Summary
Gfi1encodes a zinc-finger transcription factor essential for the development and maintenance of haematopoiesis and the inner ear. In mouse inner ear, Gfi1 expression is confined to hair cells during development and in adulthood. To construct a genetic tool for inner ear hair cell-specific gene deletion, we generated a Gfi1-Cre mouse line by knocking-in Cre coding sequences into the Gfi1 locus and inactivating the endogenous Gfi1. The specificity and efficiency of Gfi1-Cre recombinase-mediated recombination in the developing inner ear was revealed through the expression of the conditional R26R-lacZ reporter gene. The onset of lacZ expression in the Gfi1Cre/+ inner ear was first detected at E13.5 in the vestibule and at E15.5 in the cochlea, coinciding with the generation of hair cells. Throughout inner ear development, lacZ expression was detected only in hair cells. Thus, Gfi1-Cre knock-in mouse line provides a useful tool for gene manipulations specifically in inner ear hair cells.
Keywords: Gfi1, Cre recombinase, inner ear, hair cells
INTRODUCTION
The Growth Factor Independence 1 (Gfi1) gene, first reported to be involved in IL-2-independent T cell lymphoma growth (Gilks et al., 1993), is required for immune cell differentiation (Karsunky et al., 2002) and haematopoiesis (Fiolka et al., 2006). Recent studies have shown that Gfi1 also plays an essential role in the differentiation and survival of inner ear hair cells (Hertzano et al., 2004; Wallis et al., 2003), and in the development of lung and intestine (Kazanjian et al., 2004; Shroyer et al., 2005). Gfi1 expression is also detected in developing retina, brain and dorsal root ganglia (Wallis et al., 2003; Yang et al., 2003). In inner ear hair cells and intestinal secretory cells, Gfi1 functions downstream of Atoh1 and is required for cell survival (Shroyer et al., 2005; Wallis et al., 2003). Gfi1 and its paralogue Gfi1b function equivalently during haematopoiesis but Gfi1 is uniquely required for ear development (Fiolka et al., 2006). Null mutation of Gfi1 results in a complete loss of hair cells by apoptosis before birth and in a severe loss of spiral ganglion neurons observed at five months after birth (Wallis et al., 2003). Moreover, Gfi1 is identified as a downstream target of POU4F3 in ear development (Hertzano et al., 2004) and the extent of hair cell loss in the Gfi1-null mice resembles that Pou4f3-null mice (Wallis et al., 2003; Xiang et al., 1997). It is suggested that the cyclin-dependent kinase inhibitor p57Kip2 might mediate the differentiation- and survival-promoting functions of Gfi1 in hair cells (Kirjavainen et al., 2008).
Conventional knockouts of developmentally essential genes in mice often result in the lethality of embryos and newborns, thus preventing the functional study of genes during later embryonic development and in adults. The Cre-loxP-mediated tissue-specific inactivation of critical genes can bypass embryonic and neonatal lethality and allow the functional study of developmentally essential genes (Gu et al., 1994). Since inner ear hair cells are not essential for mouse survival, hair cell-specific Cre recombinase deleter mouse strains have great potential in studies of genes in developing as well as mature hair cells. Here, we have taken advantage of the hair cell-specific expression of Gfi1 (Fiolka et al., 2006; Wallis et al., 2003) and have generated a Gfi1-Cre knock-in mouse line. Using the conditional R26R-lacZ reporter mouse line, we have demonstrated that in the inner ear, Gfi1-Cre activity is confined to hair cells. Thus, the Gfi1-Cre knock-in mouse line offers a tool for cell-specific gene deletion research in inner ear hair cells.
RESULTS AND DISCUSSION
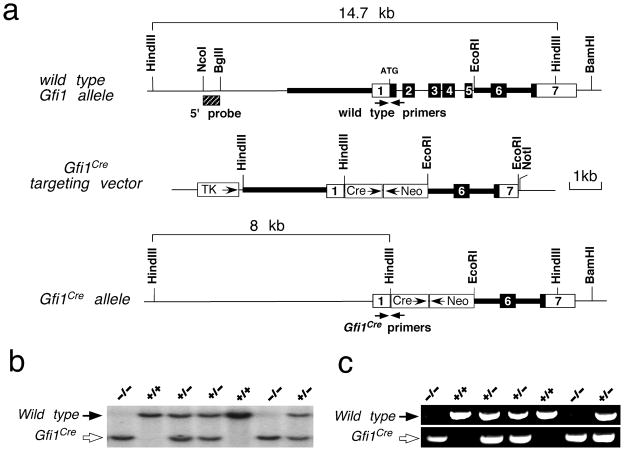
We generated the Gfi1-Cre mouse line by knocking-in Cre coding sequences into the Gfi1 locus and simultaneously inactivating the endogenous Gfi1 (Fig. 1a). The targeted event of the Gfi1-Cre mice were confirmed by Southern blotting using an external 5′ probe to identify the 14.7 kb wild type (Fig. 1b, black arrow) and 8.0 kb targeted (Fig. 1b, open arrow) DNA fragments from HindIII digested genomic DNA. Additionally, PCR genotyping was performed to determine the wild type allele (609 bp PCR product) and the Gfi1-Cre allele (672 bp PCR product) (Fig. 1c). Similar to conventional Gfi1 knockout mice (Fiolka et al., 2006; Wallis et al., 2003), heterozygous Gfi1Cre/+ mice were viable and displayed no discernible developmental defects (data not shown). Homozygous Gfi1Cre/Cre mice showed the loss of inner ear hair cells, a phenotype identical to that observed in Gfi1-null mice (Wallis et al., 2003). By crossing Gfi1-Cre with R26R-lacZ (Soriano, 1999) mice and analyzing the spatiotemporal expression of the conditional reporter lacZ gene, we determined the efficiency and specificity of Gfi1-Cre recombinase-mediated recombination in vivo.
FIG. 1.
Generation of Gfi1Cre knock-in mice. (a) Gfi1 restriction enzyme map and targeting strategy. The Gfi1 coding regions are shown as the filled boxes and the 5′ and 3′ UTR as the open boxes. Thick bars represent the DNA sequences used as the 5′ and 3′ arms for homologous recombination. The 5′ Southern probe is shown as the hatched box. Arrows indicate the approximate positions of PCR genotyping primers. Abbreviations: Cre, Cre recombinase gene and SV40 polyA cassette; Neo, PGK-neo cassette; TK, MC1-TK cassette. (b) Genotyping of a typical litter from the cross of Gfi1Cre heterozygotes using external 5′ Southern probe to identify 14.7 kb wild type and 8.0 kb targeted Gfi1Cre DNA fragments from HindIII digested genomic DNA. (c) PCR genotyping of the same litter in (b) amplifies a 609 bp product in the wide type and a 672 bp product in the Gfi1Cre knock-in mice.
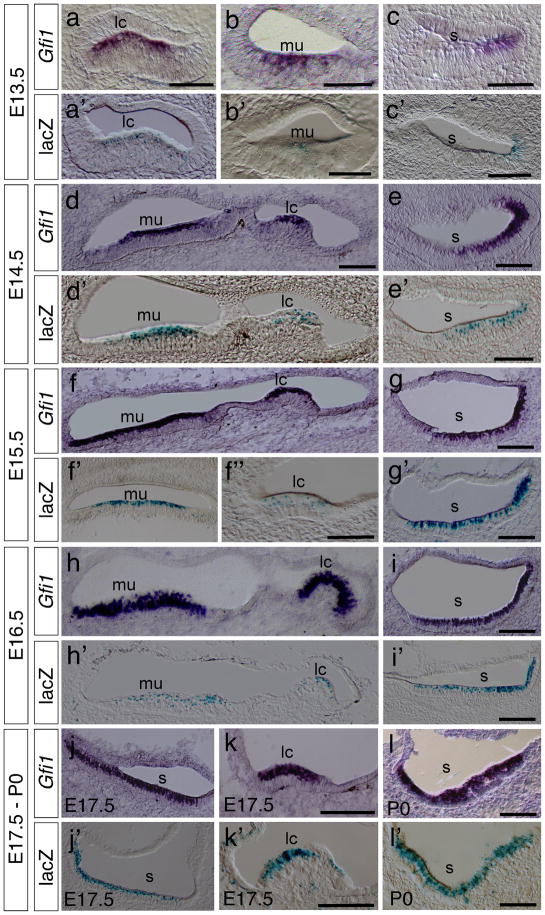
We first compared the spatiotemporal characteristics of Gfi1 expression and Cre activity in developing inner ears of Gfi1Cre/+; R26R-lacZ mice by in situ hybridization and X-Gal staining. During the development of vestibular hair cells, Gfi1 expression first appearedat E13.5 in hair cells of the cristae of the lateral, anterior and posterior semicircular canals (Fig. 2a and data not shown), the maculae of the utricle (Fig. 2b) and the saccule (Fig. 2c). At E14.5, a higher level of Gfi1 expression was seen in the cristae of semicircular canals and maculae of the utricle and saccule (Fig. 2d, e and data not shown) and was maintained in vestibular hair cells from E15.5 to P0 (Fig. 2f–l). Correspondingly, X-Gal staining revealed a similar pattern of lacZ expression in the hair cells of all cristae and maculae from E13.5 to P0 (Fig. 2a′–l′, data not shown).
FIG. 2.
Hair cell-specific expression of Gfi1-Cre in the vestibule. (a–l) In situ hybridization experiments reveals the expression pattern of Gfi1 at E13.5 (a–c), E14.5 (d,e), E15.5 (f,g), E16.5 (h,i), E17.5 (j,k) and P0 (l). (a′–l′) X-Gal staining of vestibular sections at E13.5 (a′–c′), E14.5 (d′, e′), E15.5 (f′,f″–g′), E16.5 (h′–i′), E17.5 (j′–k′) and P0 (l′) shows the Cre-recombinase-mediated activation of lacZ expression in the Gfi1Cre/+; R26R-lacZ ears. Abbreviations: lc, lateral semicircular canal; mu, macula of utricle; s, saccule. Scale bars represent 50 μm.
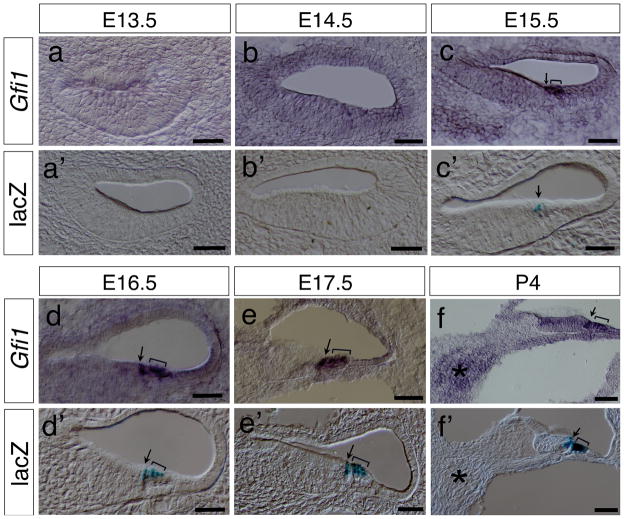
In the developing cochlea, the differentiation of hair cells starts in the basal cochlea and progresses toward the apex of cochlea. Definitive Gfi1 expression was not detected in the inner and outer hair cells in the basal cochlea until E15.5 (Fig. 3a–c) whereas lacZ expression was not detected at E13.5 to E14.5 (Fig. 3a′, b′) and was first seen only in the inner hair cells at E15.5 (Fig. 3c′), consistent with the notion that the inner hair cells arise slightly earlier than the outer hair cells. No Gfi1 or lacZ expression was detected at the middle and apical turns of cochlea at E13.5 to E15.5 (data not shown). At E16.5, both Gfi1 and lacZ were expressed in hair cells of the basal and middle cochlea but not in the apex (Fig. 3d, d′ and data not shown). Strong Gfi1 and lacZ expression was clearly identified in both the inner (arrows) and outer hair cells (brackets) at this stage. At E17.5, Gfi1 signals were seen throughout the entire cochlea including the apex (Fig. 3e and data not shown), whereas lacZ signals were observed only at the middle and basal turns but not the apex (Fig. 3e′ and data not shown). From E18.5 to P4, Gfi1 and lacZ signals were co-expressed in hair cells of the entire cochlea (Fig. 3f, f′ and data not shown). In contrast to the previously reported Gfi1 expression in inner ear neurons (Wallis et al., 2003), we did not detect the expression of Gfi1 or lacZ in the cochleovestibular ganglion (CVG) (asterisks) from E13.5 to P4 (Fig. 3f, f′, Fig. S1a, b and data not shown) although there was a relatively higher in situ hybridization background detected in the spiral ganglion cell region compared to the surrounding area (Fig. 3f and Fig. S1, asterisk).
FIG. 3.
Hair cell-specific expression of Gfi1-Cre in the cochlea. (a–c, g–i) Section in situ hybridization shows the hair cell-specific expression pattern of Gfi1 at E13.5 (a), E14.5 (b), E15.5 (c), E16.5 (d), E17.5 (e) and P4 (f). (a′–f′) X-Gal staining for lacZ activities reveals the Cre-mediated recombination events in the Gfi1Cre/+; R26R-lacZ cochleae at E13.5 (a′), E14.5 (b′), E15.5 (c′), E16.5 (d′), E17.5 (e′) and P4 (f′). Arrows indicate the inner hair cells. Brackets indicate the out hair cells. Asterisks indicate the spiral ganglion cells. Scale bars equal 50 μm.
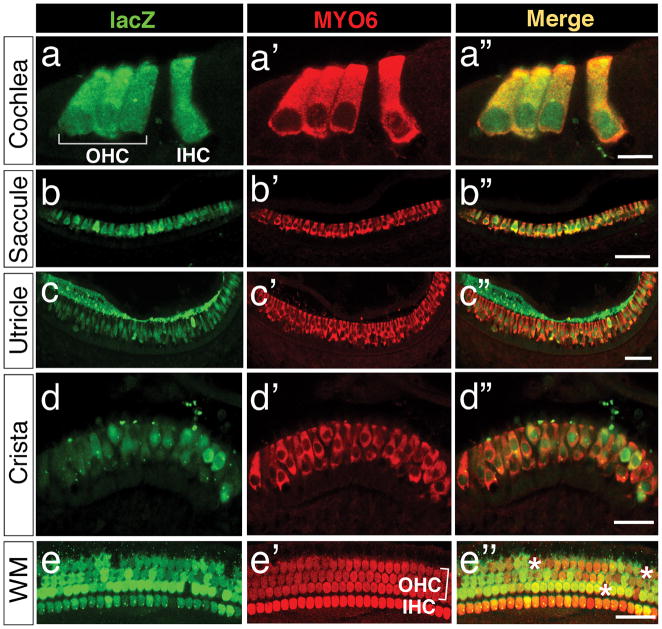
In the inner ears of Gfi1Cre/+; R26R-lacZ mice, lacZ expression was detected in both nascent and mature hair cells of the cochlea and vestibule (Fig. 2 and 3). To further determine the extent of Gfi1-Cre recombinase-mediated recombination in hair cells, we colabeled inner ear sections at P0 with anti-lacZ and anti-myosin-6 (MYO6), a hair cell-specific marker. The recombination efficiency of Gfi1-Cre, measured by the percentages of hair cells expressing lacZ, were 93.00% ± 5.35% (n=4) in the cochlea (Fig. 4a′–a″, e–e″), 90.53% ± 5.04% (n=4) in the saccule (Fig. 4b–b″), 90.50% ± 3.15% (n=4) in the utricle (Fig. 4c′c″), and 87.93% ± 7.38% (n=4) in the semicircular canal cristae (Fig. 4d′–d′). Thus, our results indicate a high efficiency of Gfi1-Cre function in hair cells.
FIG. 4.
Analysis of Gfi1-Cre activities in hair cells of the inner ear. Gfi1Cre/+; R26R-lacZ inner ears were immunolabeled with antibodies against lacZ (green) and the hair cell-specific marker myosin-6 (MYO6, red). (a–d″) Immunolabeled cryosections at P0 show lacZ expression in the hair cells of cochlea (a–a″), saccule (b–b″), utricle (c–c″), and crista (d–d″). (e–e″) Whole-mount immunostaining of the Gfi1Cre/+; R26R-lacZ cochlea at P7 reveals the widespread expression of lacZ in cochlear hair cells. Asterisks in e″ indicate the hair cells positive for MYO6 but negative for lacZ. Abbreviations: OHC, outer hair cell; IHC, inner hair cell; WM, cochlear whole-mount immunostaining. Scale bars equal 10 μm in (a–a″), 50 μm in (b′–b″, c–c″), 30 μm in (d–d″), and 50 μm in (e–e″).
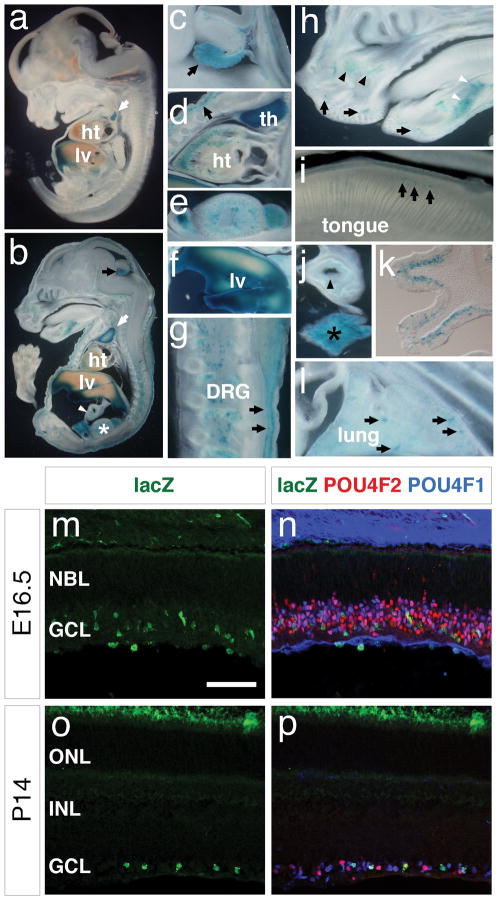
In addition to inner ear, Gfi1-Cre activities were also detected in thymus (Fig. 5a, b, d), liver (Fig. 5a, b, f), heart (Fig. 5b, d), bladder and kidney (Fig. 5b, e), dorsal root ganglia (Fig. 5b, g), choroid plexus (Fig. 5b, c), skin (Fig. 5b, d, g), upper and lower mouth areas (Fig. 5h), the maxillary and mandibular (white arrow head) areas (Fig. 5h), tongue (Fig. 5i), gut (Fig. 5j) and pancreas (Fig. 5j), olfactory epithelium (Fig. 5k), lung (Fig. 5l), and retina ganglion cells (Fig. 5m–p). Our data show Gfi1-Cre expression pattern is similar to the Gfi1 wild-type expression pattern reported previously (Wallis et al., 2003).
FIG. 5.
The expression of Gfi1-Cre in other tissues and organs. (a–l) After whole-mount X-Gal staining, Gfi1Cre/+; R26R-lacZ embryos at E13 (a), E15.5 (b–j, l), and P2 (k) were sectioned sagittally (coronally in k) and imaged. Strong lacZ activities are observed in thymus (white arrows in a and b, d), liver (a, b, f), choroid plexus (c, black arrow), dorsal root ganglia of the spinal cord (b, g), skin (b, black arrows in d and g) and heart (d). LacZ activities are also detectable in the urinary organs (asterisk in b, e), gut (white arrowhead in b; black arrowhead in j), pancreas (j, black asterisk), the dorsal epithelium of tongue (i, black arrows), the olfactory epithelia (k), the developing lung (l, arrows), the upper and lower mouth areas (h, black arrows) and in the developing maxillary (h, black arrowheads) and mandibular (h, white arrowheads) areas. (m–p) Immunostaining of the Gfi1-Cre/+; R26R-lacZ/+ retina shows Gfi1-Cre activities (lacZ, green) is confined to the retina ganglion cells marked by anti-POU4F1 (blue) and anti-POU4F2 (red) at E16.5 (m, n) and P14 (o, p). Abbreviations: DRG, dorsal root ganglia; GCL, ganglion cell layer; ht, heart; INL, inner nuclear layer; lv, liver; NBL, neuroblastic layer; ONL, outer nuclear layer; th, thymus. Scale bar equals 50 μm.
In summary, we have generated and characterized a Gfi1-Cre mouse line. Our data show that Gfi1-Cre recombination activity in the inner ear is confined to hair cells. Thus, the Gfi1-Cre knock-in mouse line provides a useful tool for gene manipulationin inner ear hair cells. In addition, this mouse line may be useful for functional study of genes in other tissues and organs where Gfi1 is expressed.
MATERIALS AND METHODS
Animals
To generate the Gfi1cre allele, genomic sequences were isolated from a mouse 129S6 (formally 129SvEvTac) BAC library (CHORI) using Gfi1 coding sequences as a probe. The Gfi1cre targeting construct was generated by inserting 3.9 kb of 5′-flanking sequences that end immediately upstream of translational initiation codon ATG and 2.7 kb of 3′-flanking sequences into the HindIII and the KpnI sites of pKII-Cre vector (L.G., unpublished), respectively (Fig. 1a). The knock-in construct removed the Gfi1 coding sequences from the translational initiation codon to Exon 5 and placed the Cre recombinase gene under the control of Gfi1 regulatory sequences. To generate Gfi1cre knock-in mice, a NotI-linearized Gfi1cre targeting construct was electroporated into AB1 embryonic stem (ES) cells (a gift from Dr. Allan Bradley, Wellcome Trust Sanger Institute) and one targeted ES clone was obtained from a total of 216 resistant ES clones. The targeted clone was confirmed by Southern blotting genotyping and injected into C57BL/6J blastocysts to generate mouse chimeras. The heterozygous Gfi1cre/+ mice were generated in a 129S6 and C57BL/6J mixed background as previously described (Gan et al., 1999; Gan et al., 1996). methods were used to genotype mice from subsequent breeding of Gfi1cre/+ heterozygotes. The PCR primers used to identify the wild type Gfi1 allele were Gfi1F (5′-GGG ATA ACG GACCAG TTG-3′) Gfi1R (5′-CCG AGG GGC GTT AGG ATA-3′). The PCR primers used to identify the Cre knock-in were Gfi1F (5′-GGG ATA ACG GAC CAG TTG-3′) and Gfi1CreR (5′-GCC CAAATG TTG CTG GAT AGT-3′). R26R-lacZ conditional reporter mice were obtained from Jackson Laboratory (Stock Number 003310) and PCR genotyping of the reporter mice was performed according to protocols provided by Jackson Laboratory. Embryonic day 0.5 (E0.5) was defined as the day when the vaginal plug was detected. University Committee of Animal Resources at the University of Rochester approved all animal procedures described here. The mouse strains were maintained in the C57BL/6J and 129S6 mixed background.
Immunohistochemistry, In Situ Hybridization and X-Gal Staining
Staged embryos and tissue samples were dissected and immediately fixed in 4% paraformaldehyde (PFA) in PBS for 1–2 hours. The samples were then embedded and frozen in OCT compound (TissueTek) for cryosections. For immunohistochemistry staining, cryosections were cut at 18 μm thickness. The working dilutions and sources of antibodies used in this study were: chicken anti-LacZ (β-galactosidase) (1:500, Abcam #ab9361-250), mouse anti-POU4F1 (BRN3A) (1:200, Santa Cruz #SC8429), goat anti-POU4F2 (BRN3B) (1:200, Santa Cruz #SC6026), and rabbit anti-MYO6 (1:500, Proteus Biosciences Inc., #25-6791). Alexa-conjugated secondary antibodies (Molecular Probes) were used at a concentration of 1:1,000. Immunofluorescence images were obtained on a Zeiss LSM 510 META confocal microscope. Whole-mount cochlear immunostaining was performed similarly to the method described previously (Ding et al., 2009). Briefly, mice were anesthetized and perfused with 4% PFA in PBS and cochleae were collected and fixed in 4% PFA in PBS for 2 hours at 4°C. After four 15-minute rinses with PBS, cochleae were blocked in 10% horse serum with 0.3% Triton-X100 in PBS for 1 hour at room temperature and incubated with primary antibodies overnight at 4°C. Then, cochleae were washed 4 times for 15 minutes each time with 0.3% Triton-X100 in PBS and incubated with Alexa-conjugated secondary antibodies in 0.3% Triton-X100 in PBS for 1 hour. After four 15-minute washes with PBS, membranous cochleae were dissected out, mounted on slides, and imaged on a Zeiss LSM 510 META confocal microscope. For in situ hybridization experiments, 20 μm thick cryosections were used as previously described (Li and Joyner, 2001). A 3′ fragment of Gfi1 cDNA (nucleotide 1873-2313) was used as the in situ hybridization probe. Detection of lacZ expression was determined by X-Gal staining (Gan et al., 1999). Briefly, cryosections were prepared at 20 μm thickness and stained at 30°C overnight in 1 mg/ml X-Gal, 4 mM K4Fe(CN)6, 4 mM K3Fe(CN)6, and 2 mM MgCl2 in PBS. For whole-mount X-Gal staining, embryos were fixed in 4% PFA in PBS overnight and stained at 30°C overnight.
To quantitate hair cells in cochlear whole-mounts, the images of immunolabeled cochlear whole-mounts were acquired and the number of labeled hair cells per 500 μm cochlear linear length at the apical, middle and basal turns were quantified. To count hair cells in cristae, saccule and utricule, immunolabeled cryosections were photographed and hair cells in ten consecutive sections were quantitated for each counting. The hair cell counts from four mice were averaged for each quantitation analysis.
Supplementary Material
In situ hybridization of the E18.5 cochlea shows Gfi1 expresson in hair cells. (a) Gfi1 antisense probe reveals Gfi1 expression specifically in cochlear hair cells. (b) The control Gfi1 sense probe shows no visible signals in cochlear hair cells and spiral ganglion cells. (c) Pou4f3 (Brn3c) probe labels cochlear hair cells specifically. The signal intensities detected in spiral ganglion cells in (a), (b) and (c) are at a similar background level. Brackets indicate the cochlear hair cells. Asterisks indicate the spiral ganglion cells. Scale bars equal 50 μm.
Acknowledgments
The authors thank Drs. Amy Kiernan and Richard Libby and the members of the Gan Laboratory for their helpful discussions and technical assistance. This work was supported by NIH Grant (DC008856) to L.G., the Research to Prevent Blindness Challenge Grant to the Department of Ophthalmology at the University of Rochester, and the Beijing Nova Program Grant (No. 2006B53) to H.Y.
LITERATURE CITED
- Ding Q, Chen H, Xie X, Libby RT, Tian N, Gan L. BARHL2 differentially regulates the development of retinal amacrine and ganglion neurons. J Neurosci. 2009;29:3992–4003. doi: 10.1523/JNEUROSCI.5237-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiolka K, Hertzano R, Vassen L, Zeng H, Hermesh O, Avraham KB, Duhrsen U, Moroy T. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep. 2006;7:326–333. doi: 10.1038/sj.embor.7400618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Wang SW, Huang Z, Klein WH. POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev Biol. 1999;210:469–480. doi: 10.1006/dbio.1999.9280. [DOI] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks CB, Bear SE, Grimes HL, Tsichlis PN. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol Cell Biol. 1993;13:1759–1768. doi: 10.1128/mcb.13.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yucel R, Frankel WN, Rechavi G, Moroy T, Friedman TB, Kelley MW, et al. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet. 2004;13:2143–2153. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- Karsunky H, Zeng H, Schmidt T, Zevnik B, Kluge R, Schmid KW, Duhrsen U, Moroy T. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet. 2002;30:295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- Kazanjian A, Wallis D, Au N, Nigam R, Venken KJ, Cagle PT, Dickey BF, Bellen HJ, Gilks CB, Grimes HL. Growth factor independence-1 is expressed in primary human neuroendocrine lung carcinomas and mediates the differentiation of murine pulmonary neuroendocrine cells. Cancer Res. 2004;64:6874–6882. doi: 10.1158/0008-5472.CAN-04-0633. [DOI] [PubMed] [Google Scholar]
- Kirjavainen A, Sulg M, Heyd F, Alitalo K, Yla-Herttuala S, Moroy T, Petrova TV, Pirvola U. Prox1 interacts with Atoh1 and Gfi1, and regulates cellular differentiation in the inner ear sensory epithelia. Dev Biol. 2008;322:33–45. doi: 10.1016/j.ydbio.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Li JY, Joyner AL. Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development. 2001;128:4979–4991. doi: 10.1242/dev.128.24.4979. [DOI] [PubMed] [Google Scholar]
- Shroyer NF, Wallis D, Venken KJ, Bellen HJ, Zoghbi HY. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain [letter] Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Wallis D, Hamblen M, Zhou Y, Venken KJ, Schumacher A, Grimes HL, Zoghbi HY, Orkin SH, Bellen HJ. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development. 2003;130:221–232. doi: 10.1242/dev.00190. [DOI] [PubMed] [Google Scholar]
- Xiang M, Gan L, Li D, Chen ZY, Zhou L, O’Malley BW, Jr, Klein W, Nathans J. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc Natl Acad Sci U S A. 1997;94:9445–9450. doi: 10.1073/pnas.94.17.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In situ hybridization of the E18.5 cochlea shows Gfi1 expresson in hair cells. (a) Gfi1 antisense probe reveals Gfi1 expression specifically in cochlear hair cells. (b) The control Gfi1 sense probe shows no visible signals in cochlear hair cells and spiral ganglion cells. (c) Pou4f3 (Brn3c) probe labels cochlear hair cells specifically. The signal intensities detected in spiral ganglion cells in (a), (b) and (c) are at a similar background level. Brackets indicate the cochlear hair cells. Asterisks indicate the spiral ganglion cells. Scale bars equal 50 μm.