated with cancer progression and poor survival rates. The widespread overexpression of PTK6 in a variety of epithelial cancers suggests that targeting it may have distinct therapeutic advantages. The 18–25% incidence of overexpression of ErbB2 in breast cancer has made it a prominent target, leading to clinical trials of both small molecule inhibitors and monoclonal antibodies [102]. The monoclonal antibody trastuzumab (Herceptin) that targets ErbB2 is an established therapeutic option, but progression is common within 1 year [103,104]. Trastuzumab's cardiotoxic effects, as well as a lack of response of some ErbB2 overexpressing tumors to treatment, underscore the importance of determining which patients may respond to therapy prior to treatment (reviewed in [105]). The correlation between PTK6 and ErbB2 overexpression in invasive human ductal breast carcinomas [43,47,106] and the finding that PTK6 cooperates with ErbB2 to promote breast tumor cell growth [43] raises the possibility that targeting PTK6 along with ErbB2 might offer a significant therapeutic advantage [107]. PTK6 is expressed in up to 86% of breast tumors [47,70], and PTK6-directed therapies could target a broad range of tumors including breast cancers that do not overexpress ErbB2.
PTK6 expression has been correlated with altered sensitivity to some targeted therapies. Introduction of both PTK6 and ErbB2 into the human MCF10a mammary gland cell line resulted in increased resistance to the ErbB1/ErbB2 kinase inhibitor lapatinib, when compared with introduction of ErbB2 alone [43]. In contrast, higher PTK6 expression correlated with increased sensitivity of squamous cell carcinoma of the head and neck cell lines to the ErbB1 kinase inhibitor gefitinib, although PTK6 knockdown did not alter sensitivity [108]. PTK6 does not respond to LFM-A13, the inhibitor of Burton's Tyrosine Kinase and Polo-Like Kinase [109]. Specific PTK6 inhibitors have not been identified.
Aberrant PTK6 expression in breast [2] and ovarian cancers [71], combined with its altered localization in prostate [60] and oral tumors [62], makes it an excellent candidate as a prognostic marker. In long-term patient studies, high PTK6 and ErbB4 protein expression was associated with a metastasis-free survival (>240 months), although ErbB2 and ErbB4 protein expression was a better prognostic marker at 60 months [35,93]. Xenograft studies in nude mice resulted in enhanced tumor growth and reduced survival when PTK6 was expressed in the MDA-MB-231 human breast adenocarcinoma cell line [32] or the mouse mammary epithelial Comma-1D cell line [43]. Further epidemiological studies and tumorigenesis/metastases studies in mouse models will clarify the contributions of PTK6 to initiation and progression of epithelial cancers.
7. Conclusions
Functions of the PTK6 family of intracellular tyrosine kinases are still, for the most part, poorly understood. PTK6 and the related kinase FRK have both been linked to differentiation, regulation of AKT, growth regulation and apoptosis (reviewed in [4]), and their ability to readily enter the nucleus sets them apart from the more intensely studied SRC family of kinases. In contrast to its functions in normal tissues, PTK6 appears to promote oncogenic signaling in epithelial tumor cells, and the majority of studies so far have focused on its roles in breast cancer cells. More than half of PTK6 substrates and associating proteins have been discovered within the last 2 years, most of which regulate proliferation and migration (Table 1). Greater efforts need to be directed towards determining the apparent context- and tissue-specific functions of PTK6 in both normal tissues as well as tumors. It is likely that different expression patterns of ligands and receptors affect the inherent functions of PTK6 in different tissues. Understanding crosstalk between PTK6 and other signaling pathways in both normal and cancer cells will be critical for evaluating its potential as a therapeutic target in cancer.
Acknowledgments
We would like to thank Dr. H.L. Palka-Hamblin, K. Weaver, A. Perekatt and Y. Zheng for their helpful comments. P.M.B. was supported by a DOD Predoctoral Traineeship Award, Army W81XWH-06-1-000. A.L.T. is supported by National Institutes of Health grants DK44525 and DK068503.
References
- 1.Lee ST, Strunk KM, Spritz RA. A survey of protein tyrosine kinase mRNAs expressed in normal human melanocytes. Oncogene. 1993;8:3403–3410. [PubMed] [Google Scholar]
- 2.Mitchell PJ, Barker KT, Martindale JE, Kamalati T, Lowe PN, Page MJ, Gusterson BA, Crompton MR. Cloning and characterisation of cDNAs encoding a novel non-receptor tyrosine kinase, brk, expressed in human breast tumours. Oncogene. 1994;9:2383–2390. [PubMed] [Google Scholar]
- 3.Siyanova EY, Serfas MS, Mazo IA, Tyner AL. Tyrosine kinase gene expression in the mouse small intestine. Oncogene. 1994;9:2053–2057. [PubMed] [Google Scholar]
- 4.Brauer PM, Tyner AL. RAKing in AKT: a tumor suppressor function for the intracellular tyrosine kinase FRK. Cell Cycle. 2009;8:2728–2732. doi: 10.4161/cc.8.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohmura N, Yagi T, Tomooka Y, Oyanagi M, Kominami R, Takeda N, Chiba J, Ikawa Y, Aizawa S. A novel nonreceptor tyrosine kinase, Srm: cloning and targeted disruption. Mol Cell Biol. 1994;14:6915–6925. doi: 10.1128/mcb.14.10.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawachi Y, Nakauchi H, Otsuka F. Isolation of a cDNA encoding a tyrosine kinase expressed in murine skin. Exp Dermatol. 1997;6:140–146. doi: 10.1111/j.1600-0625.1997.tb00161.x. [DOI] [PubMed] [Google Scholar]
- 7.Neet K, Hunter T. Vertebrate non-receptor protein-tyrosine kinase families. Genes Cells. 1996;1:147–169. doi: 10.1046/j.1365-2443.1996.d01-234.x. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell PJ, Barker KT, Shipley J, Crompton MR. Characterisation and chromosome mapping of the human non receptor tyrosine kinase gene, brk. Oncogene. 1997;15:1497–1502. doi: 10.1038/sj.onc.1201292. [DOI] [PubMed] [Google Scholar]
- 9.Serfas MS, Tyner AL. Brk, Srm, Frk, and Src42A form a distinct family of intracellular Src-like tyrosine kinases. Oncol Res. 2003;13:409–419. doi: 10.3727/096504003108748438. [DOI] [PubMed] [Google Scholar]
- 10.D'Aniello S, Irimia M, Maeso I, Pascual-Anaya J, Jimenez-Delgado S, Bertrand S, Garcia-Fernandez J. Gene expansion and retention leads to a diverse tyrosine kinase superfamily in amphioxus. Mol Biol Evol. 2008;25:1841–1854. doi: 10.1093/molbev/msn132. [DOI] [PubMed] [Google Scholar]
- 11.Park SH, Lee KH, Kim H, Lee ST. Assignment of the human PTK6 gene encoding a non-receptor protein tyrosine kinase to 20q13.3 by fluorescence in situ hybridization. Cytogenet Cell Genet. 1997;77:271–272. doi: 10.1159/000134595. [DOI] [PubMed] [Google Scholar]
- 12.Llor X, Serfas MS, Bie W, Vasioukhin V, Polonskaia M, Derry J, Abbott CM, Tyner AL. BRK/Sik expression in the gastrointestinal tract and in colon tumors. Clin Cancer Res. 1999;5:1767–1777. [PubMed] [Google Scholar]
- 13.Lee H, Kim M, Lee KH, Kang KN, Lee ST. Exon–intron structure of the human PTK6 gene demonstrates that PTK6 constitutes a distinct family of non-receptor tyrosine kinase. Mol Cells. 1998;8:401–407. [PubMed] [Google Scholar]
- 14.Kang KN, Kim M, Pae KM, Lee ST. Characterization of the 5′-flanking region of the human PTK6 gene. Biochim Biophys Acta. 2002;1574:365–369. doi: 10.1016/s0167-4781(02)00234-8. [DOI] [PubMed] [Google Scholar]
- 15.Simmen FA, Xiao R, Velarde MC, Nicholson RD, Bowman MT, Fujii-Kuriyama Y, Oh SP, Simmen RC. Dysregulation of intestinal crypt cell proliferation and villus cell migration in mice lacking Kruppel-like factor 9. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1757–G1769. doi: 10.1152/ajpgi.00013.2007. [DOI] [PubMed] [Google Scholar]
- 16.Qiu H, Miller WT. Regulation of the nonreceptor tyrosine kinase Brk by autophosphorylation and by autoinhibition. J Biol Chem. 2002;277:34634–34641. doi: 10.1074/jbc.M203877200. [DOI] [PubMed] [Google Scholar]
- 17.Kim H, Lee ST. An intramolecular interaction between SH2-kinase linker and kinase domain is essential for the catalytic activity of protein-tyrosine kinase-6. J Biol Chem. 2005;280:28973–28980. doi: 10.1074/jbc.M504568200. [DOI] [PubMed] [Google Scholar]
- 18.Derry JJ, Richard S, Valderrama Carvajal H, Ye X, Vasioukhin V, Cochrane AW, Chen T, Tyner AL. Sik (BRK) phosphorylates Sam68 in the nucleus and negatively regulates its RNA binding ability. Mol Cell Biol. 2000;20:6114–6126. doi: 10.1128/mcb.20.16.6114-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HI, Jung J, Lee ES, Kim YC, Lee WP, Lee ST. Molecular dissection of the interaction between the SH3 domain and the SH2-Kinase Linker region in PTK6. Biochem Biophys Res Commun. 2007;362:829–834. doi: 10.1016/j.bbrc.2007.08.055. [DOI] [PubMed] [Google Scholar]
- 20.Ko S, Ahn KE, Lee YM, Ahn HC, Lee W. Structural basis of the auto-inhibition mechanism of nonreceptor tyrosine kinase PTK6. Biochem Biophys Res Commun. 2009;384:236–242. doi: 10.1016/j.bbrc.2009.04.103. [DOI] [PubMed] [Google Scholar]
- 21.Koo BK, Kim MH, Lee ST, Lee W. Purification and spectroscopic characterization of the human protein tyrosine kinase-6 SH3 domain. J Biochem Mol Biol. 2002;35:343–347. doi: 10.5483/bmbrep.2002.35.3.343. [DOI] [PubMed] [Google Scholar]
- 22.Qiu H, Miller WT. Role of Brk SH3 domain in substrate recognition. Oncogene. 2004;23:2216–2223. doi: 10.1038/sj.onc.1207339. [DOI] [PubMed] [Google Scholar]
- 23.Qiu H, Zappacost F, Su W, Annan RS, Miller WT. Interaction between Brk kinase and insulin receptor substrate-4. Oncogene. 2005;24:5656–5664. doi: 10.1038/sj.onc.1208721. [DOI] [PubMed] [Google Scholar]
- 24.Lukong KE, Huot ME, Richard S. BRK phosphorylates PSF promoting its cytoplasmic localization and cell cycle arrest. Cell Signal. 2009;21:1415–1422. doi: 10.1016/j.cellsig.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 25.Hong E, Shin J, Bang E, Kim MH, Lee ST, Lee W. Complete sequence-specific 1H, 13C and 15N resonance assignments of the human PTK6 SH2 domain. J Biomol NMR. 2001;19:291–292. doi: 10.1023/a:1011221125013. [DOI] [PubMed] [Google Scholar]
- 26.Hong E, Shin J, Kim HI, Lee ST, Lee W. Solution structure and backbone dynamics of the non-receptor protein-tyrosine kinase-6 Src homology 2 domain. J Biol Chem. 2004;279:29700–29708. doi: 10.1074/jbc.M313185200. [DOI] [PubMed] [Google Scholar]
- 27.Vasioukhin V, Tyner AL. A role for the epithelial-cell-specific tyrosine kinase Sik during keratinocyte differentiation. Proc Natl Acad Sci USA. 1997;94:14477–14482. doi: 10.1073/pnas.94.26.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell PJ, Sara EA, Crompton MR. A novel adaptor-like protein which is a substrate for the non-receptor tyrosine kinase, BRK. Oncogene. 2000;19:4273–4282. doi: 10.1038/sj.onc.1203775. [DOI] [PubMed] [Google Scholar]
- 29.Zhang P, Ostrander JH, Faivre EJ, Olsen A, Fitzsimmons D, Lange CA. Regulated association of protein kinase b/akt with breast tumor kinase. J Biol Chem. 2005;280:1982–1991. doi: 10.1074/jbc.M412038200. [DOI] [PubMed] [Google Scholar]
- 30.Palka-Hamblin HL, Gierut JJ, Bie W, Brauer PM, Zheng Y, Asara JM, Tyner AL. Identification of {beta}-catenin as a target of the intracellular tyrosine kinase PTK6. J Cell Sci. 2010;123:236–245. doi: 10.1242/jcs.053264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukong KE, Richard S. Breast tumor kinase BRK requires kinesin-2 subunit KAP3A in modulation of cell migration. Cell Signal. 2008;20:432–442. doi: 10.1016/j.cellsig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Shen CH, Chen HY, Lin MS, Li FY, Chang CC, Kuo ML, Settleman J, Chen RH. Breast tumor kinase phosphorylates p190RhoGAP to regulate rho and ras and promote breast carcinoma growth, migration, and invasion. Cancer Res. 2008;68:7779–7787. doi: 10.1158/0008-5472.CAN-08-0997. [DOI] [PubMed] [Google Scholar]
- 33.Coyle JH, Guzik BW, Bor YC, Jin L, Eisner-Smerage L, Taylor SJ, Rekosh D, Hammarskjold ML. Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/BRK. Mol Cell Biol. 2003;23:92–103. doi: 10.1128/MCB.23.1.92-103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukong KE, Larocque D, Tyner AL, Richard S. Tyrosine phosphorylation of sam68 by breast tumor kinase regulates intranuclear localization and cell cycle progression. J Biol Chem. 2005;280:38639–38647. doi: 10.1074/jbc.M505802200. [DOI] [PubMed] [Google Scholar]
- 35.Aubele M, Walch AK, Ludyga N, Braselmann H, Atkinson MJ, Luber B, Auer G, Tapio S, Cooke T, Bartlett JM. Prognostic value of protein tyrosine kinase 6 (PTK6) for long-term survival of breast cancer patients. Br J Cancer. 2008;99:1089–1095. doi: 10.1038/sj.bjc.6604660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haegebarth A, Heap D, Bie W, Derry JJ, Richard S, Tyner AL. The nuclear tyrosine kinase BRK/Sik phosphorylates and inhibits the RNA-binding activities of the Sam68-like mammalian proteins SLM-1 and SLM-2. J Biol Chem. 2004;279:54398–54404. doi: 10.1074/jbc.M409579200. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda O, Miyasaka Y, Sekine Y, Mizushima A, Muromoto R, Nanbo A, Yoshimura A, Matsuda T. STAP-2 is phosphorylated at tyrosine-250 by Brk and modulates Brk-mediated STAT3 activation. Biochem Biophys Res Commun. 2009;384:71–75. doi: 10.1016/j.bbrc.2009.04.076. [DOI] [PubMed] [Google Scholar]
- 38.Liu L, Gao Y, Qiu H, Miller WT, Poli V, Reich NC. Identification of STAT3 as a specific substrate of breast tumor kinase. Oncogene. 2006;25:4904–4912. doi: 10.1038/sj.onc.1209501. [DOI] [PubMed] [Google Scholar]
- 39.Weaver AM, Silva CM. Signal transducer and activator of transcription 5b: a new target of breast tumor kinase/protein tyrosine kinase 6. Breast Cancer Res. 2007;9:R79. doi: 10.1186/bcr1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin DS, Kim YG, Kim EM, Kim M, Park HY, Kim JH, Lee BS, Kim BG, Lee YS. Solid-phase peptide library synthesis on HiCore resin for screening substrate specificity of Brk protein tyrosine kinase. J Comb Chem. 2008;10:20–23. doi: 10.1021/cc7001217. [DOI] [PubMed] [Google Scholar]
- 41.Zhong JL, Poghosyan Z, Pennington CJ, Scott X, Handsley MM, Warn A, Gavrilovic J, Honert K, Kruger A, Span PN, Sweep FC, Edwards DR. Distinct functions of natural ADAM-15 cytoplasmic domain variants in human mammary carcinoma. Mol Cancer Res. 2008;6:383–394. doi: 10.1158/1541-7786.MCR-07-2028. [DOI] [PubMed] [Google Scholar]
- 42.Kamalati T, Jolin HE, Mitchell PJ, Barker KT, Jackson LE, Dean CJ, Page MJ, Gusterson BA, Crompton MR. Brk, a breast tumor-derived non-receptor protein-tyrosine kinase, sensitizes mammary epithelial cells to epidermal growth factor. J Biol Chem. 1996;271:30956–30963. doi: 10.1074/jbc.271.48.30956. [DOI] [PubMed] [Google Scholar]
- 43.Xiang B, Chatti K, Qiu H, Lakshmi B, Krasnitz A, Hicks J, Yu M, Miller WT, Muthuswamy SK. Brk is coamplified with ErbB2 to promote proliferation in breast cancer. Proc Natl Acad Sci USA. 2008;105:12463–12468. doi: 10.1073/pnas.0805009105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mosnier JF, Jarry A, Bou-Hanna C, Denis MG, Merlin D, Laboisse CL. ADAM15 upregulation and interaction with multiple binding partners in inflammatory bowel disease. Lab Invest. 2006;86:1064–1073. doi: 10.1038/labinvest.3700465. [DOI] [PubMed] [Google Scholar]
- 45.Charrier-Hisamuddin L, Laboisse CL, Merlin D. ADAM-15: a metalloprotease that mediates inflammation. FASEB J. 2008;22:641–653. doi: 10.1096/fj.07-8876rev. [DOI] [PubMed] [Google Scholar]
- 46.Kamalati T, Jolin HE, Fry MJ, Crompton MR. Expression of the BRK tyrosine kinase in mammary epithelial cells enhances the coupling of EGF signalling to PI 3-kinase and Akt, via erbB3 phosphorylation. Oncogene. 2000;19:5471–5476. doi: 10.1038/sj.onc.1203931. [DOI] [PubMed] [Google Scholar]
- 47.Ostrander JH, Daniel AR, Lofgren K, Kleer CG, Lange CA. Breast tumor kinase (protein tyrosine kinase 6) regulates heregulin-induced activation of ERK5 and p38 MAP kinases in breast cancer cells. Cancer Res. 2007;67:4199–4209. doi: 10.1158/0008-5472.CAN-06-3409. [DOI] [PubMed] [Google Scholar]
- 48.Najy AJ, Day KC, Day ML. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. J Biol Chem. 2008;283:18393–18401. doi: 10.1074/jbc.M801329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellis C, Moran M, McCormick F, Pawson T. Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature. 1990;343:377–381. doi: 10.1038/343377a0. [DOI] [PubMed] [Google Scholar]
- 50.Moran MF, Polakis P, McCormick F, Pawson T, Ellis C. Protein-tyrosine kinases regulate the phosphorylation, protein interactions, subcellular distribution, and activity of p21ras GTPase-activating protein. Mol Cell Biol. 1991;11:1804–1812. doi: 10.1128/mcb.11.4.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Filvaroff E, Calautti E, McCormick F, Dotto GP. Specific changes of Ras GTPase-activating protein (GAP) and a GAP-associated p62 protein during calcium-induced keratinocyte differentiation. Mol Cell Biol. 1992;12:5319–5328. doi: 10.1128/mcb.12.12.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsuruzoe K, Emkey R, Kriauciunas KM, Ueki K, Kahn CR. Insulin receptor substrate 3 (IRS-3) and IRS-4 impair IRS-1- and IRS-2-mediated signaling. Mol Cell Biol. 2001;21:26–38. doi: 10.1128/MCB.21.1.26-38.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giovannone B, Scaldaferri ML, Federici M, Porzio O, Lauro D, Fusco A, Sbraccia P, Borboni P, Lauro R, Sesti G. Insulin receptor substrate (IRS) transduction system: distinct and overlapping signaling potential. Diab Metab Res Rev. 2000;16:434–441. doi: 10.1002/1520-7560(2000)9999:9999<::aid-dmrr159>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 54.Sesti G, Federici M, Hribal ML, Lauro D, Sbraccia P, Lauro R. Defects of the insulin receptor substrate (IRS) system in human metabolic disorders. FASEB J. 2001;15:2099–2111. doi: 10.1096/fj.01-0009rev. [DOI] [PubMed] [Google Scholar]
- 55.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 56.Bae CS, Lee ST. The human PTK6 interacts with a 23-kDa tyrosine-phosphorylated protein and is localized in cytoplasm in breast carcinoma T47D cells. J Biochem Mol Biol. 2000;34:33–38. [Google Scholar]
- 57.Vasioukhin V, Serfas MS, Siyanova EY, Polonskaia M, Costigan VJ, Liu B, Thomason A, Tyner AL. A novel intracellular epithelial cell tyrosine kinase is expressed in the skin and gastrointestinal tract. Oncogene. 1995;10:349–357. [PubMed] [Google Scholar]
- 58.Haegebarth A, Nunez R, Tyner AL. The intracellular tyrosine kinase Brk sensitizes non-transformed cells to inducers of apoptosis. Cell Cycle. 2005;4:1239–1246. doi: 10.4161/cc.4.9.1965. [DOI] [PubMed] [Google Scholar]
- 59.Haegebarth A, Bie W, Yang R, Crawford SE, Vasioukhin V, Fuchs E, Tyner AL. Protein tyrosine kinase 6 negatively regulates growth and promotes enterocyte differentiation in the small intestine. Mol Cell Biol. 2006;26:4949–4957. doi: 10.1128/MCB.01901-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Derry JJ, Prins GS, Ray V, Tyner AL. Altered localization and activity of the intracellular tyrosine kinase BRK/Sik in prostate tumor cells. Oncogene. 2003;22:4212–4220. doi: 10.1038/sj.onc.1206465. [DOI] [PubMed] [Google Scholar]
- 61.Wang TC, Jee SH, Tsai TF, Huang YL, Tsai WL, Chen RH. Role of breast tumour kinase in the in vitro differentiation of HaCaT cells. Br J Dermatol. 2005;153:282–289. doi: 10.1111/j.1365-2133.2005.06604.x. [DOI] [PubMed] [Google Scholar]
- 62.Petro BJ, Tan RC, Tyner AL, Lingen MW, Watanabe K. Differential expression of the non-receptor tyrosine kinase BRK in oral squamous cell carcinoma and normal oral epithelium. Oral Oncol. 2004;40:1040–1047. doi: 10.1016/j.oraloncology.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 63.Kasprzycka M, Majewski M, Wang ZJ, Ptasznik A, Wysocka M, Zhang Q, Marzec M, Gimotty P, Crompton MR, Wasik MA. Expression and oncogenic role of Brk (PTK6/Sik) protein tyrosine kinase in lymphocytes. Am J Pathol. 2006;168:1631–1641. doi: 10.2353/ajpath.2006.050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W, Li Y, Li Y, Hong A, Wang J, Lin B, Li R. NDRG3 is an androgen regulated and prostate enriched gene that promotes in vitro and in vivo prostate cancer cell growth. Int J Cancer. 2009;124:521–530. doi: 10.1002/ijc.23961. [DOI] [PubMed] [Google Scholar]
- 65.Harvell DM, Richer JK, Allred DC, Sartorius CA, Horwitz KB. Estradiol regulates different genes in human breast tumor xenografts compared with the identical cells in culture. Endocrinology. 2006;147:700–713. doi: 10.1210/en.2005-0617. [DOI] [PubMed] [Google Scholar]
- 66.Rinaudo P, Schultz RM. Effects of embryo culture on global pattern of gene expression in preimplantation mouse embryos. Reproduction. 2004;128:301–311. doi: 10.1530/rep.1.00297. [DOI] [PubMed] [Google Scholar]
- 67.Whitehead RH, Robinson PS, Williams JA, Bie W, Tyner AL, Franklin JL. Conditionally immortalized colonic epithelial cell line from a Ptk6 null mouse that polarizes and differentiates in vitro. J Gastroenterol Hepatol. 2008;23:1119–1124. doi: 10.1111/j.1440-1746.2008.05308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haegebarth A, Perekatt AO, Bie W, Gierut JJ, Tyner AL. Induction of protein tyrosine kinase 6 in mouse intestinal crypt epithelial cells promotes DNA damage-induced apoptosis. Gastroenterology. 2009;137:945–954. doi: 10.1053/j.gastro.2009.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barker KT, Jackson LE, Crompton MR. BRK tyrosine kinase expression in a high proportion of human breast carcinomas. Oncogene. 1997;15:799–805. doi: 10.1038/sj.onc.1201241. [DOI] [PubMed] [Google Scholar]
- 70.Harvey AJ, Pennington CJ, Porter S, Burmi RS, Edwards DR, Court W, Eccles SA, Crompton MR. Brk protects breast cancer cells from autophagic cell death induced by loss of anchorage. Am J Pathol. 2009;175:1226–1234. doi: 10.2353/ajpath.2009.080811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmandt RE, Bennett M, Clifford S, Thornton A, Jiang F, Broaddus RR, Sun CC, Lu KH, Sood AK, Gershenson DM. The BRK tyrosine kinase is expressed in high-grade serous carcinoma of the ovary. Cancer Biol Ther. 2006;5:1136–1141. doi: 10.4161/cbt.5.9.2953. [DOI] [PubMed] [Google Scholar]
- 72.Chen T, Boisvert FM, Bazett-Jones DP, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin HS, Berry GJ, Fee WE, Jr, Terris DJ, Sun Z. Identification of tyrosine kinases overexpressed in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2004;130:311–316. doi: 10.1001/archotol.130.3.311. [DOI] [PubMed] [Google Scholar]
- 74.Borczuk AC, Gorenstein L, Walter KL, Assaad AA, Wang L, Powell CA. Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am J Pathol. 2003;163:1949–1960. doi: 10.1016/S0002-9440(10)63553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, Pham TQ, Soriano R, Stinson J, Seshagiri S, Modrusan Z, Lin CY, O'Neill V, Amler LC. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11:8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 76.Coldren CD, Helfrich BA, Witta SE, Sugita M, Lapadat R, Zeng C, Baron A, Franklin WA, Hirsch FR, Geraci MW, Bunn PA., Jr Baseline gene expression predicts sensitivity to gefitinib in non-small cell lung cancer cell lines. Mol Cancer Res. 2006;4:521–528. doi: 10.1158/1541-7786.MCR-06-0095. [DOI] [PubMed] [Google Scholar]
- 77.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 78.Ruhe JE, Streit S, Hart S, Wong CH, Specht K, Knyazev P, Knyazeva T, Tay LS, Loo HL, Foo P, Wong W, Pok S, Lim SJ, Ong H, Luo M, Ho HK, Peng K, Lee TC, Bezler M, Mann C, Gaertner S, Hoefler H, Iacobelli S, Peter S, Tay A, Brenner S, Venkatesh B, Ullrich A. Genetic alterations in the tyrosine kinase transcriptome of human cancer cell lines. Cancer Res. 2007;67:11368–11376. doi: 10.1158/0008-5472.CAN-07-2703. [DOI] [PubMed] [Google Scholar]
- 79.Kubo T, Kuroda Y, Kokubu A, Hosoda F, Arai Y, Hiraoka N, Hirohashi S, Shibata T. Resequencing analysis of the human tyrosine kinase gene family in pancreatic cancer. Pancreas. 2009;38:e200–e206. doi: 10.1097/MPA.0b013e3181b8feb0. [DOI] [PubMed] [Google Scholar]
- 80.Kubo T, Kuroda Y, Shimizu H, Kokubu A, Okada N, Hosoda F, Arai Y, Nakamura Y, Taniguchi H, Yanagihara K, Imoto I, Inazawa J, Hirohashi S, Shibata T. Resequencing and copy number analysis of the human tyrosine kinase gene family in poorly differentiated gastric cancer. Carcinogenesis. 2009;30:1857–1864. doi: 10.1093/carcin/bgp206. [DOI] [PubMed] [Google Scholar]
- 81.Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, Cronin JC, Cruz P, Rosenberg SA, Samuels Y. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aubele M, Vidojkovic S, Braselmann H, Ritterswurden D, Auer G, Atkinson MJ, Tapio S, Hofler H, Rauser S, Bartlett JM. Overexpression of PTK6 (breast tumor kinase) protein—a prognostic factor for long-term breast cancer survival—is not due to gene amplification. Virchows Arch. 2009;455:117–123. doi: 10.1007/s00428-009-0809-8. [DOI] [PubMed] [Google Scholar]
- 83.Meric F, Lee WP, Sahin A, Zhang H, Kung HJ, Hung MC. Expression profile of tyrosine kinases in breast cancer. Clin Cancer Res. 2002;8:361–367. [PubMed] [Google Scholar]
- 84.Chen HY, Shen CH, Tsai YT, Lin FC, Huang YP, Chen RH. Brk activates rac1 and promotes cell migration and invasion by phosphorylating paxillin. Mol Cell Biol. 2004;24:10558–10572. doi: 10.1128/MCB.24.24.10558-10572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chakraborty G, Jain S, Kundu GC. Osteopontin promotes vascular endothelial growth factor-dependent breast tumor growth and angiogenesis via autocrine and paracrine mechanisms. Cancer Res. 2008;68:152–161. doi: 10.1158/0008-5472.CAN-07-2126. [DOI] [PubMed] [Google Scholar]
- 86.Lecrone V, Li W, Devoll RE, Logothetis C, Farach-Carson MC. Calcium signals in prostate cancer cells: specific activation by bone-matrix proteins. Cell Calcium. 2000;27:35–42. doi: 10.1054/ceca.1999.0083. [DOI] [PubMed] [Google Scholar]
- 87.Macri A, Versaci A, Lupo G, Trimarchi G, Tomasello C, Loddo S, Sfuncia G, Caminiti R, Teti D, Famulari C. Role of osteopontin in breast cancer patients. Tumori. 2009;95:48–52. doi: 10.1177/030089160909500109. [DOI] [PubMed] [Google Scholar]
- 88.Shevde LA, Das S, Clark DW, Samant RS. Osteopontin: an effector and an effect of tumor metastasis. Curr Mol Med (in press) doi: 10.2174/156652410791065381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ibrahim T, Flamini E, Mercantali L, Sacanna E, Serra P, Amadori D. Pathogenesis of osteoblastic bone metastases from prostate cancer. Cancer. 2010;6:1406–1418. doi: 10.1002/cncr.24896. [DOI] [PubMed] [Google Scholar]
- 90.Harvey AJ, Crompton MR. Use of RNA interference to validate Brk as a novel therapeutic target in breast cancer: Brk promotes breast carcinoma cell proliferation. Oncogene. 2003;22:5006–5010. doi: 10.1038/sj.onc.1206577. [DOI] [PubMed] [Google Scholar]
- 91.Ie Kim H, Lee ST. Oncogenic functions of PTK6 are enhanced by its targeting to plasma membrane but abolished by its targeting to nucleus. J Biochem. 2009;146:133–139. doi: 10.1093/jb/mvp050. [DOI] [PubMed] [Google Scholar]
- 92.Zhao C, Yasui K, Lee CJ, Kurioka H, Hosokawa Y, Oka T, Inazawa J. Elevated expression levels of NCOA3, TOP1, and TFAP2C in breast tumors as predictors of poor prognosis. Cancer. 2003;98:18–23. doi: 10.1002/cncr.11482. [DOI] [PubMed] [Google Scholar]
- 93.Aubele M, Auer G, Walch AK, Munro A, Atkinson MJ, Braselmann H, Fornander T, Bartlett JM. PTK (protein tyrosine kinase)-6 and HER2 and 4, but not HER1 and 3 predict long-term survival in breast carcinomas. Br J Cancer. 2007;96:801–807. doi: 10.1038/sj.bjc.6603613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Watson CJ, Neoh K. The Stat family of transcription factors have diverse roles in mammary gland development. Semin Cell Dev Biol. 2008;19:401–406. doi: 10.1016/j.semcdb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 95.Groner B, Lucks P, Borghouts C. The function of Stat3 in tumor cells and their microenvironment. Semin Cell Dev Biol. 2008;19:341–350. doi: 10.1016/j.semcdb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 96.Wei L, Laurence A, O'Shea JJ. New insights into the roles of Stat5a/b and Stat3 in T cell development and differentiation. Semin Cell Dev Biol. 2008;19:394–400. doi: 10.1016/j.semcdb.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walker SR, Nelson EA, Zou L, Chaudhury M, Signoretti S, Richardson A, Frank DA. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol Cancer Res. 2009;7:966–976. doi: 10.1158/1541-7786.MCR-08-0238. [DOI] [PubMed] [Google Scholar]
- 98.Madson JG, Lynch DT, Tinkum KL, Putta SK, Hansen LA. Erbb2 regulates inflammation and proliferation in the skin after ultraviolet irradiation. Am J Pathol. 2006;169:1402–1414. doi: 10.2353/ajpath.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yim EK, Peng G, Dai H, Hu R, Li K, Lu Y, Mills GB, Meric-Bernstam F, Hennessy BT, Craven RJ, Lin SY. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell. 2009;15:304–314. doi: 10.1016/j.ccr.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Richard S, Vogel G, Huot ME, Guo T, Muller WJ, Lukong KE. Sam68 haploinsufficiency delays onset of mammary tumorigenesis and metastasis. Oncogene. 2008;27:548–556. doi: 10.1038/sj.onc.1210652. [DOI] [PubMed] [Google Scholar]
- 101.Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 102.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 103.Nahta R, Esteva FJ. Trastuzumab: triumphs and tribulations. Oncogene. 2007;26:3637–3643. doi: 10.1038/sj.onc.1210379. [DOI] [PubMed] [Google Scholar]
- 104.Bender LM, Nahta R. Her2 cross talk and therapeutic resistance in breast cancer. Front Biosci. 2008;13:3906–3912. doi: 10.2741/2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singer CF, Kostler WJ, Hudelist G. Predicting the efficacy of trastuzumab-based therapy in breast cancer: current standards and future strategies. Biochim Biophys Acta. 2008;1786:105–113. doi: 10.1016/j.bbcan.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 106.Born M, Quintanilla-Fend L, Braselmann H, Reich U, Richter M, Hutzler P, Aubele M. Simultaneous over-expression of the Her2/neu and PTK6 tyrosine kinases in archival invasive ductal breast carcinomas. J Pathol. 2005;205:592–596. doi: 10.1002/path.1720. [DOI] [PubMed] [Google Scholar]
- 107.Harvey AJ, Crompton MR. The Brk protein tyrosine kinase as a therapeutic target in cancer: opportunities and challenges. Anticancer Drugs. 2004;15:107–111. doi: 10.1097/00001813-200402000-00002. [DOI] [PubMed] [Google Scholar]
- 108.Rogers SJ, Box C, Chambers P, Barbachano Y, Nutting CM, Rhys-Evans P, Workman P, Harrington KJ, Eccles SA. Determinants of response to epidermal growth factor receptor tyrosine kinase inhibition in squamous cell carcinoma of the head and neck. J Pathol. 2009;218:122–130. doi: 10.1002/path.2515. [DOI] [PubMed] [Google Scholar]
- 109.Uckun FM, Dibirdik I, Qazi S, Vassilev A, Ma H, Mao C, Benyumov A, Emami KH. Anti-breast cancer activity of LFM-A13, a potent inhibitor of Polo-like kinase (PLK) Bioorg Med Chem. 2007;15:800–814. doi: 10.1016/j.bmc.2006.10.050. [DOI] [PubMed] [Google Scholar]


